Abstract
Human immunodeficiency virus (HIV)'s tremendous sequence variability is a major obstacle for the development of cytotoxic-T-lymphocyte-based vaccines, especially since much of this variability is selected for by CD8+ T cells. We investigated to what extent reactivity to escape variant peptides in standard enzyme-linked immunospot (ELISPOT) assays predicts the recognition of cells infected with corresponding escape variant viruses. Most of the variant peptides tested were recognized in standard ELISPOT and intracellular cytokine stain (ICS) assays. Functional avidity of epitope-specific T cells for some of the variants was, however, markedly reduced. These mutations which reduced avidity also abrogated recognition by epitope-specific CD8+ T cells in a viral suppression assay. Our results indicate that “cross-reactive” CD8+ T-cell responses identified in ELISPOT and ICS assays using a single high concentration of variant peptide often fail to predict the recognition of cells infected with variant viruses.
Vaccines are often designed to elicit antibodies which neutralize pathogens, thereby potentially preventing infection outright or serving to blunt pathogen replication and allow control by other arms of the immune system (7). Due to the extreme difficultly in eliciting broadly neutralizing antibodies against human immunodeficiency virus (HIV), the AIDS vaccine field now also focuses on the development of vaccines that elicit effective CD8+ T-cell responses against HIV (28). This approach holds promise because CD8+ T cells are convincingly implicated in the control of HIV; rare individuals who spontaneously control HIV infection tend to have particular class I major histocompatibility complex (MHC) alleles (8, 18, 25, 33), while depleting CD8+ cells from simian immunodeficiency virus (SIV)-infected rhesus macaques leads to increased viral replication (14, 19, 22, 29). However, CD8+ T cells frequently select for variation within targeted epitopes (1, 27). The extent to which vaccine-induced CD8+ T cells recognize variants is a crucial question when measuring the ability of vaccine-induced CD8+ T cells to recognize likely challenge viruses—frequent cross-recognition of variant epitopes would suggest that a single vaccine construct might elicit responses effective against diverse viral isolates. Encouragingly, gamma interferon (IFN-γ) enzyme-linked immunospot (ELISPOT) assays indicate that CD8+ T cells generated by both vaccination (23, 32) and infection (9, 11, 12, 15, 24, 30, 38, 39) often recognize sequences derived from multiple clades of HIV. ELISPOT and intracellular cytokine stain (ICS) assays typically involve stimulating T cells with purified peptides though, bypassing processing and trafficking steps which must occur in an infected cell for a class I MHC molecule to successfully present peptides. These assays may provide an accurate picture of virus-derived peptides against which an individual makes T-cell responses, and the frequency of these responding cells, but do not necessarily provide information about CD8+ T cell' effectiveness against virus-infected target cells. Results from our laboratory (20) and others (4, 5, 37) have suggested that use of high peptide concentrations might lead to spurious stimulation of CD8+ T-cell responses in vitro, and the physiological relevance of such assays has been questioned (34, 35). Therefore, we sought to determine how effectively CD8+ T cells recognize variant sequences in ELISPOT assays and whether these “cross-reactive” CD8+ T cells can suppress the replication of variant viruses infecting primary CD4+ T cells.
We examined two previously described immunodominant CD8+ T-cell responses directed against SIVmac239 in infected rhesus macaques. The Tat28-35 SL8 epitope (amino acid sequence, STPESANL) (2, 3) is restricted by Mamu-A*01; SL8-specific CD8+ T cells typically have very high functional avidity and select for several different escape variants during the first 8 weeks of infection. We studied four variants, S1P (PTPESANL [the variant amino acid is underlined]), T2I (SIPESANL), S5L (STPELANL), and L8P (STPESANP). Binding of these variant peptides to the restricting class I MHC molecule has been described previously—the position 2 and position 8 variants affect anchor residues and reduce binding by 99% compared to the wild-type peptide, while the binding of the position 1 and position 5 mutants is reduced 67% and 85%, respectively (3). We also examined the CD8+ T-cell response directed against Nef165-173IW9 (IRYPKTFGW), which is restricted by Mamu-B*17 and is typically a low-avidity response. The most common variant selected for within the IW9 epitope in Mamu-B17+ macaques is I1T (TRYPKTFGW) (14, 26, 27), which reduces binding 75%, to a predicted 50% inhibitory concentration of 73 nM (N. J. Maness et al., unpublished data). Some animals acquire a second mutation within the epitope, often a position 6 change to methionine (TRYPKMFGW). Additionally, an I1A (ARYPKTFGW) variant has been detected in vivo (Thomas Friedrich, unpublished data).
Ex vivo IFN-γ ELISPOT assay shows robust cross-reactivity to most escape variant peptides at high peptide concentrations.
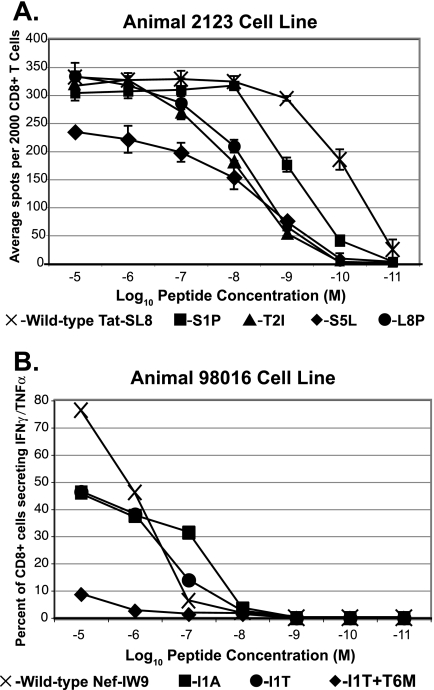
We first assessed the cross-reactivity of Tat28-35 SL8-specific cells to variant peptides with an IFN-γ ELISPOT assay at the peak of animals' CD8+ T-cell response, 17 days after intravenous infection of four Mamu-A*01+ macaques with SIVmac239. Peripheral blood mononuclear cells (PBMC) were isolated by Ficoll gradient centrifugation and resuspended in RPMI-10% fetal calf serum, and 100,000 were added to each well on IFN-γ-detecting ELISPOT assay plates (Mabtech, Nacka Strand, Sweden), which were processed by following the manufacturer's instructions. Wells were imaged with an ELISPOT assay reader (AID, Strassberg, Germany), and counted by ELISPOT Reader, version 3.2.3 (AID, Strassberg, Germany). Responses to wild-type and variant Tat-SL8 peptides were assayed in triplicate over a 7-log range of peptide concentrations, from 10 μM to 1 pM (Fig. 1A). At high concentrations of peptide, there was strong recognition of three of the variant sequences (S1P, T2I, and L8P) and detectable but significantly lower responses to the S5L variant. However, titration of the peptides revealed that the functional avidity for variants was much lower than that for the wild-type peptide. The recognition of variant peptides by ELISPOT assay was strikingly similar across the four animals tested and was confirmed again at 31 days postinfection.
FIG. 1.
Ex vivo IFN-γ ELISPOT assay responses. (A) Acute-phase responses to wild-type Tat28-35SL8 peptide (crosses) and four variant peptides, S1P (squares), T2I (triangles), S5L (diamonds), and L8P (circles). Four Mamu-A*01+ animals were infected with SIVmac239, and PBMC were isolated at 17 and 31 days postinfection for use in this assay. Each peptide was titrated over a 7-log range of concentrations, from 10 μM down to 10 pM, and assayed in triplicate. Average numbers of spots per well are shown, with error bars representing the standard deviation. Day 17 ELISPOT assay results are shown for three animals. Day 31 results are shown for a fourth animal (rh2123), as its unusually large SL8-specific response was out of the range quantifiable by the ELISPOT assay on day 17. (B) Chronic-phase responses in Mamu-B*17+ animal 98016 to wild-type Nef165-173IW9 and three variant peptides, Nef IW9 (crosses), I1T (circles), I1A (squares), and I1T+T6M (diamonds).
Ex vivo CD8+ T-cell responses directed against the Nef IW9 epitope were assessed in a chronically infected (>3 years) Mamu-B*17+ macaque. Macaque 98016's plasma virus has acquired a fixed I1T mutation. Interestingly, in the ELISPOT assay, animal 98016's responses to the wild-type and position 1 variant peptides were almost indistinguishable in terms of both magnitude and functional avidity. Its response to the peptide containing a position 6 mutation in addition to the position 1 mutation was below the limit of detection (Fig. 1B).
Recognition of variant peptides at high concentrations does not always predict recognition of escape variant viruses in vitro.
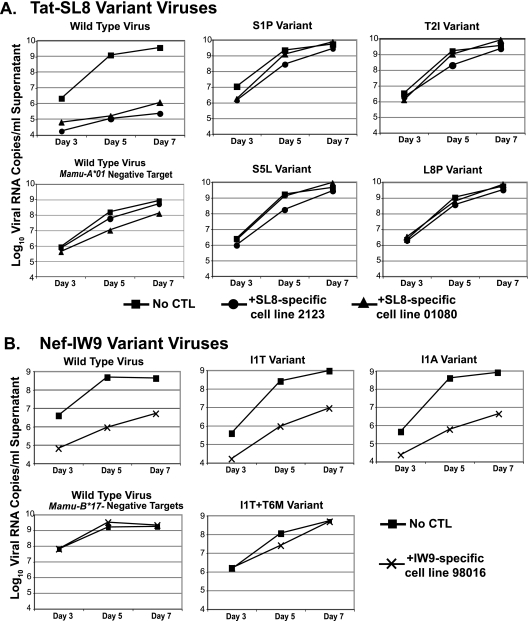
Polyclonal Tat28-35SL8-specific CD8+ T-cell lines were cultured from two of the acutely infected animals and a chronically infected animal. Cell lines were started from PBMC as described previously (31) and were stimulated every 7 to 14 days with autologous B-lymphoblastoid cell lines (B-LCLs) pulsed with wild-type SL8 peptide. Individual mutations were introduced into the SIVmac239 provirus by QuikChange PCR, and clonal viral stocks were produced by transfection of proviral DNA (13). The epitope-specific cell lines were then used in a previously described viral suppression assay (VSA) (10, 21). Briefly, we cocultured the SL8-specific cell lines with phytohemagglutinin-stimulated, CD8-depleted macaque PBMC which were infected with either SIVmac239 or viruses bearing the individual mutations. Viral replication was measured both by quantitation of viral RNA in the culture supernatant (Fig. 2) and by intracellular staining with 55-2F12 anti-Gag p27 antibody (NIH AIDS Research and Reference Reagent Program, Germantown, MD) at the end of the assay (data not shown). At an effector/target cell ratio of 1:10, SL8-specific T-cell lines suppressed wild-type viral replication 1,000- to 10,000-fold. Surprisingly, epitope-specific cell lines did not significantly suppress the replication of any of the viruses bearing individual escape mutations within the Tat28-35SL8 epitope (Fig. 2A), despite “cross-recognition” of the same variant sequences by fresh PBMC in the ELISPOT assay. Thus, these mutations conferred complete escape from suppression of viral replication in our in vitro assay, an outcome that correlated with reduced (>2 log) functional avidity for variants as measured by the ex vivo ELISPOT assay.
FIG. 2.
Suppression of viral replication by epitope-specific CD8+ T-cell lines. Primary CD8-depleted macaque PBMC were infected with wild-type SIVmac239 or a point mutant virus. (A) The infected cells were cultured in duplicate with either no CTL (squares) or SL8-specific cell lines derived from animal rh2123 (circles) or chronically infected animal r01080 (triangles). (B) The infected cells were cultured in duplicate with either no CTL (squares) or an IW9-specific cell line derived from animal 98016 (crosses). MHC mismatch cells, not expressing Mamu-A*01 or Mamu-B*17, the restricting alleles for Tat-SL8 and Nef-IW9, respectively, were infected with the wild-type virus. Supernatant was removed from cultures at days 3, 5, and 7, and the viral RNA was quantitated by PCR. Data are representative of at least two independent experiments for each epitope.
We next investigated whether cell lines cultured from the chronically infected Mamu-B17+ macaques, which recognized wild-type and position 1 variant peptides with similar magnitudes and functional avidities (<1-log difference), could inhibit the replication of these variant viruses. Cell lines from these animals were used in VSAs at an effector-to-target cell ratio of 1:5 and, indeed, inhibited the replication of both the I1A and I1T variant viruses, in addition to the wild-type virus. These epitope-specific cell lines did not inhibit the growth of the I1T+T6M variant virus (Fig. 2B).
Epitope-specific CD8+ T-cell lines secrete cytokines in response to variant peptides.
Cell lines were grown by repeated stimulation with the wild-type peptides, so we next sought to determine whether in vitro culture for 1 to 3 months had eliminated the cross-reactivity seen when ex vivo PBMC were used in the ELISPOT assay. Therefore, after they were used in VSAs, we examined the functional avidities of our cell lines for the wild-type and mutant peptides, measuring IFN-γ secretion in either the ELISPOT or the ICS assay. Our cell lines' reactivities to peptides were found to be similar to those seen with fresh PBMC, albeit with increased functional avidity. Cross-reactivity to all variants was seen when high concentrations of antigen were added (Fig. 3). The concentration of SL8 variant peptides required to trigger a half-maximal response was still at least 10-fold higher than that required to trigger a half-maximal response to the wild-type SL8 peptide in this assay (Fig. 3A), while the IW9-specific lines maintained similar avidities for the wild-type and position 1 variant peptides (Fig. 3B). Therefore, potential bias introduced by in vitro culture cannot account for the disparity between apparent recognition of the SL8 variants in ex vivo ELISPOT assays and the inability of Tat28-35SL8-specific CD8+ T cells to suppress the replication of these mutant viruses. These results are in agreement with the work of Bennett et al., who found sharp reductions in the antiviral efficacy of cytotoxic-T-lymphocyte (CTL) clones against variant viruses below epitope-dependent avidity thresholds (5).
FIG. 3.
Representative cell line functional avidity. (A) Two thousand Tat28-35SL8-specific CD8+ T cells were mixed with 10,000 autologous B-LCLs in each well and stimulated with wild-type Tat28-35SL8 peptide (crosses) or one of four variant peptides, S1P (squares), T2I (triangles), S5L (diamonds), or L8P (circles). Each peptide was titrated over a 7-log range of concentrations, from 10 μM down to 10 pM, and assayed in duplicate. The average number of spots per well is shown, with error bars representing the standard deviation. (B) Two hundred thousand Nef165-173IW9-specific CD8+ T cells were stimulated with 100,000 autologous B-LCLs pulsed with the wild-type Nef IW9 (crosses), I1T (circles), I1A (squares), and I1T+T6M (diamonds) peptides titrated over a 7-log range of concentrations, from 10 μM down to 10 pM. The percentage of CD8+ lymphocytes secreting IFN-γ and/or TNF-α in response to each peptide is shown.
Epitope-specific CD8+ T-cell line cytokine secretion in response to cells infected with variant viruses.
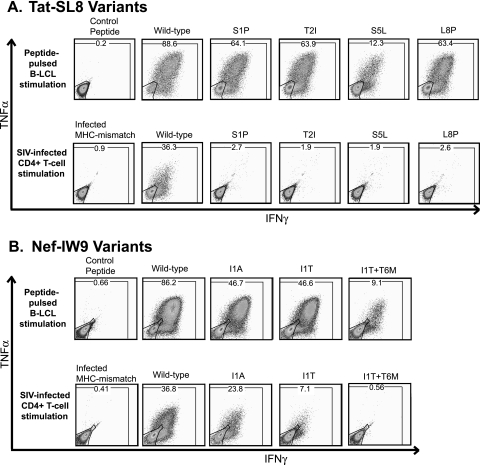
Secretion of IFN-γ and suppression of viral replication are two different readouts of CD8+ T-cell function. Betts et al. found that different T-cell functions may be stimulated by different levels of T-cell receptor occupancy (6), so we then considered whether the discrepancy between the recognition of some variant sequences at high concentrations of peptide in the IFN-γ ELISPOT assay and recognition in the VSA was a function of the different measurements of T-cell activation. To address this question, we stimulated our epitope-specific T-cell lines either with autologous B-LCLs pulsed with peptides (10 μM) or with SIVmac239-infected primary macaque cells. In this assay, 200,000 of our epitope-specific cells were stimulated with either 100,000 peptide-pulsed B-LCLs or 400,000 phytohemagglutinin-stimulated, CD8-depleted macaque PBMC which had been infected with the wild-type virus or a variant virus 6 to 8 days earlier. Intracellular cytokine staining for secretion of IFN-γ and tumor necrosis factor alpha (TNF-α) was performed as described previously (31). The infected stimulator cells were stained for SIV Gag-p27 expression to confirm that equivalent numbers of cells were infected with each of the viruses used. In agreement with the ELISPOT assay results, our cell lines recognized the variant peptides, with a large proportion secreting both IFN-γ and TNF-α (Fig. 4). Cells infected with wild-type SIVmac239 also stimulated the cell lines to secrete cytokines. However, we detected dramatically less cytokine secretion in response to cells infected with the viruses bearing different mutations within the Tat28-35SL8 epitope (Fig. 4A), although equivalent percentages of stimulator cells were infected with each of the different viruses (data not shown). Meanwhile, the Nef IW9-specific CD8+ T-cell lines recognized cells infected with both position 1 variant viruses, although a lower percentage of the T cells responded to each (Fig. 4B). These data clearly indicate that the different results of the ELISPOT assay and the VSAs cannot be attributed to the measurement of different T-cell functions. Rather, they are likely due to physiological versus nonphysiological antigen processing and presentation.
FIG. 4.
Representative cell line intracellular cytokine staining. Two hundred thousand epitope-specific CD8+ T cells were either stimulated with 100,000 autologous B-LCLs pulsed with the different peptides (10 μM) or stimulated with 400,000 primary macaque PBMC infected with the wild-type virus or a mutant virus. IFN-γ and TNF-α secretion was measured by intracellular cytokine staining and gated based on CD4− CD8+ lymphocytes. Parallel anti-Gag-p27 intracellular staining of infected cells used to stimulate the T-cell line was done to confirm that similar percentages of these cells were infected (data not shown).
Taken together, our data indicate that while CD8+ T cells may secrete cytokines in response to cells loaded with synthetic peptides, such results are not necessarily relevant to the recognition of virus-infected cells. This conclusion is in agreement with previous work examining the Mamu-A*01-restricted Gag CM9 response, in which an escape variant was recognized in the ELISPOT assay but not in a VSA (20). Many factors affect the successful presentation of peptides by class I MHC molecules, including efficiency of processing, stability of peptides in the cytoplasm, transport mediated by the transporter associated with antigen presentation, and competition between peptides for MHC binding (16, 17, 36). All of these crucial processes are bypassed by addition of high concentrations of peptides.
Whether vaccine-induced CD8+ T cells recognize variants of an epitope is a central issue for the design of CTL-based HIV vaccines. Unless there is substantial cross-recognition of variant epitopes, it is unlikely that a single vaccine construct will elicit responses effective against the many possible viruses to which vaccinees might be exposed. Several groups have shown cross-reactivity to peptides derived from multiple HIV sequences in the ELISPOT assay. Our results indicate that standard ELISPOT and ICS assays, which use a single high concentration of peptide to stimulate cells, may not provide meaningful data regarding the cross-recognition of variant SIV sequences by macaque CD8+ T cells. Indeed, challenge of two macaques making vaccine-induced responses to Tat28-35SL8 with a virus bearing the S1P mutation examined herein did not stimulate anamnestic expansion of SL8-specific cells (M. Reynolds, unpublished data), in agreement with the lack of recognition of these variant-infected cells in our in vitro assays. Therefore, caution must be used when interpreting the reactivity of HIV-specific CD8+ T cells to nonphysiological stimulation with exogenous antigen. Ideally, one should evaluate the cross-reactivity of vaccine-induced CD8+ T cells in assays with HIV-infected cells. At a minimum, the concentration of wild-type (i.e., the sequence against which an immune response was initially made) and variant HIV peptides required to trigger effector functions must be titrated. We found significant differences in the functional avidities of SL8-specific CD8+ T cells for wild-type and escape variant peptides, and this decreased functional avidity correlated with a failure of CD8+ T cells to suppress the replication of the escape variant viruses, while a response with a magnitude and avidity similar to those of the Nef IW9 epitope in the ELISPOT assay predicted the recognition of SIV-infected cells.
Acknowledgments
We thank John Loffredo for advice regarding experimental design, Alex Bean for assistance in culturing cell lines, Thomas Friedrich for virus stocks, and the animal care staff and the members of Research Support Services at the Wisconsin National Primate Research Center (WNPRC).
This research was supported by National Institutes of Health grants R01 AI049120 and R01 AI052056 to D.I.W. and by National Center of Research Resources grant P51 RR000167 to WNPRC.
Footnotes
Published ahead of print on 24 October 2007.
REFERENCES
- 1.Allen, T. M., M. Altfeld, S. C. Geer, E. T. Kalife, C. Moore, M. O'Sullivan, K. I. Desouza, M. E. Feeney, R. L. Eldridge, E. L. Maier, D. E. Kaufmann, M. P. Lahaie, L. Reyor, G. Tanzi, M. N. Johnston, C. Brander, R. Draenert, J. K. Rockstroh, H. Jessen, E. S. Rosenberg, S. A. Mallal, and B. D. Walker. 2005. Selective escape from CD8+ T-cell responses represents a major driving force of human immunodeficiency virus type 1 (HIV-1) sequence diversity and reveals constraints on HIV-1 evolution. J. Virol. 7913239-13249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allen, T. M., B. R. Mothe, J. Sidney, P. Jing, J. L. Dzuris, M. E. Liebl, T. U. Vogel, D. H. O'Connor, X. Wang, M. C. Wussow, J. A. Thomson, J. D. Altman, D. I. Watkins, and A. Sette. 2001. CD8+ lymphocytes from simian immunodeficiency virus-infected rhesus macaques recognize 14 different epitopes bound by the major histocompatibility complex class I molecule mamu-A*01: implications for vaccine design and testing. J. Virol. 75738-749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allen, T. M., D. H. O'Connor, P. Jing, J. L. Dzuris, B. R. Mothe, T. U. Vogel, E. Dunphy, M. E. Liebl, C. Emerson, N. Wilson, K. J. Kunstman, X. Wang, D. B. Allison, A. L. Hughes, R. C. Desrosiers, J. D. Altman, S. M. Wolinsky, A. Sette, and D. I. Watkins. 2000. Tat-specific cytotoxic T lymphocytes select for SIV escape variants during resolution of primary viraemia. Nature 407386-390. [DOI] [PubMed] [Google Scholar]
- 4.Belz, G. T., W. Xie, and P. C. Doherty. 2001. Diversity of epitope and cytokine profiles for primary and secondary influenza A virus-specific CD8+ T cell responses. J. Immunol. 1664627-4633. [DOI] [PubMed] [Google Scholar]
- 5.Bennett, M. S., H. L. Ng, M. Dagarag, A. Ali, and O. O. Yang. 2007. Epitope-dependent avidity thresholds for cytotoxic T lymphocyte clearance of virus-infected cells. J. Virol. 814973-4980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Betts, M. R., D. A. Price, J. M. Brenchley, K. Lore, F. J. Guenaga, A. Smed-Sorensen, D. R. Ambrozak, S. A. Migueles, M. Connors, M. Roederer, D. C. Douek, and R. A. Koup. 2004. The functional profile of primary human antiviral CD8+ T cell effector activity is dictated by cognate peptide concentration. J. Immunol. 1726407-6417. [DOI] [PubMed] [Google Scholar]
- 7.Burton, D. R. 2002. Antibodies, viruses and vaccines. Nat. Rev. Immunol. 2706-713. [DOI] [PubMed] [Google Scholar]
- 8.Carrington, M., and S. J. O'Brien. 2003. The influence of HLA genotype on AIDS. Annu. Rev. Med. 54535-551. [DOI] [PubMed] [Google Scholar]
- 9.Charini, W. A., M. J. Kuroda, J. E. Schmitz, K. R. Beaudry, W. Lin, M. A. Lifton, G. R. Krivulka, A. Necker, and N. L. Letvin. 2001. Clonally diverse CTL response to a dominant viral epitope recognizes potential epitope variants. J. Immunol. 1674996-5003. [DOI] [PubMed] [Google Scholar]
- 10.Chung, C., W. Lee, J. T. Loffredo, B. Burwitz, T. C. Friedrich, J. P. Giraldo Vela, G. Napoe, E. G. Rakasz, N. A. Wilson, D. B. Allison, and D. I. Watkins. 2007. Not all cytokine-producing CD8+ T cells suppress simian immunodeficiency virus replication. J. Virol. 811517-1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coplan, P. M., S. B. Gupta, S. A. Dubey, P. Pitisuttithum, A. Nikas, B. Mbewe, E. Vardas, M. Schechter, E. G. Kallas, D. C. Freed, T. M. Fu, C. T. Mast, P. Puthavathana, J. Kublin, K. B. Collins, J. Chisi, R. Pendame, S. J. Thaler, G. Gray, J. Mcintyre, W. L. Straus, J. H. Condra, D. V. Mehrotra, H. A. Guess, E. A. Emini, and J. W. Shiver. 2005. Cross-reactivity of anti-HIV-1 T cell immune responses among the major HIV-1 clades in HIV-1-positive individuals from 4 continents. J. Infect. Dis. 1911427-1434. [DOI] [PubMed] [Google Scholar]
- 12.Ferrari, G., J. R. Currier, M. E. Harris, S. Finkelstein, A. de Oliveira, D. Barkhan, J. H. Cox, M. Zeira, K. J. Weinhold, N. Reinsmoen, F. McCutchan, D. L. Birx, S. Osmanov, and S. Maayan. 2004. HLA-A and -B allele expression and ability to develop anti-Gag cross-clade responses in subtype C HIV-1-infected Ethiopians. Hum. Immunol. 65648-659. [DOI] [PubMed] [Google Scholar]
- 13.Friedrich, T. C., E. J. Dodds, L. J. Yant, L. Vojnov, R. Rudersdorf, C. Cullen, D. T. Evans, R. C. Desrosiers, B. R. Mothe, J. Sidney, A. Sette, K. Kunstman, S. Wolinsky, M. Piatak, J. Lifson, A. L. Hughes, N. Wilson, D. H. O'Connor, and D. I. Watkins. 2004. Reversion of CTL escape-variant immunodeficiency viruses in vivo. Nat. Med. 10275-281. [DOI] [PubMed] [Google Scholar]
- 14.Friedrich, T. C., L. E. Valentine, L. J. Yant, E. G. Rakasz, S. M. Piaskowski, J. R. Furlott, K. L. Weisgrau, B. Burwitz, G. E. May, E. J. Leon, T. Soma, G. Napoe, S. V. Capuano III, N. A. Wilson, and D. I. Watkins. 2007. Subdominant CD8+ T-cell responses are involved in durable control of AIDS virus replication. J. Virol. 813465-3476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Geels, M. J., S. A. Dubey, K. Anderson, E. Baan, M. Bakker, G. Pollakis, W. A. Paxton, J. W. Shiver, and J. Goudsmit. 2005. Broad cross-clade T-cell responses to Gag in individuals infected with human immunodeficiency virus type 1 non-B clades (A to G): importance of HLA anchor residue conservation. J. Virol. 7911247-11258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Groothuis, T. A., A. C. Griekspoor, J. J. Neijssen, C. A. Herberts, and J. J. Neefjes. 2005. MHC class I alleles and their exploration of the antigen-processing machinery. Immunol. Rev. 20760-76. [DOI] [PubMed] [Google Scholar]
- 17.Herberts, C. A., J. J. Neijssen, J. de Haan, L. Janssen, J. W. Drijfhout, E. A. Reits, and J. J. Neefjes. 2006. Cutting edge: HLA-B27 acquires many N-terminal dibasic peptides: coupling cytosolic peptide stability to antigen presentation. J. Immunol. 1762697-2701. [DOI] [PubMed] [Google Scholar]
- 18.Kelleher, A. D., C. Long, E. C. Holmes, R. L. Allen, J. Wilson, C. Conlon, C. Workman, S. Shaunak, K. Olson, P. Goulder, C. Brander, G. Ogg, J. S. Sullivan, W. Dyer, I. Jones, A. J. McMichael, S. Rowland-Jones, and R. E. Phillips. 2001. Clustered mutations in HIV-1 gag are consistently required for escape from HLA-B27-restricted cytotoxic T lymphocyte responses. J. Exp. Med. 193375-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lifson, J. D., J. L. Rossio, M. Piatak, Jr., T. Parks, L. Li, R. Kiser, V. Coalter, B. Fisher, B. M. Flynn, S. Czajak, V. M. Hirsch, K. A. Reimann, J. E. Schmitz, J. Ghrayeb, N. Bischofberger, M. A. Nowak, R. C. Desrosiers, and D. Wodarz. 2001. Role of CD8+ lymphocytes in control of simian immunodeficiency virus infection and resistance to rechallenge after transient early antiretroviral treatment. J. Virol. 7510187-10199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Loffredo, J. T., B. J. Burwitz, E. G. Rakasz, S. P. Spencer, J. J. Stephany, J. P. Giraldo Vela, S. R. Martin, J. Reed, S. M. Piaskowski, J. Furlott, K. L. Weisgrau, D. S. Rodrigues, T. Soma, G. Napoe, T. C. Friedrich, N. A. Wilson, E. G. Kallas, and D. I. Watkins. 2007. The antiviral efficacy of simian immunodeficiency virus-specific CD8+ T cells is unrelated to epitope specificity and is abrogated by viral escape. J. Virol. 812624-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Loffredo, J. T., E. G. Rakasz, J. P. Giraldo, S. P. Spencer, K. K. Grafton, S. R. Martin, G. Napoe, L. J. Yant, N. A. Wilson, and D. I. Watkins. 2005. Tat28-35SL8-specific CD8+ T lymphocytes are more effective than Gag181-189]CM9-specific CD8+ T lymphocytes at suppressing simian immunodeficiency virus replication in a functional in vitro assay. J. Virol. 7914986-14991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matano, T., R. Shibata, C. Siemon, M. Connors, H. C. Lane, and M. A. Martin. 1998. Administration of an anti-CD8 monoclonal antibody interferes with the clearance of chimeric simian/human immunodeficiency virus during primary infections of rhesus macaques. J. Virol. 72164-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McKinney, D. M., R. Skvoretz, B. D. Livingston, C. C. Wilson, M. Anders, R. W. Chesnut, A. Sette, M. Essex, V. Novitsky, and M. J. Newman. 2004. Recognition of variant HIV-1 epitopes from diverse viral subtypes by vaccine-induced CTL. J. Immunol. 1731941-1950. [DOI] [PubMed] [Google Scholar]
- 24.McKinnon, L. R., T. B. Ball, C. Wachihi, P. J. McLaren, J. L. Waruk, X. Mao, S. Ramdahin, A. O. Anzala, J. Kamene, M. Luo, K. R. Fowke, and F. A. Plummer. 2007. Epitope cross-reactivity frequently differs between central and effector memory HIV-specific CD8+ T cells. J. Immunol. 1783750-3756. [DOI] [PubMed] [Google Scholar]
- 25.Migueles, S. A., M. S. Sabbaghian, W. L. Shupert, M. P. Bettinotti, F. M. Marincola, L. Martino, C. W. Hallahan, S. M. Selig, D. Schwartz, J. Sullivan, and M. Connors. 2000. HLA B*5701 is highly associated with restriction of virus replication in a subgroup of HIV-infected long term nonprogressors. Proc. Natl. Acad. Sci. USA 972709-2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.O'Connor, D. H., T. M. Allen, T. U. Vogel, P. Jing, I. P. DeSouza, E. Dodds, E. J. Dunphy, C. Melsaether, B. Mothe, H. Yamamoto, H. Horton, N. Wilson, A. L. Hughes, and D. I. Watkins. 2002. Acute phase cytotoxic T lymphocyte escape is a hallmark of simian immunodeficiency virus infection. Nat. Med. 8493-499. [DOI] [PubMed] [Google Scholar]
- 27.O'Connor, D. H., A. B. McDermott, K. C. Krebs, E. J. Dodds, J. E. Miller, E. J. Gonzalez, T. J. Jacoby, L. Yant, H. Piontkivska, R. Pantophlet, D. R. Burton, W. M. Rehrauer, N. Wilson, A. L. Hughes, and D. I. Watkins. 2004. A dominant role for CD8+-T-lymphocyte selection in simian immunodeficiency virus sequence variation. J. Virol. 7814012-14022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Robinson, H. L., and R. R. Amara. 2005. T cell vaccines for microbial infections. Nat. Med. 11S25-S32. [DOI] [PubMed] [Google Scholar]
- 29.Schmitz, J. E., M. J. Kuroda, S. Santra, V. G. Sasseville, M. A. Simon, M. A. Lifton, P. Racz, K. Tenner-Racz, M. Dalesandro, B. J. Scallon, J. Ghrayeb, M. A. Forman, D. C. Montefiori, E. P. Rieber, N. L. Letvin, and K. A. Reimann. 1999. Control of viremia in simian immunodeficiency virus infection by CD8+ lymphocytes. Science 283857-860. [DOI] [PubMed] [Google Scholar]
- 30.Turnbull, E. L., A. R. Lopes, N. A. Jones, D. Cornforth, P. Newton, D. Aldam, P. Pellegrino, J. Turner, I. Williams, C. M. Wilson, P. A. Goepfert, M. K. Maini, and P. Borrow. 2006. HIV-1 epitope-specific CD8+ T cell responses strongly associated with delayed disease progression cross-recognize epitope variants efficiently. J. Immunol. 1766130-6146. [DOI] [PubMed] [Google Scholar]
- 31.Vogel, T. U., T. C. Friedrich, D. H. O'Connor, W. Rehrauer, E. J. Dodds, H. Hickman, W. Hildebrand, J. Sidney, A. Sette, A. Hughes, H. Horton, K. Vielhuber, R. Rudersdorf, I. P. De Souza, M. R. Reynolds, T. M. Allen, N. Wilson, and D. I. Watkins. 2002. Escape in one of two cytotoxic T-lymphocyte epitopes bound by a high-frequency major histocompatibility complex class I molecule, Mamu-A*02: a paradigm for virus evolution and persistence? J. Virol. 7611623-11636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weaver, E. A., Z. Lu, Z. T. Camacho, F. Moukdar, H. X. Liao, B. J. Ma, M. Muldoon, J. Theiler, G. J. Nabel, N. L. Letvin, B. T. Korber, B. H. Hahn, B. F. Haynes, and F. Gao. 2006. Cross-subtype T-cell immune responses induced by a human immunodeficiency virus type 1 group M consensus Env immunogen. J. Virol. 806745-6756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yant, L. J., T. C. Friedrich, R. C. Johnson, G. E. May, N. J. Maness, A. M. Enz, J. D. Lifson, D. H. O'Connor, M. Carrington, and D. I. Watkins. 2006. The high-frequency major histocompatibility complex class I allele Mamu-B*17 is associated with control of simian immunodeficiency virus SIVmac239 replication. J. Virol. 805074-5077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yewdell, J. W. 2005. The seven dirty little secrets of major histocompatibility complex class I antigen processing. Immunol. Rev. 2078-18. [DOI] [PubMed] [Google Scholar]
- 35.Yewdell, J. W., and S. M. Haeryfar. 2005. Understanding presentation of viral antigens to CD8+ T cells in vivo: the key to rational vaccine design. Annu. Rev. Immunol. 23651-682. [DOI] [PubMed] [Google Scholar]
- 36.Yewdell, J. W., E. Reits, and J. Neefjes. 2003. Making sense of mass destruction: quantitating MHC class I antigen presentation. Nat. Rev. Immunol. 3952-961. [DOI] [PubMed] [Google Scholar]
- 37.Yokomaku, Y., H. Miura, H. Tomiyama, A. Kawana-Tachikawa, M. Takiguchi, A. Kojima, Y. Nagai, A. Iwamoto, Z. Matsuda, and K. Ariyoshi. 2004. Impaired processing and presentation of cytotoxic-T-lymphocyte (CTL) epitopes are major escape mechanisms from CTL immune pressure in human immunodeficiency virus type 1 infection. J. Virol. 781324-1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yu, X. G., M. Lichterfeld, S. Chetty, K. L. Williams, S. K. Mui, T. Miura, N. Frahm, M. E. Feeney, Y. Tang, F. Pereyra, M. X. Labute, K. Pfafferott, A. Leslie, H. Crawford, R. Allgaier, W. Hildebrand, R. Kaslow, C. Brander, T. M. Allen, E. S. Rosenberg, P. Kiepiela, M. Vajpayee, P. A. Goepfert, M. Altfeld, P. J. Goulder, and B. D. Walker. 2007. Mutually exclusive T-cell receptor induction and differential susceptibility to human immunodeficiency virus type 1 mutational escape associated with a two-amino-acid difference between HLA class I subtypes. J. Virol. 811619-1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yu, X. G., M. Lichterfeld, B. Perkins, E. Kalife, S. Mui, J. Chen, M. Cheng, W. Kang, G. Alter, C. Brander, B. D. Walker, and M. Altfeld. 2005. High degree of inter-clade cross-reactivity of HIV-1-specific T cell responses at the single peptide level. AIDS 191449-1456. [DOI] [PubMed] [Google Scholar]