Abstract
Understanding why human immunodeficiency virus (HIV) preferentially infects some CD4+ CD45RO+ memory T cells has implications for antiviral immunity and pathogenesis. We report that differential expression of a novel secreted factor, ps20, previously implicated in tissue remodeling, may underlie why some CD4 T cells are preferentially targeted. We show that (i) there is a significant positive correlation between endogenous ps20 mRNA in diverse CD4 T-cell populations and in vitro infection, (ii) a ps20+ permissive cell can be made less permissive by antibody blockade- or small-interference RNA-mediated knockdown of endogenous ps20, and (iii) conversely, a ps20low cell can be more permissive by adding ps20 exogenously or engineering stable ps20 expression by retroviral transduction. ps20 expression is normally detectable in CD4 T cells after in vitro activation and interleukin-2 expansion, and such oligoclonal populations comprise ps20positive and ps20low/negative isogenic clones at an early differentiation stage (CD45RO+/CD25+/CD28+/CD57−). This pattern is altered in chronic HIV infection, where ex vivo CD4+ CD45RO+ T cells express elevated ps20. ps20 promoted HIV entry via fusion and augmented CD54 integrin expression; both of these effects were reversed by anti-ps20 antibody. We therefore propose ps20 to be a novel signature of HIV-permissive CD4 T cells that promotes infection in an autocrine and paracrine manner and that HIV has coopted a fundamental role of ps20 in promoting cell adhesion for its benefit. Disrupting the ps20 pathway may therefore provide a novel anti-HIV strategy.
Recently activated CD4 memory T cells identified by CD45RO expression are a major in vivo target of human immunodeficiency virus type 1 (HIV-1) infection, central to HIV-induced disease pathogenesis (26, 43). However, studies from our lab (46, 47, 49) and elsewhere (10, 17, 19, 38) highlight profound heterogeneity in their susceptibilities to HIV, with some CD4+ CD45RO+ T cells escaping infection despite coreceptor expression. These in vitro observations are substantiated by the fact that preferential infection of CD4 T-cell subsets in vivo influences disease outcome. Blood CD4 T cells with proliferative potential at an early differentiation stage (CD45RO+ CD57low) have higher viral burden than terminally differentiated cells (CD45RO+ CD57high); this is linked to their selective loss with ensuing impaired recall responses of the host (6, 7, 26). Indeed, in chronic HIV infection, the loss of a subset of CD28hi HIV-specific interleukin-2 (IL-2)- and gamma interferon (IFN-γ)-double-positive CD4 T cells with proliferative potential correlates with increasing viral load and disease progression (4, 26). More recently, the preferential infection of a subset of gut-homing CCR5hi CD4 T cells (8, 50) may account for severe CD4 T-cell depletion in gut tissue observed particularly in the acute stages of infection, which is not directly reflected in peripheral blood. Indeed, in the simian immunodeficiency virus macaque model, differential targeting of blood CD4 T-cell subsets by pathogenic simian immunodeficiency virus challenge is linked to divergent clinical outcomes (36).
The preferential infection of some CD4 T-cell subsets may be linked to positive-acting host factors that the virus relies on either wholly or partly to complete its replication cycle. The T-cell differentiation/activation stage (6, 39, 43) and linked cytokines (1, 23, 47) regulate HIV coreceptor expression, contributing to the preferential targeting of memory versus naïve and Th2 versus Th1 cells. Additionally, CD4 T-cell adhesiveness (44), the expression of immunomodulatory cytokines that activate the HIV-1 long terminal repeat (LTR) (1), and T-cell survival/differentiation cytokines (1, 43), notably IL-2, determine permissiveness, with HIV preferentially infecting the CD7+ IL-2hi fraction of blood CD4+ CD45RO+ T cells (49). Conversely, innate antiviral proteins can equally determine infection outcome (24, 48). One anti-HIV gene that is differentially expressed in CD4 T cells is APOBEC3G, conferring resistance to infection by Vif-defective HIV strains (42). APOBEC3G down-modulation by T-cell activation increases permissiveness (29) and upregulation with IFN-α coincident with cellular resistance (12). This leads to the hypothesis that differential expression of host-encoded HIV regulatory proteins can set the infection threshold, with the most polarized populations being highly resistant or permissive to infection, thereby contributing to cellular niches for virus replication in vivo.
To identify host proteins contributing to the signature of HIV-resistant versus HIV-permissive (P) cells, we conducted a transcriptome screen of a pair of isogenic CD4 T-cell clones markedly different in their susceptibilities to HIV infection, leading to the identification of a novel HIV regulatory protein, ps20. Human ps20, encoded by the WFDC1 gene (31, 32), is a member of the whey acidic protein (WAP) family; a highly conserved core domain comprising eight cysteines in a characteristic four-disulfide bond arrangement identifies WAPs (37). WAPs are mostly secreted factors in mucosal tissue with pleiotropic functions implicated in innate immunity; some are serine protease inhibitors with anti-infective activity (20, 27). One WAP, secretory lymphocyte protease inhibitor (SLPI), detected in macrophages but not CD4 T cells, exhibits anti-HIV activity by binding to the phospholipid binding protein annexin II, which is a cofactor for macrophage HIV infection (33, 35) and is associated with reduced HIV transmission (22, 28). ps20, a secreted WAP of approximately 23 kDa with a characteristic N-terminal signal peptide that targets it for extracellular secretion, is reported to modulate cell proliferation, migration, and the formation of multicellular spheroids (31, 32, 40) and to induce angiogenesis in cancer-associated reactive stroma in vivo (34). These functions, together with microarray data showing ps20 expression in diverse tissues (see http://symatlas.gnf.org/SymAtlas/) and regulation by transforming growth factor β1 in prostate stromal cells (34), suggest an innate immunomodulatory role for ps20. WAP domain serine protease inhibitors, including the anti-HIV gene SLPI, map to a rapidly evolving region of chromosome 20 (15), consistent with an important role in innate immunity. In contrast, ps20 maps to chromosome 16 and is highly conserved (31, 32; A. Vyakarnam, unpublished data), consistent with a biologically important factor (whose function[s] remains to be fully elucidated). WAP family proteins are likely to be important to wound repair biology, as shown to be the case with SLPI (2), and their role in viral infectivity may relate to their function in tissue homeostasis. We report here the novel HIV-regulatory function of the WAP family member ps20 in CD4 T cells and highlight a potential role for the ps20 pathway in HIV pathogenesis.
MATERIALS AND METHODS
Vectors and constructs.
The WFDC1 gene sequence (GenBank accession number, NM_021197) was cloned into the EcoRI site of the pBK-CMV mammalian expression vector (Stratagene) as previously described (32) and named H1-1pBKCMV. The human ps20 cDNA was subsequently cloned from H1-1pBKCMV into the EcoRI site of the pMH expression vector (Boehringer Mannheim) to express ps20 in mammalian cells with a hemagglutinin tag fused to the C terminus of the protein. The hps20 sequence was amplified with a T3 primer (to the pBKCMV vector) and a primer, 5′CGAATTCTCCTGAAAGTGCCTCTGTTGTCC3′, that adds an EcoRI (underlined) site to the last nucleotide before the stop codon in the ps20 sequence (shown in italics).
For recombinant expression in Drosophila melanogaster cells, human ps20 was cloned into the expression vector pMT/Bip/V5-His. The mature form of hps20 (from nucleotide 251 to 815) was amplified from H1-1pBKCMV with primer 5′ CGGCAGATCTAAGAATATCTGGAAACGGGCATTGC 3′ (BglII) and primer 5′ GCCGTTTTCGAACTGAAAGTGCCTCTGTTGTCC 3′ (BstBI) in the presence of 10% dimethyl sulfoxide by using Turbo Pfu DNA polymerase (Stratagene). After digestion with BglII and BstBI, the ps20 cDNA insert was cloned in frame into the pMT/Bip/V5-His C vector (Invitrogen), creating pMT/Bip/V 5-His/hps20. All expression vector cloning was confirmed by sequence analysis.
Human recombinant ps20 (rps20)-V5-His protein in Drosophila.
The Drosophila expression system (Invitrogen) was used to express a V5-His-tagged human ps20 protein in Schneider 2 (S2) cells (Invitrogen) in a copper-inducible manner. The pMT/Bip/V5-His/ps20 vector and pCoHYGRO (Invitrogen) at a ratio of 19:1 were transfected into S2 cells with a CaPO4 transfection kit (Invitrogen) following the manufacturer's protocol. Stably transfected S2-ps20-V5-His cells were selected in complete Drosophila expression system medium containing 300 μg/ml of hygromycin B for 3 weeks, and the culture medium was switched stepwise to ultimate insect serum-free medium containing 300 μg/ml of hygromycin B. The stable S2-ps20-V5-His cells were then cultured in suspension. When cell density reached 2 × 106 cells/ml, 500 μM CuSO4 was added to induce rps20-V5-His protein expression driven by the Drosophila metallothionein promoter, as confirmed by Western blotting with the anti-V5 antibody (Ab) (32). For human rps20-V5-His protein purification, 500 ml of condition medium (CM) from CuSO4-induced S2-ps20-V5-His cells was collected, centrifuged to remove S2 cells, and concentrated 10 times by using the Amicon stirred ultrafiltration cell model 8400 (Millipore, Bedford MA) with a 5-kDa cut-off membrane, followed by dialysis against Tris-HCl buffer (20 mM Tris-HCl, 150 mM NaCl, pH 7.6) at 4°C. The sample was then loaded slowly onto a Tris-HCl buffer-balanced Ni-nitrilotriacetic acid column. After loading, the column was washed extensively with Tris-HCl buffer with 20 mM imidazole and 40 mM imidazole. The rps20-V5-His protein was eluted with 20 mM Tris-HCl, 150 mM NaCl, pH 7.6, with 250 mM imidazole. The protein was then dialyzed against 20 mM Tris-HCl, 150 mM NaCl, pH 7.6, and concentrated to around 1 mg/ml by using Amicon Ultra-4 (Millipore). The protein concentration was measured using the Bio-Rad Dc protein assay reagents (Bio-Rad Laboratories, Hercules, CA).
Monoclonal anti-ps20 Abs.
Ten milligrams of purified human rps20 was used as an antigen. Hybridoma preparation and initial Ab screening was performed by Zymed. Primary bleeds were screened by enzyme-linked immunosorbent assay (ELISA) with 96-well plates coated with purified ps20-V5-His protein at 0.15 μg/well with standard protocols. Clone 1G7A9H5 gave the highest reading and was used in these studies.
Western blot analysis of ps20.
Samples were treated according to the manufacturer's instructions and run on a 12% NuPage Bis-Tris gel (Invitrogen). All membrane wash steps were carried out with Tris-buffered saline plus 0.1% Tween 20. The membrane blocking steps were done in 5% milk, 2% fetal calf serum (FCS) in Tris-buffered saline plus 0.1% Tween 20, and the membrane was probed with the anti-ps20 Ab IG7 at a final dilution of 1:1,000 followed by a horseradish peroxidase-conjugated goat anti-mouse Ab (Pierce) at a final dilution of 1:1,000. Western blots were developed using an ECL plus Western blotting detection system (Amersham Biosciences, United Kingdom), according to the manufacturer's protocols.
Immortalized cell lines and indicator cells.
Immortalized cell lines and indicator cells were obtained though the AIDS Repository, National Centre for Biological Standards and Controls (NIBSC) (Potters Bar, United Kingdom). The indicator T-cell line CEM.G37 was a kind gift of P. Kellam (UCL). Ghost-CCR5 and CEM.G37 indicator cells express green fluorescent protein (GFP) under the control of the HIV-2 LTR and HIV-1 LTR promoters, respectively.
Memory CD4 T cells.
Peripheral blood mononuclear cells (PBMC) were separated from blood by standard density gradient centrifugation using Lymphoprep (Axis Shield, Oslo, Norway). Ex vivo CD4 CD45RO+ memory CD4 T cells were isolated by negative immunomagnetic bead depletion (Dynal Biotech, Oslo, Norway). To obtain activated cells, PBMC were cultured with 2 μg/ml phytohemagglutinin (PHA) (Biostat Diagnostic Systems, Germany) for 48 h prior to memory cell isolation. Expanded oligoclonal populations were established from the activated memory fraction by culture for 10 to 12 days with 30 IU/ml IL-2 (Proleukin, Chiron, United Kingdom) followed by further rounds of activation conducted every 12 to 14 days with allogeneic irradiated PBMC, 2 μg/ml PHA, and 20 IU/ml IL-2. Cells were fed with 30 IU/ml fresh IL-2 every 3 or 4 days and maintained at 5 × 105 cells/ml in RPMI-HEPES medium-10% FCS-10% human serum (First Link, United Kingdom).
Clinical samples.
Clinical samples were taken from members of a well-characterized cohort to study the biological and behavioral correlates of nonprogression in HIV-1 infection (21). PBMC samples from patients who had been recruited for previous immunological studies (4) were included. Samples from seven treatment-naïve, chronically infected patients with a median time of infection of 14.6 years, a median viral load of 5,060 copies/ng (range, <50 to 437,389), and a median CD4 count of 804/μl (range, 363 to 2,002) were studied.
CD4 T-cell cloning and selection of permissive and nonpermissive (NP) clones.
Clones were generated by the limiting-dilution method from a CD4 memory fraction isolated from three donors (46). Donors included an HIV-negative blood donor (donor 8), an HIV-infected long-term nonprogressor (LTNP 134), and a progressor on antiretroviral therapy with a viral load of <50 RNA copies/ml plasma (GWS 86). Clones which exhibited good growth kinetics and were confirmed to express CD4+ and CCR5+/CXCR4+ were screened for susceptibility to HIV (X4 strains 2044 [see reference 3] and NL4-3; R5 strains BaL and YU2) infection. Clones that restricted replication of either X4 or R5 strains or both (the latter in a β-chemokine-independent manner) were selected for further study alongside permissive clones from the same individual.
ps20 identified as a candidate HIV regulatory protein by differential Affymetrix screening.
RNA isolation/characterization, Affymetrix (HG-U133A and HG-U133B) array probe synthesis, hybridization, washing, and software analysis were performed by the Human Genome Mapping Project Centre, Cambridge, United Kingdom. Data analysis was performed in collaboration with Sarah Webb and Peter Underhill of the Mammalian Genetics Unit, MRC Harwell. Differential screening of a permissive/NP clone pair was conducted under three conditions: at baseline (resting before activation), 6 days after mock infection (and PHA/IL-2 activation), and 6 days after HIV-1 infection (and PHA/IL-2 activation). Productive infection was confirmed by the accumulation of HIV-1 Gag p24 in culture supernatant. The large number of transcripts investigated necessitated a tiered approach to analysis. The data were initially filtered to remove genes that had an absent call on both arrays (filter 1). An arbitrary cutoff of 100 was then applied to remove genes whose signal intensity did not exceed 100 in at least one condition (filter 2) based on the observation that data below 100 were associated with a high coefficient of variance (S. Webb, personal communication). Only those candidate genes that were at least threefold differentially expressed (filter 3) between the constituents of the clone pair were considered as the top differentials. A total of 79 differentially expressed candidates were identified. ps20 was one of three candidates that was differentially expressed in all conditions tested.
Virus stocks.
Primary X4 HIV-1 strain 2044 and R5 strain mBaL were propagated in PHA-activated PBMC (46). Supernatant from parallel uninfected cultures served for mock infection. Full-length HIV infectious molecular clones were produced by standard transient transfection of 293T cells with purified proviral DNA encoding the HIV-1 molecular clone YU2 or NL4-3 (kind gift of M. Malim). Viral stocks were standardized by reference to HIV-1 Gag-p24 concentrations measured by ELISA (HIV-1 p24 antigen capture assay kit; NCI Frederick).
GFP-encoding HIV-1-based lentiviral vector particles for single-cycle infection.
GFP-encoding HIV-1-based lentiviral vector particles for single-cycle infection were generated through the transient transfection of 293T cells by use of Fugene 6 (Roche). The packaging construct, pCMVDR8.91 (kind gift of D. Trono), was transfected along with the HIV Gag-Pol expression vector encoding enhanced GFP, pHR′SIN-cPPT-SEW (kind gift of A. Thrasher), and a chosen envelope construct, i.e., pMD.G encoding vesicular stomatitis virus glycoprotein or HIV R5 or X4 envelopes, pYU2-SV3, or pHXB2-SV3, respectively, at an optimum ratio of 1.86:2.86:1. Virus titers were determined by assessing the level of transduction (by GFP expression) in the human CD4+ T-cell line Jurkat by flow cytometry in conjunction with a measurement of viral p24-CA (p24 ELISA; NCI).
Single-cycle infection with replication-competent HIV.
Cells (2 × 105 cells/well) were seeded in a 24-well plate and exposed to modulators overnight prior to infection with NL4-3 (10 ng p24-CA/million cells). Twenty-four hours postinfection, cells were washed three times with cold phosphate-buffered saline (PBS), trypsinized (Sigma-Aldrich) for 5 min at 37°C, and washed again three times in PBS with 5% FCS. Cell pellets were resuspended in cold lysis buffer (PBS with 1% Triton X and 1% NP-40) and stored at −80°C prior to use in p24-CA ELISA.
Spreading HIV infection.
CD4 T-cell clones were activated with irradiated allogeneic PBMC (1:1 ratio) plus 2 μg/ml PHA plus 20 IU/ml IL-2. Five days later, cells were washed and a total of 1 to 5 million cells challenged with HIV virus stocks (1 to 50 ng HIV p24-CA/million cells) in a final volume of 200 to 300 μl. Eighteen hours later, cells were washed and fresh medium was added to a final volume of 1 ml and maintained in 30 IU/ml IL-2. The HIV-1 p24-CA antigen concentration in cell-free supernatant was measured over time. Experiments were optimized to achieve reproducible virus spread; the X4 HIV-1 strain 2044 (46) was consistently more efficient than the X4 strain NL4-3 in spreading the infection of primary CD4 cells/clones, even when both virus stocks were produced in the same producer cell (PHA-stimulated PBMC); on the other hand, immortalized CD4 lines were equally susceptible to both strains irrespective of producer cell (28a). Therefore, X4 strain 2044 was generally used for spreading the infection of primary CD4 cells.
ICS assay for secreted ps20.
The intracytoplasmic staining (ICS) assay for secreted ps20 was as previously described (4). One million cells were cultured with 5 μg/ml brefeldin A (Sigma-Aldrich) for 4 h prior to staining and washed and permeabilized (An Der Grub). Permeabilized cells (1 × 105 to 2 × 105) were incubated for 20 min on ice with a 1/100-to-1/500 dilution of anti-ps20 Ab IG7, washed, and then stained with a 1/200 dilution of F(ab′)2 fluorescein isothiocyanate-conjugated goat anti-mouse immunoglobulin G (IgG) (Sigma) for 20 min on ice. Samples were then washed, fixed in PBS plus 2% FCS plus 2% formaldehyde, and acquired on a FACSCalibur (BD Biosciences) using Cell Quest software (BD Biosciences).
Nonquantitative RT-PCR of ps20 mRNA.
RNA was isolated with a one-step reverse transcription-PCR (RT-PCR) kit (Qiagen, West Sussex, United Kingdom). Q solution included in the kit was necessary to remove secondary structure in the GC-rich target sequence. The amplification of full-length ps20 mRNA was optimized for use with 0.2 μg template/0.6 μM each primer (MWG Biotech, London, United Kingdom) in a total reaction volume of 25 μl. Primers were FWD (5′ GCATGCCTTTAACCGGCGTGG 3′) and REV (5′ GCTTACTGAAAGTGCTTCTG 3′). Hypoxanthine phosphoribosyltransferase (HPRT) was used as an internal control with primers FWD (5′ ACCAGTCAACAGGGGGACAT 3′) and REV (5′ CGACCTTGACCATCTTTGGA 3′) used at 0.6 μM per reaction. All reactions were performed in an Eppendorf gradient thermocycler as follows: room temperature at 50°C for 30 min, HotStarTaq initiation at 95°C for 15 min, denaturation at 94°C for 30 s, annealing at 56°C for 30 s, and extension at 72°C for 1 min for 35 cycles, followed by a final extension at 72°C for 10 min. Products were visualized by agarose (Sigma) (1.6 to 2%) gel electrophoresis containing 0.5 mg/ml ethidium bromide for 90 min at 90 V and imaged under UV light.
Quantitative real-time PCR (qRT-PCR) for ps20 mRNA.
Total RNA from predetermined numbers of cells extracted with TRIzol (Invitrogen) was converted to cDNA (Ambion). ps20 mRNA was measured with respect to HPRT by use of a Quantitect SYBR green PCR kit (Qiagen, West Sussex, United Kingdom) on an ABI Prism 7000 (Applied Biosystems, California). Cycle parameters were as follows: 2 min at 50°C, 15 min at 95°C, and then 40 cycles (20 s at 95°C, 30 s at 60°C, 30 s at 72°C) followed by a standard dissociation stage. HPRT primer sequences were as follows: 5′-ACC AGT CAA CAG GGG GACAT-3′ (forward), and 5′-CGA CCT TGA CCA TCT TTG GA-3′ (reverse). Codon-optimized ps20 primers were from Qiagen Ltd. For absolute quantitation, an in vitro-transcribed standard curve was generated. The H1-1pBKCMV plasmid was linearized and used in a T7-transcription MEGAscript kit (Ambion). A log dilution standard of known ps20 mRNA concentrations (1,000 to 0.001 pg) spiked with a uniform 1 ng of herring sperm cDNA (Promega, Madison, WI) was included in each run along with test samples and positive, negative, and RT controls. A standard curve of ps20 mRNA versus PCR cycle number was generated, and the number of ps20 molecules per cell calculated from the known molecular weight of ps20.
Stable expression of ps20 in Jurkat cells by retroviral transduction.
The ps20 sequence was cloned into the EcoRI site of pCxCR (empty vector [EV]), a Moloney murine leukemia virus-based bicistronic retroviral vector which expresses red fluorescent protein under the control of a cytomegalovirus promoter (kind gift of G. Towers, University College London) and named pCpsCR. To generate retroviral particles, 293T cells were transiently transfected with either pCxCR or pCpsCR, along with the packaging construct pCpg (Moloney murine leukemia virus gag/pol) and an envelope construct encoding vesicular stomatitis virus glycoprotein (pMD.G). Forty-eight hours after transfection, cell-free supernatants were harvested. Jurkat cells (2 × 105 CCR5+ Jurkat cells) were cultured three times with a 50% total volume of retrovirus-containing supernatant over a 3-day period. Red fluorescent protein expression was used to sort transduced cells by flow cytometry.
ps20 knockdown using small interference RNA (siRNA).
CD4+ CCR5+ CXCR4+ adherent HeLa indicator cells that express the β-galactosidase reporter gene under the control of an HIV LTR (kind gift of J.-M. Serrano) originally obtained from the NIH AIDS repository were seeded 6 h prior to transfection at a density of 2 × 105 per 24-well plate in Dulbecco's minimal essential medium plus 10% FCS plus 20 μg/ml gentamicin. Parallel triplicate cultures were set up for HIV infection and for qRT-PCR.
The following siRNAs were purchased from Ambion: siRNA 1 against ps20 sense (5′GGUGACUCAAAGAAUGUGG 3′) and antisense (5′ CCACAUUCUUUGAGUCaCC 3′) and siRNA 2 against ps20 sense (5′GGCUCAGCAUCUUGAUAUU 3′) and antisense (5′ AAUAUCAAGAUGCUGAGCC 3′). The following mitogen-activated protein kinase (MAPK) control siRNA (Qiagen) was used to confirm the specificity of knockdown: sense, 5′UGCUGACUCCAAAGCUCUGdT 3′; antisense, 5′CAGAGCUUUGGAGUCAGCAdT 3′. Each siRNA (250 nM concentration) was diluted in a total volume of 100 μl of Dulbecco's minimal essential medium without serum and then complexed with 10 μl of HiPerFect transfection reagent (Qiagen) for 15 min and added, and cultures were topped up to 500 μl to give a final concentration of 50 nM for each siRNA. Cells were cultured with a mix of the two ps20-specific siRNAs or the MAPK-specific siRNAs. Forty-eight hours later, cells were harvested by trypsinization and washed, and viable cells were plated at 2 × 104/well in a 48-well plate. Six hours later, X4 HIV-1 NL4-3 virus stock was added at various dilutions, and cells were cultured in a final volume of 500 μl. Thirty-six hours later, cell lysates were harvested using a Tropix Galacto-Star assay system as per the manufacturer's recommendation (Applied Biosystems). Cell debris was removed by centrifugation, and lysate supernatants were stored at −80°C. To assay for β-galactosidase, 15-μl portions of lysates were added to the Galacto-Star reporter gene assay system, and the amount of chemiluminescence measured on a Victor light 1420 luminescence counter, with measurements taken at peak emission, which occurs 20 to 30 min after the beginning of the reaction. For qRT-PCR measurements, parallel cultures were harvested, counted, and processed as described in Materials and Methods.
Statistical analysis.
Statistical analysis was done in GraphPad PRISM software (PRISM 4 for Macintosh). Group differences were determined by nonparametric testing, and P values of <0.05 were considered significant.
RESULTS
Endogenous ps20 mRNA levels correlate with increased HIV spread in isogenic CD4 T-cell populations from multiple donors. (i) Clones.
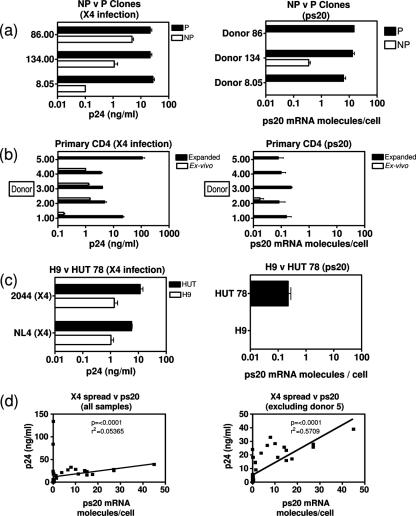
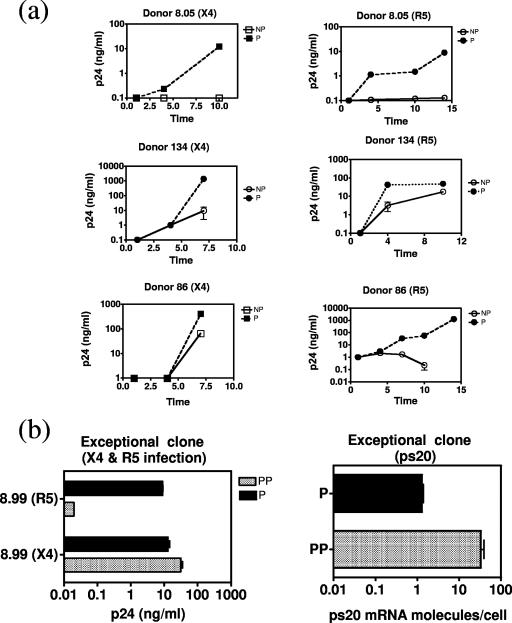
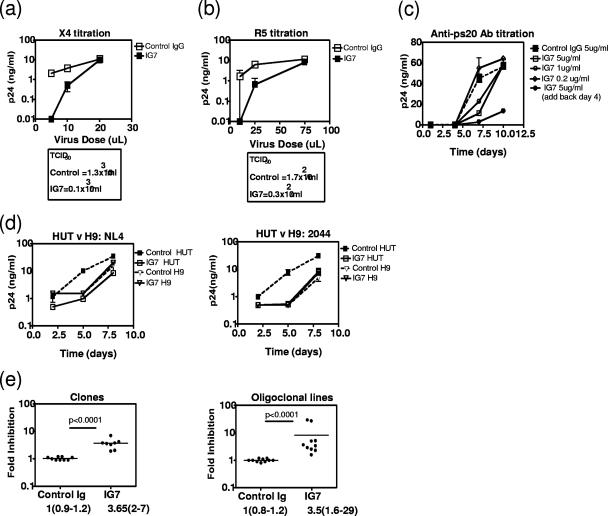
Having identified ps20 by screening a P/HIV-resistant clone pair, we first examined its expression in an expanded panel of six clones from three donors. NP clones were less permissive to both X4 HIV (Fig. 1a) and R5 HIV (Fig. 2a shows virus spread over time). Resistant clones were either refractory to infection or defined as supporting spreading infection less efficiently than the P counterpart. The mean difference in peak p24 Gag levels among the three P-versus-NP clone pairs was 99-fold for X4 HIV (range, 4.5- to 270-fold) (Fig. 1a). All P clones expressed ps20; two NP clones lacked detectable ps20, and the third had a level of ps20 41-fold lower than that seen for the P counterpart (Fig. 1a). An additional, exceptional, clone pair was identified where ps20 expression correlated with X4 but not R5 infection. Clone 8.5.7.99 was 3-fold more permissive to X4 HIV than its counterpart, consistent with previous data (46), and had 26-fold-higher ps20; however, despite high ps20, this clone was selectively resistant to R5 HIV and is therefore referred to as partially permissive (Fig. 2b).
FIG. 1.
ps20 mRNA correlates with HIV spread. One million cells (clones, primary CD4 CD45RO cells, and H9/HUT 78 cells) were infected overnight with a virus dose standardized as to HIV Gag p24-CA concentration (2 ng 2044 strain or 4 ng NL4-3 strain). Expanded CD4 cells and clones were stimulated with allogeneic allophycocyanin-PHA-IL-2 for 5 days prior to infection and cultured at 2 × 105/ml in 30 IU/ml IL-2 postinfection. Mean peak p24-CA levels (day 8) in three biological replicate cultures are shown. qRT-PCR results for ps20 per population were determined in triplicate at the time of infection, and the mean number of ps20 molecules per cell is shown. (a) Comparison of NP and P counterpart clones from donors 8, 134, and 86 infected with the 2044 (X4) strain. (b) For ex vivo results, CD4 CD45RO+ T cells were infected with 2044 and then cultured in 30 IU/ml IL-2. For expanded results, purified CD4+ CD45RO+ T cells were expanded by two rounds of stimulation with allogeneic PBMC-PHA-IL-2 (see Materials and Methods) and then infected with 2044 and maintained in 30 IU/ml IL-2 postinfection. (c) Comparison of H9 versus HUT 78 immortalized cells infected with NL4-3 or 2044 strains. (d) A statistical correlation of peak p24-CA levels versus ps20 mRNA molecules per cell for each population was derived on GraphPad PRISM software. The two-tailed P value derived by use of the nonparametric Spearman's correlation is shown. The goodness-of-fit R2 value derived by linear regression analysis is shown.
FIG. 2.
Variable pattern of spreading HIV infection in P versus NP clones. (a) Cells were challenged with either 5 ng p24-CA/million cells of X4 strain 2044 or 20 ng/million cells of R5 strain mBaL. R5 infection was conducted in the presence of a cocktail of neutralizing Abs to β-chemokines MIP-1α, MIP-1β, and RANTES (final concentration of each Ab, 1 μg/ml). Data show mean HIV-1 p24-CA levels in three to five replicate experiments over time. (b) Data on ps20 expression in an exceptional resistant clone. Mean peak p24-CA levels (day 8 for X4 virus and day 12 for R5 virus) are summarized along with qRT-PCR values for ps20 mRNA determined in triplicate at the time of infection. PP, partially permissive.
(ii) Primary cells.
Next, primary ex vivo versus activated memory CD4 H7 cells were examined. ps20 was low/undetectable in ex vivo memory cells from five donors (Fig. 1b) with no significant upregulation 48 h after PHA activation (see Fig. 7) or in time course studies at 0, 2, 6, 12, 24, and 48 h after activation with anti-CD3/28 stimulation (not shown). However IL-2 expansion of the PHA-activated cells followed by a further round of restimulation upregulated ps20 by 3- to 14-fold coincident with increased permissiveness compared to what was seen for ex vivo memory CD4 cells (Fig. 1b). One of the five donor samples was an outlier (donor 5 [expanded]) for whom p24 output was exceptionally high compared to what was seen for the others for a given virus challenge dose (>100 ng/ml) (Fig. 1b).
FIG. 7.
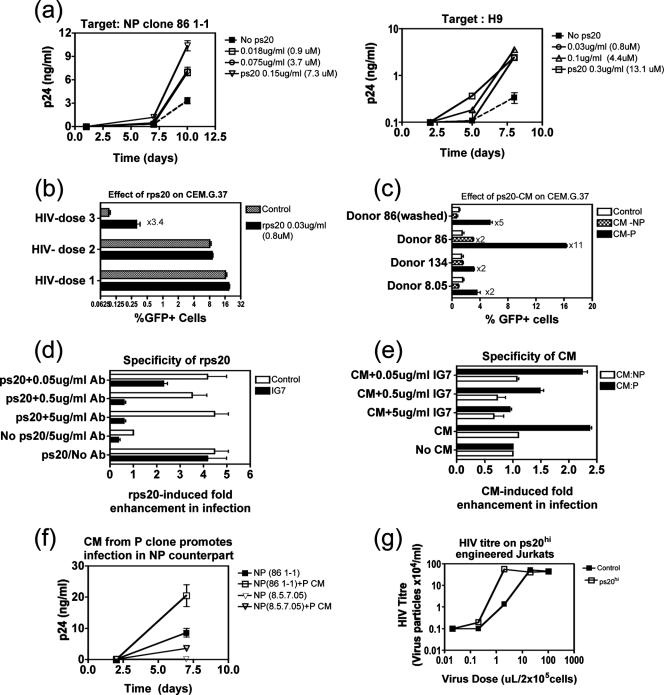
Exogenous addition or stable endogenous ps20 expression by retroviral transduction promotes HIV infection. (a) Target cells (2 × 105) (clone 86 1-1 or H9) were precultured for 18 h in the presence or absence (control) of rps20 before infection with 2044 (1 ng p24-CA virus/million 86 1-1 cells and 0.3 ng/million H9 cells). Mean p24 levels over time are shown. (b) CEM.G37 indicator cells (2 × 105) were precultured for 18 h in the presence or absence of rps20 and then infected with NL4-3 (dose 1, 3 ng p24-CA stock/million cells; dose 2, 1 ng; dose 3, 0.2 ng) followed by culturing for 4 days in a final 1-ml volume. Mean percentages of GFP+ cells are shown. (c) CEM.G37 cells (2 × 105) were precultured for 18 h in the presence or absence (control) of 10% crude CM from NP or P clones from three donors (donor 8, donor 134, and donor 86). Cells were infected with NL4-3 (0.4 ng p24-CA stock/million cells) and maintained in the same concentration of CM. Donor 86 (washed) represents cells cultured with CM prior to infection and then washed, infected, and maintained in the absence of CM. Mean percentages of GFP+ cells in triplicate cultures 4 days postinfection are shown. (d) NP clone 86 1-1 (2 × 105 cells) was precultured with 1 μM rps20 alone or in the presence of IG7 Ab/control IgG at various concentrations. Eighteen hours later, cells were infected with 2044 (1 ng p24-CA virus/million cells), and cultures were maintained in 30 IU/ml IL-2 for a further 7 days. Mean enhancements (n-fold) in p24-CA levels in triplicate cultures were calculated relative to values for control cultures infected and maintained in the absence of any treatment. ps20 and the no-Ab positive control were set up in two sets of triplicates represented by empty and filled bars, respectively. (e) CEM.G.37 cells (2 × 105) were precultured with the most potent P CM, 2% CM from P clone 86 1-3, or with the counterpart NP CM in the presence or absence of IG7 Ab at various concentrations. Eighteen hours later, cells were infected with NL4-3 (0.4 ng p24-CA stock/million cells) and cultures maintained for 4 days. The mean enhancement (n-fold) in the percentage of GFP expression in triplicate cultures was calculated relative to values for control parallel cultures infected and maintained in the absence of CM plus control mouse IgG to match the highest IG7 concentration tested, 5 μg/ml. (f) NP clones 86 1-1 and 8.5.7.05 (2 × 105 cells) were cultured for 18 h with 10% CM from P counterpart clones and then infected with 2044 (2 ng p24-CA/million cells). Cultures were maintained for 7 days. The mean p24-CA level from triplicate cultures is shown. Jurkat cells (2 × 105) transduced with EV or ps20 (G91) were infected with various dilutions of NL4-3 for 2 h, washed, and then maintained for 7 days. The HIV titer of the culture supernatant was assessed for CEM G37 indicator cells by use of GFP expression as an indication of productive infection.
(iii) Immortalized lines.
An examination of immortalized CD4 T-cell lines (a selection is shown in Fig. 2a) revealed variable ps20 levels even within isogenic subclones (H9 versus HUT78) (see reference 13 for H9/HUT78 characterization) (analogous to primary clones [Fig. 1a]). HIV infection correlated with endogenous ps20: ps20+ HUT78 was more permissive than ps20− H9 when tested with two X4 HIV-1 strains; p24 Gag output from H9 cultures was up to 1 log lower than that from HUT cultures (Fig. 1c).
The regression analysis of the ps20 mRNA molecules/cell versus the p24 Gag level in all the diverse CD4 populations represented in Fig. 1a, b, and c revealed a striking positive correlation, whether or not the outlier donor 5 sample was included in the analysis (P < 0.0001) (Fig. 1d). These data confirm ps20 to be a novel CD4 T-cell factor associated with permissiveness to HIV spread.
ps20 mRNA correlates with protein expression.
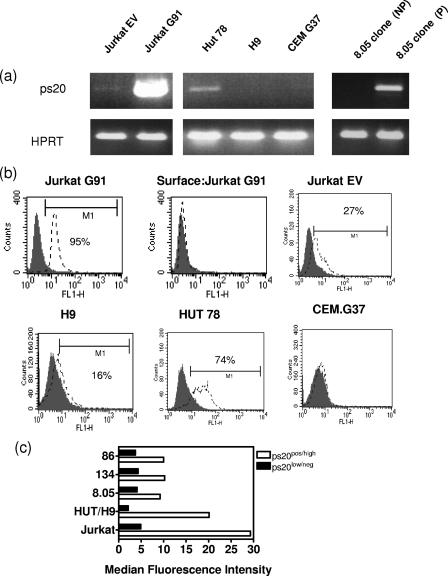
A predicted 660-bp product was seen when full-length ps20 mRNA was amplified using Jurkat cells engineered to overexpress ps20 (termed Jurkat G91) (Jurkat transduced with EV [Fig. 3a]). An ICS assay for ps20 protein corroborated the mRNA analysis (Fig. 3b), revealing low-intensity staining in the Jurkat G91 ps20hi control (median fluorescence intensity [MFI], 29) when cells were permeabilized but not upon surface staining, suggesting little retention of ps20 on the cell surface, analogous to what is seen for other T-cell secreted factors (cytokines) that, like ps20, are targeted for extracellular secretion. High MFI was noted for several P (median MFI, 10; range, 9.2 to 10.2) versus NP (median MFI, 4; range, 3.7 to 4.3) clones and for HUT (MFI, 20) versus H9 (MFI, 2) (Fig. 3c). Background staining in the ICS assay was taken as an MFI of <4 as judged by IG7 binding to ps20 mRNAneg cells.
FIG. 3.
ps20 mRNA correlates with protein expression. (a) Full-length ps20 was amplified in a one-step nonquantitative RT-PCR. Jurkat G91 refers to cells stably transduced to express full-length ps20 and Jurkat EV to Jurkat cells transduced with the EV control. (b) Immunofluorescence profiles of brefeldin A-treated permeabilized cells indirectly stained with anti-ps20 Ab IG7 or control normal mouse IgG followed by fluorescein isothiocyanate-F(ab′)2 goat anti-mouse IgG (Sigma) secondary Ab. Surface:Jurkat G91 refers to G91 stained with IG7 following culture with brefeldin A but without the prior permeabilization of cells. (c) MFIs of cells stained with IG7 determined using CellQuest software. Reference to ps20pos/high or ps20low/neg was determined by qRT-PCR levels for ps20 in the same samples. pos, positive; neg, negative.
Western blot analysis of rps20 expressed in insect cells detected a predicted band of ∼21 kDa (native folded state) and an additional band of ∼34 kDa representing the unfolded (fully reduced) form of ps20. The masses of both forms are commonly observed in reducing conditions and are identical to previously published molecular masses of native ps20 and rps20 (31, 32, 34, 40). Neither of these bands were observed when the monoclonal anti-ps20 Ab 1G7 was preabsorbed with rps20, demonstrating Ab specificity (Fig. 4a). The molecular mass of ps20 prepared in mammalian cells (293T) differed marginally from that from the insect cell preparation (molecular mass of lower band, ∼24 kDa; molecular mass of upper band, ∼33 kDa); Ab specificity for this preparation was confirmed by loss of binding following preabsorption with rps20 (Fig. 4b). The 24-kDa band was noted in Jurkat ps20hi cells and in a ps20 mRNA-positive P clone but not in a ps20 mRNA-negative NP clone (Fig. 4c). Taken together with the ps20 mRNA data, these observations demonstrate that CD4 populations from a given donor can differ inherently in terms of ps20 expression.
FIG. 4.
Western blot analysis of ps20. (a) Left lane (preabsorption), 100 ng rps20 was run on a 12% reducing gel and probed using a 1/1,000 dilution of IG7; right lane (postabsorption), 100 ng rps20 was run on a 12% reducing gel and probed with IG7 after preabsorption with 3 μg rps20 for 2 h at room temperature. (b) Left lane (preabsorption), 10 μl of culture supernatant from 293T transfected with 10 μg of a ps20-encoding expression plasmid per 1 × 105 cells was run on a 12% reducing gel and subsequently probed using a 1/1,000 dilution of anti-ps20 Ab IG7; right lane (postabsorption), 293T transfection supernatant probed with the anti-ps20 Ab preabsorbed with 3 μg rps20 for 2 h at room temperature. (c) Endogenous ps20 expression in 2 × 104 cell equivalents was assessed by immunoblotting with a 1/1,000 dilution of IG7 in cell populations as follows: left lane, ps20 mRNA-negative CD4 T-cell clone; middle lane, ps20+ CD4 T-cell clone; right lane, Jurkat cells stably transduced to overexpress ps20.
Endogenous ps20 is an HIV permissivity factor: anti-ps20 Ab blocks infection of endogenous ps20+ populations from multiple donors.
We determined if a ps20+ permissive cell could be made less permissive by blocking endogenous ps20 with a neutralizing anti-ps20 Ab. Experiments were designed to assess whether the potency of the endogenous ps20 effect was conditional on (i) virus dose and (ii) Ab concentration. As our clonal analysis showed the peripheral blood CD4 pool to be heterogeneous for ps20 (ps20hi and ps20low/neg CD4 T-cell clones isolated from a given donor's blood cells [Fig. 1]), we examined the potency of the HIV effect in a ps20-homogeneous CD4 T-cell population (clone). Anti-ps20 Ab 1G7 blocked productive infection of ps20+ permissive clone 8.16.7.05 by both an X4 HIV-1 (Fig. 5a) and an R5 HIV-1 (Fig. 5b) strain. The effect was more marked at lower virus challenge doses represented by a lower 50% tissue culture infective dose (TCID50) of each virus stock in the presence of IG7 Ab compared to what was seen for control mouse IgG at the same concentration (13-fold lower X4 titer and 5.7-fold lower R5 titer). In the same clone, a single dose of 5 μg/ml IG7 suppressed virus spread by 4-fold (Fig. 5c); suppression was augmented to 15-fold by the further addition of Ab on day 4, indicating continuous ps20 synthesis and utilization in an autocrine manner to support infection. The breadth of the ps20 regulatory effect was next examined at a single virus/Ab dose in cell populations from different donors by use of two X4 virus strains, one an infectious molecular clone (NL4-3) and the other a primary X4 strain (2044). Virus spread was suppressed by up to 10-fold by IG7 in ps20+ HUT 78 cells challenged with either the NL4-3 or the 2044 X4 strain (Fig. 5d) relative to what was seen for the IgG control. The ps20 mRNA-negative H9 cell served as an additional specificity control, as indicated by the failure of IG7 Ab (5 μg/ml) to inhibit infection (Fig. 5d) in this population despite the low-level nonspecific binding of this Ab in the ICS assay (Fig. 3b and c). As previous studies have already highlighted the tremendous inherent level of variation of the peripheral CD4 T-cell pool in susceptibility to HIV infection (17) and a number of host factors known to regulate HIV infection in CD4 T cells (19), we were keen to determine if endogenous ps20 was a permissivity factor in cells from multiple donors. A screening of ps20+ clones from three donors and ps20+ IL-2-expanded primary CD4 cells from five additional donors showed inhibition of infection in every donor's cells; the inhibitory effects of a single dose of IG7 Ab against a single high virus challenge dose ranged from 2- to 7-fold for the clones and from 2- to 29-fold for the primary cells (Fig. 5e). These experiments confirm endogenous ps20 to be an HIV permissivity factor, and the potency of its effect is determined by virus dose and Ab concentration and can be donor dependent.
FIG. 5.
Anti-ps20 Ab blocks HIV infection. ps20+ P clone 8.16.7.05 cells (2 × 105) were precultured for 18 h with 5 μg/ml of control mouse IgG or anti-ps20 Ab IG7 and then infected with various concentrations of X4 HIV-1 strain 2044 (a) or R5 HIV-1 strain YU2 (b), respectively, in triplicate cultures. After overnight infection, cells were cultured in 30 IU/ml fresh IL-2 and IG7 or control IgG in a final volume of 1 ml. p24-CA levels were measured on day 7 postinfection for 2044 infections and on day 9 for YU2 infections and TCIDs calculated based on the proportion of wells that were p24-CA positive for each virus dose (see reference 3). Mean p24-CA levels in triplicate cultures at each virus input dose is shown. (c) Spreading infection in 8.16.7.05 conducted in the presence of various anti-ps20 Ab doses. p24-CA levels in triplicate cultures are shown. (d) Spreading infection in H9 or HUT 78 cells conducted as described for panels a and b with X4 strains (4 ng p24-CA/million cells, 2044 or NL4-3). Mean p24-CA levels in triplicate cultures are shown. (e) Cells (2 × 105) (permissive clones or expanded oligoclonal CD4 lines) were precultured for 18 h with 5 μg/ml of control mouse IgG or anti-ps20 Ab IG7 or cultured in the absence of these IgGs and then infected with 2044 (2 ng p24-CA stock/million cells) for a further 18 h. Cells were then cultured in 30 IU/ml IL-2 for a further 7 days, when p24-CA levels were measured. Inhibition (n-fold) in the presence of each IgG was calculated relative to the p24-CA level in the absence of mouse IgG. Duplicate to triplicate measurements of two permissive clones and duplicate measurements of primary CD4 from five donors are shown. Group differences were determined by nonparametric Mann-Whitney testing.
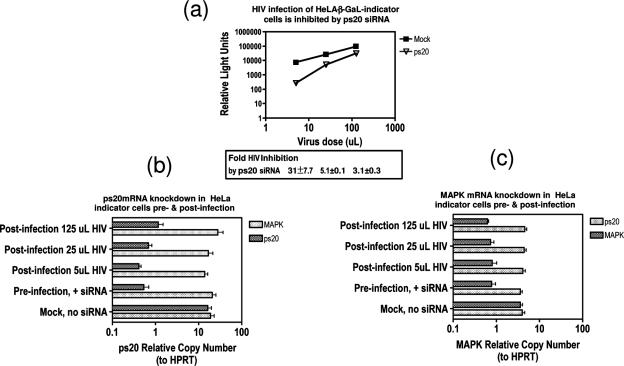
To further confirm the positive-acting effects of ps20 on HIV infection, specific siRNA was employed in knockdown experiments using a heterologous system that is amenable to transient transfection. A screen of transfectable adherent human lines identified the widely used HeLa indicator cells that express the β-galactosidase reporter gene under the control of an HIV promoter to be ps20+, providing an ideal test system. Experiments were designed to correlate ps20 knockdown efficiency on HIV infection. Specificity was controlled by including another ubiquitous host gene, MAPK mRNA. Relative to what was seen for mock transfection (transfection reagent in absence of any siRNA control), both ps20 and MAPK siRNAs were specific for their respective targets (Fig. 6b and c). ps20 knockdown over all cultures tested ranged from 24- to 39-fold and from 5- to 7-fold for MAPK siRNA, with a nonspecific effect of each siRNA on the irrelevant target restricted to less than 1.3-fold (Fig. 6b and c, respectively). Figure 6a shows a log increase (n-fold) in infection with increasing virus input in the absence of siRNA (mock control) and a significant and virus dose-dependent inhibition of HIV infection following ps20 knockdown relative to what was seen for mock transfection. A maximum HIV inhibition of 31-fold was observed at the lowest virus dose and was reduced to 3-fold inhibition at the highest virus dose despite a 24-fold ps20 knockdown. These data confirm and extend the anti-HIV effect of anti-ps20 Ab in a TCID50 assay (Fig. 5), demonstrating endogenous ps20 to be an important HIV permissivity factor that is saturated by high virus doses.
FIG. 6.
siRNA-mediated knockdown of endogenous ps20 blocks HIV infection. (a) HeLa indicator cells (2 × 105) were exposed to transfection reagent in the absence of siRNA (mock) or with 50 nM siRNA specific for ps20 or MAPK. Forty-eight hours later, adherent cells were harvested by trypsinization and washed, and viable cells were reseeded at a density of 2 × 104 cells per well and left to adhere for 6 h before the addition of virus (5 μl, 25 μl, 125 μl). Thirty-six hours later, productive HIV infection was determined for cell lysates by use of β-galactosidase levels measured as relative light units (minus background relative light units for uninfected cells) in a luminometer. (b and c) Parallel cultures as described above were set up and samples processed for ps20 mRNA or MAPK mRNA by qRT-PCR. The nonspecific effect of MAPK siRNA on ps20 knockdown is shown in panel b and vice versa (ps20 siRNA on MAPK) in panel c. Error bars represent means of three replicates.
Exogenous addition or stable endogenous expression of ps20 by retroviral transduction promotes HIV infection.
We examined if a ps20low/negative cell could be made more permissive either by exogenous addition of the factor or by stable transduction of the cells to express full-length ps20. rps20 enhanced infection in diverse ps20low/neg CD4 T-cell populations, its potency being (i) concentration dependent, (ii) virus dose dependent, and (iii) cell dependent. A 7.3 μM concentration of rps20 enhanced spreading infection by 4-fold in a ps20low NP clone, by 10-fold in ps20neg H9 cells (down to 0.8 μM) (Fig. 7a), and by 3-fold in a GFP-encoding indicator line, CEM.G.37; this effect was virus dose dependent, as shown by the enhanced infection in the presence of an optimal rps20 dose at a low but not high virus challenge dose in the CEM.G.37 assay using GFP expression as a readout (Fig. 7b). We further used the CEM.G.37 indicator cell assay to test if the native secreted factor was biologically active. Infection was increased up to 11-fold by the addition of crude CM harvested at the midpoint of the feeding cycle from ps20 mRNA+ P clones, versus 2-fold or less by CM from ps20low/neg NP clones compared to no-CM control cultures (Fig. 7c). A prior 24-h exposure to CM enhanced infection (Fig. 7c, donor 86 [washed]), but infection was higher when indicator cells were continuously exposed (Fig. 4c, donor 86, P CM). CM potencies differed between clones (donor 86 P CM potency was highest), perhaps reflecting ps20 secretion efficiency and reabsorption based on its autocrine effect. IG7 Ab completely reversed both rps20- and CM-induced enhancements in a primary clone and in the CEM.G37 assay (Fig. 7d and e, respectively). One NP clone (donor 86 NP) was an exception, as CM from this clone exhibited a modest twofold enhancing effect (Fig. 7c). Inhibition of virus spread in this clone by IG7 (Fig. 7d, no ps20/5 μg/ml Ab) suggests that this clone produces low ps20, though the level of ps20 mRNA in these cells is below the level of detection (Fig. 1a). We also show CM to be biologically active on primary cells; thus, a single addition of CM from two ps20+ P clones promoted infection by two- to fourfold in their ps20low/neg NP counterparts (Fig. 7f).
Finally, we tested whether a CD4 T cell with low endogenous ps20 could be made more permissive by engineering stable ps20 expression by retroviral transduction. ps20low Jurkat cells stably transduced to express ps20 (G91-ps20hi) were more permissive to infection than Jurkat cells transduced with EV control (EV- ps20low). The HIV titer on ps20hi G91 was 39-fold higher at a low virus dose (Fig. 7g); no differences were seen between the two populations at higher virus doses. Together, these data confirm that secreted ps20 is a paracrine-acting cofactor for HIV infection and further confirm the potency of the ps20 HIV effect to be (i) ps20 concentration dependent, (ii) virus dose dependent, and (iii) cell dependent.
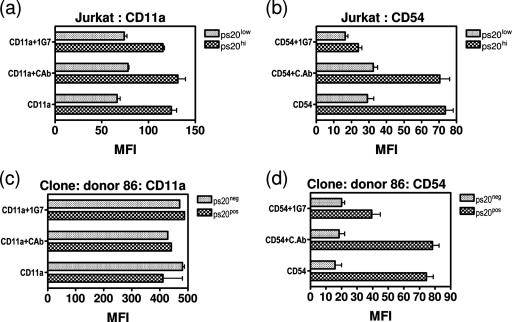
ps20 promotes infection by upregulating CD54 integrin expression.
We explored whether the HIV-regulatory effect of ps20 may be indirect, due to its known ability to promote cell piling/spheroid cell formation/migration and therefore potentially to promote cell-cell adhesion (31, 32, 34, 40). In the context of HIV, we examined if ps20 altered T-cell adhesiveness subtly, specifically by regulating the LFA-1/CD54 integrin pathway, which is well recognized to promote HIV infection of CD4+ CD45RO+ T cells (see reference 44). Experiments were conducted on populations that were homogeneous for ps20 expression using both the Jurkat cells engineered to overexpress ps20 versus the EV control (Jurkat G91 and EV) and a primary ps20pos versus ps20neg clone. LFA-1 and CD54 were two- and threefold higher, respectively, on ps20hi G91 Jurkat cells versus the EV control (Fig. 8a and b, respectively). A ps20pos clone also had 4.7-fold-higher CD54 than its ps20− counterpart (Fig. 8d), but LFA-1 was high for both P and NP clones (Fig. 8c), consistent with that seen on activated memory T cells (44). CD54 but not LFA-1 expression was reduced by 69% to the EV control level for G91 (a CD54 MFI of 73 down to 23 for G91 [Fig. 8b]) and by 50% (MFI reduced from 78 to 39) in ps20pos clone 86 (Fig. 8d) by preculturing cells for 4 days with anti-ps20 Ab, highlighting a potential role for ps20 in regulating CD54 expression.
FIG. 8.
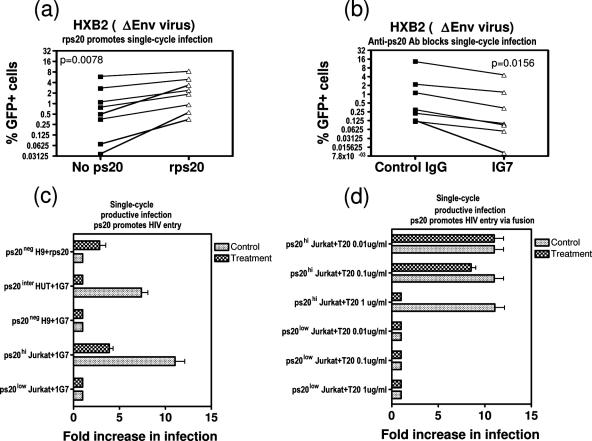
ps20 upregulates CD54. Cells (2 × 105) (ps20low EV versus ps20hi G91 Jurkat cells or ps20+ clone 86 1-3 versus ps20− NP clone 86 1-1) were directly stained for CD11a (a and c, respectively) and CD54 (b and d, respectively) by standard direct immunostaining, and MFIs from replicate cultures were determined. Cells were precultured with either 5 μg/ml control mouse IgG or IG7 for 4 days prior to staining. Untreated controls were included as indicated.
As cell adhesion factors including CD54 are known to promote HIV entry, we next explored whether ps20 promoted an early step of the virus life cycle. A single-cycle infection assay with GFP-encoding lentivirus particles pseudotyped with an HIV envelope (HXB2) prepared in a ps20low producer cell (293T) helped define the stage of the virus life cycle regulated by ps20. rps20 enhanced the infection of ps20low NP clones (median enhancement [n-fold], 2.575 [range, 1.467 to 21]) (Fig. 9a); conversely, the addition of IG7 Ab blocked infection of ps20+ permissive cells (median inhibition [n-fold], 2.4 [range, 1.4 to 3.4]) (Fig. 9b). Both the rps20 enhancing effect and the anti-ps20-mediated inhibition were more pronounced at lower challenge doses (i.e., <1% GFP+ cells), consistent with ps20 promoting an early step of the virus life cycle. This was also confirmed in a single-cycle assay with replication-competent HIV and extended to determine if HIV entry into ps20+ cells occurred via fusion and therefore was sensitive to a fusion inhibitor, especially as the expression of adhesion molecules like ICAM-3 promote virus endocytosis (41). In Fig. 9c and d, we confirm and illustrate the following. (i) ps20hi G91 cells were more susceptible to infection (by 10- to 13-fold) than the ps20low EV2 control (Fig. 9c). (ii) The infection of ps20hi G91 was completely blocked by the addition of the HIV fusion inhibitor T20 in a concentration-dependent manner (Fig. 9d). (iii) The HIV-blocking effect of the Ab was lower for a CD54hi ps20hi cell: 5 μg/ml IG7 completely blocked infection of ps20intermediate HUT 78 cells, while this amount resulted in a 60% inhibition of ps20hi G91 (ps20 mRNA molecules per cell in HUT versus G91 is equivalent to 0.34 versus 1,380, respectively) (Fig. 9c). (iv) Conversely, rps20 enhanced single-round productive infection by threefold in ps20neg H9 cells (Fig. 9c).
FIG. 9.
ps20 promotes HIV entry via fusion. (a) Cells (2 × 105) from NP clones (86 1-1 and 8.5.7.05) were precultured for 18 h with 1 μM rps20 and then infected with replication-defective GFP-encoding lentivirus particles pseudotyped with HIV-1 envelope (strain HXB2); three doses were offered (dose 1, 0.1 to 0.5% GFP+ cells; dose 2, 0.5 to 1% GFP+ cells; dose 3, >2% GFP+ cells). Cultures were maintained in 30 IU/ml IL-2. Mean percentages of GFP+ cells at 6 days postinfection in the absence and presence of rps20 are shown. Paired t testing was used to calculate statistical differences between treatments. (b) Cells (2 × 105) from two P clones (86 1-3 and 8.16.7.05) were precultured for 18 h with 5 μg/ml control mouse IgG or IG7 and then infected with replication-defective GFP-encoding lentivirus particles exactly as described for panel a. Percentages of GFP+ cells for each culture at 6 days postinfection in the presence of control IgG versus that of IG7 are shown. Paired t testing was used to calculate statistical differences between treatments. (c and d) Cells (2 × 105) were infected with NL4-3 (10 ng p24-CA/million cells) in the presence (treatment) or absence (control) of the following modulators: fusion inhibitor T20 (kind gift of B. Peters) and 0.03 μg/ml rps20. Where treatment included culturing with 5 μg/ml IG7, the control was 5 μg/ml normal mouse IgG. Twenty-four hours later, cells were washed and cell lysates harvested for p24-CA measurement. Increases (n-fold) in the infection of each sample were based on an additional sample treated in parallel, exposed to HIV for 1 min, and taken as 1.0.
Cellular profiling of a ps20+ P CD4 T-cell line shows cells at an early/intermediate stage of differentiation.
We used populations that were homogeneous for ps20 (the well-characterized primary P/NP clones [Fig. 1a]) to build a cellular profile of a ps20+ permissive CD4 T-cell line. Data summarized in Table 1 provide the following picture. (i) Cellular resistance is not associated with lower HIV receptor/coreceptor expression. Indeed, the three ps20hi permissive clones had up to twofold-lower expression of the CCR5 coreceptor. (ii) All clones were memory cells at an early/intermediate differentiation stage, as judged by being CD45RO+/CD25+/CD28+/CD57−. (iii) There were no apparent differences in the proliferative capacities of P/NP cells or in their Th1/2 cytokine profiles. Both P and NP clones could be Th2 or Th0 (Th1 clones were not examined, as previous studies have already shown that Th1 cells are less permissive to HIV than Th2 and Th0 cells [47]).
TABLE 1.
Cellular profiles of P/NP clones
| T-cell type or cytokine | % or increase (n-fold) for clone pair:
|
|||||
|---|---|---|---|---|---|---|
| 1 (HIV-negative donor)
|
2 (HIV+, long-term nonprogressor)
|
3 (HIV+, receiving highly active antiretroviral therapy)
|
||||
| NP | P | NP | P | NP | P | |
| T-cell type (%)a | ||||||
| CD3 | 91.75 | 91.72 | 72 | 84 | 76.67 | 76.8 |
| CD4 | 92.36 | 94.39 | 65 | 75 | 68.42 | 84.95 |
| CXCR4 | 15.69 | 10.4 | 65 | 39 | 42.36 | 21.73 |
| CCR5 | 12.13 | 6.39 | 41.5 | 23 | 24.37 | 16.18 |
| CD45RO | 98.37 | 98.19 | 93.9 | 74 | 86.89 | 96.49 |
| CD25 | 93.95 | 98.28 | 91.3 | 93.1 | 79.12 | 85.82 |
| CD28 | 32.53 | 46.32 | 38.3 | 17.7 | 71 | 69.98 |
| CD57 | 2.22 | 0.65 | 1.9 | 2.1 | 0.97 | 0.54 |
| Cell number (fold increase)b | 5.5 | 6 | 5.7 | 7 | 6.7 | 5.7 |
| IFN-γ+ (%) | 0.1 | 0.01 | 0.06 | 0 | 0.99 | 4.31 |
| IL-4+ (%) | 88 | 72 | 89.88 | 88.25 | 11.5 | 8.3 |
| IFN-γ+ and IL-4+ (%) | 1.3 | 1.5 | 5.97 | 2.33 | 78.46 | 66.27 |
Numbers are percentages of cells expressing a given CD marker as determined by standard flow cytometry using directly conjugated antibodies relative to the isotype control.
Numbers are increases (n-fold) in cell number 7 days after activation with PHA, allogeneic irradiated PBMC, and IL-2. Intracytoplamic cytokine staining to enumerate frequencies of IFN-γ+ and IL-4+ cells was as previously described (17). Two-color immunofluorescence was used to determine the frequencies of cells expressing IL-4 but no IFN-γ and vice versa as well as the numbers of cells expressing both cytokines.
ps20 is elevated in HIV-1 infection.
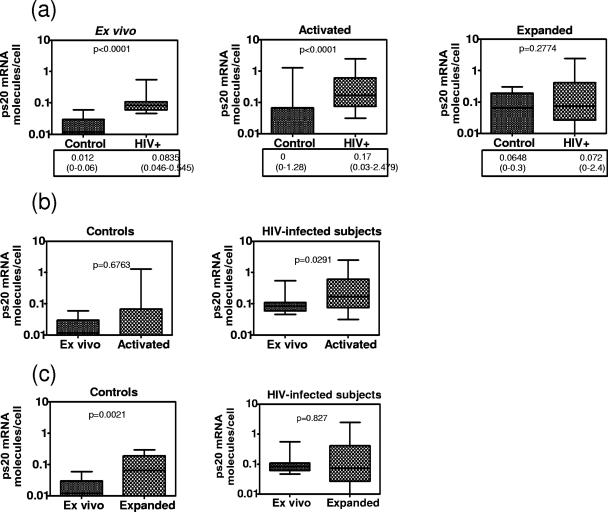
As ps20 mRNA was readily detected in activated/IL-2-expanded but rarely in ex vivo memory CD4s (Fig. 1b), we hypothesized that its expression is likely to be higher in HIV-infected subjects due to the well-recognized higher activation status of T cells in HIV infection. Samples from seven chronically infected treatment-naïve HIV+ patients showed significantly elevated ps20 mRNA levels in freshly isolated CD4 memory T cells (ex vivo) versus what was seen for control samples and similarly elevated levels in PHA/IL-2-stimulated cultures (P < 0.0001). However, no differences were noted in the IL-2-expanded cultures between patients and controls (Fig. 10a). Despite high endogenous levels, 48 h of PHA stimulation further upregulated ps20 in HIV+ but not in control samples (Fig. 10b). However, repeated restimulation of patients' cells reduced ps20 mRNA expression from 0.17 molecules/cell (median) after one round of 48-h PHA activation to 0.072 molecules/cell (median) after expansion (Fig. 10a and c). Significant ps20 expression in control cultures was observed only following IL-2 expansion, reaching levels observed for patients (Fig. 10a, right). These data suggest that ps20 is normally expressed in activated CD4 memory T cells, that ps20+ cells can be enriched by repeated restimulation, and that the propensity to find such cells is higher for blood from HIV-infected patients than that from healthy volunteers.
FIG. 10.
Higher ps20 mRNA in HIV+ than in control donors. (a) “Ex vivo” refers to CD4 CD45RO+ T cells freshly isolated from PBMC by negative selection. “Activated” refers to CD4+ CD45RO+ T cells isolated from 48-h PHA-activated PBMC. “Expanded” refers to CD4+ CD45RO+ T cells expanded with two rounds of activation with allogeneic PBMC-PHA-IL-2. Cells from six control donors and seven HIV+ subjects were examined for ps20 mRNA by qRT-PCR; mean numbers of ps20 molecules per cell from three to seven replicates for each sample are shown. Group differences were determined by nonparametric Mann-Whitney testing; median and range are shown. (b) ps20 mRNA level were analyzed by qRT-PCR as described for panel a. ps20 levels in activated versus ex vivo samples in control and HIV+ subjects are shown. (c) ps20 mRNA levels were analyzed by qRT-PCR as described for panel a. ps20 levels in expanded versus ex vivo samples in control and HIV+ subjects are shown.
DISCUSSION
Determining why HIV preferentially targets some CD4+ CD45RO+ T cells may identify novel pathways that can help preserve those CD4 T cells, whose loss is associated with impaired immunity and progression to disease (4, 6, 7, 8). Here we report that the differential expression of a novel secreted factor, ps20, can contribute to a hierarchy of susceptibility to HIV infection, with the most permissive CD4 T cells being ps20+. The ability to isolate stable clonal populations that are ps20hi versus ps20low from a given donor's blood CD4 T cells is proof that the peripheral CD4 T-cell pool is heterogeneous regarding ps20 expression, comprising of a mixture of ps20hi and ps20low/neg CD4 T-cell subsets. This is further supported by data on the two isogenic subclones of a CD4 T-cell immortalized line, H9 and HUT78, which differ markedly in ps20 expression. Taken together with the observation that ps20hi cells are more permissive to HIV, this paper highlights ps20 as a novel marker that distinguishes P and NP cells within the CD4 T-cell compartment.
Comparing ps20hi versus ps20low/neg isogenic cell populations assessed the potency of the ps20 effect. The level of virus suppression achieved in homogeneous ps20+ populations from multiple donors (primary clones, immortalized lines) by depriving cells of endogenous ps20 following infection with HIV strains prepared in different producer cells (PBMC/293T) with a neutralizing anti-ps20 Ab was significant (up to 15-fold), particularly when anti-ps20 Ab level was maintained. This was further confirmed by the up-to-31-fold HIV inhibition achieved by knocking down endogenous ps20 with specific siRNA in a heterologous HeLa system, demonstrating the importance of the HIV-ps20 effect beyond CD4 T cells. Included in the broad family of proteins that ps20 falls under are antimicrobial proteins expressed in mucosal sites as part of the innate immune response; some of these are known to regulate HIV infection (WAPs and defensins) (11, 48). The potency of the ps20/HIV effect is consistent with this class of proteins. Four experimental procedures (TCID50 determination, infection spreading, single-cycle infection, and ps20 knockdown studies) show HIV dependency on endogenous ps20 to be virus dose dependent, consistent with a number of saturable, positive-acting, and negative-acting host factors known to interact with HIV and govern the infection threshold (24, 43). Our experiments were also designed to assess the physiological relevance of the ps20 pathway. As ps20 was rarely expressed in unstimulated ex vivo CD4+ CD45RO+ T cells, we used IL-2-expanded oligoclonal populations from five donors to gauge the breadth of the ps20 effect. HIV infection in every donor's cells tested was suppressed by the addition of the anti-ps20 Ab, confirming endogenous ps20 to be a broad permissivity factor. The potency of the ps20 effect even in these heterogeneous cell populations assessed against a single high virus challenge dose varied between 2- and 29-fold; this variation in the anti-HIV effect of the IG7 Ab could reflect donor differences in (i) absolute amounts of ps20 produced, (ii) frequencies of ps20+ cells, (iii) cumulative levels of other HIV regulatory factors that impact the ps20 pathway, and (iv) ps20 binding partner(s); this is the subject of a separate ongoing investigation.
The absolute level of ps20 mRNA level in CD4 T cells is comparable to those seen for other CD4 cytokines (3). However, unlike cytokines that are rapidly induced by TCR ligation, ps20 is normally expressed only after restimulation and IL-2-expansion, similar to other late-acting factors, like the T-cell immunoglobulin proteins, expressed on some but not all CD4 memory T cells only after repetitive in vitro activation (18). Whether ps20 plays a role in CD4 T-cell differentiation is not known. Based on its expression, we predict that it is unlikely to identify CD4 T cells at either end of the differentiation spectrum: resting cells or CD57+ terminally differentiated functionally impaired cells that accumulate in HIV infection (26). Further phenotypic data are needed to ascribe ps20 to defined effector versus central memory subsets (26), but judged by proliferative capacity and some cell surface markers (Table 1), ps20 marks a subset of CD4 T cells at an early differentiation stage that have previously been described to be preferentially targeted by HIV in vivo (CD45RO+/CD28+/CD57−) and whose preservation is likely associated with nonprogression (4, 6, 7, 26). The observation that CD4 T cells from HIV patients constitutively express ps20 mRNA therefore suggests that these cells are not in a resting state, consistent with in vivo activation (26, 43). These observations suggest that ps20+ cells may be preferentially targeted in vivo and promote virus spread by autocrine or paracrine effects, thereby playing a role in disease pathogenesis. Pertinent to this issue is the question of whether HIV infection can directly regulate ps20, thereby further amplifying the ps20 HIV effect, a subject under investigation.
Clonal analysis revealed an exceptional clone in 8.5.7.99, which was resistant to R5 HIV-1 infection despite ps20 expression. β-Chemokine-independent R5 HIV resistance of CD4 memory cells (10, 17, 38, 46) may involve additional host factors. The tetraspanin protein CD63, for example, is exploited by R5 HIV but not by X4 HIV (45); cell surface annexin 11 can serve as a cofactor for HIV infection of macrophages but not lymphocytes (35). Indeed, the interaction of R5 HIV versus that of X4 HIV regulates distinct patterns of host genes (16) that in turn might regulate infection. The ability of anti-ps20 Ab to suppress both R5 and X4 HIV spread suggests that both strains exploit ps20 but that some CD4 cells possess specific restrictions to R5 HIV-1 infection that are apparently not overcome by ps20. Further clonal analysis will determine whether clone 8.5.7.99 is an exception or simply rare.
The interactions of WAPs with viruses including HIV can be diverse, and their better-known function as serine protease inhibitors is not necessarily linked to their anti-infective properties. Mutations introduced in the active site of protease inhibition of the WAP SLPI did not abrogate its anti-HIV activity (33, 35). The broad family of innate immune mediators, including antimicrobial peptides and WAPs, may regulate multiple steps of the virus life cycle, suppressing HIV infection by direct virolysis by inhibiting HIV-1 LTR transcription and blocking cell entry (20, 27, 48). Conversely, some members of this family, now including ps20, can enhance HIV infection. One mechanism of enhancement involves promoting entry-dependent steps by inducing receptor copatching (e.g., alpha-1-antitrypsin [9]) or by promoting the fusion of viral and cellular membranes via the insertion of amphipathic peptides, thereby weakening the lipid bilayer (25). Therefore, HIV exploitation of WAPs, like that of ps20, may reflect virus adaptation to their anti-infective functions (e.g., viral subversion [30]) and/or exploitation of the physiological properties of this class of proteins.
Our data are consistent with the notion that HIV has coopted, for its benefit, a fundamental role of ps20 in tissue remodeling and repair processes commonly associated with the inflammatory response and cancer progression (5). Previously, ps20 has been shown to affect cell proliferation, cell piling, and the formation of multicellular spheroids in prostate stromal cells (32, 40), indicating that this protein may affect cell adhesion and cell-cell interactions. These data suggest that ps20 may function as a secreted extracellular matrix protein. In addition, ps20 has exhibited chemokine-like properties by promoting endothelial cell migration and angiogenesis in vivo (34). Accordingly, it is possible that both the cell adhesion and chemokine-like functions of ps20 could be coopted by HIV as permissive components of infection pathways. At physiological levels, ps20 appeared to reconfigure cell phenotype by directly upregulating CD54 integrin (confirmed by CD54 downregulation by anti-ps20 Ab). Ongoing studies on the molecular signature of a ps20+ CD4 T cell line will determine whether or not CD54 integrin is one of many adhesion molecules regulated by ps20. The LFA-1/CD54 pathway is recognized to promote HIV entry into memory cells by stabilizing the virus-host fusion process (44). In addition, HIV is known to pick up a number of host factors, including LFA-1/CD54, during budding, thereby making the resultant virus more infectious in a spreading assay (14). Thus, the mechanisms through which ps20 modulates cell adhesion are likely to be important for ps20 action in promoting virus infectivity. If this is the case, depriving cells of ps20 may be therapeutically advantageous both by reducing new infections (transmission) and by ameliorating ongoing infection (virus spread).
Acknowledgments
This work was funded by grants from the Medical Research Council (G9901428) and Guy's and St Thomas' Charities (R050722) to A.V. and NIH grants R01DK45909 and R01CA58093 to D.R.
We thank M. Malim for discussions and M. Malim and P. J. Lachmann for manuscript review.
Footnotes
Published ahead of print on 17 October 2007.
REFERENCES
- 1.Alfano, M., and G. Poli. 2005. Role of cytokines and chemokines in the regulation of innate immunity and HIV infection. Mol. Immunol. 42161-182. [DOI] [PubMed] [Google Scholar]
- 2.Ashcroft, G. S., K. Lei, W. Jin, G. Longenecker, A. B. Kulkarni, T. Greenwell-Wild, H. Hale-Donze, G. McGrady, X. Y. Song, and S. M. Wahl. 2000. Secretory leukocyte protease inhibitor mediates non-redundant functions necessary for normal wound healing. Nat. Med. 61147-1153. [DOI] [PubMed] [Google Scholar]
- 3.Bas, A., G. Forsberg, S. Hammarstrom, and M. L. Hammarstrom. 2004. Utility of the housekeeping genes 18S rRNA, beta-actin and glyceraldehyde-3-phosphate-dehydrogenase for normalization in real-time quantitative reverse transcriptase-polymerase chain reaction analysis of gene expression in human T lymphocytes. Scand. J. Immunol. 59566-573. [DOI] [PubMed] [Google Scholar]
- 4.Boaz, M. J., A. Waters, S. Murad, P. J. Easterbrook, and A. Vyakarnam. 2002. Presence of HIV-1 Gag-specific IFN-gamma+IL-2+ and CD28+IL-2+ CD4 T cell responses is associated with nonprogression in HIV-1 infection. J. Immunol. 1696376-6385. [DOI] [PubMed] [Google Scholar]
- 5.Bouchard, D., D. Morisset, Y. Bourbonnais, and G. M. Tremblay. 2006. Proteins with whey-acidic-protein motifs and cancer. Lancet Oncol. 7167-174. [DOI] [PubMed] [Google Scholar]
- 6.Brenchley, J. M., B. J. Hill, D. R. Ambrozak, D. A. Price, F. J. Guenaga, J. P. Casazza, J. Kuruppu, S. A. Migueles, M. Connors, M. Roederer, D. C. Douek, and R. A. Koup. 2004. T-cell subsets that harbor human immunodeficiency virus (HIV) in vivo: implications for HIV pathogenesis. J. Virol. 781160-1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brenchley, J. M., L. E. Ruff, J. P. Casazza, R. A. Koup, D. A. Price, and D. C. Douek. 2006. Preferential infection shortens the life span of human immunodeficiency virus-specific CD4+ T cells in vivo. J. Virol. 806801-6809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brenchley, J. M., T. W. Schacker, L. E. Ruff, D. A. Price, J. H. Taylor, G. J. Beilman, P. L. Nguyen, A. Khoruts, M. Larson, A. T. Haase, and D. C. Douek. 2004. CD4+ T cell depletion during all stages of HIV disease occurs predominantly in the gastrointestinal tract. J. Exp. Med. 200749-759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bristow, C. L., D. R. Mercatante, and R. Kole. 2003. HIV-1 preferentially binds receptors co-patched with cell-surface elastase. Blood 1024479-4486. [DOI] [PubMed] [Google Scholar]
- 10.Butera, S. T., T. L. Pisell, K. Limpakarnjanarat, N. L. Young, T. W. Hodge, T. D. Mastro, and T. M. Folks. 2001. Production of a novel viral suppressive activity associated with resistance to infection among female sex workers exposed to HIV type 1. AIDS Res. Hum. Retrovir. 17735-744. [DOI] [PubMed] [Google Scholar]
- 11.Chang, T. L., and M. E. Klotman. 2004. Defensins: natural anti-HIV peptides. AIDS Rev. 6161-168. [PubMed] [Google Scholar]
- 12.Chen, K., J. Huang, C. Zhang, S. Huang, G. Nunnari, F. X. Wang, X. Tong, L. Gao, K. Nikisher, and H. Zhang. 2006. Alpha interferon potently enhances the anti-human immunodeficiency virus type 1 activity of APOBEC3G in resting primary CD4 T cells. J. Virol. 807645-7657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen, T. R. 1992. Karyotypic derivation of H9 cell line expressing human immunodeficiency virus susceptibility. J. Natl. Cancer Inst. 841922-1926. [DOI] [PubMed] [Google Scholar]
- 14.Chertova, E., O. Chertov, L. V. Coren, J. D. Roser, C. M. Trubey, J. W. Bess, Jr., R. C. Sowder II, E. Barsov, B. L. Hood, R. J. Fisher, K. Nagashima, T. P. Conrads, T. D. Veenstra, J. D. Lifson, and D. E. Ott. 2006. Proteomic and biochemical analysis of purified human immunodeficiency virus type 1 produced from infected monocyte-derived macrophages. J. Virol. 809039-9052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chimpanzee Sequencing and Analysis Consortium. 2005. Initial sequence of the chimpanzee genome and comparison with the human genome. Nature 43769-87. [DOI] [PubMed] [Google Scholar]
- 16.Cicala, C., J. Arthos, E. Martinelli, N. Censoplano, C. C. Cruz, E. Chung, S. M. Selig, D. Van Ryk, J. Yang, S. Jagannatha, T. W. Chun, P. Ren, R. A. Lempicki, and A. S. Fauci. 2006. R5 and X4 HIV envelopes induce distinct gene expression profiles in primary peripheral blood mononuclear cells. Proc. Natl. Acad. Sci. USA 1033746-3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ciuffi, A., G. Bleiber, M. Munoz, R. Martinez, C. Loeuillet, M. Rehr, M. Fischer, H. F. Gunthard, A. Oxenius, P. Meylan, S. Bonhoeffer, D. Trono, and A. Telenti. 2004. Entry and transcription as key determinants of differences in CD4 T-cell permissiveness to human immunodeficiency virus type 1 infection. J. Virol. 7810747-10754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.De Souza, A. J., T. B. Oriss, K. J. O'Malley, A. Ray, and L. P. Kane. 2005. T cell Ig and mucin 1 (TIM-1) is expressed on in vivo-activated T cells and provides a costimulatory signal for T cell activation. Proc. Natl. Acad. Sci. USA 10217113-17118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.DeVico, A. L., and R. C. Gallo. 2004. Control of HIV-1 infection by soluble factors of the immune response. Nat. Rev. Microbiol. 2401-413. [DOI] [PubMed] [Google Scholar]
- 20.Doumas, S., A. Kolokotronis, and P. Stefanopoulos. 2005. Anti-inflammatory and antimicrobial roles of secretory leukocyte protease inhibitor. Infect. Immun. 731271-1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Easterbrook, P. J. 1999. Long-term non-progression in HIV infection: definitions and epidemiological issues. J. Infect. 3871-73. [DOI] [PubMed] [Google Scholar]
- 22.Farquhar, C., T. C. Van Cott, D. A. Mbori-Ngacha, L. Horani, R. K. Bosire, J. K. Kreiss, B. A. Richardson, and G. C. John-Stewart. 2002. Salivary secretory leukocyte protease inhibitor is associated with reduced transmission of human immunodeficiency virus type 1 through breast milk. J. Infect. Dis. 1861173-1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Galli, G., F. Annunziato, C. Mavilia, P. Romagnani, L. Cosmi, R. Manetti, C. Pupilli, E. Maggi, and S. Romagnani. 1998. Enhanced HIV expression during Th2-oriented responses explained by the opposite regulatory effect of IL-4 and IFN-gamma of fusin/CXCR4. Eur. J. Immunol. 283280-3290. [DOI] [PubMed] [Google Scholar]
- 24.Goff, S. P. 2004. Retrovirus restriction factors. Mol. Cell 16849-859. [DOI] [PubMed] [Google Scholar]
- 25.Groot, F., R. W. Sanders, O. Ter Brake, K. Nazmi, E. C. Veerman, J. G. Bolscher, and B. Berkhout. 2006. Histatin 5-derived peptide with improved fungicidal properties enhances human immunodeficiency virus type 1 replication by promoting viral entry. J. Virol. 809236-9243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harari, A., V. Dutoit, C. Cellerai, P. A. Bart, R. A. Du Pasquier, and G. Pantaleo. 2006. Functional signatures of protective antiviral T-cell immunity in human virus infections. Immunol. Rev. 211236-254. [DOI] [PubMed] [Google Scholar]
- 27.Hiemstra, P. S., B. A. Fernie-King, J. McMichael, P. J. Lachmann, and J. M. Sallenave. 2004. Antimicrobial peptides: mediators of innate immunity as templates for the development of novel anti-infective and immune therapeutics. Curr. Pharm. Des. 102891-2905. [DOI] [PubMed] [Google Scholar]
- 28.Jana, N. K., L. R. Gray, and D. C. Shugars. 2005. Human immunodeficiency virus type 1 stimulates the expression and production of secretory leukocyte protease inhibitor (SLPI) in oral epithelial cells: a role for SLPI in innate mucosal immunity. J. Virol. 796432-6440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28a.King, D. F. L. 2005. PhD thesis. University of London, London, United Kingdom.
- 29.Kreisberg, J. F., W. Yonemoto, and W. C. Greene. 2006. Endogenous factors enhance HIV infection of tissue naive CD4 T cells by stimulating high molecular mass APOBEC3G complex formation. J. Exp. Med. 203865-870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lachmann, P. J. 2002. Microbial subversion of the immune response. Proc. Natl. Acad. Sci. USA 998461-8462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Larsen, M., S. J. Ressler, M. J. Gerdes, B. Lu, M. Byron, J. B. Lawrence, and D. R. Rowley. 2000. The WFDC1 gene encoding ps20 localizes to 16q24, a region of LOH in multiple cancers. Mamm. Genome 11767-773. [DOI] [PubMed] [Google Scholar]
- 32.Larsen, M., S. J. Ressler, B. Lu, M. J. Gerdes, L. McBride, T. D. Dang, and D. R. Rowley. 1998. Molecular cloning and expression of ps20 growth inhibitor. A novel WAP-type “four-disulfide core” domain protein expressed in smooth muscle. J. Biol. Chem. 2734574-4584. [DOI] [PubMed] [Google Scholar]
- 33.Ma, G., T. Greenwell-Wild, K. Lei, W. Jin, J. Swisher, N. Hardegen, C. T. Wild, and S. M. Wahl. 2004. Secretory leukocyte protease inhibitor binds to annexin II, a cofactor for macrophage HIV-1 infection. J. Exp. Med. 2001337-1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McAlhany, S. J., S. J. Ressler, M. Larsen, J. A. Tuxhorn, F. Yang, T. D. Dang, and D. R. Rowley. 2003. Promotion of angiogenesis by ps20 in the differential reactive stroma prostate cancer xenograft model. Cancer Res. 635859-5865. [PubMed] [Google Scholar]
- 35.McNeely, T. B., D. C. Shugars, M. Rosendahl, C. Tucker, S. P. Eisenberg, and S. M. Wahl. 1997. Inhibition of human immunodeficiency virus type 1 infectivity by secretory leukocyte protease inhibitor occurs prior to viral reverse transcription. Blood 901141-1149. [PubMed] [Google Scholar]
- 36.Nishimura, Y., T. Igarashi, O. K. Donau, A. Buckler-White, C. Buckler, B. A. Lafont, R. M. Goekem, S. Goldstein, V. M. Hirsch, and M. A. Martin. 2004. Highly pathogenic SHIVs and SIVs target different CD4+ T cell subsets in rhesus monkeys, explaining their divergent clinical courses. Proc. Natl. Acad. Sci. USA 10112324-12329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ranganathan, S., K. J. Simpson, D. C. Shaw, and K. R. Nicholas. 1999. The whey acidic protein family: a new signature motif and three-dimensional structure by comparative modeling. J. Mol. Graph Model. 17106-113. [DOI] [PubMed] [Google Scholar]
- 38.Riley, J. L., R. G. Carroll, B. L. Levine, W. Bernstein, D. C. St Louis, O. S. Weislow, and C. H. June. 1997. Intrinsic resistance to T cell infection with HIV type 1 induced by CD28 co-stimulation. J. Immunol. 1585545-5553. [PubMed] [Google Scholar]
- 39.Robichaud, G. A., B. Barbeau, J.-F. Fortin, D. M. Rothstein, and M. J. Tremblay. 2002. Nuclear factor of activated T cells is a driving force for preferential productive HIV-1 infection of CD45RO-expressing CD4+ T cells. J. Biol. Chem. 27723733-23741. [DOI] [PubMed] [Google Scholar]
- 40.Rowley, D. R., T. D. Dang, M. Larsen, M. J. Gerdes, L. McBride, and B. Lu. 1995. Purification of a novel protein (ps20) from urogenital sinus mesenchymal cells with growth inhibitory properties in vitro. J. Biol. Chem. 27022058-22065. [DOI] [PubMed] [Google Scholar]
- 41.Schaeffer, E., V. B. Soros, and W. C. Greene. 2004. Compensatory link between fusion and endocytosis of human immunodeficiency virus type 1 in human CD4 T lymphocytes. J. Virol. 781375-1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sheehy, A. M., N. C. Gaddis, J. D. Choi, and M. H. Malim. 2002. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature 418646-650. [DOI] [PubMed] [Google Scholar]
- 43.Stevenson, M. 2003. HIV-1 pathogenesis. Nat. Med. 9853-860. [DOI] [PubMed] [Google Scholar]
- 44.Tardif, M. R., and M. J. Tremblay. 2005. LFA-1 is a key determinant for preferential infection of memory CD4+ T cells by human immunodeficiency virus type 1. J. Virol. 7913714-13724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Von Lindern, J. J., D. Rojo, K. Grovit-Ferbas, C. Yeramian, C. Deng, G. Herbein, M. R. Ferguson, T. C. Pappas, J. M. Decker, A. Singh, R. G. Collman, and W. A. O'Brien. 2003. Potential role for CD63 in CCR5-mediated human immunodeficiency virus type 1 infection of macrophages. J. Virol. 773624-3633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vyakarnam, A., J. Eyeson, I. Teo, M. Zuckerman, K. Babaahmady, H. Schuitemaker, S. Shaunak, T. Rostron, S. Rowland-Jones, G. Simmons, and P. Clapham. 2001. Evidence for post-entry barrier to R5 HIV-1 infection of CD4 memory cells. AIDS 151613-1626. [DOI] [PubMed] [Google Scholar]
- 47.Vyakarnam, A., P. M. Matear, S. J. Martin, and M. Wagstaff. 1995. Th1 cells specific for HIV-1 gag p24 Gag are less efficient than Th0 cells in supporting HIV replication, and inhibit virus replication in Th0 cells. Immunology 8685-96. [PMC free article] [PubMed] [Google Scholar]
- 48.Wahl, S. M., T. Greenwell-Wild, and N. Vazquez. 2006. HIV accomplices and adversaries in macrophage infection. J. Leukoc. Biol. 80973-983. [DOI] [PubMed] [Google Scholar]
- 49.Wallace, D. L., P. M. Matear, D. C. Davies, R. Hicks, C. Lebosse, J. Eyeson, P. C. Beverley, and A. Vyakarnam. 2000. CD7 expression distinguishes subsets of CD4(+) T cells with distinct functional properties and ability to support replication of HIV-1. Eur. J. Immunol. 30577-585. [DOI] [PubMed] [Google Scholar]
- 50.Zaunders, J. J., S. Ip, M. L. Munier, D. E. Kaufmann, K. Suzuki, C. Brereton, S. C. Sasson, N. Seddiki, K. Koelsch, A. Landay, P. Grey, R. Finlayson, J. Kaldor, E. S. Rosenberg, B. D. Walker, B. Fazekas de St Groth, D. A. Cooper, and A. D. Kelleher. 2006. Infection of CD127+ (interleukin-7 receptor+) CD4+ cells and overexpression of CTLA-4 are linked to loss of antigen-specific CD4 T cells during primary human immunodeficiency virus type 1 infection. J. Virol. 8010162-10172. [DOI] [PMC free article] [PubMed] [Google Scholar]