Abstract
Protein sequences from multiple hepatitis B virus (HBV) isolates were analyzed for the presence of amino acid motifs characteristic of cytotoxic T-lymphocyte (CTL) and helper T-lymphocyte (HTL) epitopes with the goal of identifying conserved epitopes suitable for use in a therapeutic vaccine. Specifically, sequences bearing HLA-A1, -A2, -A3, -A24, -B7, and -DR supertype binding motifs were identified, synthesized as peptides, and tested for binding to soluble HLA. The immunogenicity of peptides that bound with moderate to high affinity subsequently was assessed using HLA transgenic mice (CTL) and HLA cross-reacting H-2bxd (BALB/c × C57BL/6J) mice (HTL). Through this process, 30 CTL and 16 HTL epitopes were selected as a set that would be the most useful for vaccine design, based on epitope conservation among HBV sequences and HLA-based predicted population coverage in diverse ethnic groups. A plasmid DNA-based vaccine encoding the epitopes as a single gene product, with each epitope separated by spacer residues to enhance appropriate epitope processing, was designed. Immunogenicity testing in mice demonstrated the induction of multiple CTL and HTL responses. Furthermore, as a complementary approach, mass spectrometry allowed the identification of correctly processed and major histocompatibility complex-presented epitopes from human cells transfected with the DNA plasmid. A heterologous prime-boost immunization with the plasmid DNA and a recombinant MVA gave further enhancement of the immune responses. Thus, a multiepitope therapeutic vaccine candidate capable of stimulating those cellular immune responses thought to be essential for controlling and clearing HBV infection was successfully designed and evaluated in vitro and in HLA transgenic mice.
Chronic hepatitis B virus (HBV) infection, which occurs in 1 to 5% of adult infections and up to 30% of pediatric infections, is characterized by high levels of viral replication averaging a daily production of about 1011 viral particles, hepatic inflammation, necrosis, and ultimately liver failure (36). An estimated 350 million individuals are classified as chronically HBV infected, with the highest concentrations of infection occurring in large parts of Asia and Africa (23).
Chronic HBV can be treated with nucleoside analogues that inhibit polymerase activity. Lamivudine was the first licensed polymerase inhibitor, and it results in significant suppression of HBV DNA levels. However, this response, similar to the loss of hepatitis B virus e antigen, is often not sustained upon discontinuation of treatment (11, 28). The emergence of viral resistance in 15 to 20% of treated patients per year clearly pinpoints the limitations of this treatment.
Newer drugs such as adefovir dipivoxil, entecavir, and telbivudine can result in less resistance, increased suppression of DNA levels, or in somewhat higher levels of hepatitis B virus e antigen loss. Real long-term treatment data with these drugs are, however, limited, and it is unclear how well these responses would be sustained if therapy were withdrawn.
Higher levels of sustained response after the cessation of therapy have been documented for treatment with alpha interferon (IFN-α) and even more so for its pegylated form. This higher efficacy may be related to the fact that interferon has not only an antiviral but also an immunomodulatory function. However, drug cost and toxicities often restrict the use of IFN (11, 28). Thus, the development of supplemental therapies, especially those aiming at an improved immune response, remain a priority, particularly as a combination of lamivudine and IFN failed to improve sustained response rates (19).
In acute HBV infection, cellular immune responses are well characterized and temporally associated with the clearance of infection. The HBV-specific cytotoxic T-lymphocyte (CTL) and helper T-lymphocyte (HTL) responses present during and immediately following resolution of acute infections are readily detected and often broadly specific, targeting epitopes from numerous viral gene products (13, 17). Chronic HBV infection is, however, only rarely resolved by the immune system. When this occurs, viral clearance is associated with increased CTL activity and increased levels of alanine transaminase (ALT), referred to as ALT flares, caused by the destruction of infected hepatocytes by the immune system (40). Viral clearance also can be induced in a significant proportion (10 to 15%) of individuals receiving IFN-α, and, similar to the effect of spontaneous clearance, the effect is correlated with increased CTL and HTL responses. Antiviral effects can be mediated through the action of cytokines, such as IFN-γ, which depress viral replication or inactivate virus particles without killing HBV-infected cells (15, 16). Similarly, in persistent HBV infection, the presence of HBV-specific CTL is associated with lower viral loads, independent from liver damage, further confirming the noncytolytic antiviral activity of CTL (4, 29). Successful vaccine immunotherapy in chronically infected individuals is expected to be characterized by similar CTL and HTL responses.
However, in the presence of active HBV replication, the immune system of chronically infected individuals no longer responds vigorously to viral epitopes. The high levels of HBV particles and HBV proteins in the circulation are thought to be responsible for this immune suppression (12, 43). This effect can be so pronounced that it develops into a generalized imbalance of HTL type 1 and 2 responses (Th1/Th2), which manifests as general peripheral tolerance (42). The rare spontaneous resolution of chronic HBV infection and the immune responses observed during IFN-α or lamivudine treatment, in which viral replication is significantly reduced, indicates that deletion of HBV-specific CTL precursors does not occur and that the disease state of immune system tolerance is reversible (7, 8).
So far, therapeutic vaccine candidates (6, 20, 25, 38, 44) for HBV have focused on the use of HBsAg and core proteins or parts thereof. However, cellular immune responses to these antigens probably are most prone to suppression, tolerance, or other dysfunctions, since these proteins circulate in very high levels in the infected host (21, 41). This was effectively demonstrated by a single core epitope lipopeptide vaccine candidate, which successfully induced CTL responses in healthy volunteers but failed to do so in chronic HBV patients (24, 25). Even under effective antiviral treatment with polymerase inhibitors or IFN-α, the levels of circulating HBsAg remain very high, possibly pointing to an explanation of why combinations of lamivudine and therapeutic vaccines based on HBsAg so far have failed to show a benefit (44). On the other hand, the polymerase antigen so far has not been as thoroughly investigated as therapeutic vaccine candidate, although the immune system may be more likely to respond to this antigen, which is produced only in minute amounts compared to production of the HBsAg and core antigen. Importantly, polymerase-specific CTL and HTL responses can be readily detected in persons resolving HBV infection (33).
The induction of broadly specific CTL and HTL responses against various HBV antigens including the polymerase thus would be a new approach to improve the efficacy of a candidate therapeutic vaccine. Such a vaccine, designed for use in the general population, needs to be based on numerous epitopes restricted by multiple HLA types in order to provide acceptable levels of population coverage. The use of synthetic peptides to deliver large numbers of CTL and HTL epitopes is not feasible, but the technology to design and synthesize genes encoding multiple epitopes for use in DNA plasmid vaccines and in viral vectors is well suited (26, 27).
The utility of using genetic constructs, such as DNA vaccines, to deliver multiple epitopes was demonstrated through the development of a vaccine product designed primarily for use with human immunodeficiency virus (HIV)-infected patients (56). This vaccine, which is designated EP HIV-1090, was successfully evaluated for safety and immunogenicity in two phase-1 clinical trials. This first-generation product proved to be safe at the dose levels tested, but immunogenicity was limited. The combined use of DNA vaccines with viral vectors in a heterologous prime-boost sequential immunization format has proved useful for augmenting response levels and rates in some clinical studies. Thus, it was decided that the combined use of DNA and modified vaccinia Ankara (MVA)-vectored vaccines encoding CTL and HTL epitopes warranted investigation.
This report describes the design, synthesis, and characterization of a multiepitope-based HBV candidate therapeutic vaccine composed of a plasmid DNA (pDNA) construct (INX102-3697) and a recombinant MVA viral vector (INX102-0557) that each contain a single gene encoding 30 HBV-derived CTL epitopes, 16 HBV-derived HTL epitopes, and the universal pan-DR epitope (PADRE) HTL epitope. These two products are designed for sequential administration using a heterologous prime-boost immunization format to maximize the cellular immune responses induced.
MATERIALS AND METHODS
Peptides.
Peptides were synthesized using an Applied Biosystems 430A peptide synthesizer (Foster City, CA) and 9-fluorenylmethyl carbamate (Fmoc) chemistry (30, 57). After synthesis, the peptides were cleaved from the resin, and the protecting groups were removed. The peptides were purified by reverse-phase high-performance liquid chromatography (HPLC) to a purity of >95% and characterized by mass spectrometry.
DNA and MVA vaccine construct.
The portion of the gene encoding CTL epitopes was designed using computer-based modeling to optimize proteasome-mediated epitope processing through the introduction of amino acid spacer sequences (26). The portion of the gene encoding the HTL epitopes was designed with Gly-Pro-Gly-Pro-Gly amino acid spacers between sequential HTL epitopes (27). The order of the HTL epitopes was selected to distribute the HLA-DR supermotif and HLA-DR3 epitopes throughout the construct.
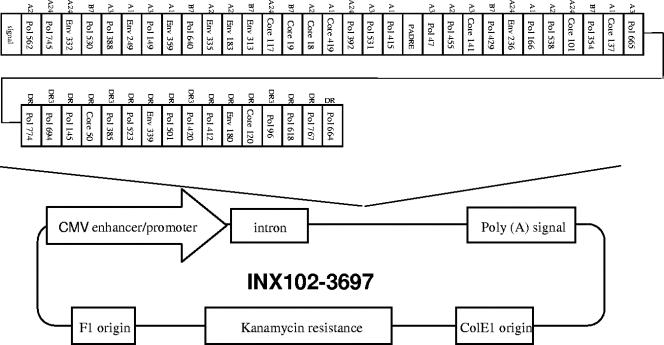
The gene construct was assembled using overlapping oligonucleotides in a PCR-based synthesis followed by subcloning into the proprietary clinically acceptable pMB75.6 (Valentis Inc., Burlingame, CA) DNA plasmid vector (56). The complete HBV pDNA vaccine INX102-3697 (6,483 bp) is depicted in Fig. 1. The kanamycin resistance gene serves for selection during production of the pDNA in bacterial cultures. Transcription of the vaccine gene in mammalian cells is driven by the cytomegalovirus immediate-early promoter, which is followed by an intron sequence to facilitate mRNA production. The 5′ end of the multiepitope gene sequence contains a consensus Kozak sequence to support translation of the gene transcript. A consensus IgKappa signal sequence was fused to the 5′ end of the multiepitope coding sequence to facilitate transport into the endoplasmic reticulum. A stop codon at the 3′ end of the open reading frame (ORF) ensures accurate termination of translation. The ORF is followed by the simian virus 40 early polyadenylation signal to ensure the production of mature and stable mRNA. The DNA sequence coding for the multiepitope gene was optimized using proprietary Gene Forge software (Aptagen Inc., Herndon, VA) to support preferred codon usage based on known codon frequencies for human genes (34). All rare and suboptimal codons were replaced, and additional codons were modified to meet the other sequence optimization criteria. These other criteria were elimination of potentially deleterious secondary RNA structures by modifying GC content, minimizing tandem repeats that may impair gene expression, and the addition of restriction sites for simple subcloning. The multiepitope gene sequence encoded by the INX102-3697 pDNA vaccine is 2,232 bp long and results in a protein product of 744 amino acids.
FIG. 1.
Schematic diagram of the HBV DNA vaccine construct INX102-3697. The orientation of the CTL and HTL epitopes in the synthetic gene is shown in the upper part; the HLA restriction of each epitope with respect to supertype also is shown. The functional elements of the DNA plasmid vector are indicated in the lower part. CMV, cytomegalovirus.
pDNA was produced by growth in Escherichia coli strain Stbl2 (Invitrogen, Carlsbad, CA) in terrific broth (54) with kanamycin (25 μg/ml) and was purified using EndoFree Plasmid Mega kit columns according to the manufacturer's directions (Qiagen, Valencia, CA). The purified pDNA HBV therapeutic vaccine construct, designated INX102-3697, was dissolved in water and stored at −70°C. For the heterologous DNA-MVA prime-boost immunizations, the pDNA was formulated in 3.4% poly(N-vinyl pyrrolidone) (PVP), 3 mg/ml ethanol, and phosphate-buffered saline (PBS), pH 7.4, at a concentration of 2 mg/ml.
Recombinant MVA was generated by homologous recombination into deletion III of the proprietary viral vector MVATGN33 (Transgene, France) using a shuttle plasmid containing the pDNA HBV vaccine gene construct functionally linked to the vaccinia virus H5R early/late promoter (9). The MVA vaccine, designated INX102-0557, was amplified and produced on chicken embryonic fibroblasts, purified by a multistep low-speed centrifugation process, and resuspended in 10 mM Tris-HCl, 5% (wt/vol) saccharose, 10 mM sodium glutamate, and 50 mM NaCl, pH 8.0, at an infectious titer of 2 × 108 PFU/ml.
Mouse strains and cell lines for immunogenicity studies.
To study the immunogenicity of HLA class I epitopes, five colonies of HLA transgenic mouse strains were used. The derivation and characterization of the HLA-A*0201/Kb, HLA-A*1101/Kb (representative of the HLA-A3 supertype), and HLA-B*0702/Kb transgenic mice were described previously (1, 2, 55). In addition, HLA-A*01/Kb and HLA-A*2402/Kb transgenic mice were produced and characterized using the same methods.
Three stably transfected cell lines were used in assays as antigen-presenting cells (APC). Jurkat (ATCC TIB-152) for HLA-A02 (55) and HLA-B07 (2) (2 × 104 cells/well) and 221 for HLA-A11 (1) (104 cells/well) were described previously. The HLA24Kb/221 cells were produced by the transfection of 721.221 cells (49) with the HLA-A*2402/Kb transgene cloned in pcDNA3.1(+) (Invitrogen, San Diego, CA). The cells (6 × 106), resuspended in cold PBS, were electroporated with 30 μg DNA at 0.2, 0.25, and 0.3 kV at 960 μF. Cells were grown in complete RPMI culture medium R10 (RPMI 1640 medium, 25 mM HEPES, pH 7.4 [Life Technologies, Grand Island, NY] supplemented with 10% FBS, 4 mM l-glutamine, 5 × 10−5 M 2-mercaptoethanol, 0.5 mM sodium pyruvate, 0.1 mM minimal essential medium nonessential amino acids, 100 μg/ml streptomycin, and 100 U/ml penicillin) for 48 to 72 h before being placed into selection medium containing 400 μg/ml Geneticin (Invitrogen). Cells were maintained under selection until a homogeneous population expressing the HLA24 transgene was obtained. Cell surface expression was confirmed by flow cytometry using a mouse anti-HLA24-specific antibody (United States Biological, Swampscott, MA).
The APC expressed a chimeric molecule composed of the α1 and α2 domains of HLA class I and the α3 domain of murine class I antigens that was identical to that of the transgenic mice. For HLA-A01 only, syngeneic spleen cells from nonimmunized mice were used as APC (2 × 105 cells/well).
Data from validation experiments demonstrated that these HLA transgenic mice responded to approximately 70% of known human epitopes. This reduced response rate probably reflects the more restricted T-cell receptor (TCR) variability in inbred mouse strains, and as such, epitope immunogenicity measurements obtained using these animals may underestimate the expected results for humans. However, the HLA transgenic mice are useful, because the immunogenicity data generated using epitopes, in the form of synthetic peptides, can provide a standard response rate and magnitude against which the responses induced using the DNA vaccine construct can be compared.
For the evaluation of HLA class II epitopes, HLA-DR transgenic mice were not available. However, H-2bxd mice (BALB/c × C57BL/6J) can be used in a limited fashion to evaluate the immunogenicity of epitopes restricted to these HLA alleles, since there is a significant degree of overlap in the binding motifs of HLA-DR and murine class II molecules (27). In these studies, irradiated syngeneic spleen cells (2 × 105 cells/well) were used as APC in vitro.
Human cell line for in vitro epitope processing and presentation studies.
The human B-lymphoblastoid cell line JY (ECACC 94022533), which is homozygous for HLA-A02, -B07, and -Cw07, was cotransfected by electroporation with plasmid INX102-3697 (coding for the multiepitope DNA vaccine construct) and a selection plasmid encoding a neomycin resistance gene. The selection of stably transfected clones was performed by culture in Iscove's modified Dulbecco's medium plus 10% fetal bovine serum supplemented with 1 mg/ml G418. Screening for recombinant protein expression was performed using insert-specific primers and reverse transcription-PCR on total RNA isolated from 106 cells. Based on reverse transcription-PCR results, a recombinant JY cell line, designated IGCL 542, was selected for banking and expansion.
Peptide binding assays.
Peptide-HLA binding measurements were completed using a standard binding competition format. Well-characterized, radiolabeled reference peptides were used as the reference for each HLA supertype, and the ability of uncharacterized competing peptides to compete for binding to HLA molecules was measured (45, 51).
Population coverage calculation.
The degree of CTL epitope conservancy was assessed on an extended (compared to the original 20 sequences used for peptide identification) data set of 698 HBV genomes (GenBank; NCBI) covering the major genotypes A, B, C, and D of HBV. The conservancy of each epitope within each HBV genotype was defined as the fraction of protein sequences containing the epitope at a 100% identity level divided by the total number of protein sequences present in the analyzed data set. Predicted population coverage for the vaccine composed of the selected epitopes was calculated using peptide binding affinities and reported gene frequencies for HLA-A, -B, and -DR alleles (10, 31). Analyses were completed by taking into account genotypic conservation and HBV genotype geographical distribution to project population coverage for Asian and Caucasian ethnicities corresponding to the major HBV genotypes.
Immunogenicity studies. (i) Immunizations.
All immunizations were performed after obtaining permission from the Local Ethics Committee for Laboratory Animal Experiments and using procedures approved by the IACUC. Eight- to 14-week-old male and female mice were used.
For peptide immunizations, individual CTL epitopes (25 μg each/mouse) were mixed with the HBV core 128-140 epitope (120 μg) to induce a T-helper response. The mixture then was emulsified in incomplete Freunds' adjuvant (IFA; Pierce Biotechnology, Rockford, IL) in a 1:1 ratio. A volume of 100 μl of the peptide emulsion was injected once subcutaneously at the base of the tail.
For DNA vaccine construct immunizations, mice were pretreated by injecting cardiotoxin (C9759; 50 μl of a 10 μM solution; Sigma) bilaterally into the tibialis anterior muscle. Five days later, mice were immunized with a single intramuscular (i.m.) injection of 100 μg pDNA INX102-3697 (50 μg bilaterally, at the same site) (18).
For heterologous DNA-MVA prime-boost immunizations, HLA-A*0201/Kb transgenic mice were immunized twice with 100 μg PVP-formulated pDNA INX102-3697 (i.m., at days 1 and 4), and boosted 3 weeks later with a single subcutaneous injection of MVA INX102-0557 (105 or 107 PFU). In a comparative homologous DNA prime-boost regimen, mice were boosted twice with 100 μg PVP-formulated pDNA (i.m., at days 26 and 29) 3 weeks after the first DNA prime immunizations.
Immunogenicity studies. (ii) Ex vivo enzyme-linked immunospot (ELISPOT) assay.
The mice were sacrificed 11 to 14 days after the last injection. The spleens were removed, and spleen cells were isolated separately from each animal. CD8+ or CD4+ cells were purified by magnetic separation using anti-CD8 or anti-CD4 antibody-coated magnetic MACS beads (Miltenyi Biotec, Bergisch Gladbach, Germany) according to the manufacturer's instructions.
Ninety-six-well ELISPOT plates (MAIP HTS plates; Millipore, Billerica, MA) were coated overnight with an anti-IFN-γ antibody (clone AN18; MabTech, Cincinnati, OH) and blocked for 2 h at room temperature with RPMI 1640 medium supplemented with 5% fetal bovine serum. APC were loaded with peptide (10 μg/ml) for 2 h at 37°C. After incubation, the excess of peptide was removed by washing the cells once. Peptide-loaded APC together with CD8+ or CD4+ spleen cells (between 105 and 2 × 105 cells/well, depending on availability) were added to the wells in triplicate. Plates then were left undisturbed overnight at 37°C. After incubation, the cells were removed and the plates were washed several times. Biotinylated anti-IFN-γ antibody (clone R46A2; MabTech) then was added to the wells, and the plates were incubated for 2 h at room temperature. After being washed, the plates were incubated with streptavidin-horseradish peroxidase (BD Biosciences, Erembodegem, Belgium) for 1 h at room temperature. After being washed, spots were visualized using 3-amino-9-ethylcarbazole (BD Biosciences) as the substrate. Finally, the plates were rinsed with tap water to stop the color reaction and were analyzed using an automated ELISPOT reader (A.EL.VIS, Hannover, Germany).
The criteria for determining the immunogenicity of individual epitopes were established using background responses measured with unloaded APC as a negative control. Results were reported as the median (of triplicate values) delta number of spot-forming cells (SFC)/106 cells after subtraction of the median background response. Any median delta response of ≥30 SFC/106 cells with a response ratio (after dividing by the median background response) of ≥2 was classified as a positive response. This was based on experience using data obtained by testing spleen cells from immunized mice with irrelevant peptides to establish background response levels. A cutoff of 30 SFC was arbitrarily selected, because this level was >2 standard deviations above the mean of background responses. This cutoff level is more conservative than the 50- to 55-SFC level commonly used for testing human peripheral blood mononuclear cells (PBMC) in clinical trials. However, background response levels observed using HLA transgenic mice and individual peptide epitopes generally are lower than those observed using peptide pools with PBMC samples.
An epitope is classified as immunogenic when a positive response is detected in at least one mouse.
In vitro epitope processing and presentation studies.
The recombinant JY cell line, IGCL 542, was expanded to 109 cells and used as the source of HLA-A02 and HLA-B07 molecules and associated peptides. Cell lysates were prepared using homogenization and freeze/thaw in PBS containing 1.0% Nonidet P-40, followed by centrifugation to remove cellular debris. HLA-peptide complexes were isolated by immunoaffinity chromatography using W6/32 monoclonal antibody (37) coated on protein A/G beads (UltraLink Immobilized Protein A/G; Pierce Biotechnology). Bound HLA-peptide complexes were eluted from the beads using three washes with 0.1% trifluoroacetic acid (TFA), pH 1.5, and then were dissociated by boiling.
Peptides were separated from the HLA molecules by subsequent filtering through a 5-kDa filter and size extraction chromatography (SEC) using a Zorbax GF-250 4.6- by 250-mm column (Agilent Technologies, Santa Clara, CA) with a mobile phase of 1% acetic acid (pH ∼3.5) plus 10% acetonitrile (ACN) in water. The peptide-containing fraction was collected and fractionated using HPLC on a 1- by 250-mm PLRP-S 100-Å polymer column with an ABI 140B LC system flowing at 50 μl/min in gradient mode with an ABI 785A UV (Applied Biosystems) detector monitoring at 214 nm. Solvent A was 2% ACN in water plus 0.1% TFA, and solvent B was 80% ACN in water plus 0.09% TFA using a gradient of 2% B to 60% B in 60 min. Fractions were collected at 1-min intervals.
Peptide identification was performed using an ion trap mass spectrometer (LCQ; ThermoFinnigan, San Jose, CA) equipped with online microcapillary HPLC (Eldex, Napa, CA) and a microspray ionization source. The microspray consisted of a microcapillary column, a 360- by 75-μm picotip, and a 15-μm tip (New Objective, Woburn, MA) self-packed with Reliasil C18 resin (Column Engineering, Ontario, CA) to a length of 10 cm. The HPLC was programmed to produce a 3-h gradient (water-ACN plus 1% acetic acid) at 30 μl/min. Prior to the loop, passive flow splitting was used to reduce the flow rate to ∼500 nl/min. The LCQ spectrometer was programmed to perform a full scan from 490 to 1,300 m/z, followed by two data-dependent scans set to select the most abundant ion species from the parent inclusion list. The parent inclusion list was filled with the singly and doubly charged m/z masses of the peptides included in the construct. This then was followed by another full scan from 490 to 1,300 m/z, followed by two more data-dependent scans set to select the most abundant ion species from the full scan. Mass spectrometry spectra were compared to those of a database of HBV proteins and the deduced protein sequence from the construct using SEQUEST (Thermofinnigan) software. SEQUEST hits with an XCorr higher than 1.2 were selected for manual verification. Expected construct peptides were found by filtering the spectra by expected m/z values with an extracted ion trace (XIC); these then were manually compared to the spectra of the synthetic peptides.
RESULTS
Vaccine epitope characterization: prediction and HLA binding properties.
Sequences from 20 HBV isolates, including adr, adw, ayr, and ayw serotypes, were analyzed using text string search software to identify amino acid sequences of 8 to 11 amino acids containing the HLA-A1, -A2, -A3, -A24, or -B7 supertype motifs and sequences of 15 or 20 amino acids containing HLA-DR supertype motifs (47, 53). Amino acid sequences that were represented in at least 45% of the HBV sequences were produced as synthetic peptides and tested for binding to purified HLA molecules.
Peptides initially were screened for binding to the prototype HLA superfamily allele(s), and those that bound with an affinity of ≤500 nM (for HLA class I) and ≤1,000 nM (for HLA class II) subsequently were tested to determine their binding affinities to the related supertype alleles. Affinity thresholds generally correlate with the capacity of a peptide to elicit an immune response (46). Accordingly, these values were utilized as a criterion for epitope selection.
A set of 30 CTL epitopes with the required moderate to high affinity binding of ≤500 nM to multiple members of the HLA-A1, -A2, -A3, -A24, or -B7 supertype finally were selected as vaccine candidates (Table 1). Similarly, a set of 16 HTL epitopes that bound HLA-DR alleles with an affinity of ≤1,000 nM were selected as vaccine candidates (Table 2).
TABLE 1.
Binding of CTL vaccine epitopes to solubilized HLA-A and HLA-B proteins
| Epitope | Sequence | Previous documentation of immunogenicitya | Conservation (%) | HLA-A2 supertype binding capacity (IC50, in nM)b
|
HLA-A2 supertype binding capacity (IC50, in nM)b
|
||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A*0201 | A*0202 | A*0203 | A*0206 | A*6802 | A*0301 | A*1101 | A*3101 | A*3301 | A*6801 | B*0702 | B*3501 | B*5101 | B*5301 | B*5401 | A*0101 | A*2902 | A*3002 | A*2301 | A*2402 | ||||
| core 18 | FLPSDFFPSV | 3, 5, 40 | 45 | 3.5 | 2.2 | 4.4 | 3.1 | 7.7 | |||||||||||||||
| env 183 | FLLTRILTI | 5, 40 | 80 | 10.4 | 106.7 | 1.3 | 18.6 | 8000 | |||||||||||||||
| env 335 | WLSLLVPFV | 5, 35, 40 | 100 | 4.4 | 252.9 | 1.8 | 11.2 | 293.5 | |||||||||||||||
| pol 455 | GLSRYVARL | 5, 40 | 55 | 46.9 | 390.9 | 18 | |||||||||||||||||
| pol 538 | YMDDVVLGV | * | 90 | 5.4 | 90.6 | 11.5 | 90.6 | 421.6 | |||||||||||||||
| pol 562 | FLLSLGIHL | 5, 40 | 95 | 7.8 | 106.3 | 988.7 | 50.6 | 2330.9 | |||||||||||||||
| core 141 | STLPETTVVRR | 5, 32 | 95 | 735 | 4.5 | 190 | 187.2 | 27.1 | |||||||||||||||
| pol 149 | HTLWKAGILYK | 5 | 100 | 15.4 | 15.6 | 536.2 | 403.4 | 48.2 | |||||||||||||||
| pol 388 | LVVDFSQFSR | 5 | 100 | 6875 | 16.7 | 706 | 142.2 | 16.9 | |||||||||||||||
| pol 47 | NVSIPWTHK | 5 | 100 | 174.7 | 214 | 283 | |||||||||||||||||
| pol 531 | SAICSVVRR | 5 | 95 | 2190 | 29.4 | 1355.7 | 461.4 | 22.9 | |||||||||||||||
| pol 665 | QAFTFSPTYKc | 95 | 249.1 | 8.4 | 18000 | 5273.6 | 7.1 | ||||||||||||||||
| core 19 | LPSDFFPSV | 5 | 45 | 3026.8 | 289.2 | 11.6 | 6.2 | ||||||||||||||||
| env 313 | IPIPSSWAF | 5 | 100 | 42.3 | 2.6 | 2.6 | 3368.5 | ||||||||||||||||
| pol 354 | TPARVTGGVF | 5 | 90 | 13.2 | 71.6 | 139.8 | |||||||||||||||||
| pol 429 | HPAAMPHLL | * | 100 | 56.6 | 297.7 | 543.5 | 160.4 | 923.3 | |||||||||||||||
| pol 530 | FPHCLAFSYM | * | 95 | 58.5 | 57.6 | 68.9 | 127.5 | 214 | |||||||||||||||
| pol 640 | YPALMPLYACI | New | 95 | 1393.4 | 119.2 | 390.4 | 111 | ||||||||||||||||
| env 359 | WMMWYWGPSLYc | New | 85 | 16.3 | 133.4 | 46.4 | |||||||||||||||||
| core 419 | DLLDTASALYc | New | 75 | 2.3 | 74.1 | 37 | |||||||||||||||||
| core 137 | LTFGRETVLEYc | 75 | 16.8 | 27.8 | 430.8 | ||||||||||||||||||
| pol 166 | ASFCGSPYc | 100 | 51.4 | 86.9 | 4.3 | ||||||||||||||||||
| pol 415 | LSLDVSAAFYc | 95 | 333.3 | 44.2 | 3.7 | ||||||||||||||||||
| env 249 | ILLLCLIFLLc | 100 | 29.9 | 1769.9 | 504.6 | ||||||||||||||||||
| env 236 | RWMCLRRFIIc | New | 95 | 7.5 | 10.2 | ||||||||||||||||||
| env 332 | RFSWLSLLVPFc | New | 100 | 12.6 | 100.8 | ||||||||||||||||||
| core 101 | LWFHISCLTFc | New | 85 | 7.7 | 6.5 | ||||||||||||||||||
| core 117 | EYLVSFGVW | 52 | 90 | 16.5 | 4.9 | ||||||||||||||||||
| pol 392 | SWPKFAVPNLc | 95 | 1 | 2.4 | |||||||||||||||||||
| pol 745 | KYTSFPWLL | 52 | 85 | 1 | 2.3 | ||||||||||||||||||
An asterisk indicates that immunogenicity was previously described for HLA transgenic mice [A02(18); B07 (2)]. New indicates that the immunogenicity of this epitope was demonstrated for the first time in this paper.
IC50, 50% inhibitory concentration.
Novel epitopes.
TABLE 2.
Binding affinity of HTL vaccine epitopes to solubilized HLA-DR proteins
| Epitope | Sequence | Previous documentation of immugenicitya | Conservation (%) | HLA-DR binding (IC50, in nM)b
|
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| DRB1* 0101 | DRB1* 1501 | DRB1* 0301 | DRB1* 0401 | DRB1* 0405 | DRB1* 1101 | DRB1* 1201 | DRB1* 1302 | DRB1* 0701 | DRB1* 0802 | DRB1* 0901 | DRB5* 0101 | DRB3* 0101 | DRB4* 0101 | ||||
| pol 412 | LQSLTNLLSSNLSWL | 33 | 90 | 2.0 | 21 | 10.0 | 47 | 303 | 397 | 143 | 173 | 598 | 791 | 1,067 | 1,837 | 4,179 | |
| pol 664 | KQAFTFSPTYKAFLC | 33 | 60 | 10 | 41 | 88 | 181 | 82 | 190 | 90 | 416 | 142 | 144 | 4,848 | 322 | ||
| env 180 | AGFFLLTRILTIPQS | 33 | 80 | 1 | 217 | 9 | 258 | 6 | 4,229 | 9 | 8 | 189 | 56 | 1,158 | 4,374 | 696 | |
| pol 774 | GTSFVYVPSALNPAD | New | 80 | 15 | 748 | 119 | 94 | 443 | 94 | 818 | 220 | 400 | |||||
| core 120 | VSFGVWIRTPPAYRPPNAPI | 14 | 90 | 27 | 43 | 58 | 220 | 11 | 817 | 565 | 78 | 76 | 1,773 | 7 | 6,454 | 395 | |
| pol 145 | RHYLHTLWKAGILYK | 33 | 100 | 17 | 4.0 | 2,271 | 1,499 | 42 | 149 | 766 | 61 | 36 | 133 | 35 | 782 | ||
| env 339 | LVPFVQWFVGLSPTV | 33 | 95 | 408 | 14 | 315 | 28 | 54 | 452 | 2,330 | 2,744 | 60 | 31 | 1,516 | 1,661 | 22 | |
| pol 501 | LHLYSHPIILGFRKI | 33 | 80 | 248 | 558 | 77 | 244 | 492 | 9,462 | 800 | 1,551 | 560 | 102 | ||||
| pol 523 | PFLLAQFTSAICSVV | New | 95 | 27 | 359 | 560 | 246 | 1,749 | 59 | 328 | 940 | 1,373 | 4,764 | 1,347 | |||
| pol 618 | KQCFRKLPVNRPIDW | 33 | 45 | 3.0 | 4,370 | 40 | 34 | 1,617 | 821 | 62 | 872 | 5,175 | 1,246 | 3,060 | |||
| pol 767 | AANWILRGTSFVYVP | 33 | 70 | 55 | 386 | 966 | 1,634 | 1,520 | 802 | 143 | 44 | 214 | 299 | 3,276 | 6,553 | ||
| core 50 | PHHTALRQAILCWGELMTLA | 14 | 90 | 810 | 8.0 | 326 | 458 | 676 | 210 | 952 | 124 | 575 | 48 | ||||
| pol 694 | LCQVFADATPTGWGL | 33 | 95 | 7,470 | 5,009 | 67 | 490 | 1,203 | 2,022 | 1,808 | 1,044 | ||||||
| pol 385 | ESRLVVDFSQFSRGN | 33 | 45 | 7,372 | 1,368 | 36 | 208 | 251 | 946 | 2,525 | 8,711 | ||||||
| pol 96 | VGPLTVNEKRRLKLI | 33 | 60 | 8,415 | 4,153 | 43 | 3,916 | 1,908 | 6,666 | 4,461 | 5,354 | 4,330 | 8,121 | ||||
| pol 420 | SSNLSWLSLDVSAAF | 33 | 85 | 38 | 3,089 | 62 | 168 | 17 | 4,923 | 1,859 | 36 | 5,063 | 1,065 | 7,126 | 5 | 7 | |
New indicates that the immunogenicity of this epitope was demonstrated for the first time in this paper.
IC50, 50% inhibitory concentration.
Vaccine epitope characterization: predicted population coverage.
In Caucasian (HBV genotypes A and D) and Asian (HBV genotypes B and C) populations, 87 and 83%, respectively, of the individuals were predicted to be capable of responding to six or more of the selected vaccine epitopes. Extending the calculation by assuming potential supertype reactivity, at least 97% of both Caucasian and Asian populations were predicted to respond to six or more of the selected epitopes (Table 3). To assess the impact of epitope variability within each of the major HBV genotypes, analysis was extended to a large data set of 698 HBV sequences sorted by genotype (compared to the 20 sequences derived from various serotypes initially used for epitope prediction). This did not, however, result in a lower level of population coverage.
TABLE 3.
Predicted population coverage of HBV vaccine epitopes
| HBV epitope | Population | Level | Population epitope recognition (%) | No. of epitopes |
|---|---|---|---|---|
| CTL | Caucasian | Allele | 87 | ≥6 |
| Supertype | 97 | ≥6 | ||
| Asian | Allele | 83 | ≥6 | |
| Supertype | 97 | ≥6 | ||
| HTL | Caucasian | Allele | 82 | ≥4 |
| DR level | 83 | ≥7 | ||
| Asian | Allele | 65 | ≥3 | |
| DR level | 86 | ≥4 |
The candidate vaccine HTL epitopes also provided a high level of predicted population coverage. Calculation of a minimal population coverage, based on only HTL epitopes with a proven allele reactivity of <100 nM, predicted that 82% of Caucasians and 65% of Asians should be genetically capable of responding to four and three HTL vaccine epitopes, respectively. Again, a less conservative calculation incorporating HLA-DR heterozygosity, gene duplication, and the highly promiscuous binding nature of the selected epitopes revealed that 83% of a Caucasian population and 86% of an Asian population may respond to at least seven or four HTL vaccine epitopes, respectively (Table 3).
Vaccine epitope characterization: immunogenicity of individual HBV-specific CTL epitopes.
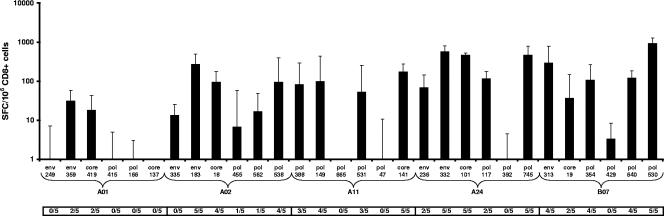
The immunogenicity of the HLA class I-restricted HBV vaccine candidate CTL epitopes was characterized using HLA transgenic mice (Fig. 2). Of the 30 CTL epitopes selected, positive responses (≥30 delta SFC/106 CD8+ cells and a response ratio of ≥2) could be shown for at least one out of five immunized mice for 21 peptides (69%) and were classified as immunogenic. All but one of the HLA-A02-, -A24-, and -B07-restricted epitopes induced significant delta CTL responses following immunization with the synthetic peptides emulsified in IFA. With the exception of epitopes pol 665 and pol 47, the HLA-A03/A11 peptides were capable of inducing positive primary CTL responses. Finally, in HLA-A01 transgenic mice, peptide immunogenicity could be demonstrated only for two out of six epitopes for two out of five mice (Fig. 2).
FIG. 2.
Median delta CTL responses induced after peptide immunization. From each HLA/Kb transgenic mouse strain, five mice were immunized once with a pool of six CTL peptides that were restricted to a specific HLA type. Direct ex vivo IFN-γ ELISPOT results are expressed as median delta SFC/106 CD8+ cells from five individual mice, with one standard deviation. In addition, the number of mice showing a positive response (≥30 SFC/106 CD8+ cells and a response ratio of ≥2) is indicated below the x axis. An epitope is considered immunogenic if a positive response is detected in at least one mouse.
Furthermore, for each HLA transgenic mouse strain, the number of responding HLA-restricted epitopes also appears to correlate with the magnitude of response: strongly immunogenic epitopes induced positive CTL responses in the majority of immunized mice. For HLA-A01, only weak median delta responses were induced towards env 359 and core 419, whereas for HLA-A24 and HLA-B07 (and, to a lesser extent, for HLA-A02 and HLA-A03/A11), strong median delta responses (>100 SFC/106 CD8+ cells) were detected, as shown in Fig. 2.
In conclusion, the identification of six new immunogenic HBV-specific CTL epitopes restricted to either HLA-A01, -A24, or -B07 demonstrates the utility of testing in transgenic animal models.
Vaccine DNA construct characterization: immunogenicity of plasmid DNA.
The pDNA construct INX102-3697 was designed to express the 30 CTL and 16 HTL vaccine candidate epitopes and the universal HTL epitope, PADRE, as a single gene product (Fig. 1). The immunogenicity of CTL epitopes in the INX102-3697 DNA vaccine candidate was characterized using the same mouse models as that for peptide immunogenicity and readily established effective expression of the vaccine antigen as well as efficient processing and presentation of the individual epitopes thereof.
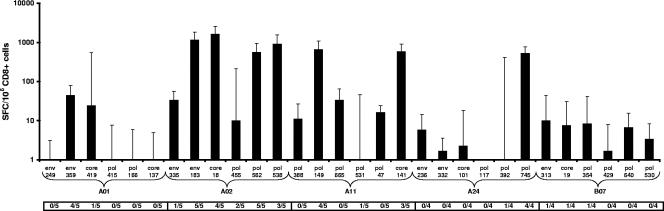
The results of the CTL immunogenicity studies are shown in Fig. 3. Experimental vaccination with 100 μg of the pDNA vaccine construct induced high-level CTL responses against all of the HLA-A02-restricted epitopes in the majority of mice. For HLA-A01, similar but rather low-level responses were measured towards two epitopes, similar to what was observed after peptide immunization. As shown in Fig. 3, for the remaining HLA types, only 3 (two HLA-A03/A11 epitopes and one HLA-A24 epitope) out of the 18 epitopes tested showed positive responses in more than one mouse. Of these 18 epitopes, several induced positive primary delta CTL responses in only one mouse, resulting in negative median delta responses (for the group of five mice).
FIG. 3.
Median delta CTL responses induced after INX102-3697 DNA construct immunization. From each HLA/Kb transgenic mouse strain, five mice were immunized once i.m. with 100 μg of pDNA after a pretreatment with cardiotoxin. Direct ex vivo IFN-γ ELISPOT results are expressed as median delta SFC/106 CD8+ cells from five individual mice, with one standard deviation. In addition, the number of responding mice (≥30 SFC/106 CD8+ cells and a response ratio of ≥2) is indicated below the x axis. An epitope is considered immunogenic if a positive response is detected in at least one mouse.
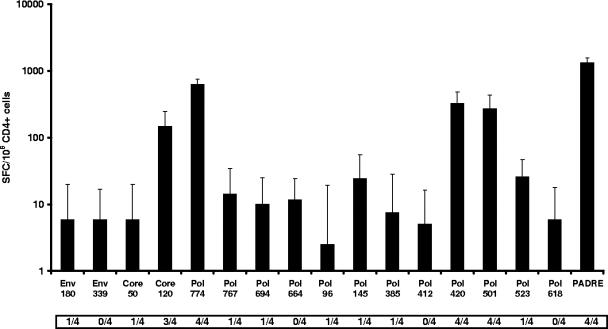
The HTL responses induced by an immunization strategy identical to that used for CTL with the INX102-3697 DNA vaccine construct were evaluated in pooled CD4+ purified spleen cells from five H-2bxd mice (Fig. 4). Not surprisingly, in all four pools of cells tested the strongest HTL responses were directed against PADRE (between 878 and 1,392 delta SFC/104 CD4+ cells). However, 12 out of 16 HBV-specific epitopes also induced positive HTL responses in at least one of the four cell pools tested and were classified as immunogenic.
FIG. 4.
Median delta HTL responses induced after INX102-3697 DNA construct immunization. H-2bxd mice were immunized once i.m. with 100 μg of pDNA after pretreatment with cardiotoxin. Direct ex vivo IFN-γ ELISPOT results are expressed as median delta SFC/106 CD4+ cells from four pools of CD4+ cells, each from five mice, with one standard deviation. In addition, the number of pools showing a positive response (≥30 SFC/106 CD8+ cells and a response ratio of ≥2) is indicated below the x axis. An epitope is considered immunogenic if a positive response is detected in at least one pool of mice.
Overall, 16 of the 30 (53%) CTL epitopes were immunogenic (in at least one out of five mice), and immunogenicity of 75% of the HTL epitopes was demonstrated in pooled cells from H-2bxd mice. Thus, the DNA vaccine format proved useful for codelivery of HTL and CTL epitopes.
Vaccine DNA construct characterization: in vitro epitope processing and presentation.
The induction of CTL responses following a single immunization with the DNA vaccine clearly indicated that some of the epitopes were processed from the single encoded multiepitope protein. The variation in results may indicate variable processing and presentation efficiency for individual epitopes. To test this directly, we compared the processing efficiency of HLA-A02 epitopes to that of HLA-B07 epitopes, measuring epitope peptides copurified with the HLA molecules.
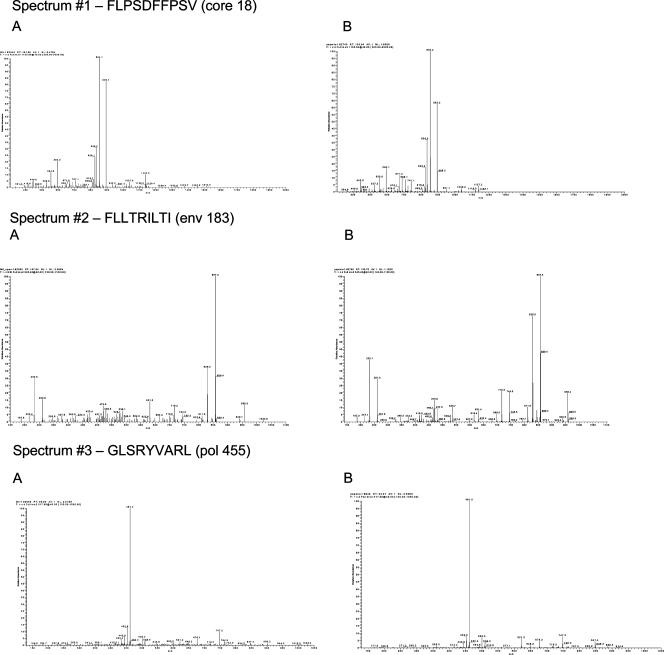
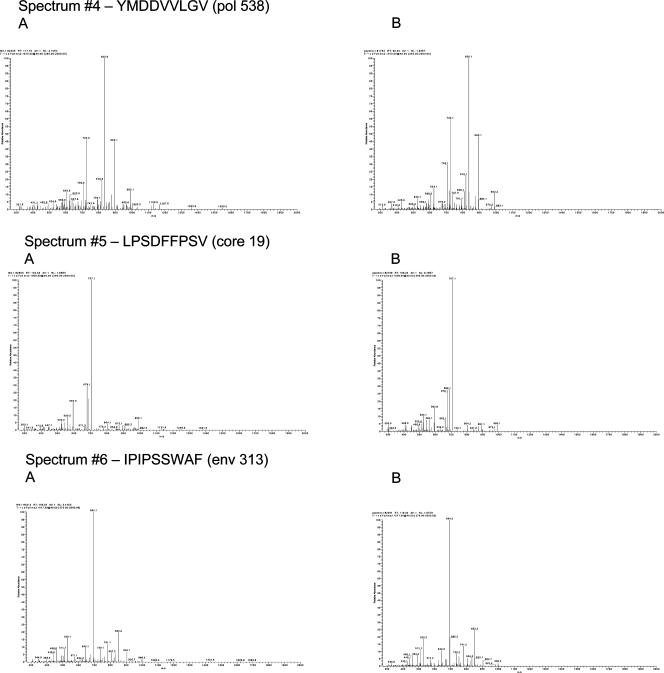
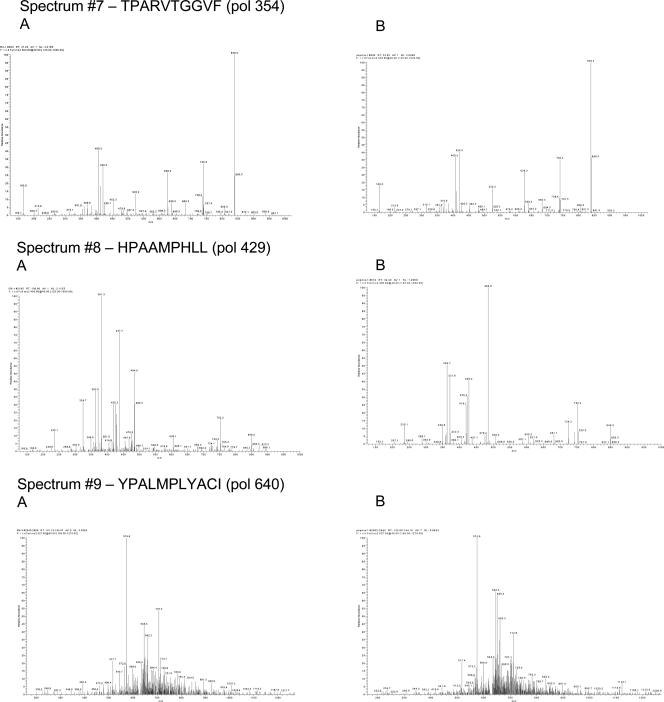
Major histocompatibility complex (MHC)-associated peptides were isolated from the recombinant JY cell line IGCL 542 (stably transfected with the multiepitope plasmid construct) and were subjected to further purification steps, consisting of a bulk elution from a SEC, followed by reverse-phase HPLC. Individual fractions were collected at 1-min intervals for 60 min. Each HPLC fraction then was subjected to mass spectrometry analysis on a high-sensitivity microspray liquid chromatography/mass spectrometry/mass spectrometry ion-trap-type mass spectrometer. Acquired fragmentation spectra first were searched against the deduced protein sequence of the DNA construct using the SEQUEST search algorithm to quickly locate potential hits. The fragmentation spectra of the peptides obtained from transfected cells then were manually compared to the fragmentation spectra of the synthetic peptides, which were run separately. Matching spectral patterns between the synthetic and cell samples validated the peptide sequences. Approximate quantification of the peptides was determined by the ratio of the total ion current (TIC) of the sample fragmentation spectra to the TIC of the synthetic fragmentation spectra (Fig. 5).
FIG. 5.
Mass spectra of the peptides from the cell sample (column A) and the mass spectra of the corresponding synthetic peptides (column B).
Of the 12 peptides expected in the DNA construct (six HLA-A02-restricted and six HLA-B07-restricted peptides), 9 were identified and validated (Table 4). Three peptides that were not found had very weak synthetic fragmentation spectra, approximately 1/50 the signal of the other peptides. In addition, two of these three peptides were the most hydrophobic peptides of the set. It is likely that these three peptides, even if displayed on the cell surface, would be difficult to detect without a very large sample size or more extensive peptide fractionation due to their hydrophobic nature and weak ionization efficiency. The spectra from the cell samples were identical to the synthetic peptide spectra, indicating that the protein was made in the cells following DNA transfection, was processed through the MHC class I pathway, and finally, appropriately cleaved individual peptides were associated with MHC molecules.
TABLE 4.
In vitro epitope processing and presentation of HBV multiepitope DNA vaccine in human cells
| HLA restriction | Epitope | Level identified by mass spectrometry (fmol) | Copies/cella |
|---|---|---|---|
| A2 | pol 538 | 15 | 75 |
| pol 562 | |||
| pol 455 | 250 | 1,250 | |
| core 18 | 6 | 30 | |
| env 183 | 3 | 15 | |
| env 335 | |||
| B7 | env 313 | 50 | 250 |
| core 19 | 75 | 350 | |
| pol 354 | 400 | 2,000 | |
| pol 429 | 700 | 3,500 | |
| pol 640 | 5 | 25 | |
| pol 530 |
Copies/cell is the approximate number of copies of the peptides per cell, calculated using the starting cell number.
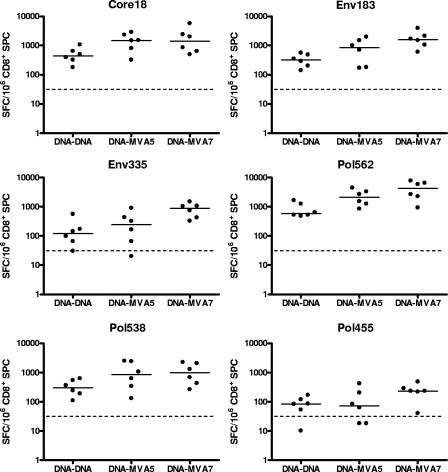
Heterologous prime-boost immunization using PVP-formulated DNA followed by MVA.
This study was designed to support the comparison of the DNA and MVA vaccine components when used to boost responses induced by DNA vaccination and gave rise to two important observations (Fig. 6). First, the use of the PVP-formulated DNA component as both the priming and boosting vaccine induced responses to all of the HLA-A02 epitopes, including env 335 and pol 455, which were minimally immunogenic when a single immunization was used (Fig. 3). Second, the use of PVP-formulated DNA and MVA in a heterologous prime-boost regimen induced CTL responses that were even further enhanced compared to those of the homologous DNA prime-boost immunization. The cumulative CTL response to all six HLA-A02-restricted epitopes as assessed by ELISPOT was 1.98 times higher when boosting with 105 PFU MVA, resulting in a trend for a difference after a Bonferroni correction for multiple testing (P = 0.1087), whereas 107 PFU MVA as the boost resulted in 3.90 times more spots, resulting in a significant difference of P = 0.0012.
FIG. 6.
Epitope-specific delta CTL responses induced after prime-boost immunization using PVP-formulated DNA construct INX102-3697 in combination with MVA construct INX102-0557. All HLA-A*02/Kb transgenic mice were primed twice with 100 μg PVP-DNA i.m. 3 to 4 days apart. Three weeks later, the first group (DNA-DNA) was boosted twice i.m. with 100 μg PVP-DNA, the second group received a single subcutaneous boost with 105 PFU MVA (DNA-MVA5), and the last group was boosted once with 107 PFU MVA subcutaneously (DNA-MVA7). Direct ex vivo IFN-γ ELISPOT results for the six HLA-A02-restricted epitopes are expressed as delta SFC/106 CD8+ cells. Each dot represents a peptide-specific delta response (from one pool of three mice), and the line represents the median response per immunization group (from six pools of three mice each).
DISCUSSION
The general acceptance and use of prophylactic HBV vaccines based on recombinant HBsAg protein has significantly reduced the risk of infection by this virus. Despite the global success of these vaccines, there are still an estimated 350 million individuals with chronic HBV infection, and these infections result in one million deaths every year (23). The limitations of the current therapies (toxicity, low level of sustained responses upon therapy cessation, and the emergence of escape variants) stress the need to develop other therapies, such as cytokine-based immunotherapy or therapeutic vaccines. We chose to address this need through the development of a therapeutic vaccine designed specifically to induce cellular immune responses to multiple CTL and HTL epitopes to ensure broad population coverage.
Comparative analysis of the immune responses raised by individuals who have recovered from acute hepatitis to those with chronic hepatitis revealed properties that were used to direct the process of designing a therapeutic vaccine as follows. (i) We identified breadth of response as a critical component of therapeutic effect and therefore elected to produce a vaccine with multiple CTL and HTL epitopes derived from different HBV proteins (13, 20). (ii) We selected epitopes restricted to the most common HLA supertypes to increase the potential recognition rate within individuals as well as to provide utility in the genetically diverse global population (47, 53). (iii) We focused on epitopes that were relatively highly conserved among multiple divergent HBV isolates. This was done not only to provide a vaccine with maximum utility against existing HBV strains but also as a strategy to combat viral mutations that could contribute to immunological escape from CTL and HTL. The reasoning behind this selection criterion is that conserved regions of the viral genome are those that have been maintained throughout evolution, since changes would affect gene product function and general viral fitness. (iv) We selected only those epitopes that bind with moderate or high affinity to HLA molecules, because high-affinity binding is one the best predictors of immunogenicity (46). (v) We included the highly immunogenic universal HTL epitope, termed PADRE, to boost the overall level of vaccine immunogenicity in order to address potential impairments to the immune system of HBV-infected individuals (42, 43, 48). (vi) The selection of a large number of polymerase-derived epitopes also is unique to our approach and may further help to overcome immune impairment, as this antigen is far less abundant than other HBV antigens. (vii) We produced the candidate vaccine using both a PVP-formulated DNA plasmid and a MVA vector for use in a heterologous prime-boost immunization schedule.
Not surprisingly, many of the selected epitopes have been confirmed to represent CTL or HTL epitopes in individuals with acute or resolving HBV infections. This is the case for five out of six HLA-A02 (3, 5, 35, 40) and all six HLA-A03/-A11 (5, 32) superfamily epitopes that are known to be immunogenic in infected individuals. Furthermore, three out of six epitopes from the HLA-B7 superfamily were positive in samples from patients with acute HBV (5). Unfortunately, the HLA-A01 and -A24 types have been much less studied. More recently, however, two of the HLA-A24 epitopes were confirmed to be dominantly recognized by HLA-A24-positive individuals with acute HBV (52). Finally, reactivity against the two core (14), the two HBsAg, and 10 out of 12 polymerase HTL epitopes was documented (33). Documentation of epitope immunogenicity as a consequence of infection is important, because these data demonstrate the generation of these epitopes from intact viral proteins through normal processing pathways. In the absence of such data, it cannot be assumed that the selected epitopes will be expressed on the surface of infected cells.
Thus, the selected 30 CTL and 16 HTL epitopes provided the basis for designing a vaccine that has the capability to induce responses against three HBV proteins. The predicted response rate and population coverage for different ethnic groups was at least 83% for the CTL epitopes, providing the required breadth of immune response. It should be noted that the estimate for the CTL epitopes was based only on the epitopes themselves and the HLA types used in the binding assays and is in fact a conservative or minimal estimate. It was previously demonstrated that numerous instances of CTL recognition of HLA-A02, -A03, and -B07 epitopes derived from HIV-1 occurred by primary blood mononuclear cells of infected donors that were not of these HLA types (56), and as such, we anticipate that the response rate could be significantly higher. In addition, several epitopes contain other (embedded) epitopes, as summarized in Table 5. This list may not be complete, and such reactivities could further add to the immunogenicity and population coverage of the vaccine construct.
TABLE 5.
Additional epitopes present in INX 102-3697
|
Epitope in construct
|
Additional HLA binding epitope
|
Reference | ||||
|---|---|---|---|---|---|---|
| Name | HLA type | Sequence | HLA type | Sequence | Immunogenicity type | |
| pol 14 | A3 | HTLWKAGILYK | A3 | TLWKAGILYK | Human acute HBV infection | 4 |
| env 249 | A1 | ILLLCLIFLL | A2 | ILLLCLIFLL | 46 | |
| A2 | LLLCLIFLL | Human acute HBV infection | 46 | |||
| A2 | ILLLCLIFL | Human acute HBV infection | 46 | |||
| pol 530 | B7 | FPHCLAFSYM | B7 | FPHCLAFSY | 50 | |
| pol 767 | DR | AANWILRGTSFVYVP | A2 | ILRGTSFVYV | Human acute HBV infection | 39 |
| env 332 | A24 | RFSWLSLLVPF | A24 | SWLSLLVPF | Human acute HBV infection | 22 |
| pol 415 | A1 | LSLDVSAAFY | A11 | LSLDVSAAFY | HLA transgenic mice | 1 |
Immunogenicity testing of the final selection of candidate epitopes focused on the CTL epitopes, as HLA transgenic mice had been developed for each of the five HLA class I (super) types. Of the 30 CTL epitopes, immunogenicity could be demonstrated in HLA transgenic mice for 21 out of 30 peptides. Large differences were observed for the different HLA types, and, especially in the case of HLA-A*01/Kb transgenic mice, induction and/or detection of CTL responses may have been suboptimal. In spite of its limitations, these results show the strength of the HLA transgenic mouse technology: this is the first demonstration of immunogenicity for three HLA-A24 (env 236, env 332, and core 101), one HLA-B07 (pol 640), and two HLA-A01 epitopes (env 359 and core 419). It should be noted that these data were generated using a single immunization. The use of booster immunizations indicates that the response rate could be much higher, and therefore more epitopes may have been detected if multiple immunizations had been used. The inclusion of a limited number of epitopes with high binding affinity but lower immunogenicity (or lower immunodominance) may be of particular interest for a therapeutic vaccine. Such epitopes may promote the induction of additional immune responses on top of the immunodominant responses typically detected in persons that resolve from acute HBV infection. Ultimately only clinical studies will be able to shed light on the relative value of different sets of epitopes.
The 30 CTL epitopes were linked by spacers designed to optimize processing (26). The PADRE epitope was located between the CTL epitopes, and the 16 selected HTL epitopes were constructed to form the C-terminal domain of the multiepitope protein. Using this DNA vaccine, 16 epitopes induced detectable CTL responses after a single DNA immunization preceded by cardiotoxin treatment. Again, large differences were noted not only between the different HLA transgenic mouse strains but also between peptide and DNA immunizations for the same HLA type. HLA-A*0201/Kb transgenic mice responded to all six epitopes with generally stronger responses than those observed after peptide vaccination. On the other hand, the HLA-B*0702/Kb transgenic mice mounted only very weak responses against epitopes that were clearly immunogenic after peptide vaccination. The nature of this difference is unknown, but a first hypothesis is that a suboptimal processing of (some of) the epitopes of the multiepitope construct result in weaker responses after DNA vaccination.
Processing of the DNA construct also was studied in vitro in human APC for the HLA-A02 and -B07 epitopes. The amount of each epitope exported by the cell as part of an HLA complex was determined by mass spectrometry, a highly sensitive in vitro analysis tool. Processing and presentation in human cells, at least for these two HLA types, seems to occur at high efficiency, demonstrating the complementarity of the mass spectrometry analysis and the assessment of in vivo immunogenicity. However, comparison of in vivo and in vitro data did not reveal a correlation between these two variables. Using mass spectrometry, higher levels of HLA-B07 peptides were identified than of HLA-A02 epitopes, yet the HLA-B07 epitopes were minimally immunogenic after DNA injection in the HLA transgenic mice. Of the two HLA-A02 epitopes (pol 562 and env 335) that could not be resolved by mass spectrometry because of their hydrophobic nature, env 335 was minimally immunogenic, but pol 562 was highly immunogenic. The pol 455 epitope, which was observed in the highest concentration for the HLA-A02 epitopes, also was minimally immunogenic. Taken together, these data indicate that epitope processing from the vaccine protein is highly efficient and that variation associated with the use of the HLA transgenic mice provided an underestimate of epitope processing. Unless processing in mouse cells proves to be significantly different from that in human cells, which seems unlikely, these data reject the hypothesis of a processing failure and point towards a presentation failure typical for some of the HLA transgenic mouse strains.
Furthermore, when comparing peptide and DNA immunogenicity, it also should be taken into account that exogenously added peptides may displace the ligands of HLA molecules already present on the membrane and therefore do not rely on intracellular processes. In contrast, antigens resulting from DNA vaccination can be displayed only if stable HLA-peptide complexes are formed in the cell and are successfully transported to the cell membrane. It cannot be excluded that the chimeric human-mouse HLA molecules that need to associate with murine β2-microglobulin have less favorable folding kinetics and/or are less stable. The proportion of HLA-epitope complexes reaching the cell membrane to display a single epitope consequently could be too small, as competition with other endogenous epitopes is always present. If this is the case, it may be that some HLA constructs are more readily affected. Alternatively, such events may be very dependent on the epitope sequence.
For the evaluation of the immunogenicity of the HTL epitopes in the DNA vaccine construct, HLA-DR transgenic mice were not available. However, the PADRE is highly immunogenic in several mouse strains, and murine MHC class II antigens are well known to overlap with human HLA-DR in terms of peptide binding motifs (27). Consequently, after immunizing H-2bxd mice only once i.m., we demonstrated that the DNA vaccine candidate induced sufficient HTL responses to support induction of a CTL response.
To conclude, 30 CTL and 16 HTL selected epitopes were combined with the universal HTL epitope PADRE in HBV DNA and MVA vaccine components. The chosen selection procedure allowed a high population and HBV variant coverage. For 24 CTL epitopes, immunogenicity has been demonstrated by others in samples from patients with acute or resolving HBV infection and/or in this study, in which we used HLA transgenic mice. For the less frequently studied HLA types A01 and A24, we identified six high-affinity binding epitopes for each HLA type, including four new ones for HLA-A24 and six for HLA-A01. In this report, immunogenicity of 5 out of these 10 novel epitopes has been demonstrated for the first time. Similarly, for all 16 HTL epitopes, immunogenicity either was previously described from studies performed on samples from HBV patients (14 epitopes) or was demonstrated by us for the first time in HLA-DR cross-reacting mice (two epitopes, pol 523 and pol 774).
The DNA candidate vaccine was able to induce significant responses in HLA transgenic mice after only a single i.m. injection after cardiotoxin pretreatment. Furthermore, we also demonstrated that the use of a combination of this DNA vaccine component with a recombinant viral vector further enhanced both the magnitude and breadth of these immune responses. The additional benefits of a heterologous prime-boost strategy also may be found in a further avidity maturation of the CTL responses. The latter may overcome the lack of antiviral efficacy noted with polymerase-specific responses, which were in contrast to HBsAg responses readily induced in HBV transgenic mice (21). The combination of this multiepitope design with a heterologous prime-boost vaccination strategy is a rational approach to counter the limitations currently experienced in therapeutic vaccination against HBV.
Acknowledgments
This work was supported in part by grant 050052 from the Instituut voor de Aanmoediging van Innovatie door Wetenschap en Technologie in Vlaanderen (IWT).
We thank Anne Farmer for editorial assistance.
Footnotes
Published ahead of print on 17 October 2007.
REFERENCES
- 1.Alexander, J., C. Oseroff, J. Sidney, P. Wentworth, E. Keogh, G. Hermanson, F. V. Chisari, R. T. Kubo, H. M. Grey, and A. Sette. 1997. Derivation of HLA-A11/Kb transgenic mice: functional CTL repertoire and recognition of human A11-restricted CTL epitopes. J. Immunol. 1594753-4761. [PubMed] [Google Scholar]
- 2.Alexander, J., C. Oseroff, J. Sidney, and A. Sette. 2003. Derivation of HLA-B*0702 transgenic mice: functional CTL repertoire and recognition of human B*0702-restricted CTL epitopes. Hum. Immunol. 64211-223. [DOI] [PubMed] [Google Scholar]
- 3.Bertoletti, A., F. V. Chisari, A. Penna, S. Guilhot, L. Galati, G. Missale, P. Fowler, H. J. Schlicht, A. Vitiello, R. C. Chesnut, F. Fiaccadori, and C. Ferrari. 1993. Definition of a minimal optimal cytotoxic T-cell epitope within the hepatitis B virus nucleocapsid protein. J. Virol. 672376-2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bertoletti, A., M. Maini, and R. Williams. 2003. Role of hepatitis B virus specific cytotoxic T cells in liver damage and viral control. Antiviral Res. 6061-66. [DOI] [PubMed] [Google Scholar]
- 5.Bertoni, R., J. Sidney, P. Fowler, R. W. Chesnut, F. V. Chisari, and A. Sette. 1997. Human histocompatibility leukocyte antigen-binding supermotifs predict broadly cross-reactive cytotoxic T lymphocyte responses in patients with acute hepatitis. J. Clin. Investig. 100503-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bienzle, U., M. Gunther, R. Neuhaus, P. Vandepapeliere, J. Vollmar, A. Lun, and P. Neuhaus. 2003. Immunization with an adjuvant hepatitis B vaccine after liver transplantation for hepatitis B-related disease. Hepatology 38811-819. [DOI] [PubMed] [Google Scholar]
- 7.Boni, C., A. Bertoletti, A. Penna, A. Cavalli, M. Pilli, S. Urbani, P. Scognamiglio, R. Boehme, R. Panebianco, F. Fiaccadori, and C. Ferrari. 1998. Lamivudine treatment can restore T cell responsiveness in chronic hepatitis B. J. Clin. Investig. 102968-975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boni, C., A. Penna, G. S. Ogg, A. Bertoletti, M. Pilli, C. Cavallo, A. Cavalli, S. Urbani, R. Boehme, R. Panebianco, F. Fiaccadori, and C. Ferrari. 2001. Lamivudine treatment can overcome cytotoxic T-cell hyporesponsiveness in chronic hepatitis B: new perspectives for immune therapy. Hepatology 33963-971. [DOI] [PubMed] [Google Scholar]
- 9.Braun, S., C. Thioudellet, P. Rodriguez, D. Ali-Hadji, F. Perraud, N. Accart, J. M. Balloul, C. Halluard, B. Acres, B. Cavallini, and A. Pavirani. 2000. Immune rejection of human dystrophin following intramuscular injections of naked DNA in mdx mice. Gene Ther. 71447-1457. [DOI] [PubMed] [Google Scholar]
- 10.Bui, H. H., J. Sidney, K. Dinh, S. Southwood, M. J. Newman, and A. Sette. 2006. Predicting population coverage of T-cell epitope-based diagnostics and vaccines. BMC Bioinformatics 7153-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Buster, E. H., and H. L. Janssen. 2006. Antiviral treatment for chronic hepatitis B virus infection-immune modulation or viral suppression? Neth. J. Med. 64175-185. [PubMed] [Google Scholar]
- 12.Chen, M., M. Sallberg, J. Hughes, J. Jones, L. G. Guidotti, F. V. Chisari, J. N. Billaud, and D. R. Milich. 2005. Immune tolerance split between hepatitis B virus precore and core proteins. J. Virol. 793016-3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chisari, F. V., and C. Ferrari. 1995. Hepatitis B virus immunopathology. Springer Semin. Immunopathol. 17261-281. [DOI] [PubMed] [Google Scholar]
- 14.Ferrari, C., A. Bertoletti, A. Penna, A. Cavalli, A. Valli, G. Missale, M. Pilli, P. Fowler, T. Giuberti, F. V. Chisari, and F. Fiaccadori. 1991. Identification of immunodominant T cell epitopes of the hepatitis B virus nucleocapsid antigen. J. Clin. Investig. 88214-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guidotti, L. G., T. Ishikawa, M. V. Hobbs, B. Matzke, R. Schreiber, and F. V. Chisari. 1996. Intracellular inactivation of the hepatitis B virus by cytotoxic T lymphocytes. Immunity 425-36. [DOI] [PubMed] [Google Scholar]
- 16.Guidotti, L. G., R. Rochford, J. Chung, M. Shapiro, R. Purcell, and F. V. Chisari. 1999. Viral clearance without destruction of infected cells during acute HBV infection. Science 284825-829. [DOI] [PubMed] [Google Scholar]
- 17.Hilleman, M. R. 2001. Overview of the pathogenesis, prophylaxis and therapeutics of viral hepatitis B, with focus on reduction to practical applications. Vaccine 191837-1848. [DOI] [PubMed] [Google Scholar]
- 18.Ishioka, G. Y., J. Fikes, G. Hermanson, B. Livingston, C. Crimi, M. Qin, M. F. del Guercio, C. Oseroff, C. Dahlberg, J. Alexander, R. W. Chesnut, and A. Sette. 1999. Utilization of MHC class I transgenic mice for development of minigene DNA vaccines encoding multiple HLA-restricted CTL epitopes. J. Immunol. 1623915-3925. [PubMed] [Google Scholar]
- 19.Janssen, H. L., M. van Zonneveld, H. Senturk, S. Zeuzem, U. S. Akarca, Y. Cakaloglu, C. Simon, T. M. So, G. Gerken, R. A. de Man, H. G. Niesters, P. Zondervan, B. Hansen, S. W. Schalm, et al. 2005. Pegylated interferon alfa-2b alone or in combination with lamivudine for HBeAg-positive chronic hepatitis B: a randomised trial. Lancet 365123-129. [DOI] [PubMed] [Google Scholar]
- 20.Jung, M. C., N. Gruner, R. Zachoval, W. Schraut, T. Gerlach, H. Diepolder, C. A. Schirren, M. Page, J. Bailey, E. Birtles, E. Whitehead, J. Trojan, S. Zeuzem, and G. R. Pape. 2002. Immunological monitoring during therapeutic vaccination as a prerequisite for the design of new effective therapies: induction of a vaccine-specific CD4+ T-cell proliferative response in chronic hepatitis B carriers. Vaccine 203598-3612. [DOI] [PubMed] [Google Scholar]
- 21.Kakimi, K., M. Isogawa, J. Chung, A. Sette, and F. V. Chisari. 2002. Immunogenicity and tolerogenicity of hepatitis B virus structural and nonstructural proteins: implications for immunotherapy of persistent viral infections. J. Virol. 768609-8620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kondo, Y., S. Asabe, K. Kobayashi, M. Shiina, H. Niitsuma, Y. Ueno, T. Kobayashi, and T. Shimosegawa. 2004. Recovery of functional cytotoxic T lymphocytes during lamivudine therapy by acquiring multi-specificity. J. Med. Virol. 74425-433. [DOI] [PubMed] [Google Scholar]
- 23.Lavanchy, D. 2004. Hepatitis B virus epidemiology, disease burden, treatment, and current and emerging prevention and control measures. J. Viral Hepat. 1197-107. [DOI] [PubMed] [Google Scholar]
- 24.Livingston, B. D., C. Crimi, H. Grey, G. Ishioka, F. V. Chisari, J. Fikes, H. Grey, R. W. Chesnut, and A. Sette. 1997. The hepatitis B virus-specific CTL responses induced in humans by lipopeptide vaccination are comparable to those elicited by acute viral infection. J. Immunol. 1591383-1392. [PubMed] [Google Scholar]
- 25.Livingston, B. D., J. Alexander, C. Crimi, C. Oseroff, E. Celis, K. Daly, L. G. Guidotti, F. V. Chisari, J. Fikes, R. W. Chesnut, and A. Sette. 1999. Altered helper T lymphocyte function associated with chronic hepatitis B virus infection and its role in response to therapeutic vaccination in humans. J. Immunol. 1623088-3095. [PubMed] [Google Scholar]
- 26.Livingston, B. D., M. Newman, C. Crimi, D. McKinney, R. Chesnut, and A. Sette. 2001. Optimization of epitope processing enhances immunogenicity of multiepitope DNA vaccines. Vaccine 194652-4660. [DOI] [PubMed] [Google Scholar]
- 27.Livingston, B., C. Crimi, M. Newman, Y. Higashimoto, E. Appella, J. Sidney, and A. Sette. 2002. A rational strategy to design multiepitope immunogens based on multiple Th lymphocyte epitopes. J. Immunol. 1685499-5506. [DOI] [PubMed] [Google Scholar]
- 28.Lok, A. S., and B. J. McMahon. 2007. Chronic hepatitis B. Hepatology 45507-539. [DOI] [PubMed] [Google Scholar]
- 29.Maini, M. K., C. Boni, C. K. Lee, J. R. Larrubia, S. Reignat, G. S. Ogg, A. S. King, J. Herberg, R. Gilson, A. Alisa, R. Williams, D. Vergani, N. V. Naoumov, C. Ferrari, and A. Bertoletti. 2000. The role of virus-specific CD8+ cells in liver damage and viral control during persistent hepatitis B virus infection. J. Exp. Med. 1911269-1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Merrifield, R. B. 1963. Solid phase peptide synthesis I: the synthesis of a tetrapeptide. J. Am. Chem. Soc. 852149-2154. [Google Scholar]
- 31.Middleton, D., L. Menchaca, H. Rood, and R. Komerofsky. 2003. New allele frequency database: http://www.allelefrequencies.net. Tissue Antigens 61403-407. [DOI] [PubMed] [Google Scholar]
- 32.Missale, G., A. Redeker, J. Person, P. Fowler, S. Guilhot, H. J. Schlicht, C. Ferrari, and F. V. Chisari. 1993. HLA-A31- and HLA-Aw68-restricted cytotoxic T cell responses to a single hepatitis B virus nucleocapsid epitope during acute viral hepatitis. J. Exp. Med. 177751-762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mizukoshi, E., J. Sidney, B. Livingston, M. Ghany, J. H. Hoofnagle, A. Sette, and B. Rehermann. 2004. Cellular immune responses to the hepatitis B virus polymerase. J. Immunol. 1735863-5871. [DOI] [PubMed] [Google Scholar]
- 34.Nakamura, Y., T. Gojobori, and T. Ikemura. 2000. Codon usage tabulated from international DNA sequence databases: status for the year 2000. Nucleic Acids Res. 28292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nayersina, R., P. Fowler, S. Guilhot, G. Missale, A. Cerny, H. J. Schlicht, A. Vitiello, R. Chesnut, J. L. Person, A. G. Redeker, and F. V. Chisari. 1993. HLA A2 restricted cytotoxic T lymphocyte responses to multiple hepatitis B surface antigen epitopes during hepatitis B virus infection. J. Immunol. 1504659-4671. [PubMed] [Google Scholar]
- 36.Nowak, M. A., S. Bonhoeffer, A. M. Hill, R. Boehme, H. C. Thomas, and H. McDade. 1996. Viral dynamics in hepatitis B virus infection. Proc. Natl. Acad. Sci. USA 934398-4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Parham, P., C. J. Barnstable, and W. F. Bodmer. 1979. Use of a monoclonal antibody (W6/32) in structural studies in HLA-A,B,C, antigens. J. Immunol. 123342-349. [PubMed] [Google Scholar]
- 38.Pol, S., B. Nalpas, F. Driss, M. L. Michel, P. Tiollais, J. Denis, C. Brechot, et al. 2001. Efficacy and limitations of a specific immunotherapy in chronic hepatitis B. J. Hepatol. 34917-921. [DOI] [PubMed] [Google Scholar]
- 39.Rehermann, B., P. Fowler, J. Sidney, J. Person, A. Redeker, M. Brown, B. Moss, A. Sette, and F. V. Chisari. 1995. The cytotoxic T lymphocyte response to multiple hepatitis B virus polymerase epitopes during and after acute viral hepatitis. J. Exp. Med. 1811047-1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rehermann, B., D. Lau, J. H. Hoofnagle, and F. V. Chisari. 1996. Cytotoxic T lymphocyte responsiveness after resolution of chronic hepatitis B virus infection. J. Clin. Investig. 971655-1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reignat, S., G. J. Webster, D. Brown, G. S. Ogg, A. King, S. L. Seneviratne, G. Dusheiko, R. Williams, M. K. Maini, and A. Bertoletti. 2002. Escaping high viral load exhaustion: CD8 cells with altered tetramer binding in chronic hepatitis B virus infection. J. Exp. Med. 1951089-1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rossol, S., G. Marinos, P. Carucci, M. V. Singer, R. Williams, and N. V. Naoumov. 1997. Interleukin-12 induction of Th1 cytokines is important for viral clearance in chronic hepatitis B. J. Clin. Investig. 993025-3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schlaak, J. F., G. Tully, H. F. Lohr, G. Gerken, and K. H. Meyer zum Buschenfelde. 1999. The presence of high amounts of HBV-DNA in serum is associated with suppressed costimulatory effects of interleukin 12 on HBV-induced immune response. J. Hepatol. 30353-358. [DOI] [PubMed] [Google Scholar]
- 44.Schneider, J., W. Halota, D. Delic, Z. Nesic, D. Prokopowicz, R. Flisiak, J. Kuydowicz, M. Jablokowski, J. Cianciara, T. Mach, R. Modrzewska, M. Fabri, D. Tomic, A. Horban, W. Krycka, and M. Cripps. 2006. A novel primeboost therapeutic vaccine induces sustained seroconversion at 52 weeks in patients with HBeAg+ chronic hepatitis B: a phase IIA clinical trial. J. Hepatol. 44(Suppl. 2)S277. [Google Scholar]
- 45.Sette, A., J. Sidney, M. F. del Guercio, S. Southwood, J. Ruppert, C. Dahlberg, H. M. Grey, and R. T. Kubo. 1994. Peptide binding to the most frequent HLA-A class I alleles measured by quantitative molecular binding assays. Mol. Immunol. 31813-822. [DOI] [PubMed] [Google Scholar]
- 46.Sette, A., A. Vitiello, B. Reherman, P. Fowler, R. Nayersina, W. M. Kast, C. J. Melief, C. Oseroff, L. Yuan, J. Ruppert, J. Sidney, M. F. del Guercio, S. Southwood, R. T. Kubo, R. W. Chesnut, H. M. Grey, and F. V. Chisari. 1994. The relationship between class I binding affinity and immunogenicity of potential cytotoxic T cell epitopes. J. Immunol. 1535586-5592. [PubMed] [Google Scholar]
- 47.Sette, A., and J. Sidney. 1999. Nine major HLA class I supertypes account for the vast preponderance of HLA-A and -B polymorphism. Immunogenetics 50201-212. [DOI] [PubMed] [Google Scholar]
- 48.Sette, A. D., C. Oseroff, J. Sidney, J. Alexander, R. W. Chesnut, K. Kakimi, L. G. Guidotti, and F. V. Chisari. 2001. Overcoming T cell tolerance to the hepatitis B virus surface antigen in hepatitis B virus-transgenic mice. J. Immunol. 1661389-1397. [DOI] [PubMed] [Google Scholar]
- 49.Shimizu, Y., and R. DeMars. 1989. Production of human cells expressing individual transferred HLA-A,-B,-C genes using an HLA-A,-B,-C null human cell line. J. Immunol. 1423320-3328. [PubMed] [Google Scholar]
- 50.Sidney, J., S. Southwood, M. F. del Guercio, H. M. Grey, R. W. Chesnut, R. T. Kubo, and A. Sette. 1996. Specificity and degeneracy in peptide binding to HLA-B7-like class I molecules. J. Immunol. 1573480-3490. [PubMed] [Google Scholar]
- 51.Sidney, J., S. Southwood, C. Oseroff, M-F. del Guercio, A. Sette, and H. M. Grey. 1998. Measurement of MHC/peptide interactions by gel filtration, p. 18.3.1-18.3.19. In J. E. Coligan et al. (ed.), Current protocols in immunology. John Wiley & Sons, New York, NY.
- 52.Sobao, Y., K. Sugi, H. Tomiyama, S. Saito, S. Fujiyama, M. Morimoto, S. Hasuike, H. Tsubouchi, K. Tanaka, and M. Takiguch. 2001. Identification of hepatitis B virus-specific CTL epitopes presented by HLA-A*2402, the most common HLA class I allele in East Asia. J. Hepatol. 34922-929. [DOI] [PubMed] [Google Scholar]
- 53.Southwood, S., J. Sidney, A. Kondo, M. F. del Guercio, E. Appella, S. Hoffman, R. T. Kubo, R. W. Chesnut, H. M. Grey, and A. Sette. 1998. Several common HLA-DR types share largely overlapping peptide binding repertoires. J. Immunol. 1603363-3373. [PubMed] [Google Scholar]
- 54.Tartof, K. D., and C. A. Hobbs. 1987. Improved media for growing plasmid and cosmid clones. Bethesda Res. Lab. Focus 912. [Google Scholar]
- 55.Vitiello, A., D. Marchesini, J. Furze, L. A. Sherman, and R. W. Chesnut. 1991. Analysis of the HLA-restricted influenza-specific cytotoxic T lymphocyte response in transgenic mice carrying a chimeric human-mouse class I major histocompatibility complex. J. Exp. Med. 1731007-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wilson, C. C., D. McKinney, M. Anders, S. MaWhinney, J. Forster, C. Crimi, S. Southwood, A. Sette, R. Chesnut, M. J. Newman, and B. D. Livingston. 2003. Development of a DNA vaccine designed to induce cytotoxic T lymphocyte responses to multiple conserved epitopes in HIV-1. J. Immunol. 1715611-5623. [DOI] [PubMed] [Google Scholar]
- 57.Wolfe, H. R., and R. R. Wilk. 1989. The RAMPS system: simplified peptide synthesis for life science researchers. Pept. Res. 2352-356. [PubMed] [Google Scholar]