Abstract
Hepatitis C virus-positive serum (HCVser, genotypes 1a to 3a) or HCV cell culture (JFH1/HCVcc) infection of primary normal human hepatocytes was assessed by measuring intracellular HCV RNA strands. Anti-CD81 antibodies and siRNA-CD81 silencing markedly inhibited (>90%) HCVser infection irrespective of HCV genotype, viral load, or liver donor, while hCD81-large intracellular loop (LEL) had no effect. However, JFH1/HCVcc infection of hepatocytes was modestly inhibited (40 to 60%) by both hCD81-LEL and anti-CD81 antibodies. In conclusion, CD81 is involved in HCVser infection of human hepatocytes, and comparative studies of HCVser versus JFH1/HCVcc infection of human hepatocytes and Huh-7.5 cells revealed that the cell-virion combination is determinant of the entry process.
We recently reported that the low-density lipoprotein receptor plays a critical role in serum-derived hepatitis C virus (HCVser) infection of human hepatocytes (26). However, several other receptor candidates have been proposed (4, 12, 24, 36, 37), including tetraspanin CD81 (32). HCV glycoprotein E2 binds with high affinity to the large extracellular loop (LEL) of human and chimpanzee CD81 in a genotype-dependent manner (29, 35, 38). In contrast, it does not bind mouse or rat CD81 and binds only weakly to African green monkey CD81 (14). A number of reports confirmed the role of CD81 in the entry of viral pseudoparticles (HCVpp) and cell culture particles (JFH1/HCVcc) in liver cell lines (6, 7, 10, 18, 22, 23, 43, 46). However, these experimental models do not mimic the natural infection process, and several lines of evidence indicate that the infectious properties of HCV may be modulated by host factors that are present in vivo (5, 27, 42).
The current study was undertaken to evaluate the role of CD81 in HCVser infection of highly differentiated normal primary human hepatocytes. Hepatocyte isolation, culture conditions, infection with serum from patients chronically infected with HCV, and treatments were performed as described elsewhere (9, 15, 26, 30). These cultures retain liver phenotypic markers (3, 8, 11, 13, 16, 28, 31) and are sensitive to HCV or hepatitis delta virus (HDV) infection (1, 9, 15, 26, 39). Intracellular accumulation of replicative and genomic HCV RNA strands, assessed by rTth-reverse transcription (RT)-PCR and quantitative RT-PCR, respectively, were used as experimental end points (9, 15, 20, 21, 26). Statistical analysis of data was performed using the nonparametric signed-rank Wilcoxon test (34). We verified that hepatocytes cultured under our standard conditions express both CD81 and CD9, a closely related tetraspanin (not shown). On average, after HCVser infection of CD81+/CD9+ hepatocytes at a genome equivalent per cell (Geq/cell) ratio of 0.025, the number of HCV genome copies per hepatocyte is 0.18 ± 0.10 (n = 13; range, 0.04 to 0.37).
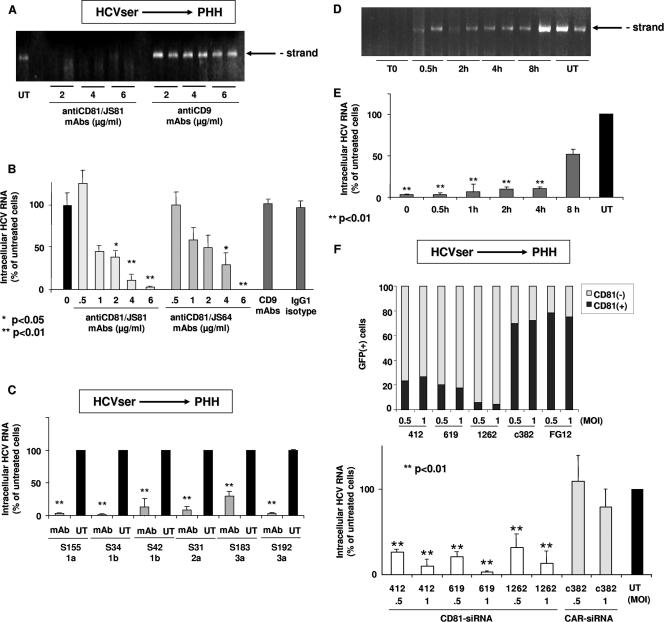
Anti-CD81 MAbs JS81 and JS64 inhibited HCVser (genotypes 1 to 3) infection of hepatocytes in a concentration-dependent manner (Fig. 1A to C). On average, MAb JS81 produced 92 ± 9% inhibition. Anti-CD9 MAb M-L13 or the mouse immunoglobulin G1 isotype control produced no inhibition. When cells were treated with MAb JS81 at the time of inoculation or shortly after (30 min), the accumulation of both negative and positive HCV RNA strands was clearly reduced (>90% on the positive strand) (Fig. 1D and E). However, as the interval between HCVser inoculation and anti-CD81 treatment increased, the inhibitory effect gradually decreased. When antibodies were added 8 h after inoculation, the inhibition was markedly reduced. Neither anti-CD81 nor anti-CD9 MAbs prevented HDV infection of hepatocytes (not shown). The inhibitory effect of anti-CD81 MAbs therefore interferes with an early step of HCVser cell entry and is specific to HCV infection.
FIG. 1.
Effect of anti-CD81 MAbs and of siRNA-mediated silencing of CD81 on HCVser infection of primary human hepatocytes (PHH). Three days postplating, hepatocytes FTBP (Biopredic) from a 52-year-old male with metastasis of a colic tumor were exposed or not (UT) to increasing concentrations (0.5 to 6 μg/ml) of anti-CD81 MAb JS81 or JS64, anti-CD9 MAb M-L13, or mouse immunoglobulin G1 isotype MOPC-21 for 30 min before inoculation with HCV-positive serum S298 (genotype 3a; 1.5 × 106 IU/ml). Following overnight exposure, cells were washed three times and the culture was continued. Five days postinoculation, cells were washed three times and total cellular RNA was extracted. (A) One μg of total RNA was analyzed by rTth-RT-PCR for the replicative RNA strand. (B) One μg of the same RNA sample was analyzed by real-time PCR for the genomic strand. RNA quality control was evaluated by performing a quantification of GAPDH mRNA in each sample. Amounts of HCV RNA were normalized to GAPDH mRNA. Results are expressed as the percent intracellular viral RNA relative to the amount obtained in in vitro-infected hepatocytes in the absence of treatment (0). Data are means from three independent assays. These results are representative of observations made with all other hepatocyte cultures from different donors used in this work. (C) Three days postplating, hepatocytes FT168 (75-year-old female with metastasis of a colic tumor) were exposed to HCV-positive serum S42 (genotype 1b, 6 × 106 IU/ml), S31 (genotype 2; 13 × 106 IU/ml), or S34 (genotype 1b; 60 × 106 IU/ml); hepatocytes FT196 (35-year-old female with metastasis of a colic tumor) were exposed to serum S155 (genotype 1a; 1.1 × 106 IU/ml); hepatocytes FT 212 (52-year-old male with metastasis of a colic tumor) were exposed to serum S183 (genotype 3a, 2.6 × 106 IU/ml); and hepatocytes FT249 (52-year-old male with metastasis of a colic tumor) were exposed to serum S192 (genotype 3a; 2.5 × 106 IU/ml) in the absence (UT) or presence of anti-CD81 MAbs JS81 at 4 μg/ml (mAb); further analysis was carried out as indicated above. Amounts of HCV RNA were normalized to GAPDH mRNA. Results are expressed as the percent intracellular viral RNA relative to the amount obtained in in vitro-infected hepatocytes in the absence of treatment (UT). Data are means from three independent assays. These results are representative of observations made with other hepatocyte cultures from different donors, FT161 (35-year-old male organ donor), 167 (57-year-old male with metastasis of a colic tumor), 176 (69-year-old female with metastasis of a colic tumor), 195 (17-year-old male organ donor), and FTBP. (D and E) Three days postplating, hepatocytes (FT196) were exposed to HCV-positive human serum S155. Cells were left untreated (UT) or treated with anti-CD81 MAbs JS81 (4 μg/ml) added either at the time of inoculation with HCV-positive serum (0), or 30 min, 1 h, 2 h, 4 h, or 8 h later, and further analysis was carried out as indicated above. (D) One μg of total RNA was analyzed by rTth-RT-PCR for the replicative RNA strand. (E) One μg of the same RNA sample was analyzed by real-time PCR for the genomic strand. Amounts of HCV RNA were normalized to GAPDH mRNA. Results are expressed as percent intracellular viral RNA relative to the amount obtained in in vitro infected hepatocytes in the absence of treatment (UT). Data are means from three independent assays. These results are representative of observations made with another hepatocyte culture from a different donor (FT195). (F) Hepatocytes FT259 (76-year-old male with hepatocellular carcinoma) were transduced overnight with suspensions of lentivirus expressing hCD81 siRNA 412, 619, or 1262, or expressing human CAR siRNA 382, at a multiplicity of infection (MOI) of 0.5 or 1. Nontransduced cells were used as the control (NT). Seven days later, GFP-positive hepatocytes were stained with anti-CD81 MAb JS-81 conjugated with Zenon and analyzed by flow cytometry. The results are expressed as the percentage of GFP-positive cells expressing (+) or not expressing (−) CD81 protein. Hepatocytes were then infected, on day 8, with HCV-positive serum sample S268 (genotype 1a; 4.9 × 106 IU/ml), and at day 3 postinoculation, the intracellular HCV RNA was analyzed by quantitative real-time RT-PCR as indicated above. All experiments were carried out in duplicate. These results are representative of those for various cell cultures and infectious serum samples S297 (genotype 3a; 1.5 × 106 IU/ml) for culture FT256 (43-year-old female with adenoma) and S268 and S301 (genotype 3a; 3.3 × 106 IU/ml) for culture FT259. Statistical analysis and P values were obtained using the Wilcoxon test.
To reduce CD81 expression, hepatocytes were transduced with FG12 lentivirus (33) expressing small interfering RNAs (siRNAs) directed against hCD81 mRNA, CD81/siRNA412 (5′-GCAGTTCTATGACCAGGCC-3′), CD81/siRNA619 (5′-GATCGATGACCTCTTCTCC-3′), or CD81/siRNA1262 (5′-GAAGGAACATCAGGCATGC-3′). Cells transduced with lentivirus expressing human constitutive androstane receptor (CAR) siRNA (5′-GCATGAGGAAAGACATGA; CAR/siRNAc382) (45) or green fluorescent protein (GFP) only were used as controls. More than 90% of cells were transduced as assessed by GFP expression, and Western blot analysis revealed that CD81 siRNA produced a >75% inhibition of CD81 expression (not shown). Levels of membrane-bound CD81 assessed by fluorescence-activated cell sorter analysis were strongly decreased in CD81 siRNA-transduced cells compared to CAR siRNA- or FG12-transduced cells (Fig. 1F). hCD81 siRNAs produced a consistent and significant inhibition (91 ± 6% at a multiplicity of infection of 1; P < 0.01) of HCVser genome replication, while CAR siRNA or empty lentivirus did not (Fig. 1F). These results confirm the role of CD81 in HCVser infection of human hepatocytes.
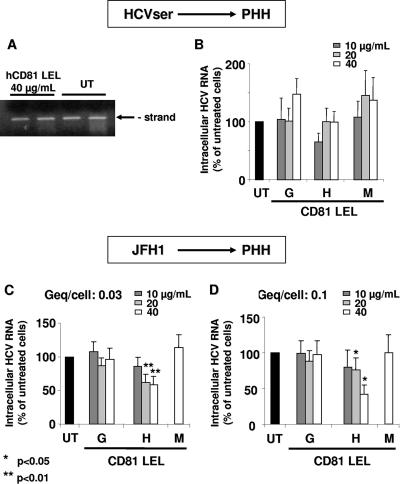
Recombinant CD81-LEL glutathione S-transferase fusion proteins from humans, African green monkeys, and mice were synthesized as described previously (17). We first verified that hCD81-LEL binds glycoprotein E2 and that LEL and anti-CD81 MAbs JS81 and JS64 markedly inhibit (>90%) JFH1/HCVcc infection of Huh-7.5 cells (not shown), as also observed by others (23, 43). Interestingly, hCD81-LEL did not inhibit HCVser infection of hepatocytes (Fig. 2A and B). As this result was rather unexpected, we compared HCVser and JFH1/HCVcc infections of human hepatocytes. We first established that under our standard conditions of infection (9, 15, 26), JFH1/HCVcc infects hepatocytes at Geq/cell ratios of 0.03 and 0.1, as assessed by both genomic and replicative HCVcc RNA strand accumulation in cells and production of infectious virions in the cell supernatant (not shown). The JFH1/HCVcc genomic strand level was 3.3 ± 1 (n = 3; range, 2.5 to 4.1) copies per hepatocyte after infection at a Geq/cell ratio of 0.03 (and 9.7 at a Geq/cell ratio of 0.1). This is an order of magnitude greater than that observed (0.18 ± 0.10) after HCVser infection at the same Geq/cell ratio. Unexpectedly, however, JFH1/HCVcc infection of human hepatocytes was inhibited significantly but only partly (40 to 60%) by both hCD81-LEL (Fig. 2C and D) and anti-CD81 MAbs (compare Fig. 3A and B and Fig. 1B), in sharp contrast with the strong inhibition (>90%) observed with JFH1/HCVcc infection of Huh-7.5 cells (not shown) and with HCVser infection of hepatocytes (Fig. 1) (92%).
FIG. 2.
Effect of recombinant mouse, green monkey, and human CD81-LEL on HCVser and JFH1/HCVcc infections of primary human hepatocytes (PHH). (A and B) HCV-positive serum S297 or S298 was preincubated for 30 min at 4°C with 10, 20, or 40 μg/ml green monkey (G), mouse (M), or human (H) CD81-LEL fusion proteins and, 3 days postplating, hepatocytes FT257 (30-year-old female with hydatid cyst) or FT259 were inoculated with these mixtures under our standard conditions. After exposure, cells were washed three times to remove excess inoculum, and the culture was continued for 3 days. Total cellular RNA was extracted, and 1 μg was analyzed by rTth-RT-PCR for the replicative RNA strand (FT257/S297) or by real-time PCR for the genomic strand. Amounts of HCV RNA were normalized to GAPDH mRNA. All experiments were carried out in duplicate. These results represent average data obtained from these four series of experiments (FT257/S297 or FT257/S298; FT259/S297 or FT259/S298). These experiments were repeated with two different lots of CD81-LEL fusion proteins with same results. (C and D) JFH1/HCVcc was preincubated or not (UT) with 10, 20, or 40 μg/ml of green monkey (G), human (H), or mouse (M; only 40 μg/ml) CD81-LEL for 30 min and used to inoculate hepatocytes FT273 (64-year-old male with metastasis of a colic tumor), FT276 (62-year-old female with metastasis of a colic tumor), and FT281 (56-year-old male with metastasis of a colic tumor) at different Geq/cell ratios (0.03 and 0.1) 3 days postplating. Three days later, cells were washed three times, and total cellular RNA was extracted. One μg of the same RNA sample was analyzed by real-time PCR for the genomic strand. RNA quality control was evaluated by performing a quantification of GAPDH mRNA in each sample. Amounts of HCV RNA were normalized to GAPDH mRNA. Results are expressed as Geq copies per cell. Data are means from three independent assays. Statistical analysis and P values were obtained using the Wilcoxon signed-rank test.
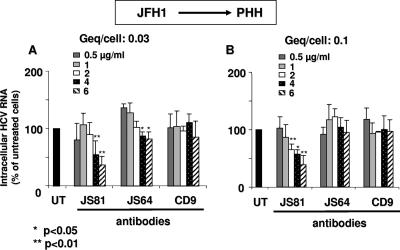
FIG. 3.
Effect of anti-CD81 MAbs on JFH1/HCVcc infection of primary human hepatocytes (PHH). Human hepatocytes (FT273, FT276, and FT281) were inoculated with JFH1/HCVcc at different Geq/cell ratios (0.03 and 0.1) in the absence (UT) or presence of 0.5 to 6 μg/ml of anti-CD81 MAb JS81 or JS64 added for 30 min before inoculation. Control experiments were performed under the same conditions with anti-CD9 MAbs M-L13. Following overnight exposure, cells were washed three times, and the culture was continued. Three days later, cells were washed three times, and total cellular RNA was extracted. One μg of total RNA was analyzed by real-time PCR for the genomic strand. RNA quality control was evaluated by performing a quantification of GAPDH mRNA in each sample. Amounts of HCV RNA were normalized to GAPDH mRNA. Results are expressed as a percentage of the control (UT). Data are means ± standard deviations for three independent assays. Statistical analysis and P values were obtained using the Wilcoxon signed-rank test.
The current observation that hCD81-LEL does not inhibit HCVser infection of hepatocytes, and only partly inhibits JFH1/HCVcc infection of hepatocytes, is puzzling on the basis that HCVser does infect human hepatocytes in a CD81-dependent manner that can be specifically inhibited with anti-CD81 MAbs or CD81-specific siRNA. Indeed, the present paper and previous studies have shown that hCD81-LEL strongly reduces HCVpp or JFH1/HVCcc infection of Huh-7 or Huh-7.5 cells (6, 7, 10, 18, 23, 43, 46). Several hypotheses may be proposed for this observation. First, it is possible that within the infectious HCVser lipoviroparticles (2, 19, 25, 41), lipoproteins prevent the interaction between glycoprotein E2 and hCD81-LEL. This idea is supported by the findings that viral particles from human plasma do not bind hCD81-LEL (44) and that some human sera have to be treated with a detergent (deoxycholate) to release HCV particles that are able to interact with CD81 in Huh-7 cells (40). Second, it is possible that the affinity of hCD81-LEL for E2 is much lower than that of membrane-bound CD81 in normal hepatocytes. Third, a primary binding of HCVser to a non-CD81 membrane molecule, for example, LDLR and SR-BI (14, 26), may trigger the virus binding to CD81 on the hepatocyte membrane so that hCD81-LEL cannot compete in the natural membrane environment.
In contrast to recombinant CD81-LEL proteins, which are expected to compete with the natural membrane receptor for binding virus particles, antibodies directly target the cellular receptor. Their efficiency is therefore expected to be greater. This is consistent with current results revealing that the efficacy of MAb JS81 is approximately one to two orders of magnitude greater than that of recombinant hCD81-LEL on a molar concentration basis (not shown). Anti-CD81 MAbs inhibited JFH1/HCVcc infection of primary hepatocytes weakly (60%) compared with HCVser infection of primary hepatocytes (92%), and JFH1/HCVcc infection of Huh-7.5 cells (>90%; not shown). It is possible that the higher input of infectious virus in JFH1/HCVcc versus HCVser infection of hepatocytes, the difference in affinity or kinetics of HCV interaction with Huh-7.5 versus hepatocyte membrane receptors, and the difference in membrane protein population in Huh-7.5 cells versus hepatocytes account for these differences in inhibition efficiency. In sum, the data obtained on recombinant CD81-LEL and anti-CD81 antibodies suggest that CD81 has a much greater impact on the entry of JFH1/HCVcc in Huh-7.5 cells than it does on the entry of HCVcc particles in primary hepatocytes. Nevertheless, other data provided here show that CD81 indeed mediates the entry of HCVser in hepatocytes. It is therefore possible that the nature of the cell-virion combination is an important determinant of the virus entry process.
It is interesting that JFH1/HCVcc particles are able to infect primary human hepatocytes as revealed here by the intracellular accumulation of both genomic and replicative strands (not shown). Indeed, although HCVser is infectious for primary hepatocytes, it has to be noted that less than 15% of the tested sera are actually infectious (15). The reason for this observation is unclear but could be related to the presence of anti-HCV antibodies in the sera of chronically infected patients. In addition, the Geq/cell ratio with HCVser can hardly be greater than 0.1 because of the limited amounts of infectious serum. Hence, the possibility to infect primary hepatocytes with unlimited amounts of JFH1/HCVcc at a much greater Geq/cell ratio will strongly facilitate future studies. The rate of genome replication of JFH1/HCVcc in human hepatocytes appears to be one order of magnitude greater than that observed with HCVser, whereas it is one order of magnitude smaller than that observed for Huh-7.5 cells at the same Geq/cell ratio. Thus, although JFH1/HCVcc particles are adapted to Huh-7.5 cells, they can replicate their genome and proliferate in primary human hepatocytes, albeit at a lower rate. This could result from the greater antiviral response of human hepatocytes than Huh-7.5 cells (L. Pichard-Garcia, S. Molina, and P. Maurel, unpublished results). On the other hand, the fact that the rate of genome replication of JFH1/HCVcc in hepatocytes is greater than that of HCVser suggests that either only a small portion of HCVser virions are infectious or antibodies present in the serum strongly affect virion infectivity.
In conclusion, CD81 plays a critical role in an early step of HCVser infection of normal primary human hepatocytes. Comparative studies on HCVser versus JFH1/HCVcc infection of human hepatocytes and on JFH1/HCVcc infection of human hepatocytes versus Huh-7.5 cells suggest that the nature of the cell-virion combination is a determinant factor for virus entry. This would be consistent with the fact that several receptor candidates have been identified for HCV and that their interactions with one another or with other membrane components might modulate the way they associate with the various virion populations.
Acknowledgments
We thank Shoshana Levy (Stanford University Medical Center) for providing the transformed Escherichia coli strains expressing recombinant CD81-LEL fusion proteins, Takaji Wakita (Tokyo Metropolitan Institute of Neuroscience) for providing the pJFH1 plasmid, Charles Rice (Rockefeller University, New York, NY) for providing the Huh-7.5 cell line, and François-Loïc Cosset, Marlène Dreux, and Dimitri Lavillette (Inserm, U412, Lyon, France) for their help in the preparation of lentivirus and their experiments on the CD81-LEL protein effect on HCVpp entry in Huh-7 cells.
This work was supported by grant 01031 from the “Agence Nationale de Recherches sur le SIDA et les hépatites virales” (ANRS) and grant 5869 C from the “Association pour la Recherche sur le Cancer” (ARC) to C.F.-W. Both V.C. and S.M. were supported by an ANRS fellowship. J.D. is an international scholar of the Howard Hughes Medical Institute.
Footnotes
Published ahead of print on 17 October 2007.
REFERENCES
- 1.Abou-Jaoudé, G., S. Molina, P. Maurel, and C. Sureau. 2007. Myristoylation signal transfer from the large to the middle or the small HBV envelope protein leads to a loss of HDV particles infectivity. Virology 365204-209. [DOI] [PubMed] [Google Scholar]
- 2.André, P., F. Komurian-Pradel, S. Deforges, M. Perret, J. L. Berland, M. Sodoyer, S. Pol, C. Bréchot, G. Paranhos-Baccalà, and V. Lotteau. 2002. Characterization of low- and very-low-density hepatitis C virus RNA-containing particles. J. Virol. 766919-6928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Assenat, E., S. Gerbal-Chaloin, D. Larrey, J. Saric, J. M. Fabre, P. Maurel, M. J. Vilarem, and J. M. Pascussi. 2004. Interleukin 1beta inhibits CAR-induced expression of hepatic genes involved in drug and bilirubin clearance. Hepatology 40951-960. [DOI] [PubMed] [Google Scholar]
- 4.Barth, H., C. Schafer, M. I. Adah, F. Zhang, R. J. Linhardt, H. Toyoda, A. Kinoshita-Toyoda, T. Toida, T. H. Van Kuppevelt, E. Depla, F. Von Weizsacker, H. E. Blum, and T. F. Baumert. 2003. Cellular binding of hepatitis C virus envelope glycoprotein E2 requires cell surface heparan sulfate. J. Biol. Chem. 27841003-41012. [DOI] [PubMed] [Google Scholar]
- 5.Barth, H., E. K. Schnober, F. Zhang, R. J. Linhardt, E. Depla, B. Boson, F. L. Cosset, A. H. Patel, H. E. Blum, and T. F. Baumert. 2006. Viral and cellular determinants of the hepatitis C virus envelope-heparan sulfate interaction. J. Virol. 8010579-10590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bartosch, B., J. Dubuisson, and F. L. Cosset. 2003. Infectious hepatitis C virus pseudo-particles containing functional E1-E2 envelope protein complexes. J. Exp. Med. 197633-642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bartosch, B., A. Vitelli, C. Granier, C. Goujon, J. Dubuisson, S. Pascale, E. Scarselli, R. Cortese, A. Nicosia, and F. L. Cosset. 2003. Cell entry of hepatitis C virus requires a set of co-receptors that include the CD81 tetraspanin and the SR-B1 scavenger receptor. J. Biol. Chem. 27841624-41630. [DOI] [PubMed] [Google Scholar]
- 8.Biron-Andreani, C., C. Bezat-Bouchahda, E. Raulet, L. Pichard-Garcia, J. M. Fabre, J. Saric, J. Baulieux, J. F. Schved, and P. Maurel. 2004. Secretion of functional plasma haemostasis proteins in long-term primary cultures of human hepatocytes. Br. J. Haematol. 125638-646. [DOI] [PubMed] [Google Scholar]
- 9.Castet, V., C. Fournier, A. Soulier, R. Brillet, J. Coste, D. Larrey, D. Dhumeaux, P. Maurel, and J. M. Pawlotsky. 2002. Alpha interferon inhibits hepatitis C virus replication in primary human hepatocytes infected in vitro. J. Virol. 768189-8199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cormier, E. G., F. Tsamis, F. Kajumo, R. J. Durso, J. P. Gardner, and T. Dragic. 2004. CD81 is an entry coreceptor for hepatitis C virus. Proc. Natl. Acad. Sci. USA 1017270-7274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Drocourt, L., J. C. Ourlin, J. M. Pascussi, P. Maurel, and M. J. Vilarem. 2002. Expression of CYP3A4, CYP2B6, and CYP2C9 is regulated by the vitamin D receptor pathway in primary human hepatocytes. J. Biol. Chem. 27725125-25132. [DOI] [PubMed] [Google Scholar]
- 12.Evans, M. J., T. von Hahn, D. M. Tscherne, A. J. Syder, M. Panis, B. Wolk, T. Hatziioannou, J. A. McKeating, P. D. Bieniasz, and C. M. Rice. 2007. Claudin-1 is a hepatitis C virus co-receptor required for a late step in entry. Nature 446801-805. [DOI] [PubMed] [Google Scholar]
- 13.Ferrini, J. B., L. Pichard, J. Domergue, and P. Maurel. 1997. Long-term primary cultures of adult human hepatocytes. Chem. Biol. Interact. 10731-45. [DOI] [PubMed] [Google Scholar]
- 14.Flint, M., T. von Hahn, J. Zhang, M. Farquhar, C. T. Jones, P. Balfe, C. M. Rice, and J. A. McKeating. 2006. Diverse CD81 proteins support hepatitis C virus infection. J. Virol. 8011331-11342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fournier, C., C. Sureau, J. Coste, J. Ducos, G. Pageaux, D. Larrey, J. Domergue, and P. Maurel. 1998. In vitro infection of adult normal human hepatocytes in primary culture by hepatitis C virus. J. Gen. Virol. 792367-2374. [DOI] [PubMed] [Google Scholar]
- 16.Gerbal-Chaloin, S., M. Daujat, J. M. Pascussi, L. Pichard-Garcia, M. J. Vilarem, and P. Maurel. 2002. Transcriptional regulation of CYP2C9 gene. Role of glucocorticoid receptor and constitutive androstane receptor. J. Biol. Chem. 277209-217. [DOI] [PubMed] [Google Scholar]
- 17.Higginbottom, A., E. R. Quinn, C. C. Kuo, M. Flint, L. H. Wilson, E. Bianchi, A. Nicosia, P. N. Monk, J. A. McKeating, and S. Levy. 2000. Identification of amino acid residues in CD81 critical for interaction with hepatitis C virus envelope glycoprotein E2. J. Virol. 743642-3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hsu, M., J. Zhang, M. Flint, C. Logvinoff, C. Cheng-Mayer, C. M. Rice, and J. A. McKeating. 2003. Hepatitis C virus glycoproteins mediate pH-dependent cell entry of pseudotyped retroviral particles. Proc. Natl. Acad. Sci. USA 1007271-7276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kanto, T., N. Hayashi, T. Takehara, H. Hagiwara, E. Mita, M. Naito, A. Kasahara, H. Fusamoto, and T. Kamada. 1995. Density analysis of hepatitis C virus particle population in the circulation of infected hosts: implications for virus neutralization or persistence. J. Hepatol. 22440-448. [DOI] [PubMed] [Google Scholar]
- 20.Lanford, R. E., D. Chavez, F. V. Chisari, and C. Sureau. 1995. Lack of detection of negative-strand hepatitis C virus RNA in peripheral blood mononuclear cells and other extrahepatic tissues by the highly strand-specific rTth reverse transcriptase PCR. J. Virol. 698079-8083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lanford, R. E., C. Sureau, J. R. Jacob, R. White, and T. R. Fuerst. 1994. Demonstration of in vitro infection of chimpanzee hepatocytes with hepatitis C virus using strand-specific RT/PCR. Virology 202606-614. [DOI] [PubMed] [Google Scholar]
- 22.Lavillette, D., A. W. Tarr, C. Voisset, P. Donot, B. Bartosch, C. Bain, A. H. Patel, J. Dubuisson, J. K. Ball, and F. L. Cosset. 2005. Characterization of host-range and cell entry properties of the major genotypes and subtypes of hepatitis C virus. Hepatology 41265-274. [DOI] [PubMed] [Google Scholar]
- 23.Lindenbach, B. D., M. J. Evans, A. J. Syder, B. Wolk, T. L. Tellinghuisen, C. C. Liu, T. Maruyama, R. O. Hynes, D. R. Burton, J. A. McKeating, and C. M. Rice. 2005. Complete replication of hepatitis C virus in cell culture. Science 309623-626. [DOI] [PubMed] [Google Scholar]
- 24.Lozach, P. Y., H. Lortat-Jacob, A. de Lacroix de Lavalette, I. Staropoli, S. Foung, A. Amara, C. Houles, F. Fieschi, O. Schwartz, J. L. Virelizier, F. Arenzana-Seisdedos, and R. Altmeyer. 2003. DC-SIGN and L-SIGN are high affinity binding receptors for hepatitis C virus glycoprotein E2. J. Biol. Chem. 27820358-20366. [DOI] [PubMed] [Google Scholar]
- 25.Miyamoto, H., H. Okamoto, K. Sato, T. Tanaka, and S. Mishiro. 1992. Extraordinarily low density of hepatitis C virus estimated by sucrose density gradient centrifugation and the polymerase chain reaction. J. Gen. Virol. 73715-718. [DOI] [PubMed] [Google Scholar]
- 26.Molina, S., V. Castet, C. Fournier-Wirth, L. Pichard-Garcia, R. Avner, D. Harats, J. Roitelman, R. Barbaras, P. Graber, P. Ghersa, M. Smolarsky, A. Funaro, F. Malavasi, D. Larrey, J. Coste, J. M. Fabre, A. Sa-Cunha, and P. Maurel. 2007. The low-density lipoprotein receptor plays a role in the infection of primary human hepatocytes by hepatitis C virus. J. Hepatol. 46411-419. [DOI] [PubMed] [Google Scholar]
- 27.Ng, T. I., H. Mo, T. Pilot-Matias, Y. He, G. Koev, P. Krishnan, R. Mondal, R. Pithawalla, W. He, T. Dekhtyar, J. Packer, M. Schurdak, and A. Molla. 2007. Identification of host genes involved in hepatitis C virus replication by small interfering RNA technology. Hepatology 451413-1421. [DOI] [PubMed] [Google Scholar]
- 28.Pascussi, J. M., A. Robert, M. Nguyen, O. Walrant-Debray, M. Garabedian, P. Martin, T. Pineau, J. Saric, F. Navarro, P. Maurel, and M. J. Vilarem. 2005. Possible involvement of pregnane X receptor-enhanced CYP24 expression in drug-induced osteomalacia. J. Clin. Investig. 115177-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Petracca, R., F. Falugi, G. Galli, N. Norais, D. Rosa, S. Campagnoli, V. Burgio, E. Di Stasio, B. Giardina, M. Houghton, S. Abrignani, and G. Grandi. 2000. Structure-function analysis of hepatitis C virus envelope-CD81 binding. J. Virol. 744824-4830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pichard, L., E. Raulet, G. Fabre, J. B. Ferrini, J. C. Ourlin, and P. Maurel. 2006. Human hepatocyte culture. Methods Mol. Biol. 320283-293. [DOI] [PubMed] [Google Scholar]
- 31.Pichard-Garcia, L., S. Gerbal-Chaloin, J. B. Ferrini, J. M. Fabre, and P. Maurel. 2002. Use of long-term cultures of human hepatocytes to study cytochrome P450 gene expression. Methods Enzymol. 357311-321. [DOI] [PubMed] [Google Scholar]
- 32.Pileri, P., Y. Uematsu, S. Campagnoli, G. Galli, F. Falugi, R. Petracca, A. J. Weiner, M. Houghton, D. Rosa, G. Grandi, and S. Abrignani. 1998. Binding of hepatitis C virus to CD81. Science 282938-941. [DOI] [PubMed] [Google Scholar]
- 33.Qin, X. F., D. S. An, I. S. Chen, and D. Baltimore. 2003. Inhibiting HIV-1 infection in human T cells by lentiviral-mediated delivery of small interfering RNA against CCR5. Proc. Natl. Acad. Sci. USA 100183-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Richardson, B. A., and J. Overbaugh. 2005. Basic statistical considerations in virological experiments. J. Virol. 79669-676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roccasecca, R., H. Ansuini, A. Vitelli, A. Meola, E. Scarselli, S. Acali, M. Pezzanera, B. B. Ercole, J. McKeating, A. Yagnik, A. Lahm, A. Tramontano, R. Cortese, and A. Nicosia. 2003. Binding of the hepatitis C virus E2 glycoprotein to CD81 is strain specific and is modulated by a complex interplay between hypervariable regions 1 and 2. J. Virol. 771856-1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saunier, B., M. Triyatni, L. Ulianich, P. Maruvada, P. Yen, and L. D. Kohn. 2003. Role of the asialoglycoprotein receptor in binding and entry of hepatitis C virus structural proteins in cultured human hepatocytes. J. Virol. 77546-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Scarselli, E., H. Ansuini, R. Cerino, R. M. Roccasecca, S. Acali, G. Filocamo, C. Traboni, A. Nicosia, R. Cortese, and A. Vitelli. 2002. The human scavenger receptor class B type I is a novel candidate receptor for the hepatitis C virus. EMBO J. 215017-5025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shaw, M. L., J. McLauchlan, P. R. Mills, A. H. Patel, and E. A. McCruden. 2003. Characterisation of the differences between hepatitis C virus genotype 3 and 1 glycoproteins. J. Med. Virol. 70361-372. [DOI] [PubMed] [Google Scholar]
- 39.Sureau, C., C. Fournier-Wirth, and P. Maurel. 2003. Role of N glycosylation of hepatitis B virus envelope proteins in morphogenesis and infectivity of hepatitis delta virus. J. Virol. 775519-5523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tan, Y. J., S. P. Lim, P. Ng, P. Y. Goh, S. G. Lim, Y. H. Tan, and W. Hong. 2003. CD81 engineered with endocytotic signals mediates HCV cell entry: implications for receptor usage by HCV in vivo. Virology 308250-269. [DOI] [PubMed] [Google Scholar]
- 41.Thomssen, R., S. Bonk, and A. Thiele. 1993. Density heterogeneities of hepatitis C virus in human sera due to the binding of beta-lipoproteins and immunoglobulins. Med. Microbiol. Immunol. 182329-334. [DOI] [PubMed] [Google Scholar]
- 42.von Hahn, T., B. D. Lindenbach, A. Boullier, O. Quehenberger, M. Paulson, C. M. Rice, and J. A. McKeating. 2006. Oxidized low-density lipoprotein inhibits hepatitis C virus cell entry in human hepatoma cells. Hepatology 43932-942. [DOI] [PubMed] [Google Scholar]
- 43.Wakita, T., T. Pietschmann, T. Kato, T. Date, M. Miyamoto, Z. Zhao, K. Murthy, A. Habermann, H. G. Krausslich, M. Mizokami, R. Bartenschlager, and T. J. Liang. 2005. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat. Med. 11791-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wünschmann, S., J. D. Medh, D. Klinzmann, W. N. Schmidt, and J. T. Stapleton. 2000. Characterization of hepatitis C virus (HCV) and HCV E2 interactions with CD81 and the low-density lipoprotein receptor. J. Virol. 7410055-10062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yamamoto, Y., T. Kawamoto, and M. Negishi. 2003. The role of the nuclear receptor CAR as a coordinate regulator of hepatic gene expression in defense against chemical toxicity. Arch. Biochem. Biophys. 409207-211. [DOI] [PubMed] [Google Scholar]
- 46.Zhong, J., P. Gastaminza, G. Cheng, S. Kapadia, T. Kato, D. R. Burton, S. F. Wieland, S. L. Uprichard, T. Wakita, and F. V. Chisari. 2005. Robust hepatitis C virus infection in vitro. Proc. Natl. Acad. Sci. USA 1029294-9299. [DOI] [PMC free article] [PubMed] [Google Scholar]