Abstract
Cereal yellow dwarf virus-RPV (CYDV-RPV) is transmitted specifically by the aphids Rhopalosiphum padi and Schizaphis graminum in a circulative nonpropagative manner. The high level of vector specificity results from the vector aphids having the functional components of the receptor-mediated endocytotic pathways to allow virus to transverse the gut and salivary tissues. Studies of F2 progeny from crosses of vector and nonvector genotypes of S. graminum showed that virus transmission efficiency is a heritable trait regulated by multiple genes acting in an additive fashion and that gut- and salivary gland-associated factors are not genetically linked. Utilizing two-dimensional difference gel electrophoresis to compare the proteomes of vector and nonvector parental and F2 genotypes, four aphid proteins (S4, S8, S29, and S405) were specifically associated with the ability of S. graminum to transmit CYDV-RPV. The four proteins were coimmunoprecipitated with purified RPV, indicating that the aphid proteins are capable of binding to virus. Analysis by mass spectrometry identified S4 as a luciferase and S29 as a cyclophilin, both of which have been implicated in macromolecular transport. Proteins S8 and S405 were not identified from available databases. Study of this unique genetic system coupled with proteomic analysis indicated that these four virus-binding aphid proteins were specifically inherited and conserved in different generations of vector genotypes and suggests that they play a major role in regulating polerovirus transmission.
Viruses in the family Luteoviridae, including Barley yellow dwarf virus (BYDV), Cereal yellow dwarf virus (CYDV), Potato leafroll virus, and Beet western yellows virus, are collectively referred to in this paper as luteovirids. They are transmitted in a circulative persistent nonpropagative manner only by aphids (Aphididae: Hemiptera) (20). Ultrastructural studies indicate that all luteovirids follow a similar pathway through their aphid vectors (12). Aphids acquire the viruses from infected phloem cells while feeding with their piercing-sucking stylets. Virions are drawn up the food canal of the stylets and into the gut lumen within the aphid. Subsequently, virions traverse the lining of the midgut, hindgut, or both (7, 8, 25) and are released into the body cavity (hemocoel) to circulate in the hemolymph. Virions suspended in hemolymph that contact the paired accessory salivary glands (ASG) are actively transported by endocytosis into the ASG cells and then transported into the salivary duct to be transmitted into potential host plants. Interestingly, each luteovirid species is transmitted most efficiently only by a specific set of aphid species or populations within an aphid species, thus demonstrating a high level of vector specificity. The cellular mechanisms responsible for vector specificity are regulated by distinct interactions between the two virus structural proteins and unknown proteins in the aphid (12).
The discovery of Gildow and Rochow (9) that competition occurred between serologically related luteovirids for transmission by a common vector supported the hypothesis that receptor-mediated endocytosis is the mechanism that regulates the vector-specific transmission of each luteovirid species. Subsequently, three cellular sites in aphids that are associated with differential virus passage that may determine vector transmission ability have been identified (reviewed in reference 12). Potential barriers to transmission occur at the gut and two distinct sites at the ASG, the basal lamina surrounding the ASG, and the basal plasmalemma of each of the four cells that comprise each gland. The collective information thus far suggests that aphids possess specific protein receptors to facilitate binding and endocytosis on the surfaces of the gut endothelial cells, on the ASG basal lamina, and on the ASG cell plasmalemma. Additional aphid proteins likely regulate the transport of virions across the cell cytoplasm and their subsequent release into the hemolymph or salivary duct.
Five proteins from the aphid vector Myzus persicae were identified that were capable of binding potato leafroll virus (32), one of which was subsequently identified as symbionin, an Escherichia coli GroEL homologue produced by the aphid endosymbiont Buchnera aphidicola. Symbionin may protect the virus from recognition by the aphid immune system (5, 33). Using similar methods, Li et al. (18) isolated a number of proteins from the head of the English grain aphid (Sitobion avenae) that were able to bind to BYDV-MAV, a virus that is transmitted specifically by this aphid. Two proteins SaM35 (molecular mass of 35 kDa, pI 4.35) and SaM50 (molecular mass of 50 kDa, pI 4.51) bound in vitro with high affinity to BYDV-MAV virions and also to an anti-idiotypic antibody that mimics an epitope on the virion surface and competes with virions in antibody-binding competition assays (15). These two proteins were not detected in the corn leaf aphid, Rhopalosiphum maidis, which is not a vector of BYDV-MAV. Symbionin was among the proteins detected, but interestingly, although it bound to the anti-idiotypic antibody, it did not bind virions. A protein with a size similar to that of SaM50 was detected in the aphids Schizaphis graminum and S. avenae that bound to BYDV-GAV virions, a virus transmitted specifically by these two aphid species (34). In situ labeling localized this protein to the aphid ASG. Furthermore, aphids fed an antibody made against this protein prior to virus acquisition were less efficient at transmitting the virus. With the exception of symbionin, none of the other virus-binding aphid proteins have been identified. A more recent study (29) identified a number of M. persicae proteins that bound beet western yellows virus, including symbionin. Other proteins were identified by mass spectrometry (MS) as homologues of an aphid cuticle protein, various actins, the Drosophila melanogaster receptor for activated C kinase (protein kinase C) (Rack-1) and the D. melanogaster glyceraldehyde-3-phosphate dehydrogenase 3 (GAPDH3). The last three proteins can function in various aspects of membrane and cytosolic transport and may be involved in some aspect of luteovirid movement into, across, or out of gut or salivary tissues. GADPH3 also functions in glycolysis, and recent evidence suggests that glycosylation of luteovirid structural proteins is required for aphid transmission (28). Although the aforementioned studies have identified luteovirid-binding proteins in aphids, they have not directly linked these proteins to the virus transmission phenotype of the aphid.
Recently we have established multiple parthenogenetic colonies of S. graminum that have a common genetic background but differ in their ability to transmit two viruses that cause barley yellow dwarf disease. Two naturally occurring S. graminum genotypes, Sg-F and Sg-SC, differ in the ability to transmit CYDV and BYDV; Sg-F is an efficient vector, while Sg-SC is a poor vector (11, 13). Sexual crosses with aphids with these genotypes coupled with transmission efficiency studies on F1 and F2 progeny indicated that the hybrids segregated not only for the ability to transmit each virus species but also for which cellular barrier (hindgut or ASG) blocked virus movement and transmission (3). Subsequent genetic analysis indicated that genetic inheritance of vector competence was a multigenic trait involving only a few major genes and several minor genes that function in an additive manner (4). Evidence that multigenetic factors in aphids regulated luteovirus transmission is consistent with other work describing transmission barriers to luteovirus transmission occurring at different sites in aphid tissues, such as the gut and salivary gland (12). Based on this information, multiple gene products (proteins) would be expected to be involved in luteovirus movement through aphid gut and salivary gland tissues, and it should be possible to identify proteins unique to or overexpressed in vector genotypes. In this study we compared the proteomes of S. graminum vector (Sg-F) and nonvector (Sg-SC) genotypes to identify aphid proteins specifically expressed by the vector phenotype. F2 genotypes derived from Sg-SC × Sg-F crosses were then used to confirm that the proteins that were either specifically expressed or significantly upregulated in Sg-F were correlated with expression levels in all vector competent F2 genotypes. Furthermore, aphid proteins involved in luteovirus transmission should bind to virus particles. To test this hypothesis, whole-aphid protein extracts were incubated in vitro with purified CYDV-RPV and then immunoprecipitated using anti-CYDV immunoglobulin G (IgG).
MATERIALS AND METHODS
Aphids and viruses.
Parthenogenetic colonies of several genotypes of the aphid S. graminum were maintained on caged barley seedlings (Hordeum vulgare) at 20 to 22°C with an 18- or 24-h photoperiod. The Sg-F genotype is an efficient vector of BYDV-SGV, BYDV-PAV, and CYDV-RPV; the Sg-SC genotype is an inefficient vector of these viruses (13). The F1 and F2 matings and subsequent testing and analyses of virus transmission efficiency were described previously (3). Six of the F2 hybrid genotypes differing in their ability to transmit CYDV were used in this study, along with the Sg-F and Sg-SC parental genotypes.
CYDV-RPV was purified from oat plants (′Coast black') inoculated 4 to 5 weeks previously using nonviruliferous aphids (Sg-F or Rhopalosiphum padi) that were allowed a 48-h acquisition feeding on detached leaves from RPV-infected oat source plants and then allowed a 5-day inoculation feeding on the 7-day-old Coast black oat seedlings. Infection was determined by obvious yellowing and dwarfing symptoms and then verified as CYDV-RPV by double antibody sandwich enzyme-linked immunosorbent assay (13). CYDV-RPV was purified from freshly harvested oat tissues or oat tissues stored at −80°C by the pectinase-cellulase enzyme-assisted protocol described by Hammond et al. (14). After purification through sucrose density gradients and high-speed pelleting, the virus was resuspended in 0.01 M phosphate buffer (pH 7) and stored at −80°C.
Extraction of aphid proteins.
For each aphid genotype (Sg-F, Sg-SC, A3, G8, G11, K2, K3, and C2), soluble proteins were extracted from aphid whole-body homogenates. Aphids previously stored frozen at −80°C were placed in a cold mortar and immediately covered with liquid nitrogen. The aphids were homogenized to a dry frozen powder with a chilled pestle. An extraction buffer, 0.1 M phosphate buffer (pH 6.7) containing 2.1% of the EDTA-free Halt protease inhibitor cocktail (Pierce, Rockford, IL), was added to the aphid powder at a ratio of 1 gram of aphids per 1 ml of buffer. The aphid extract was homogenized a second time in the extraction buffer at 4°C on ice with the pestle. The homogenates were then transferred into plastic tubes, sonicated for 4 min in an ice-water bath to disrupt aphid cells, frozen at −80°C overnight, and thawed at 4°C the next day. A total of four freeze-thaw cycles were performed to further disrupt aphid cell membranes, and vigorous vortexing (1 min) was carried out after each cycle. The homogenates were then centrifuged at 16,000 × g for 5 h at 4°C, and the supernatants were collected and frozen at −80°C in aliquots for proteomics analyses. The protein concentrations were determined with a Bio-Rad DC protein assay kit using bovine serum albumin as a standard.
Coimmunoprecipitation of aphid proteins and virus.
Aphid proteins were allowed to bind to RPV virions by mixing 3 ml of whole-body aphid protein extracts from 2 g of aphids (∼8,000 adult aphids) with 200 μg of purified RPV and incubating at 4°C for 4 h. Extracts from aphids with Sg-F and Sg-SC genotypes were incubated in parallel experiments. Following the aphid protein-RPV incubations, anti-RPV IgG antibody (20 μg in 10 μl) was added and incubated overnight with agitation at 4°C to allow binding to the RPV component of the complex. Protein A agarose (Sigma, St. Louis, MO) was then added at a 1:25 ratio (vol/vol), and this mixture was incubated for an additional 24 h with agitation at 4°C to allow binding of the protein A to the IgG component of the aphid protein-RPV-IgG complex. Finally, the mixture was centrifuged at 1,500 × g at 4°C for 5 min and the agarose bead-protein complex pellet was washed six times in 3 ml of 0.025 M phosphate-buffered saline containing 0.15 M NaCl (pH 7) to remove unattached or loosely bound proteins. Elution of the aphid proteins bound to the protein A-antibody complex was achieved by adding 125 μl of elution buffer (pH 2.8 containing 0.5 M NaCl) (ProFound pull-down kit; Pierce, Rockford, IL). This buffer separates aphid proteins from the RPV antibody-protein A-agarose complex. Two more elutions were performed, and the resulting solutions were combined. The eluted solutions were concentrated with Microcon YM-3 centrifugal filters with a 3-kDa molecular mass cutoff (Millipore, Billerica, MA). Controls included aphid proteins precipitated by the antibody-protein A complex when no purified virus was added. Thus, aphid proteins binding specifically to RPV were differentiated from proteins binding nonspecifically to anti-RPV antibody-protein A-agarose complexes.
Two-dimensional difference gel electrophoresis (2D DIGE) protein analysis.
Whole-body protein extracts from vector and nonvector genotypes or proteins from coimmunoprecipitation were compared in individual 2D gels to identify proteins unique to or significantly upregulated in either the vector or nonvector. The parent vector (Sg-F) and nonvector (Sg-SC) proteins were compared in the initial experiments, and then proteins of six F2 genotypes (three that are vectors, A3, G8, and G11, and three that are nonvectors, C2, K2, and K3) were analyzed to establish whether the presence or absence of proteins in Sg-F or Sg-SC was correlated with the vector phenotype of each aphid genotype. Proteins were labeled with CyDye DIGE Fluors (Cy2, Cy3, and Cy5) (GE Healthcare Bio-Sciences Corp., Piscataway, NJ) according to the manufacturer's recommendations. Proteins from a pair of vector and nonvector genotypes (50 μg total proteins/fluor/gel) were labeled with Cy3 or Cy5 fluor (4 pmol/μg proteins) and corun on the same gels. A dye swap was used to normalize for any bias introduced by the dyes, since different CyDye DIGE Fluors may have different efficiencies in labeling different proteins. An internal standard was prepared; this standard contained a mixture of equal amounts of proteins from each extract and from all genotypes included in the experiment. This standard was labeled with Cy2 fluor (4 pmol/μg proteins) and was included in each gel for intergel comparisons used by gel image analysis software. For the coimmunoprecipitated proteins, the nonspecific control was labeled with Cy3 and the treatment was labeled with Cy5. The protein samples were loaded onto immobilized pH gradient (IPG) strips (pH 3 to 10 nonlinear, 24 cm) during an overnight passive rehydration of the strips according to the manufacturer's specifications. The first dimension was run on the IPGphore II (GE Healthcare Bio-Sciences Corp., Piscataway, NJ) at 20°C with the following settings: step 1, 500 V, 1 h; step 2, 1,000 V, 1 h; and step3, 8,000 V, 9.3 h. Before the second dimension was run, the strips were reduced for 15 min with 64.8 mM of dithiothreitol in sodium dodecyl sulfate (SDS) equilibration buffer (50 mM Tris-HCl [pH 8.8], 6 M urea, 30% glycerol, 2% SDS, 0.002% bromophenol blue) and then alkylated for 15 min with 135.2 mM of iodoacetamide in the same equilibration buffer. The second dimension was carried out in the Ettan DALT Six system (GE Healthcare Bio-Sciences Corp., Piscataway, NJ) at 25°C in an electrode buffer (25 mM Tris, 192 mM glycine, and 0.1% [wt/vol] SDS) with the following settings: step 1, 2.5 W/gel, 25 min; step 2, 17 W/gel, 4 h. The gels used in the second dimension were 12.5% homogenous acrylamide gels cast in the laboratory. Immediately after electrophoresis, the gels were scanned with a Typhoon 9400 variable-mode imager (GE Healthcare Bio-Sciences Corp., Piscataway, NJ). Four replicate 2D gels for each pair of vector-nonvector genotypes were used, and one 2D gel of the coimmunoprecipitated proteins for each genotype was run. The gel images were initially analyzed with the Decyder 2D V 6.0 software (GE Healthcare Bio-Sciences Corp., Piscataway, NJ) and then further confirmed by Progenesis SameSpots, V2.0 (Nonlinear USA Inc., Durham, NC). To identify the conserved proteins associated with CYDV-RPV transmission with the SameSpots image analysis software, the gel images were clustered into vector and nonvector groups (i.e., parent vector and nonvector groups and F2 vector and nonvector groups). To analyze the numbers of upregulated and specific protein spots in the parent and F2 aphids, upregulated or specific spots were determined by spot-by-spot and gel-by-gel manual confirmation on all the 2D and 3D images for the group. The spots reported all had at least 1.3-fold intensity (normalized volume) difference, and all were statistically significant, which was measured with the built-in statistical tool in the SameSpots software.
For MS analyses, a new set of 2D gels was run for each genotype of the parent aphids with the same settings and procedure, except that the gels were run at a protein concentration of 500 μg/gel and without labeling the proteins with CyDye DIGE Fluors. These “picking gels” were stained by using a colloidal blue stain kit (Invitrogen, Carlsbad, CA) and scanned with a Typhoon 9400 variable-mode imager (GE Healthcare Bio-Sciences Corp., Piscataway, NJ). By manually comparing the colloidal blue images with the 2D DIGE images, we were able to identify a set of spots that were unique to or obviously upregulated in vector or nonvector genotypes. These spots were then excised from the picking gels and digested with trypsin, and the resulting peptides were extracted for analysis by MS (19). Special attention was paid to unique protein spots associated with vector or nonvector phenotypes that were conserved in both parent aphids and F2 offspring. The digestions were carried out with sequencing-grade modified porcine trypsin (Promega, Madison, WI), and the peptide extracts were desalted and concentrated with C18 ZipTips (Millipore, Billerica, MA) according to the manufacturer's instructions.
Analysis by MS.
All mass spectra were obtained using a model 4700 proteomics analyzer with tandem time of flight optics using 4000 Explorer software (version 3.0) (Applied Biosystems, Foster City, CA). The sample was reconstituted in 3 μl of 0.1% trifluoroacetic acid in 50% CH3CN prior to MS analysis. Samples of 1 μl each were applied to a target plate and mixed with 0.5 μl of matrix (10 mg/ml α-cyano-4-hydroxycinnamic acid in 50% CH3CN-0.1% trifluoroacetic acid-1 mM ammonium phosphate) using the dried droplet method (16). Prior to analysis, the mass spectrometer was calibrated, externally, using a six-peptide calibration standard available from Applied Biosystems (4700 Cal mix). Most samples were calibrated internally, using the common trypsin autolysis products (at m/z values of 842.51, 1045.5642, and 2211.1046 Da) as mass calibrants. The external calibration was used as the default if the trypsin autolysis products were not observed in the spectra of the samples. MS spectra were acquired across the mass range of 800 to 4,000 Da using the 1 kV positive ions and the reflector mode with a laser power of 4100. The signal from 1,000 laser shots was averaged to produce the final MS spectra. For tandem MS (MS-MS) experiments, the instrument was operated at a laser power of 4,900 with the collision-induced dissociation and metastable ion suppressor off. Calibration was external, using the known fragments of Glu-fibrinopeptide B as calibrants. The 10 most abundant ions not appearing on the exclusion list with a minimum signal/noise ratio of 25 were selected automatically as precursor ions for MS-MS analysis. The signal from 4,000 laser shots was averaged to produce each MS-MS spectra. All m/z values reported in this study are monoisotopic.
Protein identification.
The MS and MS-MS data collected were submitted as a combined search to Mascot (23) using the GPS Explorer software, V3.5 (Applied Biosystems, Foster City, CA). Preliminary protein identifications were obtained by comparing the experimental data to the NCBI nonredundant (nr) and Acyrthosiphon pisum expressed sequence tag (EST) databases. Multiple searches of the current NCBI databases (April 2007) were performed using following taxonomies: nr, Drosophila, Aphididae, Insecta, and viruses. The search criteria used were as follows: carbamidomenthyl-cysteine and methionine oxidation were selected as variable modifications, and one missed tryptic cleavage was allowed. The searches were done with a mass tolerance of 75 ppm for the MS mode and 0.4 Da in the MS-MS mode. The preliminary protein identifications obtained automatically from the software were inspected manually for conformation prior to acceptance. Homology to known proteins was determined by searching against protein databases in NCBI with protein BLAST programs (1).
RESULTS AND DISCUSSION
Vector competency of S. graminum genotypes.
S. graminum genotypes differ in their ability to transmit several luteovirids (13). Aphids collected in the field with the Sg-F and Sg-SC genotypes represent two extremes in their ability to act as a vector for CYDV-RPV. Similar to previously reports (11, 13), Sg-F aphids transmitted CYDV-RPV efficiently, while Sg-SC aphids rarely transmitted the virus. As previously described (3), there are multiple genetically controlled barriers to CYDV-RPV movement in aphids with the Sg-SC genotype, and these barriers can segregate independently in the sexual progeny. Sg-SC aphids are not a vector of CYDV-RPV due to an efficient barrier to virus movement through the ASG and an incomplete barrier to movement at the hindgut (3). In addition, six F2 progeny from a random mating of 13 F1 progeny of the Sg-F and Sg-SC mating were used in this proteomic analysis. Aphids with genotypes Sg-A3, Sg-G8, and Sg-G11 are efficient vectors of CYDV-RPV, and aphids with genotypes Sg-C2, Sg-K2, and Sg-K3 are inefficient vectors or nonvectors (Table 1). Aphids with the Sg-C2 genotype possess an efficient barrier to virus movement at the hindgut, since introduction of virus directly into the hemocoel by microinjection resulted in a transmission efficiency (55%) similar to that of the Sg-F parental vector. Introduction of virus into the hemocoel of aphids with Sg-K2 and Sg-K3 genotypes resulted in transmission efficiencies of 21 to 30% compared to 0% by feeding acquisition, suggesting that there were incomplete barriers to virus movement functioning at the ASG but a complete barrier at the hindgut of these two aphids (Table 1). This provides a unique genetic system to study the proteomes of vector and nonvector aphids in an attempt to identify proteins that may define the vector phenotype of an aphid genotype. Although a wide variation in luterovirus transmission efficiency exists in natural populations of S. graminum (13), the F2 progeny and the segregating transmission barriers provide a method to correlate the presence or absence of protein(s) in vector and nonvector genotypes with the same genetic background.
TABLE 1.
Transmission efficiency of CYDV-RPV by parental and selected F2 progeny genotypes of Schizaphis graminum aphidsa
| Genotype | No. of plants infected/no. of plants infested by aphids (%) following virus acquisition by:
|
|
|---|---|---|
| Feeding | Injection | |
| Sg-F | 361/479 (75) | 142/236 (60) |
| Sg-A3 | 28/32 (88) | NTb |
| Sg-G8 | 26/32 (81) | NT |
| Sg-G11 | 38/46 (83) | NT |
| Sg-SC | 1/195 (0) | 11/71 (15) |
| Sg-C2 | 3/48 (6) | 32/52 (55) |
| Sg-K2 | 0/63 (0) | 15/66 (21) |
| Sg-K3 | 0/63 (0) | 3/10 (30) |
Transmission efficiency of CYDV-RPV by parental (Sg-F and Sg-SC) and selected F2 progeny genotypes of S. graminum aphids following feeding on infected plants or by injecting virus directly into the aphid hemocoel.
NT, not tested.
Proteome comparison between S. graminum parental vector and nonvector genotypes.
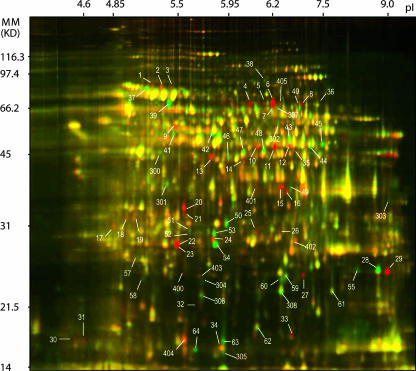
The soluble proteomes extracted from whole Sg-F (vector) and Sg-SC (nonvector) aphid bodies were directly compared by 2D DIGE (31). Four replicate gels were run with samples that included in each gel 50 μg of total protein extracts from Sg-F aphids and 50 μg of total protein extracts from Sg-SC aphids labeled with Cy3 or Cy5, respectively. Internal standards consisted of Cy2-labeled combined protein samples (25 μg from Sg-F and 25 μg from Sg-SC). This allowed for quantitative measurement of the degree of fluctuation in the spot volume ratios of image pairs from identical protein samples. Additionally, a dye swap between Sg-F and Sg-SC samples was done as a replicate analysis that controlled for any bias introduced by differences in the reactivities of the dye lots. Figure 1 shows an overlay image of Sg-F (red) and Sg-SC (green). The red spots represent the Sg-F-specific or upregulated protein spots, and the green spots represent the Sg-SC-specific or upregulated protein spots. The yellow spots indicate proteins that are expressed similarly or equally in both genotypes. A different set of 2D gels (picking gels) was used to separate proteins (500 μg/gel/aphid) extracted from the vector and nonvector aphids. These were stained with colloidal blue to provide sufficient material for isolation of individual protein spots from the gel and subsequent protein identification by MS.
FIG. 1.
Proteomic profiling of vector and nonvector genotypes as revealed by 2D DIGE analysis. Proteins extracted from whole-body protein extracts of the vector genotype Sg-F (red) and the nonvector genotype Sg-SC (green) were labeled with Cy5 and Cy3, respectively. An internal standard comprising proteins from a combination of both aphid genotypes, labeled with the Cy2 dye, was included in all gels. IPG strips (pH 3 to 10) were used for isoelectric focusing prior to standard SDS-polyacrylamide gel electrophoresis (12.5% polyacrylamide) for the second dimension. Spots marked with a number indicate proteins whose abundance was significantly different for the vector and nonvector extracts, within a 95% confidence level. The yellow-colored spots are proteins expressed equally in both genotypes. Molecular mass (MM) (in kilodaltons) is shown on the y axis, and pI is shown on the x axis.
The 2D DIGE images were clustered into vector and nonvector groups in two ways: parent vector versus parent nonvector and F2 vector versus F2 nonvector. The gel images were initially analyzed with DeCyder 2D software and then further analyzed and confirmed with SameSpots software. A total of 1,945 protein spots were detected by the SameSpots software program in the parental aphid whole-body protein extracts (Fig. 1). A comparison of the parental genotypes (Sg-F and Sg-SC) identified 120 differentially expressed protein spots. The molecular masses ranged from 15.0 to 77.3 kDa, and the pIs ranged from 4.40 to 9.25. The vector with Sg-F genotype had 68 upregulated/specific protein spots; 13 were Sg-F specific and 55 were expressed at significantly higher levels than in Sg-SC (by analysis of variance [ANOVA], 7.00 × 10−9 ≤ P ≤ 0.012) (Table 2). The nonvector Sg-SC aphids had 52 upregulated/specific protein spots; 11 were detected only in Sg-SC and 41 were significantly upregulated (by ANOVA, 2.46 × 10−8 ≤ P ≤ 0.003).
TABLE 2.
Summary of upregulated or specific protein spots in comparisons between parent vector and nonvector genotypes and between F2 vector and nonvector genotypes
| Comparison | No. of protein spots
|
Total no. of upregulated or specific protein spots | Statistical significance (P)a | |
|---|---|---|---|---|
| Specific | Upregulated | |||
| Parent aphids | ||||
| Vector | 13 | 55 | 68 | 7.00 × 10−9 ≤ P ≤ 0.012 |
| Nonvector | 11 | 41 | 52 | 2.46 × 10−8 ≤ P ≤ 0.003 |
| F2 aphids | ||||
| Vector | 2 | 12b | 14 | 8.2 × 10−5 ≤ P ≤ 0.033 |
| Nonvector | 0 | 1c | 1 | P = 0.022 |
The statistical significance of the intensity of the protein spots of vector and nonvector aphids was calculated by ANOVA.
Only 2 of these 12 spots are upregulated in the parent vector.
This protein was not upregulated in the parent nonvector.
Detection of proteins specifically linked to CYDV-RPV transmission.
Aphid proteins that may be associated with virus recognition, endocytotic and exocytotic pathways, and transcellular transport through aphid tissues resulting in circulative virus transmission were expected to be conserved in both the parental (Sg-F) and F2 (Sg-A3, Sg-G8, and Sg-G11) vector genotypes. Similarly, proteins that may be specifically associated with preventing CYDV-RPV transmission by blocking pathways and preventing transmission would be expected to be shared by both the parental (Sg-SC) and F2 (Sg-C2, Sg-K2, and Sg-K3) nonvector genotypes. We recognize that vector and nonvector genotypes may not be distinguished by the presence or absence of proteins. The difference may be more subtle such as a slightly altered amino acid sequence or posttranslational modification that could alter protein function. Some subtle changes would not be discovered using the methods described here, but many changes or alterations can be identified by slight alterations in protein spot migration in 2D gels.
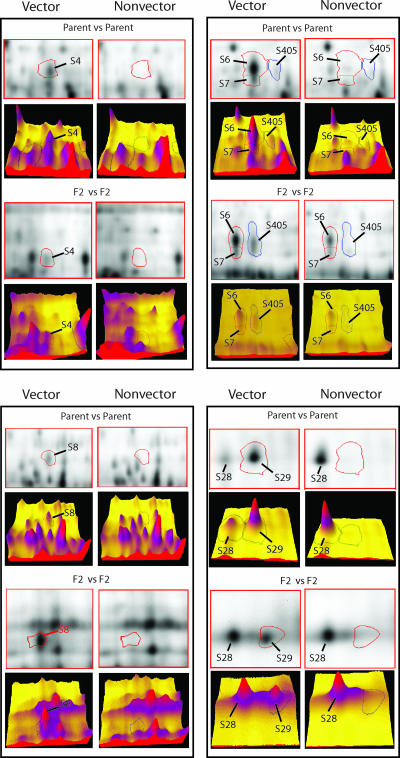
The comparison of F2 genotypes identified 14 protein spots that were specific or upregulated in the F2 vector genotypes (Table 2). Two of them, S8 and S29, were F2 vector-specific spots, and they were among the 13 parent vector-specific spots (Fig. 2). The other 12 spots were significantly upregulated (by ANOVA, 8.2 × 10−5 ≤ P ≤ 0.033), two of which (S4 and S405) were also identified in the parent vector Sg-F (Fig. 2). Therefore, four protein spots (S4, S8, S29, and S405) are conserved between the parental and F2 vectors that efficiently transmit CYDV-RPV (Table 3), indicating that these four proteins, especially the two vector-specific proteins S8 and S29, are linked to CYDV-RPV transmission. Although S4 was significantly downregulated at the group level in the F2 nonvectors, one exception was that S4 was highly expressed in the nonvector genotype Sg-C2. However, Sg-C2 is an efficient vector of BYDV-SGV (3). Thus, S4 expression in Sg-C2 may be related to BYDV-SGV transmission. The other two nonvector F2 genotypes do not transmit BYDV-SGV. With the exception of S4, the other three proteins were all significantly downregulated in Sg-C2 (Table 3). S4, S8, and S405 have similar molecular masses (68.4 to 70.2 kDa) and pIs (6.2 to 6.6), suggesting these may be related proteins; S29 is a smaller basic protein (26.6 kDa; pI 9.0).
FIG. 2.
2D and 3D images showing comparisons of key protein spots between vector and nonvector genotypes of the parent aphids or the F2 progeny. 2D DIGE analysis identified significantly increased expression of proteins S4, S8, S29, and S405 in all vector genotypes (parent Sg-F and F2 hybrids) compared to the nonvector genotypes (parent Sg-SC and F2 hybrids). Densitometric volume is shown in a 3D view generated by the SameSpots image analysis software package. Computational analysis of the images with the SameSpots software allowed for the detection of significant abundance changes (95% confidence level) based on the variance of the mean change within the cohort. The amount of the protein is proportional to the volume of the protein peak. A spot circled with a red or blue line represents the protein identified by the label. S6 and S7 are examples of nonconserved proteins. In the parent vector, the expression of S6 was significantly increased relative to the nonvector parent, and S7 was specific to the vector parent; however, no significant difference in S6 or S7 expression was found between F2 vector and nonvector groups. The size and pI of S28 are similar to those of S29, and both proteins were identified by MS analysis as cyclophilin-like proteins.
TABLE 3.
Four aphid proteins are conserved between parent and F2 vectors efficiently transmitting CYDV-RPV
| Protein | Molecular mass (kDa) | pI | Expression of protein ina:
|
Statistical significance (P)b | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Vector
|
Nonvector
|
||||||||||
| Sg-F | Sg-A3 | Sg-G8 | Sg-G11 | Sg-SC | Sg-K2 | Sg-K3 | Sg-C2 | ||||
| S4 | 68.4 | 6.1 | + | + | + | + | − | − | − | + | ≤3.40 × 10−4 |
| S8 | 70.2 | 6.6 | + | + | + | + | − | − | − | − | ≤0.004 |
| S29 | 26.6 | 9.0 | + | + | + | + | − | − | − | − | ≤0.002 |
| S405 | 70.2 | 6.3 | + | + | + | + | − | − | − | − | ≤6.78 × 10−4 |
The expression of proteins S4, S8, S29, and S405 in vector or nonvector aphids was examined. Symbols: +, specific or significantly upregulated; −, absent or significantly downregulated.
Statistical significance was determined by ANOVA done on Sg-F versus Sg-SC and F2 vectors (A3, G8, and G11) versus F2 nonvectors (K2, K3, and C2) for each spot. The greater of the two P values was reported.
The F2 nonvector group had only one upregulated protein spot (by ANOVA, P = 0.022). This spot is not among the 52 upregulated/specific spots in the nonvector parent, Sg-SC, i.e., none of these 52 proteins were conserved in F2 nonvector genotypes. Therefore, no strong association could be made with the occurrence of a specific protein and the nonvector phenotype. This suggests that the inability of vectoring CYDV-RPV in nonvectors is not due to the presence of proteins preventing transmission of CYDV-RPV in the nonvectors but more likely due to a lack of or inactivity of specific CYDV-RPV-binding proteins that facilitate transmission.
Eliminating false-positive results and the power of using genetic screens to identify target proteins.
There were 68 and 52 specific/upregulated protein spots detected in the vector and nonvector parents, respectively (Table 2). Since these parents represent two distinct genetic populations, it is difficult to conclude which proteins are actually related to virus transmission with this information alone. It can be presumed that proteins associated with CYDV-RPV transmission by the parental vector Sg-F would be represented within these 68 proteins. Naturally, one would assume that the parental vector-specific or upregulated protein spots are related to virus transmission, but this may not be true. For example, S6 is upregulated and S7 is vector specific in the parent generation; however, their levels of expression are not significantly different between F2 vectors and nonvectors (Fig. 2). In addition, any of the 52 proteins associated specifically with the Sg-SC nonvector phenotype could be involved in preventing CYDV-RPV transmission due to mechanisms like irreversible binding of the virus to certain Sg-SC proteins. The numbers of specific/upregulated protein spots consistently identified in all F2 vector and nonvector genotypes were reduced to 14 and 1, respectively, probably due to a more similar (homologous) genetic background among the F2 hybrids than between the parental types. Through the proteome comparison among parent vector and F2 vectors, only four protein spots (S4, S8, S29, and S405) were found to be conserved between generations (Fig. 2 and Tables 2 and 3). The number of protein spots of interest was reduced from 68 to 4, enabling one to focus efforts on significantly fewer proteins more likely to be involved in virus transmission. Similar comparisons between parent nonvector and F2 nonvectors eliminated the need to look further into the nonvectors for virus-binding proteins, since no protein was found to be conserved in all nonvector genotypes. Thus, coupling the proteomics data with the genetic analysis greatly reduced the number of false-positive results and thereby the number of protein spots of interest requiring further analysis. Our data demonstrated the power and usefulness of using a genetic system in proteomics research, something that may have a broad application.
Coimmunoprecipitation of CYDV-RPV-binding aphid proteins.
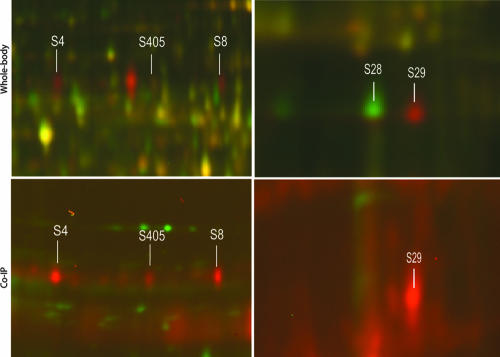
We expect that many aphid proteins involved in virus transport into and through gut or salivary tissues will bind virions due to the well-described vector-specific transmission of each virus species (12). Other aphid proteins involved in general macromolecular transport are undoubtedly involved in virus transmission, but not the specificity of virus species transmission; these proteins may not bind directly to the virus. The population of virus-binding proteins would also be expected to overlap with proteins identified in the 2D DIGE experiments if the proteins are involved in regulating virus transmission. Soluble protein extracts from whole aphid bodies were incubated with purified, transmissible CYDV-RPV. Virus-aphid protein complexes were subsequently immunoprecipitated with antibodies to whole virus. The same aphid protein extracts were also immunoprecipitated with antibodies without the previous addition of virus. Following dissociation of the immunoprecipitated protein complexes and labeling with Cy3 or Cy5 dye, proteins were separated and compared by 2D DIGE. This technique provides an advantage over far-Western blots used previously to identify virus-binding proteins (18, 29, 32, 34) in that the aphid proteins are in their native conformation, and the DIGE format provides a direct comparison of virus-precipitated proteins with proteins precipitated nonspecifically. Numerous CYDV-RPV-binding proteins were identified from the vector genotype Sg-F, including proteins with molecular masses and pIs corresponding to those of proteins S4, S8, S29, and S405 (Fig. 3). The identities of these spots were confirmed by aligning the gel images of the immunoprecipitated proteins to the gel images of the whole-body protein extracts from parent vector Sg-F using the SameSpots software. Similar proteins were not observed in controls lacking CYDV-RPV or in the nonvector Sg-SC. It should be noted that the amount of immunoprecipitated proteins was increased due to the concentrating effect of the virus antibody (Fig. 3). Our results indicated that these four proteins recognize and bind to CYDV-RPV, thus fulfilling a functional requirement expected of proteins involved in virus-specific uptake and transport through aphid cells. Additionally, numerous protein spots were observed on the immunoprecipitated gels that were not present on the gels with whole-aphid extracts (data not shown). These may represent proteins that are involved in virus recognition and transport and that are either minor components of the total proteome or proteins not resolved on 2D gels loaded with the total protein extracts. Isolation of CYDV-RPV-binding aphid proteins by virus coimmunoprecipitation further substantiates the hypothesis that these proteins are linked to CYDV-RPV recognition and binding associated with cellular transport.
FIG. 3.
Comparison of protein spots following 2D DIGE analysis of whole-aphid-body protein extracts (top) from vector (red) and nonvector (green) aphids and CYDV-RPV-binding proteins immunoprecipitated from whole-aphid-body protein extracts (bottom) (virus treatment [red]; control treatment [green]). The four proteins, S4, S8, S405 and S29, were specific to or significantly upregulated in vector genotypes, and all were confirmed to have CYDV-RPV binding properties by coimmunoprecipitation (Co-IP). S28 was a cyclophilin identified in all genotypes, S29 was a cyclophilin identified specifically in vector genotypes. Only S29 was able to bind virus and was identified in the coimmunoprecipitation experiments.
MS analysis and protein identification.
To further characterize and identify the four protein spots identified in both the analysis of whole-aphid protein extracts and coimmunoprecipitated proteins (i.e., S4, S8, S29, and S405), protein spots were cut from 2D picking gels and digested with trypsin, and the resulting peptides were analyzed by MS using a model 4700 proteomics analyzer with 4000 series Explorer software. MS-MS data from the aphid proteins were analyzed by Mascot using GPS Explorer software, and the results were compared for protein homology in various protein databases. Both peptide mass fingerprint and peptide fragmentation (MS-MS) data were used to identify proteins. The Mascot protein score judged the significance of the peptide mass fingerprint data, and the Mascot ion score judged the significance of the MS-MS data. The protein score confidence interval percentages and total ion confidence interval percentages were all 100%. For S29, the average protein score was 214, and average mass accuracy was ±6.9 ppm within the range of 1 to 16 ppm; for S4, the protein score was 122, and average mass accuracy was ±5.3 ppm within the range of 0 to 16 ppm. MS data for each of the four proteins matched ESTs from available aphid databases, but only two proteins had homology to known proteins by BLAST search. S4 was identified as a luciferase-like protein, and S29 was identified as a cyclophilin-like protein (Table 4). In both cases, the E values were <10−27 and amino acid identity was >68%. These types of proteins are associated with endoplasmic reticulum (cyclophilin) (22) or peroxisomes derived from endoplasmic reticulum (luciferase) (21).
TABLE 4.
Identification of aphid proteins conserved in vector genotypes of Schizaphis graminum transmitting CYDV based on comparative sequence analysis of mass spectral data
| Protein | Matched organism | Total no. of matchesa | Characteristic gi | Protein(s) matched by BLASTb |
|---|---|---|---|---|
| S4 | Acyrthosiphon pisum | 1 | gi 46998571 | Luciferase |
| Toxoptera citricida | 1 | gi 30049101 | Luciferase | |
| S8 | No match | 0 | ||
| S29 | Acyrthosiphon pisum | 11 | gi 82571971 | Cyclophilins |
| Aphis gossypii | 1 | gi 73612692 | Cyclophilins | |
| Myzus persicae | 3 | gi 112432157 | Cyclophilins | |
| Toxoptera citricida | 2 | gi 31365720 | Cyclophilins | |
| Drosophila melanogaster | 1 | gi 7291447 | Cyclophilins | |
| Maconellicoccus hirsutus | 1 | gi 120485537 | Cyclophilins | |
| S405 | No match | 0 |
Total number of sequence matches identified for each aphid protein sequence compared to NCBI nonredundant and Acyrthosiphon pisum EST databases using Mascot and GPS Explorer. Protein identifications were inspected for conformation prior to acceptance.
BLAST E values were between 10−99 and 10−27.
Luciferase contains a microbody targeting signal (10) that directs the protein to subcellular organelles that are bound by a single membrane (17). Luteovirids are transported across the gut and ASG cell cytoplasm in single-membrane-bound vesicles, and the transport pathway is often observed to include endosomes, lysosomes, and possibly other vesicles (2, 12). A search of the protein databases at NCBI reveals that homologues of luciferase are found in all sequenced genomes of insects. Firefly luciferase is a member of a superfamily of homologous enzymes, which includes acyl-coenzyme A (acyl-CoA) ligases and peptide synthetases. Recently, luciferase has been found to have additional activities in long-chain fatty acyl-CoA synthesis (21). Its functional roles in insects outside of the luminescence reaction in fireflies are not well characterized, and additional roles of these proteins are likely to be discovered, some of which may be applicable to luteovirus transport in aphids.
The cyclophilin present in the aphid is most closely related to cyclophilin A-like and cyclophilin B-like proteins that are localized in endoplasmic reticulum via an N-terminal signal peptide. Cyclophilin B proteins are reported to function in the secretory pathway, possibly by chaperoning of membrane proteins or having a role in receptor signaling pathways (22). Cyclophilin A is required for the attachment of human immunodeficiency virus (HIV) to host cells by binding a proline-rich loop on the surface of the HIV type 1 capsid (27, 30) and is involved in additional postentry events associated with HIV infection (26). This function of cyclophilin A may have relevance to CYDV-RPV movement in vector aphids. Also, cyclophilin A associates with the dynein/dynactin motor protein complex and has been suggested to perform a general function related to molecular binding for movement along microtubules (6). Cyclophilin B is targeted to the endoplasmic reticulum and functions within secretory pathways and in complexes on the plasma membrane (24). They chaperone plasma membrane proteins and function in receptor signaling pathways (22). Luteovirids are dependent upon receptor-mediated endocytosis pathways to enter both the gut and ASG cells and appear to be directed to specific transcytosis pathways (2, 12).
Intriguingly, cyclophilin B in Drosophila melanogaster has undergone a gene duplication event and the duplicated gene is expressed in a tissue-specific manner (22). Such a gene duplication event may have occurred in aphids also, since we detect another cyclophilin B isoform in the proteomes of both vector and nonvector aphids (S28 [Fig. 2]). Only the cyclophilin protein identified in the vectors (S29) was detected in the coimmunoprecipitation reaction. S28, the cyclophilin isoform identified in both vector and nonvector genotypes, was not detected in the coimmunoprecipitation reaction, indicating it does not bind to CYDV-RPV (Fig. 3). This result also shows the utility of this assay to determine relative expression of proteins in different aphid genotypes.
Although these analyses have not identified the same virus-binding proteins as reported by others, the common denominator is the identification of proteins involved in receptor binding or targeting and transport of macromolecules (i.e., viruses) through cell cytoplasm. We have not identified symbionin as an aphid protein involved in RPV transmission in the S. graminum experimental system, but our biological data suggest that symbionin does not play a role in preventing transmission in the nonvector genotypes (3).
The proteomics approach utilizing 2D DIGE and mass spectral data is an effective way to identify aphid proteins unique to specific aphid genotypes that differed in their ability to transmit viruses. These data alone identified a relatively large number of proteins that be involved in virus transmission; however, the different proteins may represent only genetic differences among aphid populations that could not be associated with virus transmission. Coupling these approaches with the aphid genetic system allowed us to directly correlate the presence or upregulation of specific proteins in multiple aphid genotypes with the same genetic background and the same vector competency phenotypes. This allowed the identification of four proteins out of an initial pool of 120 candidates. Furthermore, these data show that specific aphid proteins associated with CYDV-RPV vector competence are heritable. Coimmunoprecipitation was a method other than 2D DIGE to identify a different set of aphid proteins that were able to bind to virus particles, but again this provides only indirect evidence that the proteins are linked to virus transmission, and not all proteins involved in virus transmission may be involved in direct interactions with the virus. In our studies the four proteins that were conserved in different generations of only the vector competent genotypes of S. graminum were also immunoprecipitated with CYDV-RPV, indicting that these proteins specifically bind to CYDV-RPV. This strengthens the evidence that these proteins are involved in circulative virus transport, although their specific function in luteovirid transmission remains to be determined. Previous research does support a potential role of cyclophilins and luciferase in viral transmission. Our experimental system now allows us to investigate virus-aphid interactions from both the perspective of the aphid and the perspective of the virus. The results of these studies may provide a means to screen aphid populations in the future to determine their capacity to serve as efficient virus vectors. We anticipate that our methods and results will have relevance to the mechanisms underlying the transmission of both plant and animal viruses by other arthropod vectors.
Acknowledgments
We thank William Sackett in the Department of Plant Pathology at the Pennsylvania State University for assistance in the greenhouse and growth chambers for preparing and maintaining the aphid colonies, plants, and virus stocks needed to complete this research. We appreciate Yong Yang, Kevin Howe, and Tara Fish in the Functional and Comparative Proteomics Center, USDA-ARS Plant, Soil, and Nutrition Laboratory at Cornell University for assistance in the laboratory.
This project was supported in part by the National Research Initiative of the USDA Cooperative State Research, Education and Extension Service (NRI award 03-01647).
Footnotes
Published ahead of print on 24 October 2007.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. H. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 253389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brault, V., E. Herrbach, and C. Reinbold. 2007. Electron microscopy studies on luteovirid transmission by aphids. Micron 38302-312. [DOI] [PubMed] [Google Scholar]
- 3.Burrows, M. E., M. C. Caillaud, D. M. Smith, E. C. Benson, F. E. Gildow, and S. M. Gray. 2006. Genetic regulation of polerovirus and luteovirus transmission in the aphid Schizaphis graminum. Phytopathology 96828-837. [DOI] [PubMed] [Google Scholar]
- 4.Burrows, M. E., M. C. Caillaud, D. M. Smith, and S. M. Gray. 2007. Biometrical genetic analysis of luteovirus transmission in the aphid Schizaphis graminum. Heredity 98106-113. [DOI] [PubMed] [Google Scholar]
- 5.Filichkin, S. A., S. Brumfield, T. P. Filichkin, and M. J. Young. 1997. In vitro interactions of the aphid endosymbiotic SymL chaperonin with barley yellow dwarf virus. J. Virol. 71569-577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Galigniana, M. D., Y. Morishima, P. A. Gallay, and W. B. Pratt. 2004. Cyclophilin-A is bound through its peptidylprolyl isomerase domain to the cytoplasmic dynein motor protein complex. J. Biol. Chem. 27955754-55759. [DOI] [PubMed] [Google Scholar]
- 7.Garret, A., C. Kerlan, and D. Thomas. 1993. The intestine is a site of passage for potato leafroll virus from the gut lumen into the haemocoel in the aphid vector, Myzus persicae Sulz. Arch. Virol. 131377-392. [DOI] [PubMed] [Google Scholar]
- 8.Gildow, F. E. 1993. Evidence for receptor-mediated endocytosis regulating luteovirus acquisition by aphids. Phytopathology 83270-277. [Google Scholar]
- 9.Gildow, F. E., and W. F. Rochow. 1980. Transmission interference between two isolates of barley yellow dwarf virus in Macrosiphum avenae. Phytopathology 70122-126. [Google Scholar]
- 10.Gould, S. J., G. A. Keller, M. Schneider, S. H. Howell, L. J. Garrard, J. M. Goodman, B. Distel, H. Tabak, and S. Subramani. 1990. Peroxisomal protein import is conserved between yeast, plants, insects and mammals. EMBO J. 985-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gray, S. M., J. W. Chapin, D. M. Smith, N. Banerjee, and J. S. Thomas. 1998. Barley yellow dwarf luteoviruses and their predominant aphid vectors in winter wheat grown in South Carolina. Plant Dis. 821328-1333. [DOI] [PubMed] [Google Scholar]
- 12.Gray, S. M., and F. Gildow. 2003. Luteovirus-aphid interactions. Annu. Rev. Phytopathol. 41539-566. [DOI] [PubMed] [Google Scholar]
- 13.Gray, S. M., D. M. Smith, L. Barbierri, and J. Burd. 2002. Virus transmission phenotype is correlated with host adaptation among genetically diverse populations of the aphid Schizaphis graminum. Phytopathology 92970-975. [DOI] [PubMed] [Google Scholar]
- 14.Hammond, J., R. M. Lister, and J. E. Foster. 1983. Purification, identity and some properties of an isolate of barley yellow dwarf virus from Indiana. J. Gen. Virol. 64667-676. [Google Scholar]
- 15.Hu, J. S., and W. F. Rochow. 1988. Anti-idiotypic antibodies against an anti-barley yellow dwarf virus monoclonal-antibody. Phytopathology 781302-1307. [Google Scholar]
- 16.Karas, M., and F. Hillenkamp. 1988. Laser desorption ionization of proteins with molecular masses exceeding 10000 daltons. Anal. Chem. 602299-2301. [DOI] [PubMed] [Google Scholar]
- 17.Keller, G. A., S. Krisans, S. J. Gould, J. M. Sommer, C. C. Wang, W. Schliebs, W. Kunau, S. Brody, and S. Subramani. 1991. Evolutionary conservation of a microbody targeting signal that targets proteins to peroxisomes, glyoxysomes, and glycosomes. J. Cell Biol. 114893-904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li, C. Y., D. Cox-Foster, S. M. Gray, and F. Gildow. 2001. Vector specificity of barley yellow dwarf virus (BYDV) transmission: identification of potential cellular receptors binding BYDV-MAV in the aphid, Sitobion avenae. Virology 286125-133. [DOI] [PubMed] [Google Scholar]
- 19.Matsui, N. M., D. M. Smith, K. R. Clauser, J. Fichmann, L. E. Andrews, C. M. Sullivan, A. L. Burlingame, and L. B. Epstein. 1997. Immobilized pH gradient two-dimensional gel electrophoresis and mass spectrometric identification of cytokine-regulated proteins in ME-180 cervical carcinoma cells. Electrophoresis 18409-417. [DOI] [PubMed] [Google Scholar]
- 20.Mayo, M. A., and C. J. D'Arcy. 1999. Family Luteoviridae: a reclassification of luteoviruses, p. 15-22. In H. G. Smith and H. Barker (ed.), The Luteoviridae. CABI Publishing, Wallingford, United Kingdom.
- 21.Oba, Y., M. Sato, M. Ojika, and S. Inouye. 2005. Enzymatic and genetic characterization of firefly luciferase and Drosophila CG6178 as a fatty acyl-CoA synthetase. Biosci. Biotechnol. Biochem. 69819-828. [DOI] [PubMed] [Google Scholar]
- 22.Pemberton, T. J., and J. E. Kay. 2005. Identification and comparative analysis of the peptidyl-prolyl cis/trans isomerase repertoires of H. sapiens, D. melanogaster, C. elegans, S. cerevisiae and Sz. pombe. Comp. Funct. Genomics 6277-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Perkins, D. N., D. J. C. Pappin, D. M. Creasy, and J. S. Cottrell. 1999. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 203551-3567. [DOI] [PubMed] [Google Scholar]
- 24.Price, E. R., M. J. Jin, D. Lim, S. Pati, C. T. Walsh, and F. D. McKeon. 1994. Cyclophlin-B trafficking through the secretory pathway is altered by binding of cyclosporine-A. Proc. Natl. Acad. Sci. USA 913931-3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reinbold, C., E. Herrbach, and V. Brault. 2003. Posterior midgut and hindgut are both sites of acquisition of Cucurbit aphid-borne yellows virus in Myzus persicae and Aphis gossypii. J. Gen. Virol. 843473-3484. [DOI] [PubMed] [Google Scholar]
- 26.Saphire, A. C. S., M. D. Bobardt, and P. A. Gallay. 2002. Cyclophilin A plays distinct roles in human immunodeficiency virus type 1 entry and postentry events, as revealed by spinoculation. J. Virol. 764671-4677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saphire, A. C. S., M. D. Bobardt, and P. A. Gallay. 1999. Host cyclophilin A mediates HIV-1 attachment to target cells via heparans. EMBO J. 186771-6785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Seddas, P., and S. Boissinot. 2006. Glycosylation of beet western yellows virus proteins is implicated in the aphid transmission of the virus. Arch. Virol. 151967-984. [DOI] [PubMed] [Google Scholar]
- 29.Seddas, P., S. Boissinot, J. M. Strub, A. Van Dorsselaer, M. H. V. Van Regenmortel, and F. Pattus. 2004. Rack-1, GAPDH3, and actin: proteins of Myzus persicae potentially involved in the transcytosis of beet western yellows virus particles in the aphid. Virology 325399-412. [DOI] [PubMed] [Google Scholar]
- 30.Sokolskaja, E., and J. Luban. 2006. Cyclophillin, TRIM5, and innate immunity to HIV-1. Curr. Opin. Microbiol. 9404-408. [DOI] [PubMed] [Google Scholar]
- 31.Unlu, M., M. E. Morgan, and J. S. Minden. 1997. Difference gel electrophoresis: a single gel method for detecting changes in protein extracts. Electrophoresis 182071-2077. [DOI] [PubMed] [Google Scholar]
- 32.van den Heuvel, J., M. Verbeek, and F. van der Wilk. 1994. Endosymbiotic bacteria associated with circulative transmission of potato leafroll virus by Myzus persicae. J. Gen. Virol. 752559-2565. [DOI] [PubMed] [Google Scholar]
- 33.van den Heuvel, J. F. J. M., S. A. Hogenhout, and F. van der Wilk. 1999. Recognition and receptors in virus transmission by arthropods. Trends Microbiol. 771-76. [DOI] [PubMed] [Google Scholar]
- 34.Wang, X. F., and G. H. Zhou. 2003. Identification of a protein associated with circulative transmission of barley yellow dwarf virus from cereal aphids, Schizaphis graminum and Sitobion avenae. Chinese Sci. Bull. 482083-2087. [Google Scholar]