Abstract
Human T-lymphotropic virus type 1 (HTLV-1) is the etiologic agent for adult T-cell leukemia. The HTLV-1-encoded protein Tax transactivates the viral long terminal repeat and plays a critical role in virus replication and transformation. Previous work from our laboratory demonstrated that coactivator-associated arginine methytransferase 1, a protein arginine methytransferase, was important for Tax-mediated transactivation. To further investigate the role of methyltransferases in viral transcription, we utilized adenosine-2,3-dialdehyde (AdOx), an adenosine analog and S-adenosylmethionine-dependent methyltransferase inhibitor. The addition of AdOx decreased Tax transactivation in C81, Hut102, and MT-2 cells. Unexpectedly, we found that AdOx potently inhibited the growth of HTLV-1-transformed cells. Further investigation revealed that AdOx inhibited the Tax-activated NF-κB pathway, resulting in reactivation of p53 and induction of p53 target genes. Analysis of the NF-κB pathway demonstrated that AdOx treatment resulted in degradation of the IκB kinase complex and inhibition of NF-κB through stabilization of the NF-κB inhibitor IκBα. Our data further demonstrated that AdOx induced G2/M cell cycle arrest and cell death in HTLV-1-transformed but not control lymphocytes. These studies demonstrate that protein methylation plays an important role in NF-κB activation and survival of HTLV-1-transformed cells.
Human T-lymphotropic virus type 1 (HTLV-1) is a complex retrovirus that infects CD4+ T lymphocytes. It is the causative agent of adult T-cell leukemia (ATL) (42, 52) and tropical spastic paraparesis/HTLV-1 associated myelopathy (12, 32, 35), a neurodegenerative disease. Fewer than 5% of infected individuals progress to disease, which presents itself after a long latent period of 20 to 50 years (30). Once diagnosed, ATL has a poor prognosis, as most of the currently available therapies are ineffective (2, 50). The mechanism by which infected individuals develop ATL is still unknown, but it is believed to be a multistep process involving the virus-encoded protein Tax (51).
Utilizing alternative splicing and initiation codons, this complex retrovirus encodes regulatory and accessory proteins, including Tax, Rex, p12I, p27I, p13II, p30II, and HBZ, in addition to the common structural and enzymatic proteins Gag, Pol, and Env (10). The viral regulatory protein Tax functions as a transcriptional activator of the NF-κB, CREB, and SRF pathways (51) and is essential for viral gene expression, replication and transformation (1, 14, 33, 47). Tax activates NF-κB through activation of the upstream IκB kinase (IKK) complex, leading to phosphorylation and proteosome-mediated degradation of the NF-κB inhibitor IκBα (15, 17, 24). Tax activates the IKK complex, at least in part, through direct interaction with IKKγ leading to constitutive activation of the IKK kinase activity (15, 18, 45). Constitutive activation of NF-κB is considered central to HTLV-1 induced leukemia, where this pathway remains activated even in the absence of viral gene expression. Activation of NF-κB in HTLV-1-transformed cells also leads to functional inhibition of p53 (21, 38, 40, 41). By inhibiting p53, HTLV-1-infected cells evade the normal surveillance mechanisms of the cell which detect genomic instability, preventing the infected cells from undergoing apoptotic cell death.
Recent work from our laboratory demonstrated that coactivator-associated arginine methytransferase 1 (CARM1) also known as PRMT4, plays an important role in Tax transactivation (19). Small interfering RNA (siRNA)-mediated inhibition of CARM1 inhibited Tax transactivation (19). CARM1 was shown to be associated with the HTLV-1 transcription complex by chromatin immunoprecipitation assays (19). Consistent with the presence of CARM1, specific methylation of chromatin histone H3 was observed (19). CARM1 belongs to family of eight protein arginine methyltransferases, the function of which is to transfer one or two methyl groups to the guanidine nitrogen atoms of arginines, resulting in monomethylarginine or symmetric and asymmetric dimethylarginines (11). Recent advances in the understanding of arginine methyltransferases have revealed this to be an important posttranslational protein modification regulating many cellular processes, including RNA processing, transcription regulation, DNA repair, and signal transduction (reviewed in reference 3).
To further investigate the role of arginine methyltransferases in viral transcription in HTLV-1-transformed cells, we used adenosine-2,3-dialdehyde (AdOx), a potent small-molecule inhibitor of adenosylhomocysteine hydrolase that results in intracellular accumulation of adenosylhomocysteine leading to feedback inhibition of S-adenosylmethionine-dependent methyltransferases, including the protein arginine methyltransferase CARM1. Consistent with our previous studies, inhibition of arginine methyltransferases by AdOx inhibited Tax transactivation in HTLV-1-transformed cells. Unexpectedly, we also noticed that AdOx dramatically inhibited the growth of HTLV-1-transformed cells. To further characterize and investigate the mechanism of AdOx-mediated growth inhibition, we performed growth inhibition and cell cycle analysis on HTLV-1-transformed cell lines. AdOx preferentially inhibited the growth of HTLV-1-transformed cells compared to noninfected transformed T cells. We found that the decrease in viability correlated with inhibition of NF-κB. AdOx inhibited NF-κB through the degradation of the IKK complex and stabilization of the NF-κB inhibitor IκBα. Inhibition of NF-κB activity resulted in reactivation of p53 and induction of apoptosis. These studies suggest that methyltransferase inhibitors may provide a novel class of therapeutic agents for treatment of ATL.
MATERIALS AND METHODS
Cell lines and culture conditions.
The cells lines used in this study included HTLV-1-transformed cell lines C81, MT2, Hut102, C91-PL, and TLOM1 and noninfected transformed cell lines Jurkat and Molt4. All cell lines were maintained in RPMI 1640 medium supplemented with 10% heat-inactivated fetal calf serum, 2 mM l-glutamine, and penicillin-streptomycin. Peripheral blood mononuclear cells (PBMCs) were isolated from healthy donors using Ficoll-Paque Plus (GE Healthcare) according to the manufacturer's instructions. The isolated lymphocytes were washed in phosphate-buffered saline (PBS), and cultured in RPMI 1640 medium supplemented with 20% heat-inactivated fetal calf serum, and activated in the presence of 2 μg/ml phytohemagglutinin-L (PHA-L) for 24 h. Following activation, the PBMCs were cultured in serum supplemented RPMI-1640 medium in the presence of 50U/ml of interleukin-2. Cell viability studies were carried out as outlined below.
Reagents and antibodies.
AdOx was purchased from Sigma-Aldrich. β-Tubulin antibody was purchased from Boehringer Mannheim. Horseradish peroxidase-coupled secondary rabbit antibody and a mouse antibody were purchased from GE Healthcare. Anti-dimethyl arginine (anti-DMA) antibody was from Millipore. Unless otherwise mentioned, all other antibodies used in Western blot experiments were purchased from Cell Signaling.
Cell viability assay.
Cell viability assay was performed using the CellTiter-Glo luminescent cell viability assay from Promega according to the manufacturer's instructions. The CellTiter-Glo luminescence assay determines the number of viable cells based on the quantitation of ATP present, which signals the presence of metabolically active cells. Briefly, 1 × 105 to 2 × 105 cells were cultured in sterile 96-well culture plates in the presence of an appropriate concentration of AdOx (20 to 40 μM) in 100 μl of RPMI medium. The plates were then incubated for 24 to 96 h. At the desired times, 100 μl of CellTiter-Glo reagent was added to lyse the cells. The contents were mixed in an orbital shaker for 2 min and then incubated at room temperature for 10 min. The luminescence was then recorded in a luminometer with an integration time of 1 second per well. The luminescent signals for the AdOx-treated cells were normalized to the luminescent signal of cells treated with dimethyl sulfoxide (DMSO), which was arbitrarily set to 1.
Western blot analysis.
Whole-cell extracts (WCEs) were made in HEPES or radioimmunoprecipitation assay lysis buffer supplemented with 1× protease inhibitor cocktail (Roche). Cells (107) were resuspended in lysis buffer (500 μl) and incubated on ice for 15 min. After thorough vortexing, the samples were centrifuged at 14,000 RPM and supernatants were collected for analysis. Twenty-five to 50 μg of protein samples was separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred to Immobilon-P membranes (Millipore). The membranes were blocked with PBS containing 0.1% Tween and 5% nonfat dry milk or 5% bovine serum albumin and then probed with appropriate antibody. Chemiluminescence detection was performed using ECL Plus reagent according to the manufacturer's instructions (GE Healthcare).
Cell cycle analysis.
The cells (2 × 105to 5 × 105) were cultured in RPMI for appropriate time in presence or absence of the drug AdOx. The cells were harvested, washed with cold PBS, and fixed with 70% ethanol overnight at 4°C. Fixed cells were treated with 100 μl of RNase (10 mg/ml), stained with 50 μg/ml of propidium iodide, and then subjected to flow cytometry using a FACSCalibur (Becton Dickinson). The resultant data were analyzed with Modfit V3.0 software or FlowJo V8.0 software using appropriate gates.
TUNEL assay.
To quantify apoptosis, terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling (TUNEL) assay was done using an APO-bromodeoxyuridine (BrdU) TUNEL assay kit (Molecular Probes) following the manufacturer's instructions. Briefly, AdOx-treated and control cells were washed with cold PBS and fixed with 1% paraformaldehyde in PBS, followed by fixing with 70% ethanol. The cells were then labeled with BrdUTP using terminal deoxynucleotide transferase at 37°C for 60 min. At the end of the incubation, the cells were rinsed with rinse buffer (manufacturer supplied), stained with Alexa Fluor 488 dye-labeled anti-BrdU antibody for 30 min at room temperature away from light, and finally treated with propidium iodide-RNase A staining buffer for an additional 30 min at room temperature away from light. The immunostained cells were analyzed using a FACSCalibur (Becton Dickinson). TUNEL-positive cells were quantified with Cell Quest (Becton Dickinson) and FlowJoV8.0 software.
Transfection and luciferase assay.
HTLV-1-transformed cells were transfected with 1 μg of reporter constructs HTLV-1LTR-Luc, PG13-Luc, and NF-κB-Luc (kindly provided by Warner Green) using Transfast transfection reagent (Promega) according to the manufacturer's protocol. All transfections included 0.5 μg Rous sarcoma virus (RSV) β-galactosidase (β-Gal) plasmid as a control for transfection efficiency. The transfected cells were treated with AdOx at 1 h after transfection. For luciferase assay, cell lysates were made in radioimmunoprecipitation assay buffer at 24 h posttransfection, and luciferase and β-Gal activity values were determined using Promega Dual luciferase and Tropix GalactoLight assay kits following the manufacturer's instructions.
EMSA.
An electrophoretic mobility shift assay (EMSA) kit (Active Motif) was used to detect NF-κB-bound complexes according to the manufacturer's instructions. Briefly, double-stranded oligonucleotides containing the NF-κB consensus binding sequence were end labeled with [γ-32P]ATP using T4 polynucleotide kinase and used as probes for detecting NF-κB-bound complexes. Nuclear extracts were prepared using an NE-PER kit (Pierce), and the DNA-protein complexes were resolved on 5% polyacrylamide gels in 1× Tris-borate-EDTA at 4°C. The gels were subsequently exposed to a phosphor screen overnight, and the bands were visualized with a PhosphorImager (Molecular Dynamics) and quantitated using Image Quant software.
Quantitative reverse transcription-PCR (RT-PCR).
Total RNA was prepared with Trizol reagent (Invitrogen) using the manufacturer's protocol. One microgram of total cellular RNA was reverse transcribed with oligo(dT) primers in a 20-μl reaction volume using the ImprompII reverse transcription kit (Promega). The primer sequences used in the study were as follows: Tax, 5′-ACGTGATTTTTTGCCACCCCGG-3′ (forward) and 5′-TGGAAAAGGGTGGTGGGCAA-3′ (reverse); β-actin, 5′-CCAGATCATGTTTGAGACCTTCAAC-3′ (forward) and 5′-CCAGAGGCGTACAGGGATAGC-3′ (reverse). The 6-carboxyfluorescein-labeled Mdm2 and GAPDH (glyceraldehyde-3-phosphate dehydrogenase) primers were purchased as 20× stocks from Applied Biosystems. The PCR mixtures contained 1× SYBR green mix (Stratagene), 400 nM primers, and 1 μl cDNAs. Fivefold dilutions of the cDNA pool from the “untreated control” were run to generate standard curves. The values from the PCRs were normalized to the values obtained for β-actin mRNAs, and the standard curves generated from the control mRNA were used to determine the fold changes in the mRNA values. The values from the Mdm2 PCRs were similarly normalized to the values obtained for GAPDH mRNAs.
Adenovirus infection of cells.
An adenovirus-p53 siRNA construct (kindly provided by Ling-Jun Zhao, St. Louis University) and an adenovirus-green fluorescent protein (GFP) construct (Q-Biogene) were used to infect HTLV-1-transformed C81 cells (2 × 106) in RPMI medium not supplemented with serum. At 3 h postinfection, the cells were spun down and resuspended in serum-supplemented RPMI medium. The infected cells were incubated for 24 h for p53 siRNA expression, followed by AdOx treatment for different time intervals.
RESULTS
AdOx represses the transactivation function of Tax and inhibits Tax expression in HTLV-1-transformed cells.
Previous work in our laboratory has shown that CARM1, a protein arginine methyltransferase, plays an important role in Tax transactivation of the HTLV-1 long terminal repeat (LTR) (19). In these studies, we demonstrated that an siRNA against CARM1 inhibited CARM1 expression and Tax transactivation. Chromatin immunoprecipitation assays further demonstrated that CARM1 is associated with the active HTLV-1 LTR in vivo. To further investigate the role of arginine methyltransferase in Tax-mediated viral transcription in HTLV-1-transformed cells, we used AdOx, an inhibitor of enzymes belonging to a family of S-adenosylmethionine-dependent methyltransferases, including CARM1. HTLV-1-transformed cell lines C81, Hut102, and MT2 and the ATL-derived Tax-negative cell line TLOM1 were cotransfected with an HTLV-1 LTR-luciferase construct and a β-Gal reporter construct. At 1 hour posttransfection, the cells were treated with increasing concentrations of AdOx for 24 h. The cells were then harvested and luciferase activity measured. As seen in Fig. 1A, there is a dose-dependent repression of the transactivation function of Tax in Tax-expressing C81, Hut102, and MT-2 cells. The TLOM1 cell line showed basal-level luciferase activity compared to the other cell lines, consistent with it being a Tax-negative cell line (data not shown).
FIG. 1.

AdOx inhibits transcriptional activity of HTLV-1 LTR and Tax transcription. (A) HTLV-1-transformed cell lines C81, Hut102, MT2, and TLOM1 were transiently transfected using Transfast transfection reagent (Promega) with HTLV-1 LTR-Luc (1 μg) and RSV β-Gal (0.5 μg). At 1 h posttransfection, the methyltransferase inhibitor AdOx was added at increasing concentrations as indicated. After 24 h, the cells were collected and luciferase activities were measured. All luciferase values were adjusted for transfection efficiency using RSV β-Gal. The graph represents the luciferase activity from three independent experiments with standard deviation. (B) Quantitative RT-PCR was done to estimate the relative Tax message in the RNA from HTLV-1-transformed cell line C81 grown in the presence of 20 μM AdOx for increasing periods of time as indicated. The levels of Tax mRNA at different time points are shown relative to the Tax message from cells grown in the presence of medium containing control DMSO.
To determine whether AdOx could inhibit transcription from the integrated template in the HTLV-1-transformed cells, we examined the effect of AdOx on Tax mRNA expression in HTLV-1-transformed cells. C81 cells were cultured in the presence or absence of AdOx for various time intervals. Quantitative RT-PCR was done on RNA isolated from the AdOx-treated and untreated cells with Tax-specific primers. β-Tubulin primers were used as a normalization control. As shown in Fig. 1B, addition of AdOx led to a significant reduction of Tax message. Greater than 80% reduction of Tax mRNA synthesis was observed by 24 h of treatment with AdOx. Similar results were obtained with HTLV-1-transformed cell line MT2 (data not shown). In contrast to the case for Tax expression, no change in β-actin was observed (data not shown).
AdOx-induced loss of cell viability in HTLV-1-transformed cell lines.
During the course of the experiments described above, we observed that AdOx dramatically inhibited the growth of HTLV-1-transformed cells in culture. Therefore, we next tested the effect of 20 μM AdOx on the growth and survival of HTLV-1-transformed cell lines. As controls, we also tested the effect of this drug on noninfected transformed Jurkat and Molt4 T cells and PHA-L-activated PBMCs. In vitro cell viability assays using the CellTiter-Glo luminescent cell viability assay (Promega) determined the number of viable cells based on quantitation of ATP, which signals the presence of metabolically active cells (9, 22, 27). As shown in Fig. 2A, although the sensitivity to the drug varied in the different HTLV-1-transformed cell lines, they were all more susceptible to AdOx than the control lymphocyte cell lines Jurkat and Molt4. The susceptibility of the cells was not dependent on the ability of the HTLV-1-transformed cells to express Tax protein, as the ATL-derived cell line TLOM1 showed similar sensitivity to the drug (Fig. 2A). Control PBMCs also showed very little sensitivity to AdOx compared to the AdOx-treated HTLV-1-transformed cell line C81 (Fig. 2D).
FIG. 2.
AdOx inhibits the growth of HTLV-1-transformed cell lines. (A) Jurkat cells, Molt4 cells, and HTLV-1-transformed cell lines C81, C91-PL, MT2, Hut102, and TLOM1 were treated with 20 μM of AdOx. At the indicated times, CellTiter-Glo luminescent cell viability assay reagent (Promega) was added and the luminescence was recorded with a luminometer. This reagent determines the number of viable cells in culture based on quantitation of the ATP present, which signals the presence of metabolically active cells. (B) Jurkat cells, Molt4 cells, and HTLV-1-transformed cell lines C81, C91-PL, MT2, Hut102, and TLOM1 were treated for 72 h with 20 and 40 μM AdOx. CellTiter-Glo luminescent cell viability assay reagent was used as described above to determine the number of viable cells. (C) Western blot analysis was done on WCEs made from C81 and control Molt4 cells that had been pretreated with 20 μM AdOx or DMSO for 24 h. Fifty micrograms of protein extract was immunoblotted with antidimethyl antibodies. β-Tubulin was used as a loading control. (D) PBMCs were isolated from healthy donors using Ficoll-Paque Plus (GE Healthcare). The isolated lymphocytes were washed in PBS and cultured in RPMI 1640 medium supplemented with 20% heat-inactivated fetal calf serum and activated with 2 μg/ml PHA-L for 24 h. Following activation, the PBMCs and C81 cells were treated with 20 μM AdOx. At the indicated times CellTiter-Glo luminescent cell viability assay reagent (Promega) was added and the luminescence was recorded with a luminometer. This reagent determines the number of viable cells in culture based on quantitation of the ATP present, which signals the presence of metabolically active cells. Error bars indicate standard deviations.
To determine the effects of increasing concentrations of AdOx on cell survival, we added AdOx to final concentrations of 20 and 40 μM to the culture media of HTLV-1-transformed and nontransformed cells for 72 h as indicated in Fig. 2B. Concentrations above 40 μM were not used in the study because they are of general toxicity to all cells. Following treatment, the cells were assayed for cell viability using the CellTiter-Glo luminescent cell viability assay. As shown in Fig. 2B, the HTLV-1-transformed cells were in general more sensitive to AdOx and showed a concentration-dependent loss of viability. Control Molt4 and Jurkat cells showed an approximately 30% drop in cell viability, whereas the HTLV-1-transformed cells showed a 70 to 95% reduction in cell viability. MT2 cells turned out to be the least sensitive of the HTLV-1-transformed cells, with a 50% decrease in cell viability at 40 μM AdOx.
Given the difference in the sensitivities of the cells to AdOx, it was important to demonstrate that the drug was being taken up in control and HTLV-1-transformed cells. To address this point, we compared the relative levels of protein methylation in two cell lines: C81 cells, which are very sensitive to AdOx, and Molt4 cells, which are more resistant. C81 and Molt4 cells were cultured in the absence and presence of 20 μM AdOx for 24 h. Cell extracts were then prepared, and Western blot analysis was performed with an antibody which detects methylated proteins (49). The results presented in Fig. 2C demonstrate that the inhibition of protein methylation was roughly equivalent in C81 and Molt4 cells, providing indirect evidence that AdOx was taken up at roughly equal levels in the two cell lines, which were remarkably different in sensitivity to AdOx.
AdOx induces cell cycle arrest and apoptosis in HTLV-1-transformed cells.
To determine the effects of AdOx on the cell cycle, we added 20 μM AdOx to the culture media of HTLV-1-transformed and nontransformed cells and left them for 24 to 48 h. The cells were then fixed, stained with propidium iodide, and analyzed by flow cytometric analysis. As seen in Fig. 3A, AdOx treatment induced significant time-dependent changes in the cell cycle distribution in the HTLV-1-transformed cell line C81. The cells first accumulated in the G2/M phase of the cell cycle, with the G2/M population increasing from 12 to 52% in 36 h (Fig. 3A, bottom panel). With slightly slower kinetics, the G0/G1 population decreased from 73 to 36%. We also noted that the appearance of the sub-G1 population increased over time. In contrast, the noninfected HTLV-1-transformed T-cell lines Jurkat and Molt-4, when treated with the same concentration of drug, showed very little change in their cell cycle distribution (Fig. 3A). HTLV-1-transformed cell line MT2 showed similar G2/M accumulation and appearance of sub-G1 cells (Fig. 3B).
FIG. 3.
AdOx induces the G2/M checkpoint and induces apoptosis in HTLV-1-transformed cell lines. (A) HTLV-1-transformed cell line C81 and HTLV-1-negative transformed T-cell lines Molt4 and Jurkat were treated with 20 mM AdOx. At the indicated times, cells were collected, fixed with 70% ethanol, RNase treated, stained with propidium iodide, and subjected to flow cytometry. (B) HTLV-1-transformed cell line MT2 was treated similarly to the cells for panel A. At the indicated time points, the cells were fixed with 70% ethanol, RNase treated, stained with propidium iodide, and subjected to flow cytometry. (C) TUNEL analysis of C81 and Molt4 cells treated for 72 h with AdOx. C81 and Molt4 cells were treated with 20 mM AdOx for 72 h. The cells were then fixed with paraformaldehyde, followed by ethanol. DNA breaks were then labeled with terminal deoxynucleotide transferase and BrdUTP. Detection of BrdUTP incorporation was through an Alexa Fluor 488 dye-labeled anti-BrdU antibody, which was analyzed by flow cytometry. Error bars indicate standard deviations.
To more accurately quantitate the level of apoptosis induced by AdOx, the BrdU-based TUNEL assay was used to compare the level of apoptosis in HTLV-1-transformed cell line C81 and noninfected cell line Molt4. Consistent with the fluorescence-activated cell sorter (FACS) analysis, when C81 cells were treated with 20 μM AdOx for 72 h, more than 80% of the C81 cells were positive for BrdU, compared to 1.6% TUNEL-positive cells among the DMSO-treated cells (Fig. 3C). Treatment of control Molt4 cells with AdOx showed much lower levels of TUNEL-positive cells, in this case about 15%. These results provide further evidence that AdOx preferentially induces apoptosis in HTLV-1-transformed cells.
AdOx treatment stabilizes the NF-κB inhibitor IκBα and inactivates NF-κB in HTLV-1-transformed cells.
Constitutive activation of NF-κB in HTLV-1-transformed cells plays a key role in cell survival and prevents infected cells from undergoing apoptotic cell death (18, 51). To determine if the effect of AdOx was due to the effect of the drug on the NF-κB pathway, we looked at the activity and status of NF-κB in AdOx-treated C81 cells. WCEs were made from C81 cells grown in the absence or presence of 20 μM AdOx for various time intervals as indicated in Fig. 4A. Western blot analysis was done with IκBα and phospho-IκBα (ser32/36) monoclonal antibodies. As seen in Fig. 4A, a significant reduction in phospho-IκBα was observed (top panel). The reduction of the phosphorylated form of IκBα was accompanied by concomitant stabilization of the protein (middle panel). β-Tubulin was used as a loading control and demonstrates that equivalent amounts of proteins were added (bottom panel).
FIG. 4.
AdOx inhibits NF-κB through destabilization of the IKK complex in HTLV-1-transformed cells. (A) Effect of AdOx on NF-κB inhibitor IκB-α. C81 cells were treated with 20 mM AdOx. At 0, 24, 36, and 48 h posttreatment, cells were harvested and WCEs prepared. Twenty-five micrograms of protein extract was immunoblotted with IκB-α and phospho-IκB-α (Ser32/36) antibodies. β-Tubulin was used as a loading control. (B) C81 cells were transfected using Transfast transfection reagent (Promega) with NF-κB-Luc (2 μg) and RSV β-Gal (0.5 μg) plasmids. At 1 h after transfection, the cells were treated with AdOx (0, 20, 40, and 80 μM) for 24 h. Cells were then harvested, and luciferase activities were measured. Luciferase values were adjusted for transfection efficiency using RSV β-Gal activity. Error bars represent standard deviations from three independent experiments. (C) Effect of AdOx on NF-κB DNA binding activity. C81 cells were treated with AdOx for 72 h, and nuclear extracts were made. NF-κB DNA binding activity was assessed by EMSA using a consensus NF-κB oligonucleotide probe. The specificity of the reaction was evaluated by adding a 50-fold excess of cold competitor probe containing the wild-type or mutant NF-κB recognition site. (D) C81 cells were treated with 20 μM AdOx. At different times after AdOx treatment, cells were harvested and WCEs prepared. Twenty-five micrograms of protein extract was immunoblotted with Bcl-xL and Bcl-2 antibodies. β-Tubulin was used as a loading control. (E) C81 cells were treated with 20 μM AdOx for 48 h. The cells were harvested and WCEs prepared. Twenty-five micrograms of protein extract was immunoblotted with specific antibodies against IKKα, IKKβ, and IKKγ. β-Tubulin was used as a loading control. (F) One milligram of WCE was immunoprecipitated (IP) with IKKα, IKKβ, and IKKγ antibodies, and Western blot (W.B) analysis was done with anti-DMA antibody. Reciprocally, 1 mg of WCE was immunoprecipitated with DMA antibody, and Western blot analysis was done with IKKα, IKKβ, and IKKγ antibodies. Immunoprecipitation with immunoglobulin G (IgG) was used as control. (G) Ten nanograms of recombinant IKKα, IKKβ, and IKKγ proteins was immunoblotted with specific antibodies against IKKα, IKKβ, and IKKγ. Arrows indicate the molecular mass of each recombinant protein.
To analyze NF-κB transcriptional activity, C81 cells were transfected with reporter construct 4xNF-κB-Luc and subsequently treated with increasing amounts of AdOx. At 24 h posttransfection, the cells were harvested and analyzed for luciferase activity. Consistent with the results presented above, there was a significant reduction in NF-κB transcriptional activity following AdOx treatment (Fig. 4B). At 20 μM AdOx, greater than 50% inhibition of NF-κB transcription activity was observed by 24 h. At a concentration of 40 μM AdOx, an 80% reduction in NF-κB transcriptional activity was observed. We also performed EMSAs on nuclear extracts prepared from C81 cells treated with 20 μM AdOx. Using a 32P-labeled oligonucleotide probe which contained the recognition sequence for NF-κB, one predominant positive shifted complex was detected in the untreated lane (Fig. 4C, lane 1). Consistent with the transcription data, there was a significant decrease in the NF-κB gel shift complex following AdOx treatment (Fig. 4C, lane 2). The specificity of the gel shift reaction was confirmed by competition analysis. The NF-κB gel shift complex disappeared in the presence of excess (50-fold) unlabeled wild-type oligonucleotide but not in presence of excess (50-fold) unlabeled mutant oligonucleotide (Fig. 4C, lanes 3 and 4). Quantitation of the EMSA experiment is shown in the bar graph in Fig. 4C.
If NF-κB is inhibited in AdOx-treated cells, it would be expected that expression of NF-κB-responsive genes would be decreased. Bcl-xL, an antiapoptotic member of the Bcl-2 family of proteins, is constitutively expressed in HTLV-1-infected cell lines, and the expression is controlled by NF-κB (4, 28). Consistent with inhibition of NF-κB activity by AdOx, treatment of the HTLV-1-positive cell line C81 resulted in a significant reduction of Bcl-xL expression (Fig. 4D). Quantitation of the Western blots revealed that Bcl-xL protein expression was reduced by 80% compared to that in untreated cells. In contrast, AdOx treatment did not affect Bcl-2 expression, which is not regulated by NF-κB (Fig. 4D).
Our experiments thus far have demonstrated that AdOx treatment leads to stabilization of the NF-κB inhibitor IκBα and inhibition of NF-κB. To test whether this effect was due to the effect of AdOx on the upstream IKK kinase complex, WCEs were made from C81 cells grown in the absence or presence of 20 μM AdOx. Western blot analysis was done with IKKα-, IKKβ-, and IKKγ-specific antibodies. As seen in Fig. 4E, a marked reduction of IKKα, IKKβ, and ΙΚΚγ was observed in the AdOx-treated extracts (Fig. 4E). β-Tubulin was used as a loading control, which shows no effect upon AdOx treatment. To test whether the kinase subunits IKKα, IKKβ, and ΙΚΚγ were subjected to methylation in vivo, WCEs were made from C81 cells and subjected to immunoprecipitation with IKKα-, IKKβ-, and IKKγ-specific antibodies (Fig. 4G). Western blot analysis was then done with an antibody raised against asymmetric DMA. Interestingly, the IKKγ immunoprecipitate showed a reactive band corresponding to the size of IKKγ (Fig. 4F, upper panel). In contrast IKKα and IKKβ immunoprecipitates did not show reactivity in their size ranges (Fig. 4F, upper panel). To further confirm the above result, reciprocal immunoprecipitation was done on WCEs made from C81 cells with asymmetric DMA antibody and Western blot analysis was done with IKKα-, IKKβ-, and IKKγ-specific antibodies. Consistent with the above result, only IKKγ showed positive reactivity (Fig. 4F, lower panel). Western blots with IKKα and IKKβ antibodies did not yield any positive results (Fig. 4F and data not shown). Our data indicate that the IKKγ subunit is methylated, and this modification potentially plays a role in the stability of the IKK complex. The specificity of the IKK antibodies is shown in the Western blots in Fig. 4G. Using recombinant IKKα, -β, and -γ proteins, we demonstrated that under the Western blot conditions used in our assays, the antibodies show complete specificity for the respective proteins.
NF-κB inhibition results in reactivation of p53.
Earlier work from this and other labs has demonstrated that constitutive activation of the NF-κB pathway in HTLV-1-transformed cells functionally inhibited p53 (8, 20, 34, 39, 40, 43, 46). To determine if AdOx treatment had an effect on p53 protein levels and function, Western blot analysis of p53 protein was performed with HTLV-1-transformed C81 cells following AdOx treatment. As seen in Fig. 5A, there was a dramatic increase in the p53 protein level in AdOx-treated cells. To check whether the p53 protein regained functional activity, we analyzed the expression of p53-regulated Mdm2 protein. As shown in Fig. 5A, a significant increase in Mdm2 protein was observed at 24 and 36 h after treatment of the cells with AdOx. To check if there was a concomitant change in Mdm2 transcription, we analyzed Mdm2 mRNA levels by quantitative RT-PCR. As shown in Fig. 5B, at 36 h posttreatment a fivefold increase in Mdm2 RNA was observed in AdOx-treated C81 cells.
FIG. 5.
AdOx treatment results in p53 stabilization and reactivation in HTLV-1-transformed cells (A) Effect of AdOx on p53. C81 cells were treated with 20 μM AdOx. At 0, 24, 36, and 48 h posttreatment, cells were harvested and WCEs prepared. Twenty-five micrograms of protein extract was immunoblotted with p53 and Mdm2 antibodies. β-Tubulin was used as a loading control. Equal amounts of protein extract from Jurkat cells were loaded as a negative control for p53. (B) Quantitative RT-PCR was done to quantitate relative Mdm2 mRNA in HTLV-1-transformed cell line C81 in the absence or presence of 20 μM AdOx for increasing periods of time as indicated. The Mdm2 message at different time points are shown relative to the Mdm2 message from cells grown in presence of medium with control DMSO alone. GAPDH was used as a normalization control. (C) C81 cells were transfected using Transfast transfection reagent (Promega) with PG13-Luc (2 μg) and RSV β-Gal (0.5 μg) plasmids. At 1 h after transfection, the cells were treated with 20 μM AdOx for 0, 24, 36, and 48 h Cells were then harvested, and luciferase activities were measured. Luciferase values were adjusted for transfection efficiency using RSV β-Gal activity. Error bars represent standard deviations from three independent experiments.
Although the Mdm2 gene is a p53-responsive gene, it is possible that it is regulated by other effectors. To address this issue and unambiguously demonstrate that p53 transcription activity was increased, we transfected the C81 cells with a p53-responsive reporter, PG13-Luc, which contains 13 repeats of the p53 response sequence upstream of the promoter. The transfected cells were then treated with 20 μM AdOx, and samples were taken at various time points posttreatment. The results of multiple experiments demonstrate that treatment of HTLV-1-transformed C81 cells with AdOx does, in fact, result in an increase in p53 activity (Fig. 5C). At 36 h posttreatment, we observed approximately a four- to fivefold increase in p53 activity, consistent with the fold increase in p53-responsive Mdm2 promoter activity shown in Fig. 5B.
AdOx induces p53-dependent apoptosis in C81 cells.
To determine whether p53 reactivation was linked to apoptosis, we utilized an adenovirus vector encoding an siRNA to p53 (53). C81 cells were infected with adenovirus-p53 siRNA or adenovirus-GFP control virus. AdOx (20 μM) was added at 24 h postinfection, and cells were harvested 48 h after AdOx addition. Western blot analysis of the proteins showed complete disappearance of the p53 protein in the p53 siRNA-treated cells, in both AdOx-treated and untreated cells (Fig. 6A, lanes 3 and 4). To determine the effect of p53 depletion on AdOx mediated apoptosis, we performed cell cycle analysis on AdOx-treated C81 cells that were infected with either adenovirus-p53 siRNA or the control adenovirus-GFP expression vector. Following FACS analysis, the sub-G0/G1 population was calculated as an index for apoptosis. Consistent with the results presented in Fig. 3, treatment of the cells with AdOx increased the percentage of cells in the sub-G1 fraction from 2% to 16% in control adenovirus-GFP-infected cells (Fig. 6B). There was a significant reduction in apoptosis, from 16% to 6%, in adenovirus-p53 siRNA-infected cells compared to AdOx-treated cells infected with adenovirus expressing GFP. The quantitative difference in the number of apoptotic cells was not due to a difference in the adenovirus-infected cells, since a similar level of apoptotic cells was observed among the control GFP- and p53 siRNA-infected cells (1.96 versus 2.3%). These results suggest that a significant population of the C81 cells underwent p53-dependent apoptosis following AdOx treatment.
FIG. 6.
AdOx induces p53-dependent apoptosis in C81 cells. (A) C81 cells were infected with adenovirus expressing p53 siRNA or GFP. At 24 h postinfection, the cells were treated with 20 μM AdOx. After 48 h, cells were harvested and extracted with cell lysis buffer, and 25 μg of protein extract was immunoblotted with p53 and β-tubulin antibodies. (B) C81 cells were infected and treated with AdOx as described above. Cells were then harvested, fixed with 70% ethanol, RNase treated, stained with propidium iodide, and subjected to flow cytometry for evaluating DNA content. The percent sub-G1 population, which is an index of apoptosis, is indicated.
DISCUSSION
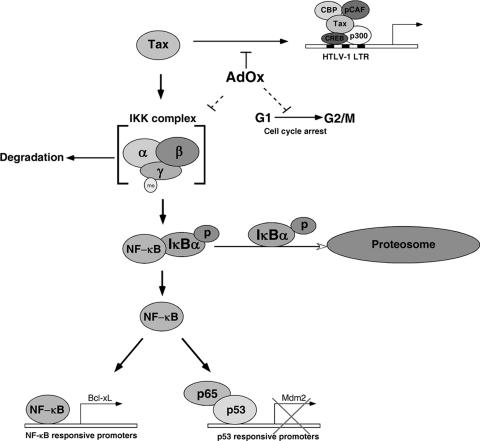
In this study, we have shown that inhibition of cellular methyltransferases by AdOx affects several critical pathways in HTLV-1-transformed cells (Fig. 7). First, AdOx blocks Tax transactivation of the HTLV-1 LTR. This observation was consistent with the positive role for CARM1 in Tax transactivation reported previously (19). Intriguingly, AdOx also induced loss of cell viability, cell cycle arrest, and apoptosis in HTLV-1-infected cells but did not significantly affect noninfected control lymphocytes. By transient transfection and gel shift assay, we have shown that AdOx treatment led to inhibition of NF-κB. Consistent with NF-κB inhibition, the expression of Bcl-xL was inhibited. Inhibition of methyltransferases led to rapid turnover of the IKK complex subunits IKKα, IKKβ, and IKKγ and reduced phosphorylation of IκBα. Studies with siRNA against p53 established that AdOx-induced apoptosis was, at least in part, regulated by p53.
FIG. 7.
AdOx affects multiple pathways in HTLV-1-transformed cells.
Posttranslational modifications of proteins play an important role in modulating protein function in diverse cellular processes. Though arginine methylation was discovered many years ago, it has only recently been recognized to play an important part in modulating protein function. It is now clear that arginine methylation plays important roles in RNA processing, transcriptional regulation, signal transduction, and DNA repair (3). For example many RNA binding proteins which play roles in processing, folding, stabilization, and localization of RNAs and mRNA translation are major targets for PRMTs and have been shown to be arginine methylated (16, 26). Similarly, methylation contributes to transcription regulation by methylation of histones (29, 36, 37). In signal transduction pathways, methylated arginines have been shown to both promote and inhibit protein-protein interactions (29). Recently, MRE11 a component of the double-strand break repair protein complex MRE11-RAD50-NBS1, was shown to be arginine methylated by PRMT1 (5).
The NF-κB pathway is intimately linked to the survival pathways of mammalian cells, especially HTLV-1-infected cells, where this pathway is constitutively activated. NF-κB activation involves degradation of the NF-κB inhibitor IκBα through phosphorylation by the IKK complex, resulting in release and migration of the p50-p65 heterodimers into the nucleus, which activates transcription of genes involved in survival and growth. In the present study, treatment of the cells with AdOx reduced IκBα phosphorylation and NF-κB-mediated transcription. A novel finding in this study is that inhibition of methyltransferases by AdOx led to rapid turnover of the IKK complex subunits IKKα, -β, and -γ, which could explain at least partly the inhibition of NF-κB. Though data on the effect of methylation on protein stability are scarce, recent work from Naeem et al. (31) has shown that the activity and stability of transcriptional coactivator p/CIP/SRC3 (p300/CBP-interacting protein) are regulated by CARM1-dependent methylation. In this case arginine methylation caused an increase in p/CIP turnover as a result of enhanced degradation. As with many posttranslational modifications having opposing functions in different targets, we speculated that methylation of one of the IKK complex proteins is necessary for protein stability. We tested this idea in the present study and showed that the IKKγ subunit is methylated in vivo. We speculate that methylation controls the stability of the complex by modulating IKKγ function and is at least partially responsible for the stability of the IKK complex. The stability of the IKK complex is also linked to the chaperone function of HSP90. The relationship of IKKγ methylation to HSP90's chaperone function is under investigation.
In HTLV-1-infected cells, constitutive activation of NF-κB has been shown to play a critical role in the inhibition of p53 transactivation function (31, 40). Inhibition of p53 is thought to be an important event in transformation of HTLV-1-infected cells. In this study we show that AdOx inhibited the NF-κB pathway. There was dramatic stabilization of p53 protein at the same time. With stabilization of p53 protein and inhibition of NF-κB, we expected that p53 might regain normal transactivation function. Consistent with this hypothesis, we observed induction of p53-responsive genes such as the Mdm2 gene upon AdOx treatment, indicating that p53 protein had overcome the functional inactivation. We also investigated whether induction of apoptosis was in part due to stabilization and reactivation of p53. We inhibited p53 expression with adenovirus-mediated siRNA prior to treatment with AdOx. Our results clearly demonstrated that inhibition of p53 expression resulted in a significant reduction of apoptosis. This result was consistent with the p53-dependent apoptosis observed in AdOx-treated Hct116 cells (44).
A role for methyltransferases in viral replication and gene regulation has been reported for human immunodeficiency virus (HIV)-, hepatitis delta virus-, and vaccinia virus-infected cells (6, 7, 13, 23, 25). Protein methylation is required to maintain optimal HIV-1 infectivity. Inhibition of protein methylation in a cell culture model increased the level of HIV virions produced in culture but, surprisingly, reduced the level of infectivity (48). Inhibition of arginine methylation also prevents the replication of hepatitis delta virus (25). These observations suggest that methylation plays an important role in replication of several human viruses and that development of methylation inhibitors should be an important consideration in antiviral therapy.
In conclusion, we have demonstrated that treatment of HTLV-1-transformed cells with AdOx reduced Tax transactivation, reduced NF-κB activity through destabilization of the IKK complex, and induced accumulation of cells in G2/M and apoptosis (Fig. 7). At least part of the AdOx-induced cell death of HTLV-1-transformed cells was due to reactivation of p53. AdOx preferentially affected HTLV-1-transformed cells and not noninfected T lymphocytes and PHA-L-activated PBMCs. The specificity of the drug towards HTLV-1-infected cells raises the possibility of development of small molecules which would specifically target the methyltransferases involved in NF-κB or p53 regulation. Further investigation to elucidate the role of arginine methylation in the regulation of NF-κB and p53 regulation is therefore important. Given the dramatic effect of AdOx on HTLV-1 growth, there might be a role for methyltransferase inhibitors in treatment of HTLV-1-associated neuropathies and ATL.
Acknowledgments
We are grateful to Cynthia Masison, Mike Radonovich, Keven Huang, and the other members of the Brady lab for discussions, suggestions, and comments on the manuscript. We thank the FACS core facility of CCR, NCI, NIH, for flow cytometry analysis.
This research was supported by the Intramural Research Program of the NIH, National Cancer Institute.
Footnotes
Published ahead of print on 17 October 2007.
REFERENCES
- 1.Akagi, T., H. Ono, and K. Shimotohno. 1995. Characterization of T cells immortalized by Tax1 of human T-cell leukemia virus type 1. Blood 864243-4249. [PubMed] [Google Scholar]
- 2.Bazarbachi, A., and O. Hermine. 2001. Treatment of adult T-cell leukaemia/lymphoma: current strategy and future perspectives. Virus Res. 7879-92. [DOI] [PubMed] [Google Scholar]
- 3.Bedford, M. T., and S. Richard. 2005. Arginine methylation an emerging regulator of protein function. Mol. Cell 18263-272. [DOI] [PubMed] [Google Scholar]
- 4.Bernal-Mizrachi, L., C. M. Lovly, and L. Ratner. 2006. The role of NF-κB-1 and NFκB-2-mediated resistance to apoptosis in lymphomas. Proc. Natl. Acad. Sci. USA 1039220-9225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boisvert, F. M., U. Dery, J. Y. Masson, and S. Richard. 2005. Arginine methylation of MRE11 by PRMT1 is required for DNA damage checkpoint control. Genes Dev. 19671-676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Borchardt, R. T., B. T. Keller, and U. Patel-Thombre. 1984. Neplanocin A. A potent inhibitor of S-adenosylhomocysteine hydrolase and of vaccinia virus multiplication in mouse L929 cells. J. Biol. Chem. 2594353-4358. [PubMed] [Google Scholar]
- 7.Boulanger, M. C., C. Liang, R. S. Russell, R. Lin, M. T. Bedford, M. A. Wainberg, and S. Richard. 2005. Methylation of Tat by PRMT6 regulates human immunodeficiency virus type 1 gene expression. J. Virol. 79124-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cereseto, A., F. Diella, J. C. Mulloy, A. Cara, P. Michieli, R. Grassmann, G. Franchini, and M. E. Klotman. 1996. p53 functional impairment and high p21waf1/cip1 expression in human T-cell lymphotropic/leukemia virus type I-transformed T cells. Blood 881551-1560. [PubMed] [Google Scholar]
- 9.Crouch, S. P., R. Kozlowski, K. J. Slater, and J. Fletcher. 1993. The use of ATP bioluminescence as a measure of cell proliferation and cytotoxicity. J. Immunol. Methods 16081-88. [DOI] [PubMed] [Google Scholar]
- 10.Franchini, G., C. Nicot, and J. M. Johnson. 2003. Seizing of T cells by human T-cell leukemia/lymphoma virus type 1. Adv. Cancer Res. 8969-132. [DOI] [PubMed] [Google Scholar]
- 11.Gary, J. D., and S. Clarke. 1998. RNA and protein interactions modulated by protein arginine methylation. Prog. Nucleic Acid Res. Mol. Biol. 6165-131. [DOI] [PubMed] [Google Scholar]
- 12.Gessain, A., F. Barin, J. C. Vernant, O. Gout, L. Maurs, A. Calender, and G. de The. 1985. Antibodies to human T-lymphotropic virus type-I in patients with tropical spastic paraparesis. Lancet ii407-410. [DOI] [PubMed] [Google Scholar]
- 13.Gordon, R. K., K. Ginalski, W. R. Rudnicki, L. Rychlewski, M. C. Pankaskie, J. M. Bujnicki, and P. K. Chiang. 2003. Anti-HIV-1 activity of 3-deaza-adenosine analogs. Inhibition of S-adenosylhomocysteine hydrolase and nucleotide congeners. Eur. J. Biochem. 2703507-3517. [DOI] [PubMed] [Google Scholar]
- 14.Grassmann, R., C. Dengler, I. Muller-Fleckenstein, B. Fleckenstein, K. McGuire, M. C. Dokhelar, J. G. Sodroski, and W. A. Haseltine. 1989. Transformation to continuous growth of primary human T lymphocytes by human T-cell leukemia virus type I X-region genes transduced by a Herpesvirus saimiri vector. Proc. Natl. Acad. Sci. USA 863351-3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harhaj, E. W., and S. C. Sun. 1999. IKKgamma serves as a docking subunit of the IkappaB kinase (IKK) and mediates interaction of IKK with the human T-cell leukemia virus Tax protein. J. Biol. Chem. 27422911-22914. [DOI] [PubMed] [Google Scholar]
- 16.Herrmann, F., M. Bossert, A. Schwander, E. Akgun, and F. O. Fackelmayer. 2004. Arginine methylation of scaffold attachment factor A by heterogeneous nuclear ribonucleoprotein particle-associated PRMT1. J. Biol. Chem. 27948774-48779. [DOI] [PubMed] [Google Scholar]
- 17.Hoyos, B., D. W. Ballard, E. Bohnlein, M. Siekevitz, and W. C. Greene. 1989. Kappa B-specific DNA binding proteins: role in the regulation of human interleukin-2 gene expression. Science 244457-460. [DOI] [PubMed] [Google Scholar]
- 18.Jeang, K. T. 2001. Functional activities of the human T-cell leukemia virus type I Tax oncoprotein: cellular signaling through NF-kappa B. Cytokine Growth Factor Rev. 12207-217. [DOI] [PubMed] [Google Scholar]
- 19.Jeong, S. J., H. Lu, W. K. Cho, H. U. Park, C. Pise-Masison, and J. N. Brady. 2006. Coactivator-associated arginine methyltransferase 1 enhances transcriptional activity of the human T-cell lymphotropic virus type 1 long terminal repeat through direct interaction with Tax. J. Virol. 8010036-10044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jeong, S. J., C. A. Pise-Masison, M. F. Radonovich, H. U. Park, and J. N. Brady. 2005. Activated AKT regulates NF-kappaB activation, p53 inhibition and cell survival in HTLV-1-transformed cells. Oncogene 246719-6728. [DOI] [PubMed] [Google Scholar]
- 21.Jeong, S. J., M. Radonovich, J. N. Brady, and C. A. Pise-Masison. 2004. HTLV-I Tax induces a novel interaction between p65/RelA and p53 that results in inhibition of p53 transcriptional activity. Blood 1041490-1497. [DOI] [PubMed] [Google Scholar]
- 22.Kangas, L., M. Gronroos, and A. L. Nieminen. 1984. Bioluminescence of cellular ATP: a new method for evaluating cytotoxic agents in vitro. Med. Biol. 62338-343. [PubMed] [Google Scholar]
- 23.Keller, B. T., and R. T. Borchardt. 1987. Adenosine dialdehyde: a potent inhibitor of vaccinia virus multiplication in mouse L929 cells. Mol. Pharmacol. 31485-492. [PubMed] [Google Scholar]
- 24.Li, X. H., K. M. Murphy, K. T. Palka, R. M. Surabhi, and R. B. Gaynor. 1999. The human T-cell leukemia virus type-1 Tax protein regulates the activity of the IkappaB kinase complex. J. Biol. Chem. 27434417-34424. [DOI] [PubMed] [Google Scholar]
- 25.Li, Y. J., M. R. Stallcup, and M. M. Lai. 2004. Hepatitis delta virus antigen is methylated at arginine residues, and methylation regulates subcellular localization and RNA replication. J. Virol. 7813325-13334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu, Q., and G. Dreyfuss. 1995. In vivo and in vitro arginine methylation of RNA-binding proteins. Mol. Cell. Biol. 152800-2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lundin, A., M. Hasenson, J. Persson, and A. Pousette. 1986. Estimation of biomass in growing cell lines by adenosine triphosphate assay. Methods Enzymol. 13327-42. [DOI] [PubMed] [Google Scholar]
- 28.Manicassamy, S., S. Gupta, Z. Huang, and Z. Sun. 2006. Protein kinase C-theta-mediated signals enhance CD4+ T cell survival by up-regulating Bcl-xL. J. Immunol. 1766709-6716. [DOI] [PubMed] [Google Scholar]
- 29.McBride, A. E., and P. A. Silver. 2001. State of the arg: protein methylation at arginine comes of age. Cell 1065-8. [DOI] [PubMed] [Google Scholar]
- 30.Murphy, E. L., B. Hanchard, J. P. Figueroa, W. N. Gibbs, W. S. Lofters, M. Campbell, J. J. Goedert, and W. A. Blattner. 1989. Modelling the risk of adult T-cell leukemia/lymphoma in persons infected with human T-lymphotropic virus type I. Int. J. Cancer 43250-253. [DOI] [PubMed] [Google Scholar]
- 31.Naeem, H., D. Cheng, Q. Zhao, C. Underhill, M. Tini, M. T. Bedford, and J. Torchia. 2007. The activity and stability of the transcriptional coactivator p/CIP/SRC-3 are regulated by CARM1-dependent methylation. Mol. Cell. Biol. 27120-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nakamura, F., H. Kajihara, M. Nakamura, H. Sasaki, T. Kumamoto, and K. Okada. 1989. HTLV-1 associated myelopathy in an HTLV-1 and HBV double carrier family: report of a case and the mode of vertical transmission of both viruses. J. Gastroenterol. Hepatol. 4387-390. [DOI] [PubMed] [Google Scholar]
- 33.Nerenberg, M., S. H. Hinrichs, R. K. Reynolds, G. Khoury, and G. Jay. 1987. The tat gene of human T-lymphotropic virus type 1 induces mesenchymal tumors in transgenic mice. Science 2371324-1329. [DOI] [PubMed] [Google Scholar]
- 34.Newcomb, E. W. 1995. P53 gene mutations in lymphoid diseases and their possible relevance to drug resistance. Leuk. Lymphoma 17211-221. [DOI] [PubMed] [Google Scholar]
- 35.Osame, M., K. Usuku, S. Izumo, N. Ijichi, H. Amitani, A. Igata, M. Matsumoto, and M. Tara. 1986. HTLV-I associated myelopathy, a new clinical entity. Lancet i1031-1032. [DOI] [PubMed] [Google Scholar]
- 36.Paik, W. K., and S. Kim. 1967. Enzymatic methylation of protein fractions from calf thymus nuclei. Biochem. Biophys. Res. Commun. 2914-20. [DOI] [PubMed] [Google Scholar]
- 37.Pal, S., S. N. Vishwanath, H. Erdjument-Bromage, P. Tempst, and S. Sif. 2004. Human SWI/SNF-associated PRMT5 methylates histone H3 arginine 8 and negatively regulates expression of ST7 and NM23 tumor suppressor genes. Mol. Cell. Biol. 249630-9645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pise-Masison, C. A., K. S. Choi, M. Radonovich, J. Dittmer, S. J. Kim, and J. N. Brady. 1998. Inhibition of p53 transactivation function by the human T-cell lymphotropic virus type 1 Tax protein. J. Virol. 721165-1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pise-Masison, C. A., S. J. Jeong, and J. N. Brady. 2005. Human T cell leukemia virus type 1: the role of Tax in leukemogenesis. Arch. Immunol. Ther. Exp. (Warsz.) 53283-296. [PubMed] [Google Scholar]
- 40.Pise-Masison, C. A., R. Mahieux, H. Jiang, M. Ashcroft, M. Radonovich, J. Duvall, C. Guillerm, and J. N. Brady. 2000. Inactivation of p53 by human T-cell lymphotropic virus type 1 Tax requires activation of the NF-κB pathway and is dependent on p53 phosphorylation. Mol. Cell. Biol. 203377-3386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pise-Masison, C. A., M. Radonovich, K. Sakaguchi, E. Appella, and J. N. Brady. 1998. Phosphorylation of p53: a novel pathway for p53 inactivation in human T-cell lymphotropic virus type 1-transformed cells. J. Virol. 726348-6355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Poiesz, B. J., F. W. Ruscetti, A. F. Gazdar, P. A. Bunn, J. D. Minna, and R. C. Gallo. 1980. Detection and isolation of type C retrovirus particles from fresh and cultured lymphocytes of a patient with cutaneous T-cell lymphoma. Proc. Natl. Acad. Sci. USA 777415-7419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reid, R. L., P. F. Lindholm, A. Mireskandari, J. Dittmer, and J. N. Brady. 1993. Stabilization of wild-type p53 in human T-lymphocytes transformed by HTLV-I. Oncogene 83029-3036. [PubMed] [Google Scholar]
- 44.Schwerk, C., and K. Schulze-Osthoff. 2005. Methyltransferase inhibition induces p53-dependent apoptosis and a novel form of cell death. Oncogene 247002-7011. [DOI] [PubMed] [Google Scholar]
- 45.Sun, S. C., E. W. Harhaj, G. Xiao, and L. Good. 2000. Activation of I-kappaB kinase by the HTLV type 1 Tax protein: mechanistic insights into the adaptor function of IKKgamma. AIDS Res. Hum. Retroviruses 161591-1596. [DOI] [PubMed] [Google Scholar]
- 46.Takemoto, S., R. Trovato, A. Cereseto, C. Nicot, T. Kislyakova, L. Casareto, T. Waldmann, G. Torelli, and G. Franchini. 2000. p53 stabilization and functional impairment in the absence of genetic mutation or the alteration of the p14(ARF)-MDM2 loop in ex vivo and cultured adult T-cell leukemia/lymphoma cells. Blood 953939-3944. [PubMed] [Google Scholar]
- 47.Tanaka, A., C. Takahashi, S. Yamaoka, T. Nosaka, M. Maki, and M. Hatanaka. 1990. Oncogenic transformation by the tax gene of human T-cell leukemia virus type I in vitro. Proc. Natl. Acad. Sci. USA 871071-1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Willemsen, N. M., E. M. Hitchen, T. J. Bodetti, A. Apolloni, D. Warrilow, S. C. Piller, and D. Harrich. 2006. Protein methylation is required to maintain optimal HIV-1 infectivity. Retrovirology 392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yadav, N., J. Lee, J. Kim, J. Shen, M. C. Hu, C. M. Aldaz, and M. T. Bedford. 2003. Specific protein methylation defects and gene expression perturbations in coactivator-associated arginine methyltransferase 1-deficient mice. Proc. Natl. Acad. Sci. USA 1006464-6468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yasunaga, J., and M. Matsuoka. 2007. Human T-cell leukemia virus type I induces adult T-cell leukemia: from clinical aspects to molecular mechanisms. Cancer Control 14133-140. [DOI] [PubMed] [Google Scholar]
- 51.Yoshida, M. 2001. Multiple viral strategies of HTLV-1 for dysregulation of cell growth control. Annu. Rev. Immunol. 19475-496. [DOI] [PubMed] [Google Scholar]
- 52.Yoshida, M., I. Miyoshi, and Y. Hinuma. 1982. Isolation and characterization of retrovirus from cell lines of human adult T-cell leukemia and its implication in the disease. Proc. Natl. Acad. Sci. USA 792031-2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhao, L. J., H. Jian, and H. Zhu. 2003. Specific gene inhibition by adenovirus-mediated expression of small interfering RNA. Gene 316137-141. [DOI] [PubMed] [Google Scholar]