Abstract
Here we describe a strategy to fluorescently label the envelope of rabies virus (RV), of the Rhabdoviridae family, in order to track the transport of single enveloped viruses in living cells. Red fluorescent proteins (tm-RFP) were engineered to comprise the N-terminal signal sequence and C-terminal transmembrane spanning and cytoplasmic domain sequences of the RV glycoprotein (G). Two variants of tm-RFP were transported to and anchored in the cell surface membrane, independent of glycosylation. As shown by confocal microscopy, tm-RFP colocalized at the cell surface with the RV matrix and G protein and was incorporated into G gene-deficient virus particles. Recombinant RV expressing the membrane-anchored tm-RFP in addition to G yielded infectious viruses with mosaic envelopes containing both tm-RFP and G. Viable double-labeled virus particles comprising a red fluorescent envelope and a green fluorescent ribonucleoprotein were generated by expressing in addition an enhanced green fluorescent protein-phosphoprotein fusion construct (S. Finke, K. Brzozka, and K. K. Conzelmann, J. Virol. 78:12333-12343, 2004). Individual enveloped virus particles were observed under live cell conditions as extracellular particles and inside endosomal vesicles. Importantly, double-labeled RVs were transported in the retrograde direction over long distances in neurites of in vitro-differentiated NS20Y neuroblastoma cells. This indicates that the typical retrograde axonal transport of RV to the central nervous system involves neuronal transport vesicles in which complete enveloped RV particles are carried as a cargo.
Rabies virus (RV) is a highly neurotropic and prototypic member of the Rhabdoviridae family within the Lyssavirus genus. The viral 12-kb, negative-strand RNA genome encodes five proteins which all are essential for virus replication and virus particle formation. The viral RNA is wrapped with nucleoprotein (N) and associated with the phosphoprotein (P) and the large RNA-dependent RNA polymerase (L) in a transcriptionally active ribonucleoprotein (RNP) complex. During virus budding at the cell surface, RNPs are condensed and wrapped by a lipid bilayer envelope containing an internal matrix protein (M) layer and the transmembrane spike glycoprotein (G) (26), which is exposed on the virus surface as a trimeric spike (14, 46).
As is well established in cell culture, entry of RV into cells involves receptor binding by the G protein, endocytosis, pH-dependent membrane fusion (13, 37), and, as demonstrated for vesicular stomatitis virus (VSV) rhabdovirus, release of M and the RNP into the cytoplasm (34). RVs lacking the G gene must be complemented in trans with G to allow infection of cells and are not able to spread further in cell culture (25) or in mice in vivo (7). In vitro, a broad spectrum of nonneuronal cells are effectively infected by RV (32), indicating the existence of ubiquitous, non-neuron-specific receptors. In addition, three cellular proteins have been proposed as high-affinity neuron-specific receptors, namely, nicotinic acetylcholine receptor, neuronal adhesion molecule, and p75 neurotrophin receptor (20, 44, 45). The RV G protein sequence and differential usage of receptors by G proteins appear to be a prime virulence factor determining the spread of virus and outcome of infection. Indeed, attenuation of a recombinant laboratory strain adapted to cell culture could be overcome by exchanging the G gene with that of a highly pathogenic challenge virus strain, CVS (10, 28).
A hallmark of RV infection in vivo is peripheral infection and swift transport of virus to the central nervous system (CNS). Crossing such long distances in a reasonable time requires that viruses hijack the cellular motor-driven cargo transport systems (for a review, see reference 15). In the case of RV, this involves microtubule-dependent dynein-driven retrograde cargo transport of axons. Intriguingly, interaction of a component of the dynein motor complex, dynein light chain (DLC), with the RV P protein has recently been demonstrated (17, 31), but a contribution to transport of viral RNP complexes is questionable (10, 22, 30, 43). There is more direct evidence in favor of the envelope G protein being involved in retrograde axonal transport of viruses, since retroviruses pseudotyped with RV G are transported in a retrograde way and can gain access to the nervous system after peripheral delivery (21, 27). Binding of G to cell surface proteins may therefore be important not only for membrane fusion and cell infection but also for long-distance transport of enveloped viruses along axons within nonacidified transport vesicles.
The availability of recombinant RV expressing fluorescent proteins such as green fluorescent protein (GFP) has in the past greatly facilitated the identification and analysis of living virus-infected cells, including neurons in situ (47). In combination with RV pseudotyping approaches, the first monosynaptic viral tracers that allow determination of the connectivity of individual cells have been developed (48). In addition, studies on the motility of virus particles and subunits have benefited greatly from imaging of fluorescent proteins fused to virus structural proteins (for a review, see reference 15). Fluorescent labeling of rhabdovirus and paramyxovirus RNPs has been achieved by generating fluorescent N (18), P (5, 8), and L (6) proteins. Single fluorescent RV particles expressing an enhanced GFP (eGFP)-P fusion protein could be tracked by fluorescence microscopy (8), and for the first time this allowed live observation of the release of eGFP-labeled RNPs from late endosomal structures into the cytoplasm (10).
Here, we report the generation of replicating RV particles with a stable red fluorescent envelope, allowing live discrimination of naked RNPs from enveloped virus particles. Since initial approaches involving fluorescent labeling of RV M failed (unpublished results) or fusion of GFP to the cytoplasmic domain of VSV G resulted in rapid loss of GFP expression by mutations (4), we followed a novel strategy involving pseudotyping of RV particles (for a review, see reference 11) with a transmembrane-anchored red fluorescent protein (tm-RFP). This was achieved by equipping the pH-stable RFP derivative tdtomato with the signal sequence and the membrane-spanning domains of RV G. tm-RFPs with or without an N-glycosylation site were transported to the cell surface and colocalized to RV M and G proteins. Expression of tm-RFP from recombinant RV resulted in infectious mosaic viruses carrying both G and tm-RFP in their envelopes, which could be tracked by fluorescence microscopy in cell-free solution and in the endosomes of living cells. Long-distance transport of particles containing both GFP-labeled RNPs and tm-RFP in neuroblastoma cell axon-like outgrowths confirmed that intact RV virus particles can be transported.
MATERIALS AND METHODS
Cells and viruses.
Viruses were grown on BHK-21 (clone BSR) cell monolayers maintained in Glasgow minimal essential medium supplemented with 10% newborn calf serum. BSR T7/5 cells constitutively expressing bacteriophage T7 RNA polymerase (2) were used for recovery of RV from transfected cDNA as described previously (9).
Recombinant RV lacking G expression (SAD ΔG) was generated from pSAD ΔG (25), which is a cDNA derivative of pSAD L16 (38). Infectious virus stocks of SAD ΔG were prepared by in trans complementation in MGon cells, expressing both RV matrix and glycoprotein after induction with doxycycline (12). In SAD eGFP-P, the authentic P coding sequence was replaced with the eGFP-P fusion protein-encoding sequence described previously (8).
Murine neuroblastoma NS20Y cells (1) were differentiated to nondividing cells with outgrowing neurites as described previously (16) with some modifications. Briefly, NS20Y cells were seeded into μ-slide VI or μ-slide I (Ibidi, Munich, Germany) and were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 5% fetal calf serum (FCS). When the cells were 70% confluent, the medium was changed to DMEM supplemented with 0.5% FCS and 1 mM butyryl-cyclic AMP (butyryl-cAMP) (Sigma-Aldrich). The medium was replaced every 3 days. After 7 days of cultivation, the medium was changed to DMEM supplemented with 5% FCS, 50 μM AraC (Sigma-Aldrich), and 300 μM uridine (Sigma-Aldrich) and again was replaced every 3 days. After a further 7 days of cultivation, the NS20Y cells were used for infection and live cell imaging experiments.
Construction of RFP/RV G chimera-encoding cDNAs.
To generate RFP/RV G chimeras, the N-terminal signal sequence of RV G (amino acids [aa]1 to 29) was PCR amplified (using primers 5′-CTCGTCAAGCTTGGGAAAGATGGTTCCTCAGG-3′ [forward] and 5′-GCTGTCGGATCCCTTGTCTGGTATCGTGTA-3′ [reverse]) and was inserted into the multiple cloning site of the expression plasmid pCR3 (Invitrogen) using the HindIII and BamHI restriction sites, resulting in pCR3ssRVG. A C-terminal sequence of RV G containing the cytoplasmic tail as well as the transmembrane domain of RV G (aa 410 to 524) was PCR amplified (using primers 5′-CTCGTCCTCGAGTCGGTTATCCCCCTTGTG-3′ [forward] and 5′-GCTGTCTCTAGATTACAGTCTGGTCTCACC-3′ [reverse]) and was cloned into pCR3ssRVG (XhoI/XbaI), giving rise to pCR3sstmRVG. A DNA fragment encoding tdtomato (40) (aa 2 to 476) was PCR amplified (using primers 5′-CTCGTCGAATTCTGGTGAGCAAGGGCGAGGAG-3′ [forward] and 5′-GCTGTCGATATCCTAGCTTGTACAGCTCGTCCATGC-3′ [reverse]), EcoRI/EcoRV digested, and inserted between the signal sequence and the transmembrane domain of the RV G tail sequence. The resulting pCR3 tm-RFP encodes the RFP/RV G chimeric protein under the control of a cytomegalovirus promoter. The tdtomato open reading frame encodes two copies of the tomato variant of Discosoma sp. fluorescent protein DsRed in tandem orientation (40).
To introduce larger C-terminal parts of RV G with an RV G glycosylation site included, a sequence coding for aa 316 to 524 of RV G was introduced into pCR3sstmRVG, giving rise to the expression plasmid pCR3 tm-RFP Asn319. A synthetic glycosylation site was introduced into pCR3 tm-RFP by site-directed mutagenesis to produce an amino acid exchange from Asn-Gly-Arg to Asn-Gly-Thr at positions 34 to 36 of tm-RFP, resulting in tm-RFP(Asn).
cDNA rescue experiments.
cDNA plasmids were transfected into cells after CaPO4 precipitation (mammalian transfection kit; Stratagene) according to the supplier's instructions. Vaccinia virus-free rescue of recombinant RV was performed as described previously (9) by transfection of 10 μg of full-length cDNA and plasmids pTIT-N (5 μg), pTIT-P (2.5 μg), and pTIT-L (2.5 μg) in 106 BSR T7/5 cells grown in 8-cm2 culture dishes.
Density gradient centrifugation.
Supernatants from 3 × 106 virus-infected cells were harvested at day 2 postinfection at a multiplicity of infection of 1, and the virions were concentrated by centrifugation on size exclusion columns (Centricon Plus-20, polyethersulfone 100,000 MVV CO; Millipore). The concentrated virus suspension was diluted in TEN buffer (50 mM Tris-HCl, 1 mM EDTA, 150 mM NaCl [pH 7.4]) and was loaded on a 12-ml 10% to 35% iodixanol (Optiprep; Axis-Shield) density gradient. After 18 h of centrifugation (Beckman SW28, 27,000 rpm, 4°C), 12 fractions of 1 ml each were collected. After sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Western Blotting, the fractions were analyzed with RV N, P, and M protein-specific sera or with a serum recognizing DsRed and its derivatives (Clontech, BD Biosciences).
Immobilization of supernatant virions on glass coverslips.
Immobilization of supernatant virions was performed as described previously (8). Briefly, 25 μl of density-purified virus from cell culture supernatants was loaded onto glass coverslips and incubated for 2 h at 4°C. After binding of virions to the glass surface, the virions either were directly analyzed by confocal laser scanning microscopy or were fixed with 3% paraformaldehyde (PFA) and subsequently immunostained.
Immunostaining.
Cells and virions were incubated at room temperature for 30 min with 3% PFA in phosphate-buffered saline (PBS). After membrane permeabilization with 0.5% Triton X-100 (15 min, room temperature), cells were incubated with specific antibodies directed against RV G (E559) or RV M (M2D4) for 2 h at 37°C. The cells were then washed with PBS and incubated with fluorescence-labeled secondary antibodies (Alexa 488- or 633-conjugated antibodies directed against mouse or rabbit immunoglobulin G) for a further 30 min at 37°C. After two washing steps with PBS and one with double-distilled water, the cells were mounted and analyzed on a fluorescence microscope.
Fluorescence microscopy.
Confocal laser scanning microscopy was performed with a Zeiss LSM510 Meta system using a Zeiss Axiovert 200 microscope with 63× and 40× objectives. eGFP and tdtomato proteins were excited at laser wavelengths of 488 nm and 543 nm. Fluorescence emission was detected using a 505/530-nm band pass filter for eGFP. tdtomato fluorescence was detected using long pass filters (>585 and >560 nm) or an appropriate band pass filter (585/615 nm). Live cell imaging was performed with cells growing in μ-dishes and μ-slides (Ibidi, Munich, Germany). During acquisition, the cells were incubated in a heated chamber at 37°C with 5% CO2. Images were processed and analyzed using ImageJ software.
Western blotting.
After sodium dodecyl sulfate-polyacrylamide gel electrophoresis, proteins were transferred to a nitrocellulose membrane (Schleicher and Schuell) using a semidry transfer apparatus (OWL Scientific). After incubation with blocking buffer (2.5% dry milk and 0.05% Tween 20 in PBS) at room temperature for 1 h, membranes were incubated overnight with the primary antibodies against RV-M (M1B3), RV-Gtail (HCA/05), and RV-N (S86). Western Lightning chemiluminescence reagent (Perkin-Elmer) was used for the detection of peroxidase-conjugated secondary antibodies.
RESULTS
Construction of transmembrane tdtomato protein.
In order to generate infectious RV particles with fluorescent membrane surface proteins, we sought to incorporate a red fluorescent transmembrane surface protein along with the viral G protein to create viruses with a mosaic envelope. The DsRed variant tdtomato (40) was considered a suitable fluorescent protein because of an increased resistance against low pH compared to GFP and the lack of intermolecular aggregation as observed with the conventional DsRed. To achieve expression of a fluorescent type I transmembrane tdtomato protein, we constructed an expression plasmid to encode the N-terminal RV G signal sequence, the tdtomato coding sequence and the C-terminal sequence of the RV G protein, including 49 aa of the ectodomain, the transmembrane anchor, and the cytoplasmic tail (Fig. 1A). The resulting tm-RFP was further modified by insertion of an artificial N-glycosylation signal at the N terminus of tdtomato to possibly support transport to the cellular surface [tm-RFP(Asn)]. In a further construct, tm-RFP(Asn319), the amino acid sequence derived from the membrane-proximal region of RV G was elongated by 75 aa to include an authentic RV glycosylation site at position 319 of RV G.
FIG. 1.
(A) Schematic presentation of chimeric RFP/RV G constructs. To ensure cell surface expression and incorporation into RV particles, the RFP derivative tdtomato (40) was N-terminally fused to the signal peptide (black boxes) of RV G and C-terminally fused to a C-terminal part of RV G comprising the intracellular cytoplasmic domain, the hydrophobic transmembrane domain (tm), and a membrane-proximal part of the RV G ectodomain (hatched boxes). Predicted glycosylation sites are indicated by diamonds. tm-RFP, no glycosylation site; tm-RFP(Asn), a synthetic glycosylation site was inserted between the signal peptide and RFP; tm-RFP(Asn319), RV ectodomain to aa 316 comprising one authentic glycosylation site (Asn319). (B) Surface expression of RFP/RV G chimeras. Plasmids encoding the indicated RFP constructs under the control of the cytomegalovirus promoter were transfected into BSR T7/5 and neuroblastoma (NA) cells, and 24 h later, RFP autofluorescence in PFA-fixed cells was observed by confocal laser scanning microscopy.
Surface expression of tm-RFP and colocalization with RV M and G proteins.
Expression and transport of the tm-RFP constructs was assayed after transfection of plasmids into BSR T7/5 and neuroblastoma (NA) cells. In both cell types tm-RFP lacking an N-glycosylation site was efficiently expressed and localized to the cell periphery, as observed by confocal fluorescence microscopy of PFA-fixed and permeabilized cells (Fig. 1B). A similar pattern of RFP fluorescence was observed for tm-RFP(Asn), harboring the artificial N-glycosylation site at the tdtomato coding sequence. Interestingly, although expressed efficiently, the tm-RFP(Asn319) comprising an elongated RV G ectodomain with an authentic N-glycosylation site was transported poorly, if at all, to the cell periphery.
Incorporation into viruses requires the expression of proteins at the sites of virus budding. To investigate whether tm-RFP and RV G are transported to the same areas of the cytoplasmic membrane, colocalization of coexpressed RV G and tm-RFP was investigated by confocal fluorescence microscopy. After immunostaining of RV G protein with a monoclonal antibody recognizing a part of the ectodomain not present in tm-RFP, colocalization of RV G and tm-RFP at the cytoplasmic membrane and within the cytoplasm of transfected cells was confirmed (Fig. 2C). No G staining was detected in cells transfected only with tm-RFP-expressing plasmid (Fig. 2B), confirming the lack of antibody cross-reaction, and expression of G (Fig. 2A) or M (Fig. 2E) alone did not yield red fluorescence. Since the RV M protein plays a major role in recruitment of RNPs and G protein to the viral budding site, colocalization of tm-RFP with plasmid-derived M protein was also checked. Perfect colocalization was observed at the plasma membrane, whereas unequivocal colocalization at internal membranes was not apparent (Fig. 2D). In terms of transport and association with membranes and RV M protein, the recombinant tm-RFP proteins were similar to authentic G protein.
FIG. 2.
tm-RFP colocalizes with RV envelope proteins G and M. tm-RFP and RV proteins G and M were expressed in BSR T7/5 cells from transfected cDNAs. After 24 h of incubation, the cells were fixed and permeabilized for immunostainings against RV G or M proteins. (A) After transfection of RV G cDNA only, no tm-RFP autofluorescence was detectable. (B) After transfection of tm-RFP cDNA only, no G-specific immunostaining was detectable. (C) Colocalization of RV G protein and tm-RFP mainly at the cytoplasmic membrane. (D) Colocalization of RV M protein with tm-RFP at the cytoplasmic membrane. (E) After transfection of RV M cDNA only, no G-specific autofluorescence was detectable. All images were acquired at identical microscope settings.
Complementation of G-deficient RV.
Incorporation of tm-RFP as a type I surface envelope protein in the absence of G was analyzed by trans-complementation of a G gene-deleted RV, SAD ΔG (25). Two days after infection of NA cells constitutively expressing the unglycosylated tm-RFP, cell culture supernatants were collected and virions were purified by density gradient centrifugation. A mixture of purified virions from tm-RFP expressing cells and eGFP-labeled RV virions (8) was immobilized by binding to glass coverslips for confocal fluorescence microscopy (Fig. 3). Detection of either red or green fluorescent particulate structures of similar size indicated that tm-RFP was incorporated into virus particles at levels sufficient for detection by fluorescence microscopy.
FIG. 3.
Incorporation of tm-RFP into SAD ΔG virions. A cell line constitutively expressing tm-RFP was infected with G gene-deleted SAD ΔG. After 2 days of infection, the cell culture supernatants were collected and virions were purified by density gradient centrifugation. Purified virions were mixed with virions from SAD eGFP-P-infected cells, which are known to produce green fluorescent virions (8). After immobilization of virus particles to glass coverslips, red fluorescent particles were detected by laser scanning microscopy. The red and green fluorescent particles were similar in size but were clearly distinguishable by fluorescence from eGFP-labeled virions. Bar, 1 μm.
Recombinant infectious viruses: coincorporation of tm-RFP and RV G into virions.
Whereas tm-RFP-labeled SAD ΔG virions are not infectious, due to the lack of receptor binding G protein (not shown), virus particles comprising tm-RFP in addition to G might be infectious and may be useful for virus tracking studies. In order to see whether viable virus with tm-RFP coincorporated can be rescued, recombinant RVs were generated in which the different tm-RFPs described above (Fig. 1) were expressed from an extra gene located between the RV G and L transcription units (Fig. 4A). All viruses were successfully rescued from cDNA, indicating that tm-RFP expression did not interfere critically with virus function. Indeed, RFP was expressed from all recombinant viruses, as determined after infection of BSR T7/5 cells (Fig. 4B). As previously observed, tm-RFP and tm-RFP(Asn) showed colocalization with RV G at the cytoplasmic membrane, in contrast to the tm-RFP(Asn319) variant (Fig. 4B, right).
FIG. 4.
Colocalization of tm-RFP and glycoprotein G in infected cells and in virions. (A) Schematic presentation of recombinant RV expressing the indicated tm-RFP constructs. The different tm-RFPs were inserted into the RV genome as an additional transcription unit between the G and L genes. (B) Surface expression of RFP/RV G chimeras in virus-infected cells. After 24 h of infection of BSR-T7/5 cells with the indicated viruses, the cells were fixed and RV G was immunostained with G-specific monoclonal antibody E559. RV G protein and RFP fluorescence colocalized at the cytoplasmic membrane in SAD tm-RFP- and in SAD tm-RFP(Asn)-infected cells. In contrast, no colocalization was detectable in SAD tm-RFP(Asn319)-infected cells. (C) Incorporation of tm-RFP into RV particles. Density gradient-purified supernatant virions were immobilized on glass coverslips, and after fixation with 3% PFA, the virions were immunostained with G-specific antibody E559. Virus particles were detected by confocal laser scanning microscopy.
The incorporation of tm-RFP and G into virus particles was analyzed by confocal microscopy of purified virus particles on glass coverslips. Viruses encoding tm-RFP and tm-RFP(Asn) displayed efficient fluorescence of virions, demonstrating efficient incorporation into virus envelopes even in the presence of RV G protein. Moreover, immunostaining of the individual virions with a G-specific antibody revealed that tm-RFP and tm-RFP(Asn) were coincorporated with RV G into the same virus particles, resulting in infectious red fluorescent mosaic virus particles. In addition to G- and RFP-containing particles, virus particles with low or nondetectable levels of RFP fluorescence were observed (Fig. 4C, left and middle), indicating some variability of the composition of envelope proteins. In contrast to these results, viruses expressing tm-RFP(Asn319) extracellular virions immobilized to glass coverslips did not yield particles with detectable fluorescence, supporting the view that surface expression is required for efficient incorporation.
Infectivity of tm-RFP-containing viruses.
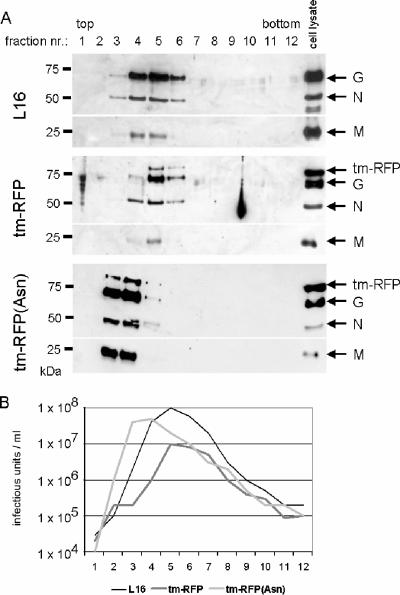
To further analyze the composition of tm-RFP- and tm-RFP(Asn)-expressing RV particles, purified supernatant virions from infected BSR T7/5 cells were analyzed by Western blotting (Fig. 5A). With a G-specific antibody directed against the common cytoplasmic tail of G and tm-RFP, specific signals for tm-RFP and tm-RFP(Asn) were detected in purified virions and cell extracts. In both tm-RFP- and tm-RFP(Asn)-expressing viruses, the signal peaks colocalized with the virus peaks identified by the presence of RV G, N, and M proteins. Whereas in cell lysates comparable levels of G protein and tm-RFP were present, purified viruses contained less tm-RFP than G, corroborating some advantage of G in incorporation into virus envelopes. Nevertheless, tm-RFP comigrated with the virus particle peaks identified by the presence of RV G, N, and M proteins and by the infectivity profiles of gradient fractions (Fig. 5B), which were determined by titration of the infectious virus titers in BSR T7/5 cells.
FIG. 5.
Incorporation of RV G and tm-RFP into virions. (A) Density gradient-purified supernatant virions were analyzed by Western blotting using the RV N, M, and G protein-specific antisera. tm-RFP was detected through a G-specific antibody which recognizes the transmembrane domain of RV G protein. The specificity of the tm-RFP signal was also tested with an antibody recognizing DsRed and its derivatives (not shown). (B) The infectious virus titer of each density gradient fraction was determined on BSR T7/5 cells and compared with the virus protein content.
As additional incorporation of heterologous transmembrane proteins into the virus membrane may interfere with receptor binding and subsequent fusion events, we determined the virus protein contents of gradient fractions by Western blotting and quantitative fluorescent imaging (not shown) and compared the measured protein levels with the infectious titers of the same fractions. Indeed, an approximately ∼10-fold-decreased infectivity compared to wild-type SAD L16 could be determined for viruses containing tm-RFP and tm-RFP(Asn), as suspected from the infectivity profiles of the nonquantitative Western blot (Fig. 5). Similarly, growth curves of parallel infections (Fig. 6) indicated a reduced production of infectious tm-RFP-encoding viruses, with approximately 10-fold-lower maximum titers.
FIG. 6.
Growth kinetics of tm-RFP-expressing RVs. The infectious virus titers (IE) in supernatants from virus-infected BSR T7/5 cells (multiplicity of infection = 0.01) were determined at the indicated time points postinfection. In comparison to those of SAD L16, the infectious virus titers of SAD tm-RFP and SAD tm-RFP(Asn) were ∼10-fold decreased.
Double-labeled RVs.
In order to allow discrimination between viral RNP and envelope in the fluorescence microscope, tm-RFP labeling was combined with RNP labeling by expression of an eGFP-P fusion protein as previously described (8). The recombinant virus SAD eGFP-P tm-RFP (Fig. 7A) was recovered from cDNA and grew to titers comparable to those of SAD eGFP-P (not shown). Supernatant virions were purified, and the glass-immobilized virions were analyzed by confocal laser scanning microscopy. Double-labeled particles with perfectly colocalizing RFP and eGFP fluorescence emissions (yellow) were readily detectable (Fig. 7B), demonstrating coincorporation of both fluorescent fusion proteins into virions. However, 27% (n = 100) green particles with nondetectable RFP fluorescence could be detected, indicating variable levels of RFP. In addition, 16% red-only signals potentially representing empty membrane vesicles were also detected. Quantification of the fluorescence of a total of 93 eGFP-positive particles revealed rather homogenous intensities of eGFP, whereas RFP fluorescence intensities were more variable (Fig. 7C), most likely reflecting competition of G and tm-RFP in incorporation.
FIG. 7.
Generation of double-labeled RV. (A) Genome organization of control virus SAD L16, SAD tm-RFP, SAD eGFP-P, and the double-labeled SAD eGFP-P tm-RFP. Recombinant SAD eGFP-P tm-RFP virus was generated by combining cDNA plasmids encoding SAD tm-RFP and the previously described SAD eGFP-P. (B) Density gradient-purified supernatant virions from SAD eGFP-P tm-RFP-infected cells were immobilized on glass coverslips. Virus particles were detectable by confocal laser scanning microscopy. (C) Quantification of the fluorescence of a total of 93 eGFP-positive particles. Error bars indicate standard deviations. (D) Double-labeled SAD eGFP-P tm-RFP virions were detectable in endosomes of living BSR T7/5 cells. Confocal fluorescence images with the individual fluorescence channels (top three rows, left and middle panels), a fluorescence merge (top three rows, right panels), and an overlay with nonconfocal DIC images (bottom panels) are shown.
To see whether detection of particles was also satisfactory under live cell conditions, BSR T7/5 cells were incubated with SAD eGFP-P tm-RFP (Fig. 7D). Virus particles were readily detected at the cytoplasmic membrane (not shown). After 3 h of incubation, multiple yellow particles were observed in large vesicular structures. Blocking of acidification with NH4Cl led to an accumulation of particles in these structures (not shown), indicating that they represent endosomal compartments which are traversed by the virus during entry (not shown). As described previously for SAD eGFP-P (10), virus particles were moving inside the vesicular compartment but always remained connected to the vesicle membrane, even after longer observation periods. Most importantly, yellow particles were observed in the vesicles, indicating that numerous enveloped virus particles accumulate in the endosomal compartment.
Enveloped viruses are transported in differentiated NS20Y cells.
To address whether double-labeled enveloped virus particles are transported over substantial distances within vesicles prior to membrane fusion and cell entry, the neuroblastoma cell line NS20Y was used for differentiation into neuron-like cells with butyryl-cAMP. Cultures of NS20Y cells in which long axon-like structures indicated successful differentiation were incubated with a double-labeled virus in which the glycoprotein gene was derived from the highly pathogenic RV strain CVS. For virus tracking in neuroblastoma cells, CVS G protein-expressing virus was used, since decreased pathogenicity of attenuated SAD L16 virus in peripheral infections of mice (10) may be caused by G-dependent defects in long-distance transport.
After addition of virus, the cells were incubated on the microscope stage at 37°C with 5% CO2, and image acquisition was performed with a confocal laser scanning system that allows simultaneous detection of GFP and tdtomato autofluorescence. Differential interference contrast (DIC) image acquisition was used for visualization of the complete cells to determine the retro- and anterograde directions in individual cells by localization of the cell body. Subsequently, fast image acquisition was performed in areas of axon-like neurites to detect transported virus particles.
Different types of movements were recorded in the shafts of the neurites. Directly after incubation, almost all detectable virus particles were floating in the extracellular space between the neuroblastoma cells (not shown). Although the majority of labeled viruses kept sticking to the surfaces of the neuroblastoma cells (Fig. 8A), three different types of virus motion were observed. Most of the particles performed only slight and nondirectional movements (Fig. 8B) and therefore seemed not to be attached to the motor transport machineries. Another portion of particles were moving in the anterograde direction yet showed only short-range movements that were interrupted by many stops (Fig. 8C). In contrast, particles that were transported in the retrograde direction showed rather continuous movement at constant speed of 0.1 μm/s (average for five particles; standard error of the mean, 0.0022 μm/s) with little pausing. Thus, long-range distances of on average 45 μm (standard error of the mean, 10.10 μm/s) could be determined (Fig. 8D). Most importantly, all moving particles emitted fluorescence derived from both RNP-associated GFP and membrane-anchored RFP, demonstrating the retrograde transport of complete, enveloped viruses. The dual-color labeling strategy developed here is thus suitable to determine the composition of intracellularly transported RV and may further contribute to the investigation of the mechanisms involved in the retrograde axonal transport of RV and other neurotropic viruses.
FIG. 8.
RV particle movement in axon-like structures of NS20Y cells differentiated by butyryl-cAMP-treatment. The differentiated cells were incubated with double-labeled virus SAD eGFP-P tm-RFP expressing the glycoprotein of the pathogenic RV strain CVS. (A) Attachment of virions to neuronal outgrowths with no or marginal movement. (B) Virus particles which showed slight movement in different directions. (C) Movement of enveloped virus in the anterograde direction (from the top to the bottom). (D) Retrograde transport of double-labeled RV in NS20Y cells at 6 h postinfection. Fluorescent confocal laser scanning microscopy images were merged with DIC images of the neuroblastoma cell. In panel D, the image acquisition was for 218 s at 6.4 s/frame. A video file is provided in the supplemental material.
DISCUSSION
Infection at the periphery, invasion of motoneurons or sensory neurons, and rapid retrograde transport to the CNS, including transsynaptic spread, are long-known key features of RV neuropathogenesis. As has been shown with RV gene deletion mutants, the G protein is an absolute requirement for RV entry into cells and RV spread between neurons (7, 48). In addition, the sequence of G proteins determines the neuroinvasiveness of RV variants (10, 28), which may be related to differential receptor usage of viruses (3) and to differential induction of apoptosis after expression of G in infected cells, affecting the integrity of neuronal network connectivity (reviewed in reference 19). The work presented here in addition demonstrates a role of RV G in virus particle transport in neuronal cells.
The study of intracellular virus trafficking has greatly benefited in the past from the ability to fluorescently label structural virus proteins (for a comprehensive review, see reference 15). In the case of enveloped viruses, which infect cells by membrane fusion, the loss of the viral envelope and release of the naked core (or RNP) at the cell surface or in endosomes is the hallmark of virus entry and infection of cells. Distinction between free enveloped and naked intracellular RNPs in live tracing approaches therefore requires differential labeling of envelope and RNP. Fluorescence-labeling strategies have been successfully applied to RV (8, 18) and other Mononegavirales (5, 6) to track intracellular RNPs. Here, we report the generation of viable RV with a red fluorescent envelope and a green fluorescent RNP to allow discrimination of enveloped virus and free RNPs by live fluorescence microscopy.
In analogy to previous successful approaches of envelope switching and retargeting (23, 24; for a review, see reference 11), we generated fluorescent type I transmembrane proteins that were incorporated, similarly to authentic G, in the envelope of budding virus. As predicted from previous work with RV and VSV G (35), a short G “stem” region comprising the membrane-proximal 49 aa of the ectodomain, the transmembrane region, and the cytoplasmic tail of G was sufficient to target tm-RFP to the cell membrane for association with the matrix protein layer and incorporation into budding virus, probably as trimers. Notably, the presence of an artificial N-glycosylation signal in tm-RFP(Asn) was not required and did not improve trafficking of the tm-RFP. Moreover, an elongated G ectodomain, including the authentic membrane-proximal N-glycosylation site (Asn319) of RV G, which has been shown to support transport of the full-length protein (39), was not transported to the cell surface, indicating that the elongated stem contains inhibitory structures. Interestingly, a similar effect was observed with a chimeric RV G carrying the anthrax protective antigen for immunization purposes (42). The autonomous incorporation of tm-RFP and tm-RFP(Asn) and their coincorporation with different G proteins, including VSV G (not shown), predict that this system is applicable to the study RV pseudotypes with heterologous spike proteins as well.
Indeed, RV expressing G and tm-RFP(Asn) contained substantial amounts of tm-RFP protein, as indicated by Western blots of gradient-purified virion preparations with a serum recognizing the common cytoplasmic tail of tm-RFP and RV G (Fig. 5A), suggesting little if any advantage of G for incorporation. As observed by fluorescence microscopy of single virus particles, the tm-RFP content of individual virions showed a more variable distribution than GFP fluorescence of RNPs (Fig. 7C), as expected from random distribution and incorporation of G and tm-RFP at the cell surface budding sites and a limited incorporation capacity of spike proteins. In contrast, fluorescence of the eGFP-P fusion protein in individual virus particles showed less variability, reflecting the incorporation of a defined number of P proteins into the RNP. This also supported the presence of a single RNP within the observed rhabdovirus particles, in contrast to the case for paramyxoviruses, which often may contain multiple RNPs (29).
The incorporation of extra nonfunctional spike protein into the viral envelope may be expected to result in a decreased infectivity of virions because of a reduced concentration of G protein, restricted access of G to receptors, or interference with G functions during membrane fusion and RNP entry. Indeed, compared to wild-type RV, an approximately 10-fold reduction of the specific infectivity was observed for RV expressing G and tm-RFP. Although reduced receptor binding of viruses cannot be excluded, the identification of double-labeled, membrane-associated virus particles in endosomal vesicles (Fig. 7D) indicated that this step was not severely hampered. Another possibility for reduced infectivity is attenuation of the mechanism of membrane fusion. A certain number of G trimers, probably in a defined arrangement, are required to form a fusion pore (36). Both the arrangement of the G layer and conformational changes could be affected by intermingled tm-RFP proteins. In addition, the formation of heterotrimeric spikes containing G and tm-RFP proteins, which potentially are no longer able to perform the conformational rearrangements required for pH-dependent membrane fusion, cannot be excluded. In any case, the moderate loss of infectivity indicated that neither step was critically affected.
To investigate the transport of enveloped virus particles in neurons, transport of double-labeled RV particles in in vitro-differentiated NS20Y neuroblastoma cells was analyzed. Since recombinant RV SAD L16 is attenuated in mice after peripheral infection, which might be due to altered receptor usage in the original SAD B19 vaccine virus (10), the G protein of the highly pathogenic RV CVS-11 (10) was initially used to analyze intraneuronal virus transport. Examination of NS20Y cells incubated with double-labeled CVS G-encoding RV revealed that enveloped virus particles enter a long-distance transport pathway. Apart from virus particles showing poor (Fig. 8A) or slight (Fig. 8B) back and forward movement, enveloped particles that moved in the anterograde direction were detected (Fig. 8C). In contrast to motions in the anterograde direction, which were interrupted by many stops, the movement in the retrograde direction (Fig. 8D) occurred at a rather constant speed of 0.1 μm/s and a run length of ∼45 μm. The velocity of transported enveloped RV was thus ∼10-fold lower than the velocities of other viral capsids, e.g., those from herpesviruses, entering the microtubule-dependent retrograde transport (41). Since differences in transport velocities strongly depend on the cell type and cultivation conditions, a direct comparison may not be possible. Furthermore, due to the image acquisition time of 6.4 seconds per frame, we cannot exclude repeated pausing during acquisition, possibly leading to an decreased average velocity.
Similar to the case for double-labeled virus particles, virions in which only the RNP was labeled by eGFP-P (although the differentiation of enveloped virions and RNP is not possible in this case) showed identical patterns of motion, indicating that the presence of tm-RFP is not instrumental in transport (not shown).
These results are in agreement with the previous observation that RV G protein can confer retrograde axonal transport to pseudotyped lentivirus vectors (21, 27), and they indicate that G-dependent retrograde transport is an intrinsic feature of authentic RV infection. In contrast, the existence of a direct role of the binding of cytosolic P protein or RNPs to DLC in motor-driven transport has been challenged by the finding that RV mutants deficient in DLC binding are still transported to the CNS after peripheral inoculation (22, 43). Rather, an indirect role could be considered, since the lack of the DLC binding site in P appears to affect intraneuronal virus replication by decreasing primary transcription of RV (43).
Our observations of RV enveloped by G protein being transported in neuronal cultures support a model in which transport of complete virions inside neuronal transport vesicles directly contributes to the spread of RV in vivo. Although it appears that productive infection of neurons is required for further spread of RV to connected neurons (7), the preceding initial transport of RV to the cell body may save time and prevent premature damage of the neuronal network and help RV to reach its final destination. Further experiments with primary neurons may further reveal the nature of neuronal transport vesicles and of receptors selected by RV G to take the correct vesicles. For instance, one of the potential neuronal receptors for RV, p75 neurotrophin receptor, is known to be internalized in vesicles entering the retrograde axonal transport (33). These studies might also reveal differences in the receptor use of different RV G proteins and help to clarify differential neuroinvasiveness of RV strains and serotypes.
Supplementary Material
Acknowledgments
We thank Nadin Hagendorf for perfect technical assistance.
This work was supported by the Deutsche Forschungsgemeinschaft through SFB 455 and SPP1175.
Footnotes
Published ahead of print on 10 October 2007.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Amano, T., E. Richelson, and M. Nirenberg. 1972. Neurotransmitter synthesis by neuroblastoma clones. Proc. Natl. Acad. Sci. USA 69258-263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Buchholz, U. J., S. Finke, and K. K. Conzelmann. 1999. Generation of bovine respiratory syncytial virus (BRSV) from cDNA: BRSV NS2 is not essential for virus replication in tissue culture, and the human RSV leader region acts as a functional BRSV genome promoter. J. Virol. 73251-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Coulon, P., J. P. Ternaux, A. Flamand, and C. Tuffereau. 1998. An avirulent mutant of rabies virus is unable to infect motoneurons in vivo and in vitro. J. Virol. 72273-278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dalton, K. P., and J. K. Rose. 2001. Vesicular stomatitis virus glycoprotein containing the entire green fluorescent protein on its cytoplasmic domain is incorporated efficiently into virus particles. Virology 279414-421. [DOI] [PubMed] [Google Scholar]
- 5.Das, S. C., D. Nayak, Y. Zhou, and A. K. Pattnaik. 2006. Visualization of intracellular transport of vesicular stomatitis virus nucleocapsids in living cells. J. Virol. 806368-6377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duprex, W. P., F. M. Collins, and B. K. Rima. 2002. Modulating the function of the measles virus RNA-dependent RNA polymerase by insertion of green fluorescent protein into the open reading frame. J. Virol. 767322-7328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Etessami, R., K. K. Conzelmann, B. Fadai-Ghotbi, B. Natelson, H. Tsiang, and P. E. Ceccaldi. 2000. Spread and pathogenic characteristics of a G-deficient rabies virus recombinant: an in vitro and in vivo study. J. Gen. Virol. 812147-2153. [DOI] [PubMed] [Google Scholar]
- 8.Finke, S., K. Brzozka, and K. K. Conzelmann. 2004. Tracking fluorescence-labeled rabies virus: enhanced green fluorescent protein-tagged phosphoprotein P supports virus gene expression and formation of infectious particles. J. Virol. 7812333-12343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Finke, S., and K. K. Conzelmann. 1999. Virus promoters determine interference by defective RNAs: selective amplification of mini-RNA vectors and rescue from cDNA by a 3′ copy-back ambisense rabies virus. J. Virol. 733818-3825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Finke, S., and K. K. Conzelmann. 2005. Replication strategies of rabies virus. Virus Res. 111120-131. [DOI] [PubMed] [Google Scholar]
- 11.Finke, S., and K. K. Conzelmann. 2005. Recombinant rhabdoviruses: vectors for vaccine development and gene therapy. Curr. Top Microbiol. Immunol. 292165-200. [DOI] [PubMed] [Google Scholar]
- 12.Finke, S., R. Mueller-Waldeck, and K. K. Conzelmann. 2003. Rabies virus matrix protein regulates the balance of virus transcription and replication. J. Gen. Virol. 841613-1621. [DOI] [PubMed] [Google Scholar]
- 13.Gaudin, Y. 2000. Rabies virus-induced membrane fusion pathway. J. Cell Biol. 150601-612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gaudin, Y., R. W. Ruigrok, C. Tuffereau, M. Knossow, and A. Flamand. 1992. Rabies virus glycoprotein is a trimer. Virology 187627-632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Greber, U. F., and M. Way. 2006. A superhighway to virus infection. Cell 124741-754. [DOI] [PubMed] [Google Scholar]
- 16.Grouse, L. D., B. K. Schrier, C. H. Letendre, M. Y. Zubairi, and P. G. Nelson. 1980. Neuroblastoma differentiation involves both the disappearance of old and the appearance of new poly(A)+ messenger RNA sequences in polyribosomes. J. Biol. Chem. 2553871-3877. [PubMed] [Google Scholar]
- 17.Jacob, Y., H. Badrane, P. E. Ceccaldi, and N. Tordo. 2000. Cytoplasmic dynein LC8 interacts with lyssavirus phosphoprotein. J. Virol. 7410217-10222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koser, M. L., J. P. McGettigan, G. S. Tan, M. E. Smith, H. Koprowski, B. Dietzschold, and M. J. Schnell. 2004. Rabies virus nucleoprotein as a carrier for foreign antigens. Proc. Natl. Acad. Sci. USA 1019405-9410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lafon, M. 2005. Modulation of the immune response in the nervous system by rabies virus. Curr. Top Microbiol. Immunol. 289239-258. [DOI] [PubMed] [Google Scholar]
- 20.Lentz, T. L., T. G. Burrage, A. L. Smith, J. Crick, and G. H. Tignor. 1982. Is the acetylcholine receptor a rabies virus receptor? Science 215182-184. [DOI] [PubMed] [Google Scholar]
- 21.Mazarakis, N. D., M. Azzouz, J. B. Rohll, F. M. Ellard, F. J. Wilkes, A. L. Olsen, E. E. Carter, R. D. Barber, D. F. Baban, S. M. Kingsman, A. J. Kingsman, K. O'Malley, and K. A. Mitrophanous. 2001. Rabies virus glycoprotein pseudotyping of lentiviral vectors enables retrograde axonal transport and access to the nervous system after peripheral delivery. Hum. Mol. Genet. 102109-2121. [DOI] [PubMed] [Google Scholar]
- 22.Mebatsion, T. 2001. Extensive attenuation of rabies virus by simultaneously modifying the dynein light chain binding site in the P protein and replacing Arg333 in the G protein. J. Virol. 7511496-11502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mebatsion, T., and K. K. Conzelmann. 1996. Specific infection of CD4+ target cells by recombinant rabies virus pseudotypes carrying the HIV-1 envelope spike protein. Proc. Natl. Acad. Sci. USA 9311366-11370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mebatsion, T., S. Finke, F. Weiland, and K. K. Conzelmann. 1997. A CXCR4/CD4 pseudotype rhabdovirus that selectively infects HIV-1 envelope protein-expressing cells. Cell 90841-847. [DOI] [PubMed] [Google Scholar]
- 25.Mebatsion, T., M. Konig, and K. K. Conzelmann. 1996. Budding of rabies virus particles in the absence of the spike glycoprotein. Cell 84941-951. [DOI] [PubMed] [Google Scholar]
- 26.Mebatsion, T., F. Weiland, and K. K. Conzelmann. 1999. Matrix protein of rabies virus is responsible for the assembly and budding of bullet-shaped particles and interacts with the transmembrane spike glycoprotein G. J. Virol. 73242-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mentis, G. Z., M. Gravell, R. Hamilton, N. A. Shneider, M. J. O'Donovan, and M. Schubert. 2006. Transduction of motor neurons and muscle fibers by intramuscular injection of HIV-1-based vectors pseudotyped with select rabies virus glycoproteins. J. Neurosci. Methods 157208-217. [DOI] [PubMed] [Google Scholar]
- 28.Morimoto, K., H. D. Foley, J. P. McGettigan, M. J. Schnell, and B. Dietzschold. 2000. Reinvestigation of the role of the rabies virus glycoprotein in viral pathogenesis using a reverse genetics approach. J. Neurovirol. 6373-381. [DOI] [PubMed] [Google Scholar]
- 29.Rager, M., S. Vongpunsawad, W. P. Duprex, and R. Cattaneo. 2002. Polyploid measles virus with hexameric genome length. EMBO J. 212364-2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rasalingam, P., J. P. Rossiter, T. Mebatsion, and A. C. Jackson. 2005. Comparative pathogenesis of the SAD-L16 strain of rabies virus and a mutant modifying the dynein light chain binding site of the rabies virus phosphoprotein in young mice. Virus Res. 11155-60. [DOI] [PubMed] [Google Scholar]
- 31.Raux, H., A. Flamand, and D. Blondel. 2000. Interaction of the rabies virus P protein with the LC8 dynein light chain. J. Virol. 7410212-10216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reagan, K. J., and W. H. Wunner. 1985. Rabies virus interaction with various cell lines is independent of the acetylcholine receptor. Arch. Virol. 84277-282. [DOI] [PubMed] [Google Scholar]
- 33.Riccio, A., B. A. Pierchala, C. L. Ciarallo, and D. D. Ginty. 1997. An NGF-TrkA-mediated retrograde signal to transcription factor CREB in sympathetic neurons. Science 2771097-1100. [DOI] [PubMed] [Google Scholar]
- 34.Rigaut, K. D., D. E. Birk, and J. Lenard. 1991. Intracellular distribution of input vesicular stomatitis virus proteins after uncoating. J. Virol. 652622-2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Robison, C. S., and M. A. Whitt. 2000. The membrane-proximal stem region of vesicular stomatitis virus G protein confers efficient virus assembly. J. Virol. 742239-2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roche, S., and Y. Gaudin. 2002. Characterization of the equilibrium between the native and fusion-inactive conformation of rabies virus glycoprotein indicates that the fusion complex is made of several trimers. Virology 297128-135. [DOI] [PubMed] [Google Scholar]
- 37.Roche, S., and Y. Gaudin. 2004. Evidence that rabies virus forms different kinds of fusion machines with different pH thresholds for fusion. J. Virol. 788746-8752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schnell, M. J., T. Mebatsion, and K. K. Conzelmann. 1994. Infectious rabies viruses from cloned cDNA. EMBO J. 134195-4203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shakin-Eshleman, S. H., A. T. Remaley, J. R. Eshleman, W. H. Wunner, and S. L. Spitalnik. 1992. N-linked glycosylation of rabies virus glycoprotein. Individual sequons differ in their glycosylation efficiencies and influence on cell surface expression. J. Biol. Chem. 26710690-10698. [PubMed] [Google Scholar]
- 40.Shaner, N. C., R. E. Campbell, P. A. Steinbach, B. N. Giepmans, A. E. Palmer, and R. Y. Tsien. 2004. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotechnol. 221567-1572. [DOI] [PubMed] [Google Scholar]
- 41.Smith, G. A., L. Pomeranz, S. P. Gross, and L. W. Enquist. 2004. Local modulation of plus-end transport targets herpesvirus entry and egress in sensory axons. Proc. Natl. Acad. Sci. USA 10116034-16039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Smith, M. E., M. Koser, S. Xiao, C. Siler, J. P. McGettigan, C. Calkins, R. J. Pomerantz, B. Dietzschold, and M. J. Schnell. 2006. Rabies virus glycoprotein as a carrier for anthrax protective antigen. Virology 353344-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tan, G. S., M. A. Preuss, J. C. Williams, and M. J. Schnell. 2007. The dynein light chain 8 binding motif of rabies virus phosphoprotein promotes efficient viral transcription. Proc. Natl. Acad. Sci. USA 1047229-7234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thoulouze, M. I., M. Lafage, M. Schachner, U. Hartmann, H. Cremer, and M. Lafon. 1998. The neural cell adhesion molecule is a receptor for rabies virus. J. Virol. 727181-7190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tuffereau, C., J. Benejean, D. Blondel, B. Kieffer, and A. Flamand. 1998. Low-affinity nerve-growth factor receptor (P75NTR) can serve as a receptor for rabies virus. EMBO J. 177250-7259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Whitt, M. A., L. Buonocore, C. Prehaud, and J. K. Rose. 1991. Membrane fusion activity, oligomerization, and assembly of the rabies virus glycoprotein. Virology 185681-688. [DOI] [PubMed] [Google Scholar]
- 47.Wickersham, I. R., S. Finke, K. K. Conzelmann, and E. M. Callaway. 2007. Retrograde neuronal tracing with a deletion-mutant rabies virus. Nat. Methods 447-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wickersham, I. R., D. C. Lyon, R. J. Barnard, T. Mori, S. Finke, K. K. Conzelmann, J. A. Young, and E. M. Callaway. 2007. Monosynaptic restriction of transsynaptic tracing from single, genetically targeted neurons. Neuron 53639-647. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.