Abstract
In 2001 and 2003, we isolated two H5N1 viruses, A/swine/Fujian/1/01 (SW/FJ/01) and A/swine/Fujian/1/03 (SW/FJ/03), from pigs in Fujian Province, southern China. Genetically, these two viruses are similar, although the NS gene of the SW/FJ/03 virus has a 15-nucleotide deletion at coding positions 612 to 626. The SW/FJ/01 virus is highly lethal for chickens, whereas the SW/FJ/03 virus is nonpathogenic for chickens when administrated intravenously or intranasally. To understand the molecular basis for the difference in virulence, we used reverse genetics to create a series of single-gene recombinants of both viruses. We found that a recombinant virus containing the mutated NS gene from the SW/FJ/03 virus in the SW/FJ/01 virus background was completely attenuated in chickens. We also found that viruses expressing the mutant NS1 protein of SW/FJ/03 did not antagonize the induction of interferon (IFN) protein. Conversely, only the recombinant virus containing the wild-type SW/FJ/01 NS gene in the SW/FJ/03 background was lethal in chickens and antagonized IFN protein levels. Further, we proved that the NS1 genes of the two viruses differ in their stabilities in the host cells and in their abilities to interact with the chicken cleavage and polyadenylation specificity factor. These results indicate that the deletion of amino acids 191 to 195 of the NS1 protein is critical for the attenuation of the SW/FJ/03 virus in chickens and that this deletion affects the ability of the virus to antagonize IFN induction in host cells.
In addition to their natural hosts, i.e., wild birds, influenza A viruses can infect numerous animal species, including humans, pigs, and domestic fowl. Based on antigenic differences in the two surface glycoproteins, hemagglutinin (HA) and neuraminidase (NA), the influenza viruses are divided into 16 HA subtypes and 9 NA subtypes (7). All of the subtypes have been detected in wild birds, although only a few subtypes have crossed the species barrier and spread widely among domestic poultry. Most avian influenza viruses display low pathogenicity in chickens; however, some of the H5 and H7 subtypes have caused significant outbreaks in poultry and wild birds (5) and have posed a significant threat for human public health. The elucidation of factors that determine influenza virus host range and virulence is therefore an area of research that has important implications for public health and agriculture.
When influenza viruses are transmitted to a novel host, mutations occur in the genome to enable the virus to adapt. When lowly pathogenic avian influenza viruses circulate in domestic poultry, they acquire additional basic amino acids in their cleavage sites (14, 19), which are associated with increased pathogenicity for H5 and H7 subtypes. The amino acid at position 627 of the PB2 gene changed from serine (E) to lysine (K) when an H7 virus was passaged in mammalian cell lines (35) or after replication in mice (13, 22). This change in PB2 amino acid sequence influenced the outcome of the virus infection in mice (13). The amino acid at position 701 of the PB2 gene also plays a crucial role in the replication and lethality of H5 and H7 subtype viruses in mice (9, 22). Recent studies demonstrated that the NS1 gene is important for the virulence of several subtypes of influenza virus in mice and pigs (30, 34). The amino acid at position 149 of NS1 correlates with the replication phenotype of a goose H5N1 avian influenza virus in chickens (23).
For this study, we performed extensive genetic and biological analyses of two H5N1 viruses that were isolated from pigs in the Fujian province of southern China. Our data indicate that these two viruses are closely related to each other and to a duck virus, A/duck/Zhejiang/52/00 (DK/ZJ/00). These two swine viruses exhibit different virulence properties in chickens, and we used reverse genetics to determine the genetic basis for this difference.
MATERIALS AND METHODS
Cells and viruses.
Chicken embryo fibroblasts (CEFs) were prepared from 10-day-old specific-pathogen-free (SPF) chicken embryos. The CEFs and human embryonic kidney (293T) cells were maintained in minimum essential medium containing 10% fetal bovine serum (Invitrogen Corp., CA).
The two H5N1 swine influenza viruses, A/swine/Fujian/1/01 (SW/FJ/01) and A/swine/Fujian/1/03 (SW/FJ/01), were isolated during routine surveillance in the Fujian province of southern China in 2001 and 2003, respectively (20). Virus stocks were propagated in 10-day-old SPF embryonated chicken eggs and stored at −70°C until they were used for RNA extraction and animal studies. Recombinant vesicular stomatitis virus (VSV) expressing green fluorescent protein (GFP) was generated as described previously by inserting the G protein gene of VSV into the VSVΔG*GFP vector (36), using reverse genetics (18).
Construction of plasmids.
We used an eight-plasmid reverse genetics system for virus rescue. We inserted the cDNA derived from the SW/FJ/01 or SW/FJ/03 viral genes between the ribozyme and promoter sequences of polymerase I of the mRNA-viral RNA bidirectional transcription vector pBD as described previously (22), Briefly, we used a set of primers with two extra nucleotides (CC and TT) at the 5′ ends of the forward and reverse primers to amplify the full-length cDNAs of the viruses. The primer sequences are listed in Table 1. We then treated the PCR products with T4 polymerase (New England Biolabs, Beverly, MA) in the presence of 100 mM dTTP and 100 mM dCTP for 10 min at 12°C to generate a CC and a TT overhang at the two ends of the viral insert. We cut plasmid pBD with SapI (New England Biolabs) and then partially filled it in by treatment with Klenow fragment in the presence of 30 mM dATP and 30 mM dGTP at 25°C for 30 min to create 5′ GG and 5′ AA ends to match the ends of the PCR products.
TABLE 1.
Primers used in this study
| Purpose | Primer sequence (5′-3′)a
|
|
|---|---|---|
| Forward | Reverse | |
| PB2 amplification | CCAGCAAAAGCAGGTCAAATATATTC | TTAGTAGAAACAAGGTCGTTTTTAAACAAT |
| PB1 amplification | CCAGCAAAAGCAGGCAAACCATT | TTAGTAGAAACAAGGCATTTTTTCAT |
| PA amplification | CCAGCAAAAGCAGGTACTGATCCAAAAT | TTAGTAGAAACAAGGTACTTTTTTGGAC |
| HA amplification | CCAGCAAAAGCAGGGGTCTAATTTGCC | TTAGTAGAAACAAGGGTGTTTTTAACTAC |
| NP amplification | CCAGCAAAAGCAGGGTAGATAATCACTC | TTAGTAGAAACAAGGGTATTTTTCTTTA |
| NA amplification | CCAGCAAAAGCAGGAGTTCAAAATGAATC | TTAGTAGAAACAAGGAGTTTTTTGAACA |
| M amplification | CCAGCAAAAGCAGGTAGATGTTGAAAG | TTAGTAGAAACAAGGTAGTTTTTTAC |
| NS amplification | CCAGCAAAAGCAGGGTGACAAAAAC | TTAGTAGAAACAAGGGTGTTTTTTATC |
| NS1 expression | GCCGCCACCATGGATTCCAACACTGTGTCAAGC | GCTATCAAACTTCTGACTCAATTGTTCTC |
| NS2 splice acceptor site mutation | ATGAGGATGTGAAAAATGCAATTGG | CCAATTGCATTTTTCACATCCTCAT |
| NS1 HA tag introduction | GCCACCATGACCAGCTACCCATACGATGTTCCAGATTACGCTGCAGATTCCAACACTGTGTCAAGC | TTAGTAGAAACAAGGGTGTTTTTTATC |
| Chicken CPSF amplification | GCCACCATGCAGGAGCTCAT | CTTTCACTGTCCACTGAG |
| CPSF Flag tag introduction | GCTGCCACCATGCAGGAGCTCATCG | GACTTACTACTTGTCATCGTCATCCTTGTAGTCGATGTCATGATCTTTATAATCACCGTCATGGTCATGGTCTTTGTAGTCAGCCTGTCCACTGAGAAAGGCCAG |
| Chicken IFN-α amplification | GGGTACGACATCCTGTTGCTC | CGGCTGATCCGGTTGAGGAG |
| Chicken IFN-β amplification | GCCACAGCCTCCTCAACCAGAT | CAACGTCCCAGGTACAAGCACT |
| Chicken β-actin amplification | AGACAGCTACGTTGGTGATGAAGCCCAG | GTTGAAGGTAGTTTCATGGATACCACAG |
Nucleotides that have been changed are shown in italics.
To generate the NS1 expression constructs, cDNAs encoding the NS1 proteins of SW/FJ/01 and SW/FJ/03 were amplified using the primers listed in Table 1 and were cloned into pBlueScript II KS (Clontech) under the control of the T7 promoter, creating pBlueNS1-01 and pBlueNS1-03, respectively. The NS1-encoding cDNAs contained silent mutations in the splice acceptor site to avoid splicing events and expression of NS2. To generate N-terminally HA-tagged full-length NS1 expression constructs of SW/FJ/01 and SW/FJ/03, the open reading frames encoding NS1 were amplified using primers that contained the HA tag sequence (listed in Table 1). The PCR products were cloned into pBluescript, creating pBlueNS1-01-HA and pBlueNS1-03-HA, using the SmaI restriction site. The 30-kDa subunit of the chicken cleavage and polyadenylation specificity factor (CPSF) gene from CEFs was cloned by PCR, using a primer pair specific for the chicken cDNA (GenBank accession no. XM-414800) (primer sequences are listed in Table 1). The product was cloned into pBlueScript II KS plasmid, creating pBlue-CPSF, by using the SmaI restriction site. To generate a C-terminally Flag-tagged CPSF construct, the CPSF gene was amplified from pBlue-CPSF by using the pair of primers listed in Table 1, blunt inserted into the pBlueScript II KS plasmid, and then subcloned into the pCAGGS plasmid, using the SmaI and XhoI sites. All constructs were confirmed by sequence analysis.
Virus rescue.
Monolayers of 293T cells at 80% to 90% confluence in six-well plates were transfected with 5 μg of the eight plasmids (about 0.6 μg/plasmid), using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. Briefly, DNA and transfection reagent were mixed (2 μl of Lipofectamine 2000 per μg of DNA), incubated at room temperature for 30 min, and added to the cells. Sixteen hours later, the DNA-transfection reagent mixture was replaced by Opti-MEM (Invitrogen) containing 0.3% bovine serum albumin and 0.01% fetal calf serum. Forty-eight hours after the transfection, the supernatants were harvested and inoculated into embryonated eggs for virus propagation. Viruses were detected by using a hemagglutination assay and were fully sequenced to ensure the absence of unwanted mutations.
Sequence analyses.
Viral RNA was extracted with an RNeasy mini kit (Qiagen, Valencia, CA) and was reverse transcribed. PCR amplification was performed by using segment-specific primers (primer sequences are available upon request). The PCR products were purified with a QIAquick PCR purification kit (Qiagen) and sequenced by using a CEQ DTCS quick-start kit on a CEQ 8000 DNA sequencer (Beckman Coulter). Sequence data were compiled using the SEQMAN program of the DNASTAR package.
Determination of IFN secretion.
Levels of interferon (IFN) secreted by virus-infected cells were determined as previously described (6, 23, 29, 34). Monolayers of CEFs at 80% confluence were infected at a multiplicity of infection (MOI) of 1 with the test viruses. The supernatants were harvested at 20 h postinfection (p.i.). Viruses present in the supernatant were UV inactivated by placing samples on ice 70 cm below a 30-W UV lamp for 20 min, with constant stirring. The UV-inactivated supernatant was added to fresh confluent CEFs and incubated for 24 h. The cells were then infected at an MOI of 0.01 with VSV-GFP. At 20 h p.i., the cells were examined for GFP expression under a fluorescence microscope (Leica, Germany).
Detection of IFN-α/β mRNA in influenza virus-infected cells by RT-PCR.
CEFs were infected at an MOI of 1 with different H5N1 swine influenza viruses. Total RNA was extracted using an RNeasy mini kit (Qiagen, Valencia, CA). Reverse transcription-PCR (RT-PCR) was conducted by using an Access RT-PCR system (Promega, Madison, WI) with specific pairs of primers (Table 1) for chicken IFN-α and chicken IFN-β mRNAs (GenBank accession numbers AB021153 and AY831397). As a control, we used a pair of specific primers (Table 1) to amplify a 660-bp fragment of the chicken β-actin gene. The products were sequenced and confirmed to be derived from the expected mRNAs.
Antisera.
To generate antibodies to the SW/FJ/01 NS1 protein, we produced the protein in Escherichia coli BL21 (Invitrogen Corp.) cells from the plasmid pET-32a (+) NS1. This plasmid encodes a truncated SW/FJ/01 NS1 protein that spans amino acids 28 to 221, with histidine tags fused to both the N and C termini. Once it was expressed, the truncated NS1 protein was purified using a Ni-nitrilotriacetic acid purification system (Invitrogen) and then injected into rabbits (Harbin Experimental Animal Center). Chicken antisera raised against the SW/FJ/03 virus were used to detect the other influenza virus proteins.
Western blot analysis.
Cell extracts were made in lysis buffer containing 0.5% NP-40, 150 mM NaCl, 20 mM HEPES, pH 7.4, 1 mM EDTA, 1 mM EGTA, 10% glycerol, and Complete protease inhibitor cocktail (Roche). The protein lysate (10 or 20 μl) was loaded onto a 10% sodium dodecyl sulfate (SDS)-polyacrylamide gel. Separated proteins were probed for the NS1 protein with a rabbit polyclonal antibody. As controls, the M1 protein of influenza virus and the β-actin protein were also detected by using monoclonal antibodies directed against these molecules. Antibody binding was visualized with DAB (3,3′-diaminobenzidine) reagent after incubation with peroxidase-conjugated secondary antibodies.
Pulse-chase radioactive labeling and immunoprecipitation.
NS1 proteins were expressed by using a vaccinia virus T7 transient expression system as described previously (2, 42). Briefly, HeLa cells seeded in 35-mm dishes were infected with vTF7-3, a recombinant vaccinia virus expressing the T7 polymerase, at a multiplicity of 10 for 1 h. After infection, the cells were transfected with plasmid DNA constructs in mixture with Lipofectin. At 10 h p.i., cells were starved in Met- and Cys-deficient Eagle's medium for 30 min and then pulse labeled with 200 μCi of a mixture of 35S-labeled methionine and cysteine ([35S]Met-Cys) (Amersham) in 600 ml of Met- and Cys-deficient Eagle's medium for 30 min. After being pulse labeled, cells were lysed with lysis buffer (150 mM NaCl, 50 mM Tris-HCl, 1 mM EDTA, 1% Triton X-100, 1% sodium deoxycholate [pH 7.5]) or chased in complete medium (Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum) for 4 or 7 h, followed by lysis. The cell lysates collected after pulse labeling or at different chase time points were immunoprecipitated with rabbit antibodies against NS1 and with protein A-agarose beads (Pierce) at 4°C overnight. Protein samples were prepared with sample buffer (125 mM Tris-HCl [pH 7.5], 4% SDS, and 20% glycerol plus 10% β-mercaptoethanol) and heated at 95°C for 5 min, followed by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) analysis.
NS1-CPSF coimmunoprecipitation.
To monitor the interaction of the different NS1 constructs with Flag-CPSF, HA-tagged NS1 cDNAs in pBluescript under the control of the T7 promoter were expressed using a TNT transcription/translation kit (Promega). The cDNA coding for Flag-CPSF was expressed by transfection into 293T cells. At 40 h posttransfection, cells were lysed in 50 mM Tris (pH 7.5), 200 mM NaCl, 0.5% NP-40, 0.2 mM EDTA, and 10% glycerol. The cleared cell lysates were incubated with the NS1 proteins in the presence of 1 μg of a monoclonal anti-Flag antibody (Sigma) and 25 μl of protein A-Sepharose beads (Amersham) for 2 h at 4°C. The beads were then washed three times in lysis buffer and incubated in SDS sample buffer for 5 min at 95°C. The proteins were subjected to 12% SDS-PAGE and transferred to membranes for Western blotting. The precipitated NS1 proteins were detected using a monoclonal antibody specific to the HA tag (Upstate Biotechnology, CA), and the precipitated Flag-CPSF was detected using a monoclonal antibody specific to Flag (Sigma).
Mouse experiments.
Groups of eight 6-week-old female BALB/c mice (Beijing Experimental Animal Center) were lightly anesthetized with CO2 and inoculated intranasally (i.n.) with 105.0, 106.0, or 107.0 50% egg infectious doses (EID50) of H5N1 influenza virus in a volume of 50 μl. Three mice in each group were euthanized on day 3 p.i., and the lungs, kidneys, spleens, and brains were collected for virus titration in eggs. The lower limit of virus detection was 0.5 log10 EID50 per ml of tissue homogenate. The remaining five mice in each group were monitored daily for 14 days for weight loss and mortality. To determine the 50% mouse lethal dose (MLD50) of viruses, groups of five mice were inoculated i.n. with 10-fold serial dilutions containing 103 to 107 EID50 of virus in a 50-μl volume. The MLD50 was calculated by the method of Reed and Muench (31).
Chicken experiments.
To determine the pathogenicity of the viruses, the intravenous pathogenicity index (IVPI) was determined according to the recommendations of the Office International Des Epizooties (27). Groups of 10 6-week-old SPF White Leghorn chickens housed in isolator cages were inoculated intravenously (i.v.) with 0.2 ml of a 1:10 dilution of bacterium-free allantoic fluid containing virus. Serum conversion of the surviving birds on day 10 p.i. was confirmed by the hemagglutination inhibition test. Ten additional chickens were inoculated i.n. with 106 EID50 of each virus in a 0.1-ml volume. On day 3 p.i., three birds in each group were euthanized, and the lungs, bursae, kidneys, brains, pancreases, and spleens were collected for virus isolation. Oropharyngeal and cloacal swabs were collected on day 3 p.i. from all birds for detection of virus shedding, and the remaining seven chickens were observed for signs of disease or death for 14 days.
RESULTS
Genetic analysis of the two H5N1 swine influenza viruses.
The H5N1 influenza viruses SW/FJ/01 and SW/FJ/03, which were isolated from pigs in Fujian Province, China, were sequenced and compared with available sequences of other H5N1 avian influenza viruses. Five amino acid differences between the two viruses were identified and mapped to the PB2, HA, and M2 genes. Both viruses have six basic amino acids in the connecting peptide of HA. The PB1, PA, NP, and NA genes of the two viruses are identical at the amino acid level, although a few silent mutations were detected. No deletions were detected in the region of the gene that encodes the NA stalk. There is a 15-nucleotide deletion spanning positions 263 to 277 in the NS genes of both viruses that has also been observed in other H5N1 viruses. However, there is a unique deletion of 15 nucleotides at positions 612 to 626 in the SW/FJ/03 virus, which results in a five-amino-acid deletion in both the NS1 and NS2 proteins (Table 2). We found that the two swine viruses are very closely related to the previously reported H5N1 duck virus A/duck/Zhejiang/52/2000 (DK/ZJ/00) (Table 2). There are fewer than 30 amino acid differences between the H5N1 swine influenza viruses and the DK/ZJ/00 virus, suggesting that DK/ZJ/00 is a close ancestor of these H5N1 swine influenza viruses.
TABLE 2.
Amino acid differences between two H5N1 swine influenza viruses and the duck virus DK/ZJ/00
| Protein | Amino acid position | Amino acid in virusa
|
||
|---|---|---|---|---|
| DK/ZJ/00 | SW/FJ/01 | SW/FJ/03 | ||
| PB2 | 51 | V | M | M |
| 92 | P | S | S | |
| 164 | L | M | M | |
| 562 | I | I | S | |
| 679 | L | P | P | |
| 725 | P | L | L | |
| PB1 | 456 | L | H | H |
| 486 | L | R | R | |
| PA | 54 | V | I | I |
| 330 | I | V | V | |
| 384 | Y | C | C | |
| 459 | V | I | I | |
| HA | 16 | S | G | G |
| 100 | N | S | S | |
| 168 | K | K | E | |
| 254 | S | A | A | |
| 444 | L | I | L | |
| NP | 65 | S | R | R |
| 257 | T | I | I | |
| 284 | V | A | A | |
| NA | 34 | V | I | I |
| M2 | 26 | L | S | L |
| 65 | T | T | A | |
| NS1 | 18 | V | I | I |
| 69 | P | L | L | |
| 88 | I | M | M | |
| 191-195 | EALQR | EALQR | ||
| NS2 | 39-43 | KLYRD | KLYRD | |
The amino acids that are different between the two swine influenza viruses are shown in italics.
Biological properties of the two H5N1 swine influenza viruses.
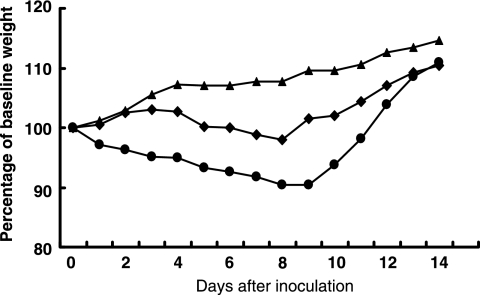
Our previous study demonstrated that H5N1 viruses isolated from healthy ducks in southern China have a clear temporal pattern of increasing pathogenicity in mice (4). Several early isolates, including DK/ZJ/00, cannot replicate in mice (4). Here we investigated the replication and virulence of the two H5N1 swine influenza viruses and DK/ZJ/00, using the previously described mouse model (4). When 6-week-old BALB/c mice were inoculated i.n. with different doses (105 to 107 EID50) of virus, the two swine influenza viruses replicated well in the lungs without prior adaptation, with titers ranging from 3.8 to 5.5 log EID50. The viruses were not detected in other tested organs, including the brain, spleen, and kidneys (Table 3). The SW/FJ/01 virus caused about 10% weight loss and killed the mice at high doses, with an MLD50 of 5.7 log EID50 (Fig. 1). Interestingly, the SW/FJ/03 virus killed only one of the five mice at the high dose of 107 EID50. As reported previously (4), DK/ZJ/00 did not cause disease or death and was not detected in any organs tested (Fig. 1; Table 3).
TABLE 3.
Replication and lethality of H5N1 swine influenza viruses and a duck virus in mice
| Virus | Virus titer in mouse lungs on day 3 p.i.a
|
MLD50 (log EID50) | ||
|---|---|---|---|---|
| 105 EID50 | 106 EID50 | 107 EID50 | ||
| DK/ZJ/00 | <0.5 | <0.5 | <0.5 | >7.5 |
| SW/FJ/01 | 4.7 ± 0.4 | 5.5 ± 0.3 | 4.6 ± 0.3 | 5.7 |
| SW/FJ/03 | 3.8 ± 0.3 | 4.8 ± 0.5 | 4.5 ± 0.7 | 7.3 |
Data are means ± standard deviations.
FIG. 1.
Weight changes in mice inoculated with H5N1 swine influenza viruses and a duck influenza virus. Groups of five mice were inoculated i.n. with 106 EID50 (in 50 μl) of DK/ZJ/00 (▴), SW/FJ/01(•), or SW/FJ/03 (⧫) and weighed daily for 14 days.
SW/FJ/01 and SW/FJ/03 have multiple basic amino acids in the connecting peptide of HA, which has been proven to be a marker for lethality in chickens. To confirm the pathogenicity of these two viruses, we tested their IVPI in SPF chickens. The IVPI for SW/FJ/01 was 2.5, with 10 inoculated chickens dying within 3 days (Table 4). In contrast, SW/FJ/03 caused only mild disease in the chickens inoculated from days 2 to 4 p.i., and all of the chickens survived over the observation period. The IVPI for SW/FJ/03 was 0.3, and all of the chickens seroconverted (Table 4).
TABLE 4.
Disease and death caused by H5N1 viruses in chickens after i.v. inoculationa
| Virus | Titer (log EID50/ml) | No. of chickens with manifestation
|
IVPI | |
|---|---|---|---|---|
| S/D/totalb | SC/totalc | |||
| SW/FJ/01 | 7.7 | 0/10/10 | / | 2.5 |
| SW/FJ/03 | 8.0 | 10/0/10 | 10/10 | 0.3 |
| R-SW/FJ/01 | 8.0 | 0/10/10 | / | 2.2 |
| R-SW/FJ/03 | 8.3 | 10/0/10 | 10/10 | 0.2 |
| SW/FJ/01-03PB2 | 8.3 | 0/10/10 | / | 2.1 |
| SW/FJ/01-03HA | 8.3 | 0/10/10 | / | 2.5 |
| SW/FJ/01-03M | 8.0 | 0/10/10 | / | 1.9 |
| SW/FJ/01-03NS | 8.3 | 4/0/10 | 10/10 | 0.2 |
| SW/FJ/03-01PB2 | 7.8 | 10/0/10 | 10/10 | 0.2 |
| SW/FJ/03-01HA | 8.3 | 10/0/10 | 10/10 | 0.3 |
| SW/FJ/03-01M | 7.8 | 10/0/10 | 10/10 | 0.2 |
| SW/FJ/03-01NS | 8.0 | 0/10/10 | / | 1.9 |
Six-week-old White Leghorn chickens housed in high-efficiency particulate air-filtered isolators were inoculated i.v. with 0.2 ml of a 1:10 dilution of bacterium-free allantoic fluid containing virus for IVPI testing, based on the Office International Des Epizooties recommendations.
S/D/total, numbers of chickens that were sick (S) and died (D) as well as total number of chickens during the observation period. Birds that showed disease signs, such as depression and ruffled feathers, but recovered at the end of the observation were counted as sick animals.
SC/total, number of chickens that seroconverted/total number of chickens at the end of the observation period. /, all birds died by the end of the observation period.
The SW/FJ/01 virus replicated systemically in chickens inoculated i.n. with 106 EID50 and could be detected on day 3 p.i. in any organ tested, including the lungs, bursa, kidneys, brain, pancreas, and spleen (Table 5). Virus shedding was detected in both pharyngeal and cloacal swabs. By 6 days p.i., all of the birds had died. In the SW/FJ/03-inoculated chickens, virus was also detected in any organ tested; however, the virus titers were significantly lower than those observed in chickens inoculated with SW/FJ/01 (Table 5). Virus shedding was not detected for any of the chickens. None of the chickens showed disease signs, and all survived during the observation period (Table 5). These results indicate that the two swine viruses are genetically close but have different phenotypes with respect to their replication and lethality in chickens.
TABLE 5.
Replication and lethality of H5N1 swine influenza viruses in chickens after i.n. inoculationa
| Virus | No. of chickens with manifestation
|
Virus replication in organ on day 3 p.i. (log10 EID50/g)
|
Virus shedding on day 3 (log10 EID50/ml)
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| S/D/totalb | SC/totalc | Lung | Brain | Kidney | Spleen | Pancreas | Bursa | Pharynx | Cloaca | |
| SW/FJ/01 | 0/7/7 | / | 7.5 ± 0.7 | 7.3 ± 1.1 | 6.8 ± 0.1 | 6.6 ± 0.6 | 7.7 ± 0.5 | 6.7 ± 0.8 | 2.3 ± 1.5 | 1.1 ± 1.1 |
| SW/FJ/03 | 0/0/7 | 7/7 | 3.5 ± 0.5d | 1.2 ± 0.3d | 1.5 ± 0.7d | 1.3 ± 0.5d | 1.8 ± 0.5d | 1.1 ± 0.8d | <0.5 | <0.5 |
| R-SW/FJ/01 | 0/7/7 | / | 6.1 ± 0.3 | 6.0 ± 1.1 | 5.0 ± 0.4 | 5.1 ± 0.3 | 5.3 ± 0.5 | 5.7 ± 0.7 | 2.4 ± 0.7 | 1.3 ± 0.9 |
| R-SW/FJ/03 | 0/0/7 | 7/7 | 3.5 ± 0.3d | 0.8 ± 0.3d | 1.7 ± 0.9d | 1.3 ± 0.1d | 1.2 ± 0.8 | 0.8 ± 0.3d | <0.5 | <0.5 |
| SW/FJ/01-03PB2 | 1/6/7 | 1/7 | 6.8 ± 0.7 | 6.1 ± 1.0 | 5.3 ± 1.0 | 4.8 ± 1.2 | 4.6 ± 1.0 | 4.1 ± 1.0 | 1.9 ± 1.1 | 0.7 ± 0.5 |
| SW/FJ/01-03HA | 0/7/7 | / | 7.4 ± 0.1 | 7.0 ± 0.4 | 6.8 ± 0.6 | 4.9 ± 0.5 | 6.4 ± 1.1 | 6.0 ± 1.6 | 2.1 ± 0.9 | 1.0 ± 0.8 |
| SW/FJ/01-03M | 0/7/7 | / | 6.2 ± 0.4 | 5.4 ± 1.4 | 5.9 ± 0.3 | 5.6 ± 0.6 | 5.1 ± 0.8 | 5.9 ± 0.3 | 2.3 ± 0.6 | 1.4 ± 0.9 |
| SW/FJ/01-03NS | 0/0/7 | 7/7 | 2.7 ± 0.1d | <0.5 | 2.2 ± 0.2d | 2.0 ± 0.1d | 0.8 ± 0.3d | <0.5 | <0.5 | <0.5 |
| SW/FJ/03-01PB2 | 0/0/7 | 7/7 | 2.4 ± 0.4 | 0.7 ± 0.3 | 2.0 ± 0.1 | 1.9 ± 0.4 | 0.8 ± 0.3 | <0.5 | <0.5 | <0.5 |
| SW/FJ/03-01HA | 0/0/7 | 7/7 | 2.6 ± 0.4 | <0.5 | 1.9 ± 0.4 | 2.1 ± 0.2 | 0.7 ± 0.3 | <0.5 | <0.5 | <0.5 |
| SW/FJ/03-01M | 0/0/7 | 7/7 | 2.8 ± 0.4 | 1.3 ± 0.8 | 2.3 ± 0.1 | 2.4 ± 0.5 | 1.8 ± 0.3 | 0.7 ± 0.7 | <0.5 | <0.5 |
| SW/FJ/03-01NS | 2/5/7 | 2/7 | 4.3 ± 0.3 | 2.4 ± 0.1e | 3.7 ± 0.3e | 3.8 ± 0.3e | 2.6 ± 0.6 | 1.8 ± 1.1 | 1.2 ± 1.0 | 1.2 ± 0.9 |
Six-week-old SPF chickens were inoculated i.n. with 106 EID50 of each virus in a 0.1-ml volume; on day 3, three birds in each group were euthanized, and virus was titrated in samples of lung, bursa, kidney, brain, pancreas, and spleen in eggs. The lower limit of virus detection was 0.5 log10 EID50 per gram of tissue. Pharyngeal and cloacal swabs were collected from all birds on day 3. <0.5, virus was not detected in that sample. Data are means ± standard deviations.
S/D/total, numbers of chickens that were sick (S) and died (D) as well as total number of chickens during the observation period. Birds that showed disease signs, such as depression and ruffled feathers, but recovered at the end of the observation were counted as sick animals.
SC/total, number of chickens that seroconverted/total number of chickens at the end of the observation period. /, all birds died by the end of the observation period.
P < 0.01 compared with the titers in the corresponding organs of the SW/01- or R-SW/01-inoculated chickens.
P < 0.05 compared with the titers in the corresponding organs of the SW/03- or R-SW/03-inoculated chickens.
The NS gene is crucial for the difference in virulence between the H5N1 swine influenza viruses in chickens.
To explore the genetic basis for the different virulence levels of the two viruses in chickens, we used a reverse genetics system to generate single-gene reassortant viruses. To rescue the parent, wild-type recombinant viruses, we inserted the cDNA of each full-length RNA segment of the SW/FJ/01 and SW/FJ/03 viruses into the viral RNA-mRNA bidirectional expression plasmid pBD. The rescued SW/FJ/01 and SW/FJ/03 wild-type viruses were designated R-SW/FJ/01 and R-SW/FJ/03, respectively. The rescued R-SW/FJ/01 virus, like its wild-type counterpart, was highly pathogenic for chickens, and the IVPI was 2.2 (Table 4). The virus replicated systemically in i.n. inoculated chickens and killed all of the animals before day 4 p.i. (Table 5). The rescued R-SW/FJ/03 virus, also like its wild-type counterpart, caused only mild clinical signs in i.v. inoculated chickens (Table 4). Although it replicated systemically in the organs tested after i.n. inoculation, the virus titers were significantly lower than those in the R-SW/FJ/01 virus-inoculated chickens (Table 5). The rescued H5N1 viruses therefore possessed the biological characteristics of the original wild-type viruses.
We then generated four single-gene reassortants. First, we tested the effects of genes derived from SW/FJ/03 on the replication and virulence of SW/FJ/01. Each recombinant contained the PB2, HA, M, or NS gene from the lowly pathogenic SW/FJ/03 and the remaining seven genes from the highly pathogenic SW/FJ/01. The pathogenicity of the recombinant viruses was tested in chickens. As shown in Table 4, i.v. inoculation of the recombinant viruses containing the M (SW/FJ/01-03M), HA (SW/FJ/01-03HA), and PB2 (SW/FJ/01-03PB2) genes from the SW/FJ/03 virus in the SW/FJ/01 background led to the death of all 10 chickens by day 4 p.i., with the IVPI ranging from 1.9 to 2.5 (Table 4). However, the recombinant virus that contained the NS1 gene of SW/FJ/03 (SW/FJ/01-03NS) induced mild disease signs in the inoculated chickens, and all of the birds survived the observation period. The IVPI for SW/FJ/01-03NS was 0.2 (Table 4). When these viruses were inoculated into chickens i.n., the SW/FJ/01-03PB2, SW/FJ/01-03HA, and SW/FJ/01-03M viruses were detected in the lungs, kidneys, brains, and pancreases of the chickens killed on day 3 p.i., and the titers were similar to those in SW/FJ/01-inoculated chickens (Table 5). Low-titer virus shedding was detected for all of the chickens on day 3 p.i., and all of the chickens died by 4 days p.i. SW/FJ/01-03NS also replicated in all of the tested organs, but the virus titers were significantly lower than those for the SW/FJ/01- or R-SW/FJ/01-inoculated chickens.
We then tested the effects of genes derived from SW/FJ/01 on the replication and virulence of the SW/FJ/03 virus. We again generated four single-gene recombinant viruses, each containing the NS, HA, M, or PB2 gene from SW/FJ/1/01 and the remaining seven genes from SW/FJ/03. The viruses that carried the PB2 (SW/FJ/03-01PB2), HA (SW/FJ/03-01HA), and M (SW/FJ/03-01M) genes of SW/FJ/01 caused transient, mild disease in chickens, and the IVPI ranged from 0.2 to 0.3 (Table 2). The recombinant virus bearing the NS1 gene from SW/FJ/01 (SW/FJ/03-01NS), however, caused severe disease and killed all of the inoculated chickens by day 5 p.i. (Table 4). The IVPI was 1.9. When the chickens were inoculated i.n., SW/FJ/03-01PB2, SW/FJ/03-01HA, and SW/FJ/03-01M were detected in the tested organs, with titers similar to those in the SW/FJ/03-inoculated chickens, but virus shedding was not detected. All of the chickens survived during the observation period. The SW/FJ/03-01NS virus also replicated in all of the tested organs, but the titers were significantly higher than those for the SW/FJ/03-inoculated chickens. The virus killed five of seven chickens during the observation period (Table 5).
These results indicate that the NS gene plays an important role in determining the virulence of SW/FJ/01 and SW/FJ/03 in chickens. There is a 15-nucleotide deletion in the NS gene of the SW/FJ/03 virus, which results in a five-amino-acid deletion in both the NS1 and NS2 proteins (Table 2). These data suggest that the amino acid deletion in the NS1 and/or NS2 protein of SW/FJ/03 is crucial for the attenuation of this virus in chickens.
The deleted amino acids at positions 191 to 195 of the NS1 gene affect the ability of SW/FJ/01 and SW/FJ/03 to antagonize IFN-α/β production.
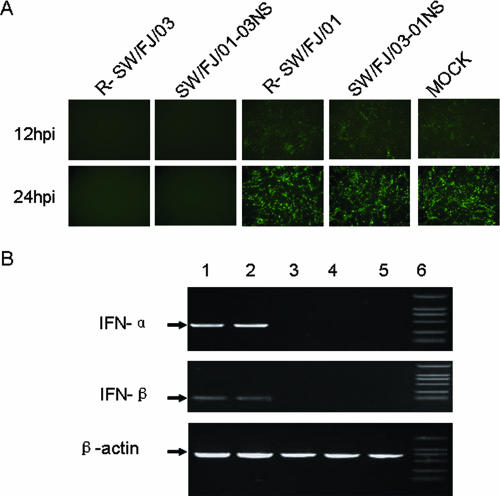
The NS1 protein of influenza virus has previously been shown to act as an IFN-α/β antagonist (10, 37). To examine whether the differences in the replication and virulence of SW/FJ/01 and SW/FJ/03 were directly correlated with their respective abilities to inhibit the IFN-α/β system, the production of IFN in cells infected with the different wild-type and reassortant viruses was assessed. To assay IFN production, supernatants from influenza virus-infected cells were tested for the ability to inhibit GFP expression and viral replication of recombinant VSV-GFP. VSV growth is very sensitive to type I IFN (6, 29, 34). Supernatants from mock-infected CEFs and R-SW/FJ/01- and SW/FJ/03-01NS-infected CEFs did not inhibit GFP expression (Fig. 2A), indicating that R-SW/FJ/01 and SW/FJ/03-01NS antagonized IFN production. However, the supernatants from CEFs infected with R-SW/FJ/03 and SW/FJ/01-03NS completely prevented the replication of VSV-GFP (Fig. 2A), indicating that viruses bearing the SW/FJ/03-like NS1 gene were unable to antagonize IFN production in these cells.
FIG. 2.
Induction of IFN-α/β in CEFs infected with different influenza viruses. (A) CEFs were infected with the indicated influenza viruses, and the UV-treated supernatants were used to examine IFN-mediated inhibition of VSV-GFP replication in CEFs as described in Materials and Methods. The cells were visualized by fluorescence microscopy at 12 and 24 h p.i. (B) RT-PCR analysis of IFN-α/β- and chicken β-actin-specific mRNAs from CEFs infected with R-SW/FJ/03 (lane 1), SW/FJ/01-03NS (lane 2), R-SW/FJ/01 (lane 3), or SW/FJ/03-01NS (lane 4) or from mock-infected CEFs (lane 5). Lane 6, marker.
To examine the effect of influenza virus infection on the synthesis of IFN-α/β mRNA, CEFs were infected with various influenza viruses and their RNAs were extracted 20 h after infection. The cDNAs of chicken IFN-α/β were amplified by using primers specific to either the IFN-α or the IFN-β gene. As shown in Fig. 2B, the cDNAs of both the IFN-α and IFN-β genes were amplified from the SW/FJ/03- and SW/FJ/01-03NS-inoculated CEFs but not from the CEFs inoculated with the SW/FJ/01 and SW/FJ/03-01NS viruses. The effects of these viruses on IFN-α/β mRNA production are consistent with their effects in the IFN-α/β bioassay. Western blot analysis revealed that the levels of the NS1 protein in the cells infected with virus containing the SW/FJ/01 NS gene were considerably higher than those in cells infected with virus containing the SW/FJ/03 NS gene (Fig. 3). However, no significant difference in the levels of M protein and β-actin were detected among the different virus-infected cells (Fig. 3).
FIG. 3.
Virus protein levels determined by Western blotting. Lysates of mock-infected cells (lane 1) or cells infected with R-SW/FJ/03 (lane 2), SW/FJ/01-03NS (lane 3), R-SW/FJ/01 (lane 4), or SW/FJ/03-01NS (lane 5) were incubated with rabbit anti-NS1 antiserum, a mouse monoclonal antibody to the viral M1 protein, or β-actin antiserum.
These results indicate that viruses bearing the NS1 gene of SW/FJ/01 are better able to antagonize the IFN-α/β response in CEFs than are viruses bearing the NS1 gene of SW/FJ/03. Since the NS1 genes of these two viruses differ only in the deletion of the amino acids at positions 191 to 195, this indicates that the five-amino-acid deletion in the NS1 gene is crucial for the reduced ability of the SW/FJ/03 virus to antagonize the host IFN-α/β response and for its attenuation in chickens.
The deleted amino acids at position 191 to 195 affect the stability of the NS1 protein and impair its interaction with chicken CPSF.
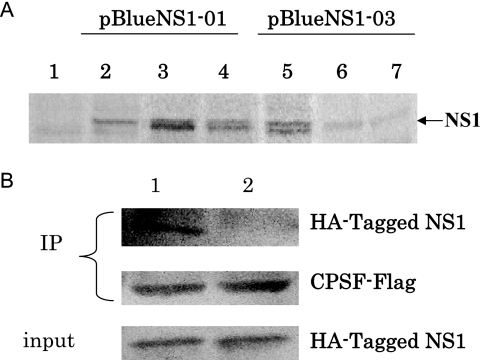
As shown in Fig. 3, the SW/FJ/01 NS1 protein was expressed at significantly higher levels than the SW/FJ/03 NS1 protein in virus-infected cells. To determine whether this was due to a reduced stability of the SW/FJ/03 NS1 protein, we carried out a pulse-chase radioactive labeling and immunoprecipitation analysis. The genes for the SW/FJ/01 and SW/FJ/03 NS1 proteins were cloned into the plasmid vector pBlueScript II KS under the control of the T7 promoter, and the NS1 proteins were expressed in HeLa cells by using the recombinant vaccinia virus T7 transient expression system (8). As shown in Fig. 4A, similar levels of labeled SW/FJ/01 and SW/FJ/03 NS1 proteins were detected in cells after pulse labeling. However, while the SW/FJ/01 NS1 protein largely maintained its level after 4- and 7-h chases, the amount of labeled SW/FJ/03 NS1 protein was significantly reduced after 4- and 7-h chases. This result indicates that the SW/FJ/03 NS1 protein, which contains a five-amino-acid deletion, is less stable than the SW/FJ/01 NS1 protein.
FIG. 4.
Stability of NS1 proteins and their interaction with chicken CPSF. (A) Analysis of NS1 expression by pulse-chase radioactive labeling and immunoprecipitation. The SW/FJ/01 and SW/FJ/03 NS1 proteins were expressed and labeled as described in Materials and Methods. The cells were lysed after being pulse labeled or at different chase time points, and cell lysates were immunoprecipitated with antibodies against NS1, followed by SDS-PAGE analysis. Lane 1, control (empty plasmid DNA vector-transfected cells); lanes 2 to 4, pBlue-NS1-01-transfected samples; lanes 5 to 7, pBlue-NS1-03-transfected samples. Lanes 2 and 5, samples prepared after being pulse labeled; lanes 3 and 6, samples prepared after a 4-h chase; lanes 4 and 7, samples prepared after a 7-h chase. The arrow indicates the NS1 protein band. (B) Analysis of CPSF binding. HA-tagged NS1 constructs were expressed by in vitro translation. Flag-tagged CPSF was expressed in transfected 293T cells. The cell lysate was mixed with the NS1 proteins and subjected to coimmunoprecipitation (IP) using a monoclonal anti-Flag antibody coupled to protein A-Sepharose. The precipitated proteins were analyzed for the HA-tagged NS1 protein and Flag-CPSF by Western blot analysis using HA tag- and Flag-specific monoclonal antibodies, respectively. Lane 1, SW/FJ/01 NS1; lane 2, SW/ FJ/03 NS1.
The effector domain of the NS1 protein of influenza A virus has been reported to bind to distinct cellular proteins and to affect their function (17). NS1 interacts with the 30-kDa subunit of CPSF (25, 26), leading to a block of cellular mRNA processing in influenza A virus-infected cells. To investigate if the five-amino-acid deletion in the NS1 gene of the SW/FJ/03 virus affects its interaction with chicken CPSF, we tested and compared the two NS1 proteins for their interaction with CPSF in a coimmunoprecipitation assay. HA-tagged NS1 constructs were expressed by in vitro transcription and translation. The NS1 proteins were mixed with Flag-tagged CPSF and subjected to immunoprecipitation using a Flag-specific antibody. We found almost no CPSF binding for SW/FJ/03 NS1 but a strong association for SW/FJ/01 NS1 (Fig. 4B). These results indicate that the deleted amino acids at positions 191 to 195 of NS1 not only affect the stability of the NS1 protein in cells but also affect its ability to interact with the chicken cellular protein CPSF.
DISCUSSION
We report an extensive characterization of two H5N1 influenza viruses, SW/FJ/01 and SW/FJ/03, that were isolated from pigs in Fujian Province, southern China. The two swine viruses share high homology with a duck virus, DK/ZJ/00, that has been characterized previously. The virus SW/FJ/03 has a unique five-amino-acid deletion in the NS gene. The two swine influenza viruses replicated in the lungs of mice without prior adaptation, but they displayed low pathogenicity in mice. Interestingly, the viruses displayed differing levels of pathogenicity in chickens. The SW/FJ/03 virus was less pathogenic than SW/FJ/01, even though the HA protein of SW/FJ/03 bears multiple basic amino acids in its cleavage site, a known marker for high pathogenicity. Using reverse genetics, we generated a series of single-gene recombinants by switching the genes of the SW/FJ/01 and SW/FJ/03 viruses, and we then tested their replication and lethality in chickens. We demonstrated that the five-amino-acid deletion at positions 191 to 195 in the NS1 protein contributes to the attenuation of the SW/FJ/03 virus in chickens and that the attenuation phenotype of this virus in chickens may be associated with its decreased ability to antagonize IFN-α/β production in host cells.
Our data also support the hypothesis that pigs can serve as an intermediate host for influenza viruses to evolve in a direction toward being able to replicate more efficiently in animals other than their natural hosts. We found that there are only 23 amino acid differences between the genomes of the SW/FJ/01 and DK/ZJ/00 viruses, but the replication phenotypes of these two viruses in mice are quite different. DK/ZJ/00 does not replicate in mice (4), whereas SW/FJ/01 replicates well in the mouse lung and kills mice at high doses (105 to 106 EID50). SW/FJ/01 does not, however, contain the markers 627K and 701N in the PB2 protein, which are associated with the replication and virulence of H5N1 influenza viruses in mice (9, 13, 22). This suggests the existence of additional mutations that contribute to the replication of H5N1 viruses in mammals. The SW/FJ/01 and SW/FJ/03 viruses were isolated from the same region of Fujian Province, and besides the deletion in the NS gene of SW/FJ/03, there are only five amino acid differences between the two isolates. Two of these five amino acids (HA 444L and M2 26L) in SW/FJ/03 are the same as those in DK/ZJ/00, implying that the virus may have circulated in this region and been transmitted back and forth between waterfowl and pigs.
NS1 has multiple regulatory functions during influenza virus infection (11, 17). Here we confirmed that the deletion of the amino acids at positions 191 to 195 in the NS1 protein plays an important role in the attenuation of the SW/FJ/03 or R-SW/FJ/01-03 virus in chickens and in its ability to antagonize host cell IFN-α/β production. There are several possible explanations for how this deletion could contribute to the low-pathogenicity phenotype and the inability to antagonize IFN. First, the deletion is located within the effector domain, whose main function is the stabilization and/or facilitation of NS1 dimerization (41). Interestingly, it was shown that truncation of the C-terminal domain significantly decreased the stability of an NS1 protein from a swine influenza virus (34) but not that of NS1 from the influenza virus WSN (41). We found that the amount of NS1 protein in cells infected with R-SW/FJ/03 or SW/FJ/01-03NS was significantly lower than that in cells infected with R-SW/FJ/01 or SW/FJ/03-01NS (Fig. 3) and that the SW/FJ/03 NS1 protein exhibited decreased stability compared to the SW/FJ/01 NS1 protein (Fig. 4A). It is thus possible that the deletion in the effector domain affects NS1 protein stabilization, resulting in reduced amounts of the NS1 protein in infected cells. The NS1 protein is responsible for IFN antagonist activity in mammalian (6, 34, 37) and chicken (23) cells. Decreased NS1 protein levels could therefore lead to a decreased ability to antagonize IFN production. Second, the deletion in the effector domain could lead to impaired multimerization of the NS1 protein, which would weaken NS1 binding to RNA (40). The NS1 protein of influenza A virus binds to double-stranded RNA and prevents double-stranded RNA-mediated activation of protein kinase R (1, 12, 24). It is this kinase that induces IFN-α/β synthesis (6, 37). Therefore, the NS1 deletion could lead to the attenuated phenotype through a decreased NS1 protein level and/or a reduction in NS1's RNA binding ability.
The deletion of amino acids 191 to 195 of the NS1 protein of SW/FJ/03 negatively affects the ability of NS1 to bind the cellular 30-kDa subunit CPSF. CPSF is an essential component of the 3′-end processing machinery of cellular pre-mRNAs, and the influenza virus NS1 protein is known to interact with the 30-kDa subunit and to inhibit 3′-end formation of cellular pre-mRNAs (21). The inhibition of cellular pre-mRNA processing and subsequent protein translation are among the antiviral mechanisms employed by influenza virus (17). Two distinct domains in the C-terminal effector domain, namely, the well-characterized region around amino acid 186 (39) and the recently identified middle region around positions 103 and 106 (16), have been proved to be required for binding to the host factor CPSF, resulting in an inhibition of cellular mRNA processing. In the present study, we proved that the amino acid deletion at positions 191 to 195 of NS1 of the SW/FJ/03 virus impairs NS1 binding to the 30-kDa CPSF subunit and may lead to more cellular pre-mRNA, including antiviral IFN-α/β pre-mRNA and proteins.
The nucleotide deletion in the SW/FJ/03 NS gene also results in a five-amino-acid deletion at positions 39 to 43 of the NS2 protein. The NS2 protein of influenza A virus mediates the nuclear export of viral ribonucleoproteins (28), and mutations within the nuclear export signal motif (amino acids 12 to 21) of NS2 affect the export of viral ribonucleoproteins and limit viral growth in cell culture and in mice (15). Although the function, if any, of the NS2 region affected by the deletion at positions 39 to 43 is unknown, we believe that it is unlikely that this deletion in NS2 affects the attenuation of the SW/FJ/03 virus in chickens.
Several reports suggest that mutations or deletions in the NS1 protein contribute to the virulence of influenza viruses in different hosts (3, 23, 32, 33, 38). In this study, we found that the amino acid deletion at positions 191 to 195 of the NS1 protein attenuated the SW/FJ/03 virus in chickens and was critical for the virus to antagonize the host cell IFN response. Our results further strengthen the view that the NS1 protein is an important virulence factor for influenza viruses and that multiple domains within this protein may be suitable targets for the development of antiviral drugs and attenuated vaccines.
Acknowledgments
We thank Gloria Kelly for editing the manuscript, Michael Whitt for providing the reverse genetics system for generating recombinant VSV-GFP, and Nancy Cox and Kanta Subbarao for providing the plasmid pBD.
This work was supported by Chinese National S&T Plan grant 2004BA519A-57; by the Chinese National Key Basic Research Program (973) (2005CB523005 and 2005CB523200); by grants-in-aid and a contract research fund from the Ministry of Education, Culture, Sports, Science and Technology, Japan, for a Program of Founding Research Centers for Emerging and Reemerging Infectious Diseases; by National Institute of Allergy and Infectious Diseases Public Health Service research grants; and by funding from the Emory Global Health Institute.
Footnotes
Published ahead of print on 17 October 2007.
REFERENCES
- 1.Bergmann, M., A. Garcia-Sastre, E. Carnero, H. Pehamberger, K. Wolff, P. Palese, and T. Muster. 2000. Influenza virus NS1 protein counteracts PKR-mediated inhibition of replication. J. Virol. 746203-6206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bu, Z., L. Ye, R. W. Compans, and C. Yang. 2003. Enhanced cellular immune response against SIV Gag induced by immunization with DNA vaccines expressing assembly and release-defective SIV Gag proteins. Virology 309272-281. [DOI] [PubMed] [Google Scholar]
- 3.Cauthen, A. N., D. E. Swayne, M. J. Sekellick, P. I. Marcus, and D. L. Suarez. 2007. Amelioration of influenza virus pathogenesis in chickens attributed to the enhanced interferon-inducing capacity of a virus with a truncated NS1 gene. J. Virol. 811838-1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen, H., G. Deng, Z. Li, G. Tian, Y. Li, P. Jiao, L. Zhang, Z. Liu, R. G. Webster, and K. Yu. 2004. The evolution of H5N1 influenza viruses in ducks in southern China. Proc. Natl. Acad. Sci. USA 10110452-10457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen, H., Y. Li, Z. Li, J. Shi, K. Shinya, G. Deng, Q. Qi, G. Tian, S. Fan, H. Zhao, Y. Sun, and Y. Kawaoka. 2006. Properties and dissemination of H5N1 viruses isolated during an influenza outbreak in migratory waterfowl in western China. J. Virol. 805976-5983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Donelan, N., C. F. Basler, and A. Garcia-Sastre. 2003. A recombinant influenza A virus expressing an RNA-binding defective NS1 protein induces high levels of beta interferon and is attenuated in mice. J. Virol. 7713257-13266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fouchier, R., V. Munster, A. Wallensten, T. Bestebroer, S. Herfst, D. Smith, G. Rimmelzwaan, B. Olsen, and A. Osterhaus. 2004. Characterization of a novel influenza A virus hemagglutinin subtype (H16) obtained from blackheaded gulls. J. Virol. 792814-2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fuerst, T. R., E. G. Niles, F. W. Studier, and B. Moss. 1986. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc. Natl. Acad. Sci. USA 838122-8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gabriel, G., B. Dauber, T. Wolff, O. Planz, H.-D. Klenk, and J. Stech. 2005. The viral polymerase mediates adaptation of an avian influenza virus to a mammalian host. Proc. Natl. Acad. Sci. USA 10218590-18595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garcia-Sastre, A., A. Egorov, D. Matassov, S. Brandt, D. E. Levy, J. E. Durbin, P. Palese, and T. Muster. 1998. Influenza A virus lacking the NS1 gene replicates in interferon-deficient systems. Virology 252324-330. [DOI] [PubMed] [Google Scholar]
- 11.Garcia-Sastre, A. 2006. Antiviral response in pandemic influenza viruses. Emerg. Infect. Dis. 1244-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hatada, E., S. Saito, and R. Fukuda. 1999. Mutant influenza viruses with a defective NS1 protein cannot block the activation of PKR in infected cells. J. Virol. 732425-2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hatta, M., P. Gao, P. Halfmann, and Y. Kawaoka. 2001. Molecular basis for high virulence of H5N1 Hong Kong influenza A viruses. Science 2931840-1842. [DOI] [PubMed] [Google Scholar]
- 14.Horimoto, T., E. Rivera, J. Pearson, D. Senne, S. Krauss, Y. Kawaoka, and R. Webster. 1995. Origin and molecular changes associated with emergence of a highly pathogenic H5N2 influenza virus in Mexico. Virology 213223-230. [DOI] [PubMed] [Google Scholar]
- 15.Iwatsuki-Horimoto, K., T. Horimoto, Y. Fujii, and Y. Kawaoka. 2004. Generation of influenza A virus NS2 (NEP) mutants with an altered nuclear export signal sequence. J. Virol. 7810149-10155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kochs, G., A. García-Sastre, and L. Martínez-Sobrido. 2007. Multiple anti-interferon actions of the influenza A virus NS1 protein. J. Virol. 817011-7021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krug, R. M., W. Yuan, D. L. Noah, and A. G. Latham. 2003. Intracellular warfare between human influenza viruses and human cells: the roles of the viral NS1 protein. Virology 309181-189. [DOI] [PubMed] [Google Scholar]
- 18.Lawson, N., E. Stillmans, M. Whit, and J. K. Rose. 1995. Recombinant vesicular stomatitis viruses from DNA. Proc. Natl. Acad. Sci. USA 924477-4481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee, C. W., Y. J. Lee, D. A. Senne, and D. L. Suarez. 2006. Pathogenic potential of North American H7N2 avian influenza virus: a mutagenesis study using reverse genetics. Virology 353388-395. [DOI] [PubMed] [Google Scholar]
- 20.Li, H., K. Yu, H. Yang, X. Xin, J. Chen, P. Zhao, Y. Bi, and H. Chen. 2004. Isolation and characterization of H5N1 and H9N2 influenza viruses from pigs in China. Chin. J. Prev. Vet. Med. 261-6. (In Chinese.) [Google Scholar]
- 21.Li, Y., Z. Y. Chen, W. Wang, C. C. Baker, and R. M. Krug. 2001. The 3′-end-processing factor CPSF is required for the splicing of single-intron pre-mRNAs in vivo. RNA 7920-931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li, Z., H. Chen, P. Jiao, G. Deng, G. Tian, Y. Li, E. Hoffmann, R. G. Webster, Y. Matsuoka, and K. Yu. 2005. Molecular basis of replication of duck H5N1 influenza viruses in a mammalian mouse model. J. Virol. 7912058-12064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li, Z., Y. Jiang, P. Jiao, A. Wang, F. Zhao, G. Tian, X. Wang, K. Yu, Z. Bu, and H. Chen. 2006. The NS1 gene contributes to the virulence of H5N1 avian influenza viruses. J. Virol. 8011115-11123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lu, Y., M. Wambach, M. G. Katze, and R. M. Krug. 1995. Binding of the influenza virus NS1 protein to double-stranded RNA inhibits the activation of the protein kinase that phosphorylates the eIF-2 translation initiation factor. Virology 214222-228. [DOI] [PubMed] [Google Scholar]
- 25.Nemeroff, M. E., S. M. Barabino, Y. Li, W. Keller, and R. M. Krug. 1998. Influenza virus NS1 protein interacts with the cellular 30 kDa subunit of CPSF and inhibits 3′ end formation of cellular pre-mRNAs. Mol. Cell 1991-1000. [DOI] [PubMed] [Google Scholar]
- 26.Noah, D. L., K. Y. Twu, and R. M. Krug. 2003. Cellular antiviral responses against influenza A virus are countered at the posttranscriptional level by the viral NS1A protein via its binding to a cellular protein required for the 3′ end processing of cellular pre-mRNAs. Virology 307386-395. [DOI] [PubMed] [Google Scholar]
- 27.Office International Des Epizooties. 2004. Manual of diagnostic tests and vaccines for terrestrial animals. Office International des Epizooties, Paris, France.
- 28.O'Neill, R. E., J. Talon, and P. Palese. 1998. The influenza virus NEP (NS2 protein) mediates the nuclear export of viral ribonucleoproteins. EMBO J. 17288-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Park, M. S., A. Garcia-Sastre, J. F. Cros, C. F. Basler, and P. Palese. 2003. Newcastle disease virus V protein is a determinant of host range restriction. J. Virol. 779522-9532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Quinlivan, M., D. Zamarin, A. Garcia-Sastre, A. Cullinane, T. Chambers, and P. Palese. 2005. Attenuation of equine influenza viruses through truncations of the NS1 protein. J. Virol. 798431-8439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reed, L. J., and H. Muench. 1938. A simple method of estimating fifty percent endpoints. Am. J. Hyg. 27493-497. [Google Scholar]
- 32.Richt, J. A., P. Lekcharoensuk, K. M. Lager, A. L. Vincent, C. M. Loiacono, B. H. Janke, W.-H. Wu, K.-J. Yoon, R. J. Webby, A. Solórzano, and A. García-Sastre. 2006. Vaccination of pigs against swine influenza viruses using an NS1-truncated modified live virus vaccine. J. Virol. 8011009-11018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Seo, S. E., E. Hoffmann, and R. G. Webster. 2002. Lethal H5N1 influenza viruses escape host antiviral cytokine responses. Nat. Med. 8950-954. [DOI] [PubMed] [Google Scholar]
- 34.Solorzano, A., R. J. Webby, K. M. Lager, B. H. Janke, A. Garcia-Sastre, and J. A. Richt. 2005. Mutations in the NS1 protein of swine influenza virus impair anti-interferon activity and confer attenuation in pigs. J. Virol. 797535-7543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Subbarao, K., W. London, and B. R. Murphy. 1993. A single amino acid in the PB2 gene of influenza A virus is a determinant of host range. J. Virol. 671761-1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Takada, A., C. Robison, H. Goto, A. Sanchez, K. Murti, M. Whitt, and Y. Kawaoka. 1997. A system for functional analysis of Ebola virus glycoprotein. Proc. Natl. Acad. Sci. USA 9414764-14769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Talon, J., C. Horvath, R. Polley, C. F. Basler, T. Muster, P. Palese, and A. Garcia-Sastre. 2000. Activation of interferon regulatory factor 3 is inhibited by the influenza A virus NS1 protein. J. Virol. 747989-7996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Talon, J., M. Salvatore, R. E. O'Neill, Y. Nakaya, H. Zheng, T. Muster, A. Garcia-Sastre, and P. Palese. 2000. Influenza A and B viruses expressing altered NS1 proteins: a vaccine approach. Proc. Natl. Acad. Sci. USA 974309-4314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Twu, K. Y., D. L. Noah, P. Rao, R. L. Kuo, and R. M. Krug. 2006. The CPSF30 binding site on the NS1A protein of influenza A virus is a potential antiviral target. J. Virol. 803957-3965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang, W., K. Riedel, P. Lynch, C. Y. Chien, G. T. Montelione, and R. M. Krug. 1999. RNA binding by the novel helical domain of the influenza virus NS1 protein requires its dimer structure and a small number of specific basic amino acids. RNA 5195-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang, X., C. F. Basler, B. R. Williams, R. H. Silverman, P. Palese, and A. Garcia-Sastre. 2002. Functional replacement of the carboxy-terminal two-thirds of the influenza A virus NS1 protein with short heterologous dimerization domains. J. Virol. 7612951-12962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ye, L., Z. Bu, A. Vzorov, D. Taylor, R. W. Compans, and C. Yang. 2004. Surface stability and immunogenicity of the human immunodeficiency virus envelope glycoprotein: role of the cytoplasmic domain. J. Virol. 7813409-13419. [DOI] [PMC free article] [PubMed] [Google Scholar]