Abstract
Alpha/beta interferon immune defenses are essential for resistance to viruses and can be triggered through the actions of the cytoplasmic helicases retinoic acid-inducible gene I (RIG-I) and melanoma differentiation-associated gene 5 (MDA5). Signaling by each is initiated by the recognition of viral products such as RNA and occurs through downstream interaction with the IPS-1 adaptor protein. We directly compared the innate immune signaling requirements of representative viruses of the Flaviviridae, Orthomyxoviridae, Paramyxoviridae, and Reoviridae for RIG-I, MDA5, and interferon promoter-stimulating factor 1 (IPS-1). In cultured fibroblasts, IPS-1 was essential for innate immune signaling of downstream interferon regulatory factor 3 activation and interferon-stimulated gene expression, but the requirements for RIG-I and MDA5 were variable. Each was individually dispensable for signaling triggered by reovirus and dengue virus, whereas RIG-I was essential for signaling by influenza A virus, influenza B virus, and human respiratory syncytial virus. Functional genomics analyses identified cellular genes triggered during influenza A virus infection whose expression was strictly dependent on RIG-I and which are involved in processes of innate or adaptive immunity, apoptosis, cytokine signaling, and inflammation associated with the host response to contemporary and pandemic strains of influenza virus. These results define IPS-1-dependent signaling as an essential feature of host immunity to RNA virus infection. Our observations further demonstrate differential and redundant roles for RIG-I and MDA5 in pathogen recognition and innate immune signaling that may reflect unique and shared biologic properties of RNA viruses whose differential triggering and control of gene expression may impact pathogenesis and infection.
RNA viruses comprise approximately 80% of all viruses and in humans are etiologic agents of infectious diseases that pose major public health concerns (20). Despite differences in genomic features and replication strategies, RNA viruses can trigger common cellular components during infection to elicit an innate immune response that serves as a first line of protection against infection (18, 38, 41). The rapid production of alpha/beta interferon (IFN-α/β) is a central and essential component of this response, leading to induced expression of hundreds of interferon-stimulated genes (ISGs) whose products direct antiviral and immunomodulatory actions that can limit infection (38).
Retinoic acid-inducible gene I (RIG-I) and melanoma differentiation-associated gene 5 (MDA5) are cytoplasmic DEx(D/H) box helicases that can detect intracellular viral products such as genomic RNA (vRNA) to signal IFN-α/β production in infected cells (41). Signaling by each occurs through homotypic caspase activation and recruitment domain (CARD) interactions with the interferon promoter-stimulating factor 1 (IPS-1) adaptor protein (19, 27, 35, 39), which recruits RIG-I and MDA5 to the outer membrane of the mitochondria as part of a macromolecular signaling complex that serves to activate downstream interferon-regulatory factors (IRFs) and other transcription factors that induce IFN-α/β and ISG expression (14). Although RIG-I and MDA5 may share similar signaling features (42) and structural homology (43), accumulating evidence suggests that the two helicases may discriminate among different ligands to trigger the innate immune response to RNA viruses. Signaling by RIG-I is triggered during infection by a number of RNA viruses and by the presence of synthetic RNA transcribed in vitro (17, 24, 33, 37). More recently, RIG-I has been implicated in the recognition of RNA moieties that harbor 5′ triphosphate ends (13, 31, 32) or of RNAs that assume complex secondary structures (33, 37). In contrast, signaling by MDA5 is uniquely triggered during picornavirus infections or in the presence of a synthetic RNA polymer consisting of annealing strands of inosine and cytosine, poly(I:C) (10, 17). To investigate how viruses from distinct genera initiate the innate immune response, we assessed the requirement for RIG-I, MDA5, and IPS-1 during infection with a panel of RNA viruses. Through functional genomic analyses, we further characterize host genes whose expression is dependent on RIG-I during influenza virus infection. Our results define an essential role for IPS-1 and demonstrate unique and redundant roles for RIG-I and MDA5 in RNA virus-triggered signaling involved in the innate immune response to infection. We provide the first identification of a RIG-I-responsive gene bioset whose expression has been linked with immunity and disease during influenza A virus infection.
MATERIALS AND METHODS
Cells, viruses, and reagents.
RIG-I−/−, MDA5−/−, and IPS-1−/− and corresponding wild-type (wt) mouse embryo fibroblast (MEF) cell lines were generated from age-matched knockout and wt mice with a C57BL/6 background as described previously (10, 17, 22). RIG-I MDA5 double-knockout cells were generated by infecting RIG-I−/− MEFs with lentivirus particles that expressed an MDA5-targeting set of short hairpin RNAs (shRNA). As a negative control, RIG-I−/− MEFs were infected with lentivirus particles that expressed a nontargeting control shRNA. Briefly, HEK293T cells were transfected with individual clones from the Sigma Mission shRNA targeting set NM_027835 or the nontargeting control shRNA plasmid along with a lentivirus packaging plasmid according to the manufacturer's recommendations. Cell culture supernatants containing lentivirus particles were collected at 24 and 48 h posttransfection, filtered, and used to infect RIG-I−/− MEFs. The following shRNA were used for silencing MDA5 expression in RIG-I−/− MEFs in this study (the targeting sequences are boldfaced): MDA5-998, 5′-CCGGCCACAGAATCAGACACAAGTTCTCGAGAACTTGTGTCTGATTCTGTGGTTTTTG-3′; MDA5-2067, 5′-CCGGCCTACAAATCAACGACACGATCTCGAGATCGTGTCGTTGATTTGTAGGTTTTTG-3′; MDA5-2911, 5′-CCGGGCAAAGCAATACAACGACAATCTCGAGATTGTCGTTGTATTGCTTTGCTTTTTG-3′; MDA5-3265, 5′-CCGGCCTGATCTTGACTACTCAGAACTCGAGTTCTGAGTAGTCAAGATCAGGTTTTTG-3′; MDA5-3547, 5′-CCGGCCCATGAGGTATTGTCCTAAACTCGAGTTTAGGACAATACCTCATGGGTTTTTG-3′. The shRNA used as a nontargeting control in this study was 5′-CCGGCAACAAGATGAAGAGCACCAACTCGAGTTGGTGCTCTTCATCTTGTTGTTTTT-3′. The human hepatoma cell lines Huh7 and Huh7.5 have been described previously (37). All cell lines were propagated in Dulbecco's modified Eagle medium supplemented with 10% fetal bovine serum, 2 mM l-glutamine, 1 mM sodium pyruvate, antibiotic-antimycotic solution, and 1× nonessential amino acids.
Stocks of red fluorescent protein (RFP)-tagged Newcastle disease virus (NDV) were produced by injecting 10-day-old specific-pathogen-free embryonated chicken eggs (Charles River Laboratory) with stocks of virus (obtained from L. Martinez-Sobrido, Mount Sinai School of Medicine) that were diluted in phosphate-buffered saline (PBS) supplemented with 0.2% (wt/vol) bovine serum albumin, 1.1 mM MgCl2, and 1.2 mM CaCl2. Infected eggs were incubated at 38°C for 48 h and then cooled to 4°C for 2 h before allantoic fluid was collected for virus stock. The virus was titrated on Madin-Darby canine kidney (MDCK) epithelial cells by assaying for focus-forming units (FFU) at 24 to 48 h postinfection. Stocks of green fluorescent protein (GFP)-tagged respiratory syncytial virus (RSV) were prepared from HeLa cells using a virus stock obtained from M. E. Peeples (Ohio State University). Infected HeLa cells and culture medium were collected at 48 h postinfection. The cell pellet was resuspended in PBS supplemented with 2 mM EDTA and was lysed by successive cycles of freezing and thawing. Cellular debris was removed by centrifugation, and the supernatant was pooled with the previously collected culture medium. The virus was concentrated with the addition of 50% (vol/vol) polyethylene glycol 8000 in NTE buffer (150 mM NaCl, 50 mM Tris base [pH 7.2], 10 mM EDTA) to a final concentration of 10% (vol/vol) polyethylene glycol, followed by centrifugation at 10,000 rpm. The virus pellet was reconstituted in 20% sucrose in NTE buffer and was further purified by sedimentation using a discontinuous gradient of 35 and 60% (wt/vol) sucrose in NTE buffer that was centrifuged at 37,000 rpm for 1 h at 4°C. The purified virus stock was titrated on HeLa cells and assayed for FFU at 48 h postinfection. Stocks of the human RSV strain A-2 were prepared from HeLa cells by using a virus stock purchased from the ATCC (VR-1540), and the virus was purified as described above for the GFP-tagged RSV.
Sendai virus (SenV) strain Cantell was obtained from Charles River Laboratory. The recombinant influenza virus with an NS1 deletion, its wt counterpart A/Puerto Rico/8/34, influenza virus strains A/Udorn/1972 (Udorn) and B/Yamagata, and rabbit polyclonal antibodies (pAb) to the nucleoprotein (NP) and NS1 protein of influenza A and B viruses were from A. García-Sastre (Mount Sinai School of Medicine) and R. M. Krug (University of Texas at Austin). Reovirus type 3 Dearing (T3D) and type 1 Lang (T1L) and rabbit pAb to reoviruses were gifts from B. Sherry (North Carolina State University). Dengue virus type 2 (DEN2) was a gift from L. Gehrke (Massachusetts Institute of Technology and Harvard Medical School).
Rabbit pAb to RIG-I and Mx-1 were produced in rabbits by repeated injections of purified recombinant RIG-I protein or synthetic peptides derived from Mx-1, respectively, at the University of Texas Southwestern Antibody Core facility. Rabbit pAb to ISG54 and ISG56 were a gift from G. Sen (Cleveland Clinic); a rabbit pAb to ISG15 was a gift from A. Haas (Louisiana State University). The remaining antisera described in this paper were obtained from commercial sources: Abcam (a mouse monoclonal antibody to DEN NS1), Charles River Laboratory (a chicken pAb to NDV), Molecular Probes (a rabbit pAb to GFP), Santa Cruz (a goat pAb to actin), Biodesign International (a goat pAb to SenV), Chemicon International (a chicken pAb to RFP), Zymed (a rabbit pAb to IRF-3), and Axxora (rabbit pAb to IPS-1 and MDA5).
Renilla luciferase and IFN-β-luciferase constructs have been described elsewhere (8); small interfering RNAs (siRNA) for RIG-I (Genome SMARTpool targeting human DDX58) and IPS-1 (Genome SMARTpool targeting human KIAA1271) and a nontargeting control siRNA were obtained from Ambion. The sequences of the siRNA to IPS-1 and the nontargeting control siRNA have been described previously (26). The set of siRNA duplexes used to silence RIG-I expression in this study included the following: (i) 5′-CAGAAGAUCUUGAGGAUAAUU-3′, (ii) 5′-GCACAGAAGUGUAUAUUGGUU-3′, (iii) 5′-AGACAUGGGUAUAGAGUUAUU-3′, and (iv) 5′-CAACCGAUAUCAUUUCUGAUU-3′.
Protein expression analysis.
Protein extracts from infected cells were prepared by lysing cells in extraction buffer (50 mM Tris HCl [pH 7.5], 150 mM NaCl, 0.5% sodium deoxycholate, 1% NP-40, 1 mM EDTA, and 0.1% sodium dodecyl sulfate, supplemented with 1 mM phenylmethylsulfonyl fluoride, 1 mg/ml aprotonin, leupeptin, and pepstatin, 1 mM sodium vanadate, and 1 mM sodium fluoride) followed by 4°C centrifugation at 16,000 × g for 15 min. The supernatant was separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and analyzed by immunoblotting. We note that ISG56 expression levels typically ranged from undetectable to a low basal level in primary MEFs (see Results), likely a reflection of subtle differences in culture conditions. Differences in viral growth were also noted in the different mouse fibroblast cell lines. We attribute these potential differences to cell population differences in phenotype, which are often observed in in vitro studies and/or which could be linked to other, unknown features of MDA5 or IPS-1 function.
For promoter luciferase reporter assays, cells were cotransfected with Renilla luciferase and IFN-β-luciferase constructs and simultaneously transfected with the appropriate pool of siRNA or a nontargeting siRNA by using Lipofectamine 2000 (Invitrogen) according to the manufacturer's recommendations. Cells were either infected with virus or mock infected 24 h after transfections. At the indicated times, cell extracts were collected and analyzed for dual luciferase activity as described elsewhere (8).
Immunostaining and fluorescent microscopy.
Cells were seeded in chamber slides and either infected with virus or mock infected as indicated below. At the indicated times postinfection, cells were fixed in 3% (wt/vol) paraformaldehyde, incubated in 0.1% (vol/vol) Triton X-100 in PBS, and blocked with 10% (vol/vol) normal goat serum. The slides were then sequentially stained with the appropriate dilutions of primary and secondary antibodies and washed before Vectashield mounting medium (Vector Laboratories) was applied. Slides were sealed under coverslips and examined by immunofluorescent microscopy using an Axiovert phase-contrast microscope in the University of Texas Southwestern Pathogen Imaging Facility.
Expression microarray format and data analysis.
Microarray formats and protocols for probe labeling and array hybridization are described at http://expression.viromics.washington.edu. Briefly, a single experiment comparing two mRNA samples was performed with four replicate mouse (version 2) 22,000-oligonucleotide expression arrays (Agilent Technologies) by using the dye label reverse technique. This allows for the calculation of mean ratios between expression levels of each gene in the sample pair analyzed, standard deviations, and P values for each experiment. Spot quantitation, normalization, and application of a platform-specific error model were performed using Agilent's Feature Extractor software, and all data were then entered into a custom-designed database, Expression Array Manager, and uploaded into the Rosetta Resolver system (version 6.0.0.2.8) for TaqMan (Rosetta Biosoftware, Kirkland, WA) and into the Spotfire DecisionSite for Functional Genomics (version 8.1; Spotfire, Somerville, MA). Data normalization and the resolver error model are described at the Public Microarray Data Download Site of the University of Washington Department of Microbiology (http://expression.viromics.washington.edu). This website is also used to publish all primary data in accordance with the proposed MIAME (minimum information about a microarray experiment) standards (3). Selection of genes for data analysis was based on a >99% probability of their being differentially expressed (P ≤ 0.01) and a change of twofold or more in at least two experiments.
qPCR.
Quantitative real-time PCR (qPCR) was used to validate the gene expression changes and measure influenza virus A/PR/8/34 hemagglutinin (HA) and NP RNA expression in infected MEFs. The results of the qPCR for selected genes are presented in Fig. 5C; additional data are available upon request. Total-RNA samples were treated with DNA-free DNase Treatment and Removal Reagents (Ambion, Austin, TX). Reverse transcription was performed using random hexamer primers and TaqMan reverse transcription reagents (Applied Biosystems, Foster City, CA). Real-time PCR was performed using an ABI 7500 real-time PCR system and TaqMan chemistry. Each target was run in triplicate with TaqMan 2× PCR Universal Master Mix and a 20-μl total reaction volume. Primer and probe sets for relative quantification were selected from the Assays-on-Demand product list (Applied Biosystems), including endogenous controls and 18S rRNA. Each gene was quantified, relative to the calibrator, according to the method of Pfaffl (30). Briefly, expression values were calculated using the following equation: log ratio = 2ΔΔCP as defined by manufacturer's instructions. Probes used for analysis (Applied Biosystems) were as follows (product numbers for probes are in parentheses): the human gene eukaryotic 18S rRNA (Hs99999901_s1) and mouse genes Ifit1 (Mm00515153_m1), Ifit2 (Mm00492606_m1), Stat1 (Mm00439518_m1), Isgf3g (Mm0049267_m1), Ifih1 (Mm00459183_m1), Tlr3 (Mm00556577_m1), Irf7 (Mm00516788_m1), Ifna6 (Mm02524285_g1), Ifnb1 (Mm00439546_s1), Ddx58_C11 (Mm01216863_m1), and Ddx58_C12 (Mm01216864_m1).
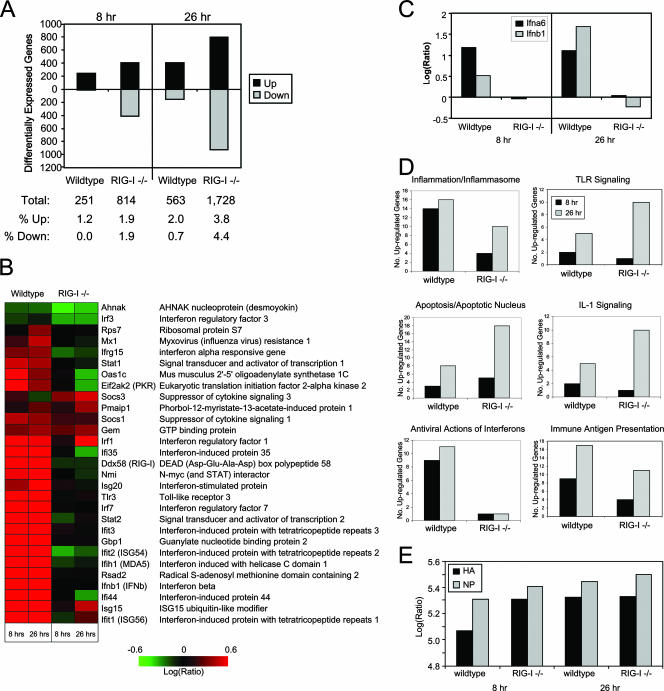
FIG. 5.
Identification of RIG-I-responsive genes by functional genomics analyses. wt or RIG-I−/− mouse fibroblasts were either mock infected or infected with influenza virus A/PR/8/34 at a multiplicity of infection of 5. At 8 and 26 h postinfection, cells were collected and cellular RNA extracted for DNA microarray or qPCR analyses as described in the text. (A) (Top) Numbers of differentially expressed genes (up- or down-regulated more than twofold) as identified by DNA microarray analyses in wt and RIG-I−/− cells following influenza virus A/PR/8/34 infection at 8 and 26 h relative to expression in mock-infected cells. (Bottom) Table showing the total numbers of differentially expressed genes and the breakdown of genes that were differentially expressed as a percentage of total genes analyzed. (B) Heat map showing the differential expression of a bioset of RIG-I-responsive genes following influenza virus A/PR/8/34 infection. (C) Gene expression of IFN-α6 and IFN-β was verified by qualitative real-time PCR. (D) Numbers of genes, segregated by their known functions, whose expression was determined by a DNA microarray to be induced more than twofold following influenza virus A/PR/8/34 infection for 8 and 26 h. Additional data are available at http://expression.viromics.washington.edu. (E) Qualitative real-time PCR analysis for viral HA protein and NP in RNA extracted from wt or RIG-I−/− cells that were either mock infected or infected with influenza virus A/PR/8/34 for 8 or 26 h.
RESULTS
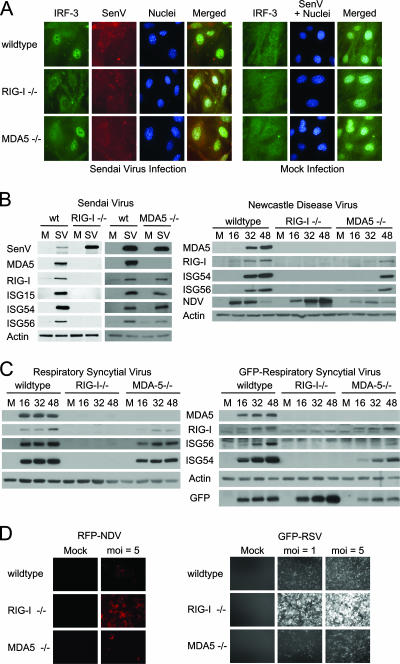
We conducted functional studies to directly compare the roles of RIG-I, MDA5, and IPS-1 in innate immunity against members of the paramyxovirus, flavivirus, reovirus, and orthomyxovirus families. Studies from knockout mice have shown that RIG-I is essential for the innate immune response to the nonhuman paramyxoviruses SenV and NDV (17, 42, 43). We therefore conducted initial experiments to compare innate immune signaling and response in cells infected with SenV, NDV, or RSV, a human pathogen of public health importance (5, 23, 36). Paramyxoviruses encode a monopartite genome of negative-sense single-stranded RNA (20). Infection of primary MEFs by SenV triggered accumulation of the active, nuclear isoforms of IRF-3 in wt and MDA5−/− cells, but infection failed to induce IRF-3 nuclear translocation in RIG-I−/− cells (Fig. 1A). IRF-3 distribution was also predominantly cytoplasmic in uninfected cells. In side-by-side immunoblot analyses, SenV, NDV, and RSV infection induced the expression of IRF-3-dependent genes including ISG15, ISG54, and ISG56 in wt and MDA5−/− cells but not in RIG-I−/− cells (Fig. 1B and C). In the case of RSV, RIG-I was essential for signaling innate immune defenses against both a clinical strain and a recombinant virus that expresses GFP (Fig. 1C). This is consistent with a recent study showing RIG-I-dependent cytokine and Toll-like receptor 3 (TLR3) expression in RSV-infected airway epithelial cells (24). Thus, RIG-I but not MDA5 is essential for triggering innate immune defenses against RSV and other paramyxoviruses. Cells lacking RIG-I were overall more permissive to RSV and NDV infection than were wt or MDA5−/− cells (Fig. 1D), indicating that RIG-I actions restrict initial infection. We found that while MDA5 is not essential for innate immune activation by paramyxoviruses, virus-induced gene expression in MDA5−/− cells was relatively attenuated or delayed compared to that in wt cells (Fig. 1B and C), suggesting that MDA5 may play an auxiliary role in amplifying innate immune signaling initiated by RIG-I during paramyxovirus infection. It should be noted that SenV, NDV, and RSV infections also led to the accumulation of RIG-I and MDA5 proteins in wt cells. MDA5 expression was not readily detectable above background levels in the absence of RIG-I during paramyxovirus infection.
FIG. 1.
RIG-I-dependent signaling of the innate immune response during paramyxovirus infection. (A) wt, RIG-I−/−, or MDA5−/− mouse fibroblasts were either mock infected (right panels) or infected with SenV at 100 HA units/ml of medium (left panels) for 12 h. Cells were fixed and stained with primary antibodies specific for IRF-3 and SenV, followed by fluorescent dye-conjugated secondary antibodies. Cellular distribution of IRF-3 (green), SenV gene products (red), and 4′,6′-diamidino-2-phenylindole (DAPI)-stained nuclei (blue) was visualized by immunofluorescence microscopy. (B) wt, RIG-I−/−, or MDA5−/− mouse fibroblasts were either mock infected (M) or infected with SenV (SV) at 100 HA units/ml of medium for 24 h (left) or with NDV at a multiplicity of infection (MOI) of 5 for the indicated times (hours) (right). (C) wt, RIG-I−/−, or MDA5−/− mouse fibroblasts were either mock infected or infected with RSV (left) or GFP-RSV (right) at an MOI of 5 for the indicated times (hours). Cells were collected and analyzed by immunoblotting for expression of ISG15, ISG54, ISG56, RIG-I, MDA5, viral proteins, and actin (used as a control). (D) wt, RIG-I−/−, or MDA5−/− mouse fibroblasts were either mock infected or infected with either RFP-NDV (left) or GFP-RSV (right) at the indicated MOI for 48 h; then they were fixed, and virus replication was analyzed using fluorescence microscopy.
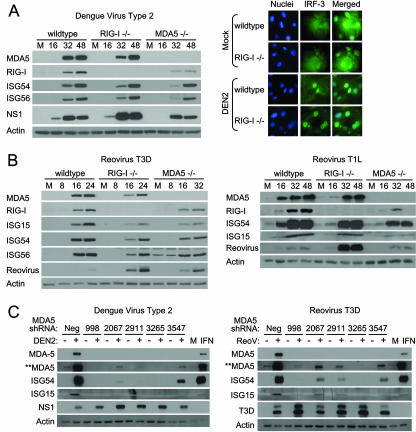
We further examined the roles of RIG-I and MDA5 in the innate immune response to infection by Dengue virus type 2 (DEN2), a flavivirus encoding a monopartite positive-sense single-stranded RNA genome, and prototypic reoviruses, encoding a multipartite double-stranded RNA genome, of two distinct serotypes: T1L and T3D (20). Whereas reovirus T3D exhibits severe neurovirulence and high fatality when injected into infant mice, infection with reovirus T1L is rarely fatal and exhibits considerably milder symptoms (1). Infection by DEN2 triggered the expression of IRF-3-responsive genes in wt, RIG-I−/−, and MDA5−/− cells and concomitant accumulation of IRF-3 in the nuclei of infected cells (Fig. 2A; also data not shown). Reovirus infection of MEFs similarly triggered ISG expression, albeit to different degrees, and this response occurred in the absence of either RIG-I or MDA5 (Fig. 2B). However, neither RIG-I−/− nor MDA5−/− cells attained wt levels of virus-induced gene expression during DEN2 or reovirus infection. Furthermore, although RIG-I and MDA5 expression increased in each case following virus infection, only wt cells attained the highest levels of expression. These results indicate that RIG-I and MDA5 are independently dispensable for innate immune signaling during DEN or reovirus infection. Alternatively, our results may indicate the involvement of a yet to be defined cytoplasmic receptor other than RIG-I or MDA5 in innate immune signaling during DEN2 or reovirus infection. To examine this possibility, MDA5 expression in RIG-I−/− MEFs was silenced by shRNA to generate double-knockout cells lacking both RIG-I and MDA5. DEN2 and reovirus T3D both triggered robust ISG expression in RIG-I−/− cells that had been treated with a nontargeting shRNA control (Fig. 2C, Neg). In contrast, DEN2 or reovirus T3D infection triggered little or no ISG expression (which directly correlated with MDA5 expression levels) in the double-knockout cells, despite robust viral replication. These results indicate that DEN2 and reoviruses trigger both RIG-I- and MDA5-dependent innate immune responses in mouse fibroblasts.
FIG. 2.
DEN and reoviruses trigger the innate immune response independently of RIG-I or MDA5. (A) wt, RIG-I−/−, or MDA5−/− mouse fibroblasts were either mock infected or infected with DEN2 at a multiplicity of infection of 1. (Left) Extracts derived from cells that were mock infected (M) or infected with DEN2 for the indicated times (in hours) were analyzed by immunoblotting for the abundance of IRF-3-responsive genes, RIG-I, MDA5, DEN2 NS1, and actin (used as a control). (Right) At 16 h postinfection, cells were fixed and stained with primary antibodies specific for IRF-3, followed by fluorescent-dye-conjugated secondary antibodies. Cellular distribution of IRF-3 (green) and 4′,6′-diamidino-2-phenylindole (DAPI)-stained nuclei (blue) was analyzed by immunofluorescence microscopy. (B) wt, RIG-I−/−, or MDA5−/− mouse fibroblasts were either mock infected (M) or infected with either reovirus T3D or reovirus T1L at a multiplicity of infection of 25. At the indicated times postinfection (in hours), cells were harvested and the extracts analyzed by immunoblotting. We note that a significantly longer exposure time was necessary for the detection of ISG54 and ISG56 in reovirus T1L-infected cells. (C) RIG-I−/− mouse fibroblasts were infected with lentivirus particles expressing either a shRNA to MDA5 (shRNA 998, 2067, 2911, 3265, or 3547) or a nontargeting shRNA control (Neg). At 36 h postinfection, cells were either mock infected (−) or infected (+) with either DEN (left) or reovirus T3D (right) at multiplicities of infection of 1 and 25, respectively. Cells were harvested 48 h postinfection (84 h after infection with lentivirus) and the extracts analyzed by immunoblotting. For controls, extracts of RIG-I−/− cells that were mock treated (M) or treated with exogenous IFN-β were also subjected to immunoblot analysis. **MDA5 indicates an image of MDA5 protein expression taken after a significantly longer exposure time.
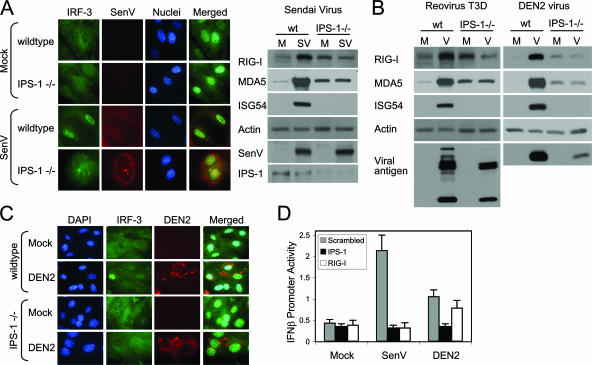
RIG-I and MDA5 signal downstream immune actions through the essential adaptor protein, IPS-1 (14). To determine the requirement for signaling by either RIG-I or MDA5 in the innate immune response to DEN2 and reovirus infection, we examined whether IPS-1 was required for triggering virus-induced gene expression. In control experiments, cells lacking IPS-1 failed to signal IRF-3 nuclear translocation and lacked ISG expression upon SenV infection (Fig. 3A). Similarly, when infected with either reovirus T3D or DEN2, IPS-1−/− cells failed to induce IRF-3-dependent gene expression above basal background levels (Fig. 3B) or to trigger IRF-3 nuclear translocation following DEN2 (Fig. 3C) or reovirus T3D (data not shown) infection. To verify these results, we measured IFN-β promoter signaling in HeLa cells transfected with siRNA targeted to IPS-1 or RIG-I. Cells were then either mock infected or infected with SenV or DEN2 and assessed for IFN-β promoter activation. As expected, knockdown of RIG-I expression abrogated IFN-β promoter activation by SenV but had only a partial impact on promoter signaling by DEN2 (Fig. 3D). Importantly, knockdown of IPS-1 completely abrogated signaling to the IFN-β promoter induced by SenV or DEN2. Thus, IPS-1 is essential for triggering innate immune defenses during infection with SenV, DEN, or reovirus. Taken together, our results suggest that reovirus and DEN signal the innate defenses through an IPS-1-regulated pathway that is likely initiated by either RIG-I or MDA5.
FIG. 3.
IPS-1 is essential for triggering of the innate immune response during paramyxovirus, reovirus, or DEN infection. (A) wt or IPS-1−/− mouse fibroblasts were either mock infected or infected with SenV at 100 HA units/ml. (Left) Cells were fixed and stained with primary and fluorescent-dye-conjugated secondary antibodies at 12 h postinfection. The cellular distribution of IRF-3 (green), SenV-infected cells (red), and nuclei of cells (blue) (stained with 4′,6′-diamidino-2-phenylindole [DAPI]) was detected by fluorescence microscopy. (Right) At 24 h, mock-infected (M) and SenV-infected (SV) cells were collected and analyzed by immunoblotting for the abundances of ISG54, RIG-I, MDA5, IPS-1, viral antigen, and actin (used as a control). (B) wt or IPS-1−/− mouse fibroblasts were either mock infected or infected (V) with either reovirus T3D at a multiplicity of infection of 25 or DEN2 at a multiplicity of infection of 1. At 24 h postinfection, cells were collected and analyzed for the abundances of RIG-I, MDA5, ISG54, viral antigen, and actin (used as a control). (C) wt or IPS-1−/− mouse fibroblasts were either mock infected or infected with DEN2 at a multiplicity of infection of 1 for 24 h. Cells were fixed, stained with primary and fluorescent-dye-conjugated secondary antibodies, and analyzed by immunofluorescent microscopy for the cellular distribution of IRF-3 (green) and the detection of DEN2-infected cells (red) and DAPI-stained nuclei (blue). (D) HeLa cells were transfected with IFN-β promoter-luciferase and Renilla luciferase reporter plasmids and either a siRNA pool that is known to suppress IPS-1 or RIG-I expression or a nonspecific control siRNA (scrambled). At 24 h posttransfection, cells were either mock infected or infected with either SenV at 100 HA units/ml or DEN2 at a multiplicity of infection of 1. Cells were collected at 24 h postinfection and analyzed for relative luciferase activity. Error bars, standard deviations calculated from three independent experiments.
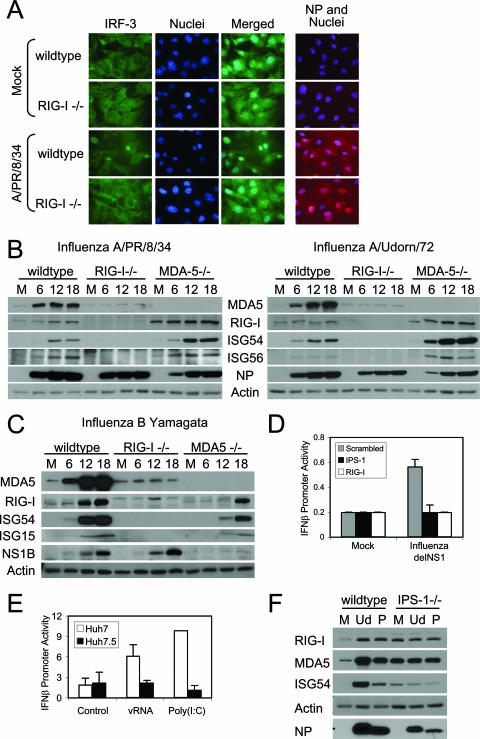
We also directly compared and assessed the roles of RIG-I and MDA5 in triggering innate immune actions during orthomyxovirus infection. Influenza A and B viruses are orthomyxoviruses, each encoding a multipartite genome comprised of single-stranded, negative-sense RNA (20). We found that the A/PR/8/34 (Fig. 4A) or A/Udorn (data not shown) strain of influenza virus triggered IRF-3 nuclear accumulation in a low frequency of cells in cultures of wt (Fig. 4A) or MDA5−/− (data not shown) MEFs. Uninfected cells show typical cytoplasmic staining of IRF-3 (Fig. 4A). In parallel experiments, we found that nuclear accumulation of IRF-3 occurred within a larger frequency of cells during infection of cultures with an isogenic mutant virus, A/PR/8delNS1, that expresses a truncated NS1 gene (data not shown). This supports previous findings that the influenza virus A/PR/8/34 NS1 protein can attenuate RIG-I signaling of IRF-3 activation (11, 28, 29, 31). Importantly, IRF-3 was never observed to accumulate in the nuclei of influenza virus-infected RIG-I−/− cells (Fig. 4A). Immunoblot analysis revealed that each strain of influenza A virus (Fig. 4B) as well as the B/Yamagata strain of influenza B virus (Fig. 4C) triggered innate immune response gene expression and concomitant increases in RIG-I and MDA5 protein levels in a manner that was completely dependent on RIG-I. We note that influenza B virus triggered a reduced response in MDA5−/− cells, which suggests that MDA5 may play a supportive role in signaling the innate immune response to influenza B virus infection. Alternatively, the reduced response may be a direct reflection of low levels of viral growth observed in MDA5−/− cells.
FIG. 4.
RIG-I- and IPS-1-dependent innate immune signaling by orthomyxoviruses. (A) (Left panels) wt or RIG-I−/− mouse fibroblasts were either mock infected or infected with influenza virus A/PR/8/34 at a multiplicity of infection of 5. At 12 h postinfection, cells were fixed and analyzed for host response activation by staining for IRF-3 (green) and nuclei (blue). (Right panel) In parallel experiments, cells were fixed and analyzed for the frequency of virus infection by staining for influenza A virus NP (red) and nuclei (blue). (B and C) wt, RIG-I−/−, or MDA5−/− mouse fibroblasts were either mock infected (M) or infected with influenza virus A/PR/8/34 or A/Udorn/72 (B) or with influenza virus B/Yamagata (C) at a multiplicity of infection of 5. At the indicated hours postinfection, cells were collected and analyzed by immunoblotting for the abundances of IRF-3-responsive genes, MDA5, RIG-I, influenza virus NP or NS1B protein, and actin (used as a control). (D) HeLa cells were transfected with IFN-β promoter-luciferase and Renilla luciferase reporter plasmids and either a siRNA pool that has been shown to suppress IPS-1 or RIG-I expression or a control siRNA with no known targets (Scrambled). At 24 h posttransfection, cells were either mock infected or infected with influenza virus A/PR/8delNS1 at a multiplicity of infection of 5. Cells were collected 18 h postinfection and analyzed for dual luciferase activity(expressed as relative IFN-β promoter activity). Error bars, standard deviations calculated from three independent experiments. (E) Huh7 or Huh7.5 cells were transfected with IFN-β promoter-luciferase and Renilla luciferase reporter plasmids for 24 h and then were either mock transfected or transfected with either vRNA or a synthetic double-stranded RNA control [poly(I:C)]. At 12 h after RNA transfection (36 h after plasmid DNA transfection), cells were collected and analyzed for dual luciferase activity. The results are expressed as relative IFN-β promoter activity. Error bars, standard deviations calculated from three independent experiments. (F) wt or IPS-1−/− mouse fibroblasts were either mock infected (M) or infected with influenza virus A/Udorn/72 (Ud) or A/PR/8/34 (P) at a multiplicity of infection of 5. At 18 h postinfection, cells were collected and the extracts analyzed for the abundances of RIG-I, MDA5, ISG54, actin (used as a control), and viral NP by immunoblotting.
To assess the mechanisms of RIG-I signaling during influenza virus infection, we conducted IFN-β promoter signaling experiments in HeLa cells and in Huh7 cell clones. HeLa cells were first transfected with siRNA to knock down RIG-I or IPS-1 expression. As shown in Fig. 4D, RIG-I or IPS-1 knockdown abrogated signaling of IFN-β promoter activation by the influenza A virus NS1 truncation mutant. In further experiments we transfected Huh7 cells or Huh7.5 cells with a synthetic double-stranded RNA polymer consisting of annealing strands of inosine and cytosine [poly(I:C)] or with vRNA isolated from influenza virus A/PR/8/34. Huh7 cells are competent for signaling through the RIG-I pathway and trigger innate immune activation in response to poly(I:C), whereas Huh7.5 cells encode defective RIG-I and cannot mediate RIG-I-dependent signaling (37). Influenza virus vRNA and poly(I:C) control RNA both activated the IFN-β promoter reporter in Huh7 cells (Fig. 4E). However, neither the vRNA nor the poly(I:C) control was able to trigger IFN-β promoter signaling in the absence of functional RIG-I in Huh7.5 cells. Furthermore, when infected with influenza virus A/PR/8/34 or A/Udorn/1972, cells lacking IPS-1 failed to induce expression of ISG above basal background levels (Fig. 4F). Taken together, our results demonstrate that RIG-I is essential for triggering the innate immune response to influenza viruses A and B, and they indicate that the orthomyxoviruses signal innate defenses through the IPS-1 adaptor protein. Our observations validate influenza A virus vRNA as a trigger of RIG-I signaling (31) and indicate that vRNA contains signatures or specific motifs that present a pathogen-associated molecular pattern that is recognized by RIG-I and sufficient to trigger the innate immune response to infection.
To further define the role of RIG-I in signaling host gene expression, we conducted functional genomics analyses to define the cellular genes whose expression is controlled by RIG-I during influenza A virus infection. We designed our bioinformatics analysis to identify RIG-I-responsive genes in cells by comparing wt and RIG-I−/− cells that had been either mock infected or infected with influenza A/PR/8/34 virus for 8 or 26 h. Overall, the number of differentially expressed genes in RIG-I−/− cells far exceeded that in wt cells after influenza virus A/PR/8/34 infection at both time points (Fig. 5A). Our data show that infection with influenza virus A/PR/8/34 triggered the expression of a number of immune system-related genes that are not expressed, or whose expression is altered, in the absence of RIG-I (Fig. 5B). In particular, RIG-I−/− cells show a profound lack of expression of a bioset of genes whose products are involved in innate defenses during influenza A virus infection. In agreement with the immunoblot results, we found that IFN-α/β gene expression is significantly attenuated in the absence of RIG-I (expression of IFN-α6 and IFN-β [Fig. 5B and C]). There was also a lack of expression of genes whose products are involved in IFN-α/β production and signaling (such as IRF3, IRF7, Stat1, and Stat2) and of ISGs with direct antiviral actions, including PKR, OAS, Mx-1, ISG54, ISG56, and Viperin, in RIG-I−/− cells (Fig. 5B and data not shown). Moreover, TLR3 and MDA5 (Ifih1) expression was attenuated overall in RIG-I−/− cells.
Expression profiling also showed a lack of expression of several genes involved in antigen presentation and the secretion of proinflammatory cytokines in RIG-I−/− cells (Fig. 5D), suggesting that RIG-I signaling may further impact those immune processes during influenza A virus infection. The absence of RIG-I also resulted in reduced expression of a subset of genes following influenza virus A/PR/8/34 infection (Fig. 5A), suggesting that RIG-I may further regulate the maintenance of basal-level expression of these genes. In contrast, the expression of genes involved in TLR signaling of the proinflammatory response, apoptosis, and interleukin-1 signaling of chemokine secretion was elevated in the absence of RIG-I (Fig. 5D); the primary microarray data for these genes are available at http://expression.viromics.washington.edu. Viral gene expression was further verified by qPCR, and the analyses showed that at all times tested, viral HA and NP genes were transcribed to higher levels in RIG-I−/− cells than in wt cells (Fig. 5E). Taken together, our results indicate that influenza viruses can engage RIG-I during infection, resulting in innate immune signaling and induction of genes broadly involved in immunity and inflammation.
DISCUSSION
The results of this study show that IPS-1 is essential for signaling the innate immune response to a wide range of RNA viruses from diverse genera with distinct genome and replication characteristics. The universal requirement for IPS-1 in these examples is unusual given the typical redundancy observed in most immune signaling processes. By playing a central role in innate immune signaling during RNA virus infections, IPS-1 presents an attractive target for virus control of host defenses, where viral strategies to disrupt IPS-1 function can provide effective means of evading the innate immune response. Indeed, hepatitis C virus targets IPS-1 through the actions of the viral NS3/4A protease, which cleaves IPS-1 to inactivate its signaling properties (15, 26, 27). Similarly, pathogenic pestiviruses and hepatitis A virus proteolytically target and inactivate IPS-1 (7, 40). IPS-1 signaling is initiated by ligand-induced CARD-CARD interaction with either RIG-I or MDA5 (19, 26, 33), which facilitates the recruitment of RIG-I and MDA5 to the mitochondria as part of a macromolecular signaling complex. The interaction with IPS-1 is thought to stimulate the actions of this signaling complex to activate IRFs and NF-κB (12, 44). Importantly, the mitochondrial localization of IPS-1 must be maintained in order to support these signaling functions (26, 35). The requirement for mitochondrial placement of IPS-1 indicates that its function requires mitochondrial cofactors that cooperate in factor recruitment and/or IPS-1 signaling actions. By extension, our results imply that mitochondrial cofactors of IPS-1 signaling play an essential role in innate immunity against the genera of RNA viruses examined in the current study.
Progress has been made in discerning the molecular signatures recognized by RIG-I during virus infection, but it is still unclear what minimal features are required for virus recognition by MDA5. In other work and in this study, RIG-I has been shown to trigger innate immune defenses during infection by paramyxoviruses, orthomyxoviruses, and the rhabdovirus vesicular stomatitis virus (17, 41), and it is suggested to play a role in recognizing the single-stranded RNA genome of the filovirus Ebola virus (4) as well as EBER RNA, carried by Epstein-Barr virus (34) (Fig. 6). In contrast, MDA5 uniquely triggers innate immune defenses during picornavirus infections (10, 17). Despite similarities in genomic features and replication strategies, members of the Flaviviridae exhibit differential triggering of the innate immune response. Whereas Japanese encephalitis virus and hepatitis C virus initiated a primarily RIG-I dependent response (6, 17, 26, 37), strains of West Nile virus (9) and DEN triggered RIG-I- and MDA5-dependent signaling, suggesting that flaviviruses may have genomic features that can be discerned by both RIG-I and MDA5. The results of our study therefore substantiate accumulating evidence for unique and redundant roles of RIG-I and MDA5 in signaling that converges on IPS-1 to direct innate immune defenses during virus infection. Identification of the specific molecular patterns or features that trigger RIG-I or MDA5 signaling may lead to targeted and specific antiviral strategies against infection.
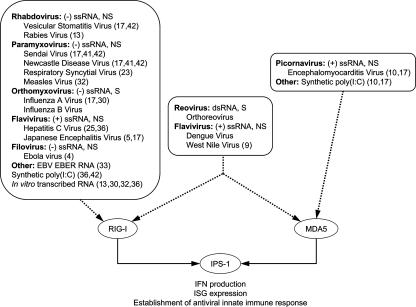
FIG. 6.
Model of viral nucleic acid pathogen-associated molecular pattern recognition and signaling to IFN-β by RIG-I and MDA5 during infection by different RNA viruses. Shown is a summary of published results (with reference numbers in parentheses) demonstrating virus-induced RIG-I and MDA5 signaling of the innate immune response. Genomic features of viruses are described in brief: positive sense (+) versus negative sense (−); single-stranded RNA (ssRNA) versus double-stranded RNA (dsRNA); segmented (S) versus nonsegmented (NS) genomes. In fibroblasts, epithelial cells, and certain immune cells, signaling by RIG-I and MDA5 converges on IPS-1 to rapidly trigger IFN-α/β production and ISG expression. Of the viruses listed, reovirus, DEN2, and West Nile virus are unique in being able to signal both RIG-I-dependent and RIG-I-independent responses.
Functional genomic analyses show that infection with contemporary strains of influenza A virus typically triggers the rapid expression of immune system-related genes responsible for mediating transient immune responses that effectively resolve infection (2). Recent studies by Kash et al. and Kobasa et al. provide evidence that infection with the 1918 pandemic strain of influenza A virus triggers the persistent expression of an overlapping pool of immune system-related genes (16, 21). Excessive triggering of these genes leads to induction of a “cytokine storm” that not only is ineffectual in facilitating viral clearance but may further contribute to disease (25). Importantly, many of these disease-related genes induced by pandemic influenza virus infection are present in the bioset of RIG-I-responsive genes identified in our functional genomics studies. Thus, the processes that trigger and control RIG-I-signaling during virus infection and the products of RIG-I-responsive genes may be important determinants of viral pathogenesis and disease outcome during influenza A virus infection.
Acknowledgments
We thank Robert M. Krug, Marco Colonna, Lee Gerhke, Barbara Sherry, Masato Hatta, and Yoshihiro Kawaoka for providing various viruses and materials for this study. We also thank Robert I. Greene and Matthew J. Thomas for technical assistance.
This study was supported by the University of Washington; NIH grants R01 AI22646, R01 AI46954, and R01AI060389 to M.G.K., A.G.-S., and M.G., respectively; an NIH-funded center to investigate viral immunity and antagonism (CIVIA, U19 AI62623; to A.G.-S.); and an NIH-funded training grant (CA09229-28; to J.F.). M.G. is additionally supported by funding from the Burroughs-Wellcome Fund and a gift from R. and R. Batcheldor.
Footnotes
Published ahead of print on 17 October 2007.
REFERENCES
- 1.Barthold, S. W., A. L. Smith, and P. N. Bhatt. 1993. Infectivity, disease patterns, and serologic profiles of reovirus serotypes 1, 2, and 3 in infant and weanling mice. Lab. Anim. Sci. 43425-430. [PubMed] [Google Scholar]
- 2.Baskin, C. R., A. García-Sastre, T. M. Tumpey, H. Bielefeldt-Ohmann, V. S. Carter, E. Nistal-Villan, and M. G. Katze. 2004. Integration of clinical data, pathology, and cDNA microarrays in influenza virus-infected pigtailed macaques (Macaca nemestrina). J. Virol. 7810420-10432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brazma, A., P. Hingamp, J. Quackenbush, G. Sherlock, P. Spellman, C. Stoeckert, J. Aach, W. Ansorge, C. A. Ball, H. C. Causton, T. Gaasterland, P. Glenisson, F. C. Holstege, I. F. Kim, V. Markowitz, J. C. Matese, H. Parkinson, A. Robinson, U. Sarkans, S. Schulze-Kremer, J. Stewart, R. Taylor, J. Vilo, and M. Vingron. 2001. Minimum information about a microarray experiment (MIAME)—toward standards for microarray data. Nat. Genet. 29365-371. [DOI] [PubMed] [Google Scholar]
- 4.Cárdenas, W. B., Y. M. Loo, M. Gale, Jr., A. L. Hartman, C. R. Kimberlin, L. Martinez-Sobrido, E. O. Saphire, and C. F. Basler. 2006. Ebola virus VP35 protein binds double-stranded RNA and inhibits alpha/beta interferon production induced by RIG-I signaling. J. Virol. 805168-5178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.CDC. 2006. Respiratory syncytial virus activity—United States, 2005-2006. Morb. Mortal. Wkly. Rep. 551277-1279. [PubMed] [Google Scholar]
- 6.Chang, T. H., C. L. Liao, and Y. L. Lin. 2006. Flavivirus induces interferon-beta gene expression through a pathway involving RIG-I-dependent IRF-3 and PI3K-dependent NF-κB activation. Microbes Infect. 8157-171. [DOI] [PubMed] [Google Scholar]
- 7.Chen, Z., Y. Benureau, R. Rijnbrand, J. Yi, T. Wang, L. Warter, R. E. Lanford, S. A. Weinman, S. M. Lemon, A. Martin, and K. Li. 2007. GB virus B disrupts RIG-I signaling by NS3/4A-mediated cleavage of the adaptor protein MAVS. J. Virol. 81964-976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Foy, E., K. Li, C. Wang, R. Sumpter, Jr., M. Ikeda, S. M. Lemon, and M. Gale, Jr. 2003. Regulation of interferon regulatory factor-3 by the hepatitis C virus serine protease. Science 3001145-1148. [DOI] [PubMed] [Google Scholar]
- 9.Fredericksen, B. L., and M. Gale, Jr. 2006. West Nile virus evades activation of interferon regulatory factor 3 through RIG-I-dependent and -independent pathways without antagonizing host defense signaling. J. Virol. 802913-2923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gitlin, L., W. Barchet, S. Gilfillan, M. Cella, B. Beutler, R. A. Flavell, M. S. Diamond, and M. Colonna. 2006. Essential role of MDA-5 in type I IFN responses to polyriboinosinic:polyribocytidylic acid and encephalomyocarditis picornavirus. Proc. Natl. Acad. Sci. USA 1038459-8464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guo, Z., L. M. Chen, H. Zeng, J. A. Gomez, J. Plowden, T. Fujita, J. M. Katz, R. O. Donis, and S. Sambhara. 2007. NS1 protein of influenza A virus inhibits the function of intracytoplasmic pathogen sensor, RIG-I. Am. J. Respir. Cell Mol. Biol. 36263-269. [DOI] [PubMed] [Google Scholar]
- 12.Hiscott, J. 29 March 2007. Triggering the innate antiviral response through IRF-3 activation. J. Biol. Chem. doi: 10.1074/jbc.R700002200. [DOI] [PubMed]
- 13.Hornung, V., J. Ellegast, S. Kim, K. Brzozka, A. Jung, H. Kato, H. Poeck, S. Akira, K. K. Conzelmann, M. Schlee, S. Endres, and G. Hartmann. 2006. 5′-Triphosphate RNA is the ligand for RIG-I. Science 314994-997. [DOI] [PubMed] [Google Scholar]
- 14.Johnson, C. L., and M. Gale, Jr. 2006. CARD games between virus and host get a new player. Trends Immunol. 271-4. [DOI] [PubMed] [Google Scholar]
- 15.Johnson, C. L., D. M. Owen, and M. Gale, Jr. 2007. Functional and therapeutic analysis of hepatitis C virus NS3.4A protease control of antiviral immune defense. J. Biol. Chem. 28210792-10803. [DOI] [PubMed] [Google Scholar]
- 16.Kash, J. C., T. M. Tumpey, S. C. Proll, V. Carter, O. Perwitasari, M. J. Thomas, C. F. Basler, P. Palese, J. K. Taubenberger, A. García-Sastre, D. E. Swayne, and M. G. Katze. 2006. Genomic analysis of increased host immune and cell death responses induced by 1918 influenza virus. Nature 443578-581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kato, H., O. Takeuchi, S. Sato, M. Yoneyama, M. Yamamoto, K. Matsui, S. Uematsu, A. Jung, T. Kawai, K. J. Ishii, O. Yamaguchi, K. Otsu, T. Tsujimura, C. S. Koh, C. Reis e Sousa, Y. Matsuura, T. Fujita, and S. Akira. 2006. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature 441101-105. [DOI] [PubMed] [Google Scholar]
- 18.Kawai, T., and S. Akira. 2006. Innate immune recognition of viral infection. Nat. Immunol. 7131-137. [DOI] [PubMed] [Google Scholar]
- 19.Kawai, T., K. Takahashi, S. Sato, C. Coban, H. Kumar, H. Kato, K. J. Ishii, O. Takeuchi, and S. Akira. 2005. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat. Immunol. 6981-988. [DOI] [PubMed] [Google Scholar]
- 20.Knipe, D. M., P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus. 2001. Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, PA.
- 21.Kobasa, D., S. M. Jones, K. Shinya, J. C. Kash, J. Copps, H. Ebihara, Y. Hatta, J. H. Kim, P. Halfmann, M. Hatta, F. Feldmann, J. B. Alimonti, L. Fernando, Y. Li, M. G. Katze, H. Feldmann, and Y. Kawaoka. 2007. Aberrant innate immune response in lethal infection of macaques with the 1918 influenza virus. Nature 445319-323. [DOI] [PubMed] [Google Scholar]
- 22.Kumar, H., T. Kawai, H. Kato, S. Sato, K. Takahashi, C. Coban, M. Yamamoto, S. Uematsu, K. J. Ishii, O. Takeuchi, and S. Akira. 2006. Essential role of IPS-1 in innate immune responses against RNA viruses. J. Exp. Med. 2031795-1803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leader, S., and K. Kohlhase. 2002. Respiratory syncytial virus-coded pediatric hospitalizations, 1997 to 1999. Pediatr. Infect. Dis. J. 21629-632. [DOI] [PubMed] [Google Scholar]
- 24.Liu, P., M. Jamaluddin, K. Li, R. P. Garofalo, A. Casola, and A. R. Brasier. 2007. Retinoic acid-inducible gene I mediates early antiviral response and Toll-like receptor 3 expression in respiratory syncytial virus-infected airway epithelial cells. J. Virol. 811401-1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Loo, Y. M., and M. Gale, Jr. 2007. Influenza: fatal immunity and the 1918 virus. Nature 445267-268. [DOI] [PubMed] [Google Scholar]
- 26.Loo, Y. M., D. M. Owen, K. Li, A. K. Erickson, C. L. Johnson, P. M. Fish, D. S. Carney, T. Wang, H. Ishida, M. Yoneyama, T. Fujita, T. Saito, W. M. Lee, C. H. Hagedorn, D. T. Lau, S. A. Weinman, S. M. Lemon, and M. Gale, Jr. 2006. Viral and therapeutic control of IFN-β promoter stimulator 1 during hepatitis C virus infection. Proc. Natl. Acad. Sci. USA 1036001-6006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meylan, E., J. Curran, K. Hofmann, D. Moradpour, M. Binder, R. Bartenschlager, and J. Tschopp. 2005. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature 4371167-1172. [DOI] [PubMed] [Google Scholar]
- 28.Mibayashi, M., L. Martinez-Sobrido, Y. M. Loo, W. B. Cardenas, M. Gale, Jr., and A. García-Sastre. 2007. Inhibition of retinoic acid-inducible gene I-mediated induction of beta interferon by the NS1 protein of influenza A virus. J. Virol. 81514-524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Opitz, B., A. Rejaibi, B. Dauber, J. Eckhard, M. Vinzing, B. Schmeck, S. Hippenstiel, N. Suttorp, and T. Wolff. 2007. IFNβ induction by influenza A virus is mediated by RIG-I which is regulated by the viral NS1 protein. Cell. Microbiol. 9930-938. [DOI] [PubMed] [Google Scholar]
- 30.Pfaffl, M. W. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pichlmair, A., O. Schulz, C. P. Tan, T. I. Naslund, P. Liljestrom, F. Weber, and C. Reis e Sousa. 2006. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science 314997-1001. [DOI] [PubMed] [Google Scholar]
- 32.Plumet, S., F. Herschke, J. M. Bourhis, H. Valentin, S. Longhi, and D. Gerlier. 2007. Cytosolic 5′-triphosphate ended viral leader transcript of measles virus as activator of the RIG I-mediated interferon response. PLoS ONE 2e279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saito, T., R. Hirai, Y. M. Loo, D. Owen, C. L. Johnson, S. C. Sinha, S. Akira, T. Fujita, and M. Gale, Jr. 2007. Regulation of innate antiviral defenses through a shared repressor domain in RIG-I and LGP2. Proc. Natl. Acad. Sci. USA 104582-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Samanta, M., D. Iwakiri, T. Kanda, T. Imaizumi, and K. Takada. 2006. EB virus-encoded RNAs are recognized by RIG-I and activate signaling to induce type I IFN. EMBO J. 254207-4214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Seth, R. B., L. Sun, C. K. Ea, and Z. J. Chen. 2005. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-κB and IRF 3. Cell 122669-682. [DOI] [PubMed] [Google Scholar]
- 36.Shay, D. K., R. C. Holman, G. E. Roosevelt, M. J. Clarke, and L. J. Anderson. 2001. Bronchiolitis-associated mortality and estimates of respiratory syncytial virus-associated deaths among US children, 1979-1997. J. Infect. Dis. 18316-22. [DOI] [PubMed] [Google Scholar]
- 37.Sumpter, R., Jr., Y. M. Loo, E. Foy, K. Li, M. Yoneyama, T. Fujita, S. M. Lemon, and M. Gale, Jr. 2005. Regulating intracellular antiviral defense and permissiveness to hepatitis C virus RNA replication through a cellular RNA helicase, RIG-I. J. Virol. 792689-2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Takaoka, A., and H. Yanai. 2006. Interferon signalling network in innate defence. Cell. Microbiol. 8907-922. [DOI] [PubMed] [Google Scholar]
- 39.Xu, L. G., Y. Y. Wang, K. J. Han, L. Y. Li, Z. Zhai, and H. B. Shu. 2005. VISA is an adapter protein required for virus-triggered IFN-β signaling. Mol. Cell 19727-740. [DOI] [PubMed] [Google Scholar]
- 40.Yang, Y., Y. Liang, L. Qu, Z. Chen, M. Yi, K. Li, and S. M. Lemon. 2007. Disruption of innate immunity due to mitochondrial targeting of a picornaviral protease precursor. Proc. Natl. Acad. Sci. USA 1047253-7258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yoneyama, M., and T. Fujita. 2007. Function of RIG-I-like receptors in antiviral innate immunity. J. Biol. Chem. 28215315-15318. [DOI] [PubMed] [Google Scholar]
- 42.Yoneyama, M., M. Kikuchi, K. Matsumoto, T. Imaizumi, M. Miyagishi, K. Taira, E. Foy, Y. M. Loo, M. Gale, Jr., S. Akira, S. Yonehara, A. Kato, and T. Fujita. 2005. Shared and unique functions of the DExD/H-box helicases RIG-I, MDA5, and LGP2 in antiviral innate immunity. J. Immunol. 1752851-2858. [DOI] [PubMed] [Google Scholar]
- 43.Yoneyama, M., M. Kikuchi, T. Natsukawa, N. Shinobu, T. Imaizumi, M. Miyagishi, K. Taira, S. Akira, and T. Fujita. 2004. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat. Immunol. 5730-737. [DOI] [PubMed] [Google Scholar]
- 44.Zhao, T., L. Yang, Q. Sun, M. Arguello, D. W. Ballard, J. Hiscott, and R. Lin. 29 April 2007. The NEMO adaptor bridges the nuclear factor-κB and interferon regulatory factor signaling pathways. Nat. Immunol. doi: 10.1038/ni1465. [DOI] [PubMed]