Abstract
RIG-I senses intracellular virus-specific nucleic acid structures and initiates an antiviral response that induces interferon (IFN) production, which, in turn, activates the transcription of RIG-I to increase RIG-I protein levels. Upon intracellular poly(I:C) stimulation, however, the levels of RIG-I protein did not correlate with the expression patterns of RIG-I transcripts. When the ISG15 conjugation system was overexpressed, ISG15 was conjugated to RIG-I and cellular levels of the unconjugated form of RIG-I decreased. The ISGylation of RIG-I reduced levels of both basal and virus-induced IFN promoter activity. Levels of unconjugated RIG-I also decreased when 26S proteasome activity was blocked by treatment with MG132, ALLN, or Lactacystin. In the presence of MG132, ISG15 conjugation to RIG-I increased, and hence, the unconjugated form of RIG-I was reduced. In Ube1L−/− cells, which lack the ability to conjugate ISG15, basal levels of both RIG-I protein and transcripts were increased compared to those in wild-type cells. As a result, enhanced production of ISGs and enhanced IFN promoter activity in Ube1L−/− cells were observed, and the phenotype was restored to that of wild-type cells by the overexpression of Ube1L. Based on these results, we propose a novel negative feedback loop which adjusts the strength of the RIG-I-mediated antiviral response and IFN production through the regulation of RIG-I protein by IFN-induced ISG15 conjugation.
Mammalian cells are equipped with two distinct innate immune machineries that detect viral infections and trigger the induction of type I interferons (IFNs) and proinflammatory cytokines (17). Extracellular viral nucleic acids are recognized by Toll-like receptors (3, 7, 8, and 9) in the endosome, while intracellular nucleic acids, such as double-stranded RNA (dsRNA) and 5′-triphosphate RNA, are recognized by the retinoic acid-inducible gene I (RIG-I)/melanoma differentiation-associated gene 5 (MDA5, or Helicard) protein in the cytoplasm (2, 11-13, 31, 34). Extracellular dsRNA activates NF-κB and IFN regulatory factor 3 (IRF3)/IRF7 via Toll-like receptor 3 and the adaptor protein TRIF, and intracellular dsRNA activates NF-κB and IRF3/IRF7 through RIG-I and the mitochondrial adaptor protein MAVS/VISA/Cardif/IPS-1; both pathways induce type I IFN production (1, 18, 28, 35, 40).
RIG-I protein is a cytosolic DEXD/H box RNA helicase with dsRNA or 5′-triphosphate RNA binding properties (13, 31, 42). RIG-I activates type I IFN production in response to the presence of an intracellular viral RNA genome, such as that of the Sendai virus or hepatitis C virus (38, 42), and plays central roles in the regulation of IFN production in response to viral infection in fibroblasts and conventional dendritic cells (15). The antiviral response to RNA virus infection is abolished in RIG-I-deficient murine embryonic fibroblast (MEF) cells, and conversely, viral replication is restricted in cells overexpressing RIG-I (42). Additionally, RIG-I/MDA5-mediated antiviral signaling can be disrupted by various viral proteins derived from hepatitis C virus, paramyxoviruses, or the influenza virus (3, 8, 29).
Type I IFN plays a central role in mediating antiviral innate immunity in mammals. IFN-α and IFN-β signaling is initiated by IFN-α/β binding to the IFN receptor, followed by the activation of Janus-activated kinase (JAK)-STAT signaling pathways and the induction of IFN-stimulated genes (ISGs). The ability of IFN to confer antiviral immunity depends largely on the production of ISG proteins, including RIG-I protein, dsRNA-dependent protein kinase (PKR), the 2′-5′-oligoadenylate (2-5A) system, the MxA proteins, and ISG15 (22, 37, 42). The expression of ISG15, one of the earliest ISG proteins to be identified (7), is strongly induced by IFN-α/β or lipopolysaccharide (LPS) stimulation, as well as by the presence of dsRNA or viral or bacterial infections (5, 7, 10). ISG15 is a ubiquitin (Ub)-like protein which is conjugated to intracellular proteins via an isopeptide bond (25). Similar to ubiquitination, the conjugation of ISG15 (ISGylation) requires a three-step process, involving an E1 activating enzyme (UBE1L), an E2 conjugating enzyme (UbcM8/H8), and an E3 ligase (19). Recently, HERC5/Ceb1 has been identified as an IFN-inducible ISG15-specific E3 ligase (6, 39) and UBP43 (USP18) has been identified as an IFN-inducible ISG15-specific deconjugating protease (27). The components of the ISGylation system identified thus far, including UBE1L, UbcM8/H8, HERC5/Ceb1, UBP43, and ISG15, are all IFN inducible (6, 19, 22, 27, 39).
Although ISG15-deficient mice were initially reported to exhibit normal IFN signaling and resistance to vesicular stomatitis virus (VSV) and lymphocytotic choriomeningitis virus (LCMV) infections, these mice were recently shown to have increased susceptibility to influenza, herpes, and Sindbis viral infections (23, 30). The antiviral role of ISG15 has also been supported by the observations that the overexpression of ISG15 represses Sindbis virus replication in multiple organs of IFN-α/β receptor-deficient mice and protects the host against virus-induced lethality (22). Among the recently identified cellular targets of ISG15 are several IFN-induced antiviral proteins, including PKR, MxA, HuP56, and RIG-I, suggesting that ISG15 conjugation may play an important regulatory role in IFN-mediated antiviral responses (26, 43). However, Ube1L-deficient mice that fail to produce ISG15 conjugates exhibited normal IFN-α/β signaling and antiviral responses to VSV and LCMV infections (20).
RIG-I protein senses intracellular viral components and initiates an antiviral response, which stimulates the production of IFN. IFN, in turn, activates the transcription of RIG-I, thus generating a positive feedback loop leading to the accumulation of RIG-I protein during the antiviral immune response. At the same time, the IFN-inducible Lgp2 interferes with the recognition of dsRNA by RIG-I protein and, hence, functions as a negative regulator (33, 41, 42). Here, we report an additional negative feedback loop that controls cellular levels of RIG-I protein through protein ISGylation, which is induced by antiviral or IFN-α/β signals. These results have revealed a mechanism by which RIG-I-mediated signaling strength can be fine-tuned to maintain a balance between innate defense and hypersensitivity during antiviral responses.
MATERIALS AND METHODS
Plasmids.
Full-length mouse ISG15 cDNA was obtained from livers of LPS-treated mice by reverse transcriptase PCR (RT-PCR) and subsequently cloned into the EcoRI/XhoI sites of the pCS2-MT plasmid. PCR primer sequences were as follows: 5′-CCGGAATTCGCAGCAATGGCCTGGGAC-3′ and 5′-CCGCTCGAGGGCACACTGGTCCCCTCC-3′. Full-length human RIG-I cDNA was isolated from pEF-BOS-Flag-RIG-I (provided by Takashi Fujita) and subcloned into the pcDNA3.1/Hygro plasmid (Invitrogen). A Flag-RIG-I (K172R) point mutation was introduced with the QuikChange site-directed-mutagenesis kit according to the instructions of the manufacturer (Stratagene). The pCAGGS-HA-UBE1L and pFlag-CMV-UbcM8 plasmids were kindly provided by Dong-Er Zhang (The Scripps Research Institute), and the hemagglutinin (HA)-Ub, Flag-human ISG15, and Flag-UBP43 plasmids were provided by Chin Ha Chung (Seoul National University, Korea). The pEGFP-N (Clonetech) and pISRE-luc (Stratagene) plasmids were purchased, and the pPRDIII-I luciferase plasmid was kindly provided by Katherine A. Fitzgerald (University of Massachusetts).
Cell culture and transfection.
All cell lines, with the exception of HepG2, were maintained in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum (FBS; HyClone, Logan, UT) and penicillin-streptomycin (Invitrogen). HepG2 cells were maintained in minimum essential medium containing 10% FBS and penicillin-streptomycin. For cytokine stimulation, 1,000-U/ml human IFN-α2a or human IFN-β (R&D Systems) was used. For dsRNA stimulation, cells were transiently transfected with poly(I:C) (Amersham Biosciences) by using Lipofectamine 2000 (Invitrogen) or Metafectene (Biontex, Germany) according to the manufacturer's instructions. For transient transfection, Lipofectamine 2000 was used. To make the total amounts of plasmids for transfection equal under every condition, cells were always cotransfected accordingly with empty vector. Cells were treated with MG132 (Sigma), ALLN (Calbiochem), or Lactacystin (Calbiochem) under the conditions indicated below. Newcastle disease virus (NDV; the La Sota strain) was obtained from the National Veterinary Research Quarantine Service, South Korea.
Antibodies and Western blotting.
Cells were lysed in lysis buffer (150 mM NaCl, 1% Triton X-100, 0.1% sodium dodecyl sulfate [SDS], 0.5% deoxycholate, 25 mM Tris-HCl, pH 7.5) containing 1 mM dithiothreitol, 0.57 mM phenylmethylsulfonyl fluoride, 5 μg of leupeptin/ml, 2 μg of pepstatin A/ml, 5 μg of aprotinin/ml, and 1 mM benzamidine. All chemicals were purchased from Sigma. Total cell lysates were analyzed using denaturing (SDS) polyacrylamide gels. Antibodies against Flag (M2; Sigma), HA (Roche), Myc (Roche), actin (Santa Cruz Biotechnology), glyceraldehyde-3-phosphate dehydrogenase (GAPDH; Chemicon), green fluorescent protein (GFP; Santa Cruz Biotechnology), and RIG-I protein (R37; Immuno-Biological Laboratories Co.) were purchased, and the anti-human RIG-I antibody was described previously (14). Immunoreactive signals were developed using the Super Signal reagent (Pierce).
Immunoprecipitation.
Total cells lysates (1 mg) were precleared with mouse immunoglobulin G (IgG) and protein G/protein A-agarose beads (Calbiochem, La Jolla, CA) and then incubated with anti-Flag or mouse IgG overnight at 4°C. The beads were washed with wash buffer (150 mM NaCl, 1% NP-40, 0.1% SDS, 0.5% deoxycholate, 1 mM EDTA, 50 mM Tris-HCl, pH 8.0) and analyzed on SDS-polyacrylamide gels. Input controls consisted of 5% of the lysates (whole-cell extracts).
Luciferase reporter assays.
COS-7, HepG2, and MEF cells were cotransfected with the pISRE or pPRDIII-I luciferase reporter plasmid and appropriate expression vectors. To normalize transfection efficiency, cells were cotransfected with pRL-CMV. At 48 h posttransfection, a dual luciferase assay (Promega) was performed. For stimulation, cells were treated with poly(I:C) (10 μg/ml) or NDV (105 50% egg infective doses/ml) at 42 to 48 h posttransfection for the times indicated in the figures.
Generation of stable cell lines.
HEK293 cells were transfected with pcDNA3.1/Hygro or pcDNA3.1/Hygro-Flag-RIG-I expression vectors, and stable transfectants were selected in 0.2 mg of hygromycin (Invitrogen)/ml for 3 weeks.
RNA isolation and analysis.
Total RNA was extracted from HepG2 or MEF cells using the TRIzol reagent (Invitrogen) and subjected to real-time or conventional RT-PCR. Total RNA (1 μg) was reverse transcribed using the Improm-II reverse transcription system (Promega). The primer sequences used for RT-PCR were as follows: mouse UbelL, 5′-CGGATGCAGCTTC TGAGGATG and 5′-CGTCCAGGGTTTCCTGCAGTT; mouse RIG-I, 5′-CGGTCGCTGATGAAGGCA and 5′-TACGGACATTTCTGCAGG; mouse ISG15, 5′-CCGGAATTCGCAGCAATGGCCTGGGAC; human LGP2, 5′-GATCCTGTGGTCATCATCAACA and 5′-TCAGTCCAGGGAGAGGTC-3′; and mouse IFN-β, 5′-CAGCTCCAGCTCCAAG and 5′-CTGAAGATCTCTGCTC-3′. Primer sequences for real-time PCR were as follows: RIG-I, 5′-GCCATTACACTGTGCTTGGAGA and 5′-CCAGTTGCAATATCCTCCACCA; IFN-β, 5′-TGCTCTCCTGTTGTGCTTCTCC and 5′-CATCTCATAGATGGTCAATGCGG; the oligoadenylate synthetase (OAS) gene, 5′-GCAGCGCCCAACCAAGCT and 5′-CCAGTTCCAAGACGGTCC; ISG15, 5′-CCTCTGAGCATCCTGGT and 5′-AGGCCGTACTCCCCCAG; MxA, 5′-CCTGGAAGAGTCTGCTGTG and 5′-AACCTGGTCCTGGCAGTAG; mouse IFN-β1, 5′-GGAATGAGACTATTGTT and 5′-CTGAAGATCTCTGCTC; mouse RIG-I, 5′-TCCCAGCAATGAGAATCCT and 5′-GTCAATGCCTTCATCAGC; mouse Ube1L, 5′-CGGATGCAGCTTCTGAGGATG and 5′-CACGCCACACAGTCTTGCCAG; LGP2, 5′-GATCCTGTGGTCATCAACA and 5′-TCAGTCCAGGGAGAGGTC; and the actin gene, 5′-TCATGAAGTGTGACGTTGACATCCGT and 5′-CCTAGAAGCATTTGCGGTGCACGATG.
Pulse-chase experiment.
For 35S metabolic labeling, Flag-RIG-I-transfected cells were preincubated in Dulbecco's modified Eagle's medium (containing 10% FBS) lacking methionine (Sigma) for 1 h at 37°C. Cells were pulse-labeled with [35S]methionine (PerkinElmer) for 90 min and were chased with complete culture medium for the times indicated below. Poly(I:C) was added during the chase period only. After the indicated times, cells were lysed in radioimmunoprecipitation assay lysis buffer and subjected to immunoprecipitation. Metabolically labeled RIG-I protein was analyzed by using autoradiography.
NDV replication analysis.
After infection with NDV (105 50% egg infective doses/ml) for various times, total RNA was prepared from infected MEF cells using TRIzol reagent. Total RNA (5 μg) was reverse transcribed using the NDV-specific primer (5′-GCTGATCATGAGGTTACCTC-3′) (21), and the presence of the NDV genome was assessed by PCR using NDV F gene-specific primers 5′-TTGATGGCAGGCCTCTTGC-3′ and 5′-GGAGGATGTTGGCAGCATT-3′ (36). For the control of equal loading levels, total RNA (1 μg) was reverse transcribed using oligo(dT) and subjected to PCR using actin gene primers at the same time.
RESULTS
Levels of RIG-I protein oscillate in cells stimulated with poly(I:C), a synthetic dsRNA.
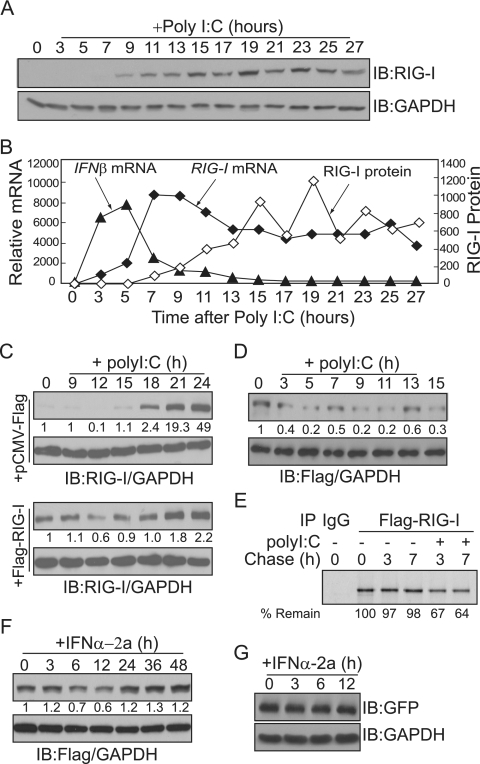
RIG-I protein senses intracellular dsRNA and activates IFN production. IFN, in turn, activates the transcription of RIG-I and increases RIG-I protein levels. The presence of this positive feedback loop predicts that intracellular levels of RIG-I protein will increase exponentially, as long as the viral signal exists. We examined the levels of endogenous RIG-I protein over time during the antiviral response. We transfected cells with poly(I:C) to mimic intracellular dsRNA infection, and levels of RIG-I protein were examined every 2 h over 24 h (Fig. 1A). A single dose of poly(I:C) was sufficient to induce RIG-I protein production, which was sustained over 24 h. However, RIG-I protein levels did not increase continuously, as initially expected; instead, levels declined slightly at 17 and 21 h after poly(I:C) treatment. We then examined IFN-β and RIG-I mRNA levels over 24 h following poly(I:C) transfection (Fig. 1B). Duplicate sets of cells were treated with poly(I:C), and samples from each culture were prepared at the same time points. Total RNA was isolated every 2 h over 24 h, and transcript levels were quantified using real-time RT-PCR. IFN-β mRNA reached a maximum level by approximately 5 h after poly(I:C) stimulation. In contrast, RIG-I mRNA began to increase by 5 h posttransfection, reached peak levels by approximately 7 h, and then declined. Thus, the overall pattern of RIG-I mRNA induction did not correlate with the pattern of RIG-I protein induction. To confirm this observation, separate sets of cells were transfected with poly(I:C), and RIG-I protein and mRNA levels were measured every 6 h over a 72-h time period. Again, the induction patterns of RIG-I protein did not agree with the mRNA expression pattern (data not shown). We repeated these assays at least three times, and the fluctuating patterns of RIG-I protein production were repeatedly observed.
FIG. 1.
Cellular levels of RIG-I protein oscillate in response to intracellular poly(I:C). (A) HepG2 cells were harvested at the indicated times after transfection with 10 μg of poly(I:C)/ml, and total cell extracts were probed with antibodies against RIG-I protein and GAPDH. Levels of RIG-I protein were quantified by measuring the immunoblot (IB) band intensity using ImageJ (NIH) and normalized to GAPDH protein levels. A line representing the degree of induction (n-fold) of RIG-I protein, compared to the protein level in untreated cells (time zero), is superimposed over the line representing RIG-I transcript levels in panel B. (B) Total RNA was isolated from HepG2 cells at the indicated times after transfection with 10 μg of poly(I:C)/ml. RIG-I and IFN-β transcripts were quantified by real-time RT-PCR, and the results were normalized to levels of β-actin gene transcripts. The degree of induction (n-fold) compared to the level in untreated cells (time zero) is shown. Duplicate sets of cells were treated at the same time for panels A and B. (C) HepG2 cells were transiently transfected with empty or Flag-RIG-I expression vectors and harvested at the indicated times after transfection with 10 μg of poly(I:C)/ml. Total cell extracts were probed with antibodies to RIG-I protein (top panel) and GAPDH (bottom panel). Relative amounts of RIG-I protein are shown below the RIG-I immunoblot. The experiment was repeated twice, with similar results. (D and F) HEK293 cells stably expressing Flag-RIG-I were stimulated with 10 μg of poly(I:C)/ml (D) or 1,000 U of IFN-α2a/ml (F) for the indicated times. Levels of exogenous Flag-RIG-I protein were quantified with antibodies against Flag (top panels) and GAPDH (bottom panels). (E) Pulse-chase analysis of Flag-RIG-I. Cells were metabolically labeled with [35S]methionine for 90 min. For poly(I:C) stimulation, cells were transfected with poly(I:C) at the beginning of the chase. Cells were harvested at the chase end points, and RIG-I protein was immunoprecipitated (IP) with Flag antibody. Percentages of remaining RIG-I protein relative to the amount at time zero (100%) are shown at the bottom. −, absent; +, present. (G) HEK293 cells stably expressing Flag-RIG-I were transfected with pCMV-GFP. After 24 h, cells were stimulated with 1,000 U of IFN-α2a/ml for the indicated times. Levels of GFP were analyzed with antibodies against GFP (top panel) and GAPDH (bottom panel).
These data suggest that cellular levels of RIG-I protein are controlled posttranscriptionally during antiviral responses. To test this hypothesis, HepG2 cells were transiently transfected with Flag-RIG-I and the level of Flag-RIG-I after poly(I:C) stimulation was examined (Fig. 1C). Since the expression of Flag-RIG-I is under the control of the cytomegalovirus promoter, it should overwhelm the transcriptional contribution by poly(I:C) or IFN stimulation. Stimulating Flag-RIG-I-expressing cells with intracellular poly(I:C) had an effect similar to that observed with endogenous RIG-I; the levels of Flag-RIG-I declined at 12 h after poly(I:C) transfection. We also used HEK293 cells stably expressing Flag-RIG-I and observed similar patterns of Flag-RIG-I protein levels after poly(I:C) stimulation (Fig. 1D). Furthermore, to examine whether the stability of RIG-I protein is altered after poly(I:C) treatment, we metabolically labeled cells with [35S]methionine and performed a pulse-chase experiment with the cells treated with or without poly(I:C) (Fig. 1E). RIG-I protein was stable, as little or no degradation at 3 and 7 h of chase was observed. However, with poly(I:C) stimulation, levels of 35S-labeled RIG-I were decreased, and about 67 and 64% of the protein remained at the end of 3 and 7 h of chase, respectively. This result indicates that stimulation with poly(I:C) leads to the degradation of the RIG-I protein. We then tested whether treatment with IFN-α can elicit a similar effect in Flag-RIG-I-expressing HEK293 cells (Fig. 1F). Flag-RIG-I levels decreased slightly by 6 to 12 h after IFN-α treatment, suggesting that IFN-α-mediated signaling pathways may be responsible for the negative regulation of RIG-I protein stability. To confirm that the cytomegalovirus promoter itself was not responsive to the IFN-α signal, levels of GFP in the cells transiently transfected with pCMV-GFP were examined after IFN-α treatment (Fig. 1G).
Cellular levels of RIG-I are diminished by overexpression of the ISG15 conjugating system.
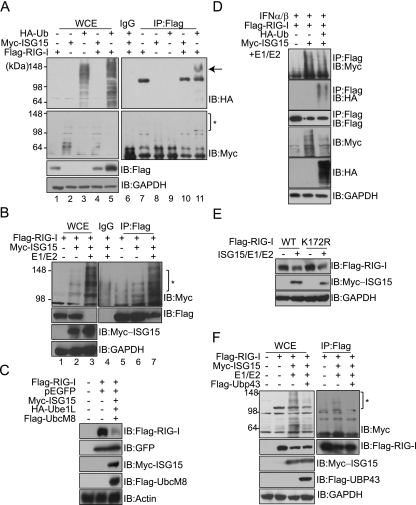
To investigate the molecular mechanism of the negative regulation of RIG-I, we specifically examined the effect of posttranslational modifications that are induced by IFN signaling. We examined ISGylation, which is strongly induced by IFN stimulation, and ubiquitination, since ISGylation utilizes a similar enzymatic cascade. Moreover, RIG-I protein has been identified as a cellular target of ubiquitination (4, 9). Cells were transfected with either HA-Ub or Myc-ISG15, together with the Flag-RIG-I expression plasmid, and the ubiquitination or ISGylation of RIG-I protein was assayed by immunoprecipitation (Fig. 2A). Although the overexpression of HA-Ub alone was sufficient to induce ubiquitin conjugation to RIG-I protein (Fig. 2A, lane 11), cellular levels of unconjugated RIG-I protein did not decrease but rather were increased by the overexpression of ubiquitin. In contrast, ISG15 conjugation to RIG-I protein was barely detectable when ISG15 alone was overexpressed (Fig. 2A, lane 10), even though RIG-I protein was previously reported to be a cellular target of ISG15 (43). We reasoned that the ISG15 conjugating system may be limiting in the cell lines assayed. Thus, we transfected cells with UBE1L and UbcM8 expression plasmids to provide the ISG15 E1 activating enzyme and the E2 conjugating enzyme and subsequently subjected total cellular extracts to immunoprecipitation with an anti-Flag antibody (Fig. 2B). In the presence of the ISG15 conjugating system, the ISGylated form of RIG-I protein clearly increased (Fig. 2B, lane 7). Interestingly, cellular levels of RIG-I protein, presumably the unconjugated form, dramatically decreased as the ISG15-conjugated form of RIG-I protein increased (Fig. 2B, lanes 3 and 7). The negative effect of ISGylation was specific to RIG-I protein, as levels of GFP in cotransfected cells were not altered (Fig. 2C). These data indicate that ISGylation may be one of the mechanisms responsible for controlling cellular levels of free (or unconjugated) RIG-I protein.
FIG. 2.
Cellular levels of RIG-I protein are decreased by ISG15 conjugation. (A) COS-7 cells were transfected with the indicated plasmids. Cells were harvested at 48 h posttransfection and subjected to immunoprecipitation (IP) with anti-Flag (lanes 7 to 11) or with mouse IgG (lane 6), followed by immunoblotting (IB) with anti-HA (top panel) or anti-Myc (bottom panel). The arrow and asterisk indicate the ubiquitinylated and ISGylated forms of RIG-I protein, respectively. Lanes 1 to 5, whole-cell extracts (WCE). −, absent; +, present. (B) COS-7 cells were transfected with 1 μg each of E1/E2, HA-Ube1L, and Flag-UbcM8 expression vectors, as indicated. To make the total amounts (4 μg/transfection) of plasmids for transfection equal under every condition, cells were cotransfected accordingly with empty vector. Total cell extracts were analyzed by immunoprecipitation with anti-Flag or with mouse IgG, followed by immunoblotting with anti-Myc, anti-Flag, or anti-GAPDH. The asterisk indicates the ISGylated form of RIG-I protein. The experiment was repeated at least three times, with similar results. (C) COS-7 cells were transfected with the indicated expression vectors. At 48 h posttransfection, total cell extracts were subjected to immunoblotting with anti-Flag, anti-GFP, anti-Myc, or antiactin. (D) COS-7 cells were transfected with the indicated plasmids. At 36 h posttransfection, cells were stimulated with IFN-α2a (1,000 U) for 12 h. Cells were harvested and subjected to immunoprecipitation with anti-Flag, followed by immunoblotting with the appropriate antibodies. (E) COS-7 cells were transfected with expression constructs, as indicated, harvested at 48 h posttransfection, and analyzed by immunoblotting with anti-Myc and anti-Flag. WT, wild type. (F) COS-7 cells were transfected with the indicated plasmids. At 48 h posttransfection, cells were harvested and subjected to immunoprecipitation with anti-Flag, followed by immunoblotting with the appropriate antibodies.
Levels of ISGylated RIG-I protein were slightly reduced with increasing HA-Ub expression in the absence (data not shown) or presence (Fig. 2D) of IFN treatment, indicating that ubiquitin and ISG15 may compete for RIG-I protein. Since a K172R mutation in the caspase recruitment domain of RIG-I protein causes the near-complete loss of ubiquitination of the protein (9), we constructed a RIG-I K172R mutant protein to test whether ISG15 uses the same site as ubiquitin for conjugation (Fig. 2E). However, in the presence of the ISGylation conjugating system, cellular levels of the unconjugated K172R mutant form decreased to levels comparable to those of the wild type, indicating that K172 is not a site for ISGylation. Lastly, we examined the effect of UBP43. UBP43 is an isopeptidase that is able to deconjugate ISG15 from target proteins. When cells were cotransfected with a UBP43 expression plasmid, ISG15 conjugation to RIG-I protein was reduced and the overall levels of cellular ISGylation decreased. However, levels of unconjugated RIG-I protein did not recover when UBP43 was overexpressed (Fig. 2F).
ISGylation negatively controls RIG-I-mediated signaling.
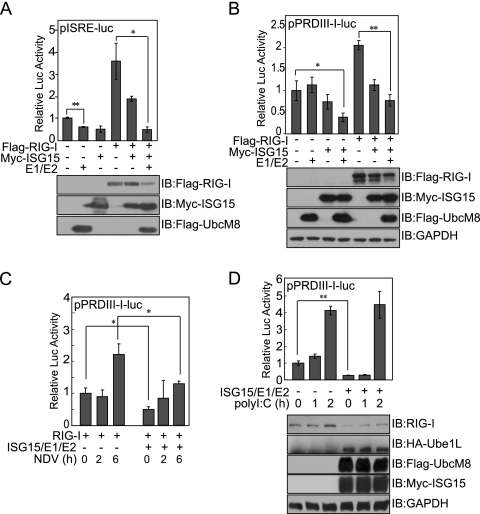
We next addressed the molecular function of ISGylated RIG-I in RIG-I-mediated signaling events. We performed luciferase reporter assays using constructs under the control of IFN-stimulated response elements (pISRE-luc) or the IFN promoter (pPRDIII-I-luc) in cells cotransfected with RIG-I, ISG15, UBE1L (E1), and UbcM8 (E2) expression vectors. RIG-I-mediated signaling was positively correlated with cellular levels of unconjugated RIG-I protein, as luciferase activity elicited in the presence of RIG-I overexpression was significantly reduced by the coexpression of the ISG15 conjugating proteins (Fig. 3A and B). We then stimulated cells with NDV, and RIG-I-mediated IFN promoter activity was measured using the pPRDIII-I-luc reporter construct (Fig. 3C). Both basal and induced levels of IFN promoter activity were lower in cells overexpressing the ISG15 conjugating system. We also used HepG2 cells, which express a relatively high level of endogenous RIG-I protein, to confirm the effect of ISGylation (Fig. 3D). Both pPRDIII-I luciferase activity and the level of unconjugated endogenous RIG-I protein were clearly reduced by the coexpression of the ISG15 conjugating system, as observed earlier. When stimulated with poly(I:C), IFN promoter activity began to increase as early as 1 h poststimulation and reached approximately fourfold induction at 2 h poststimulation. In contrast, when cells were cotransfected with the ISG15 conjugating system, the basal level of IFN promoter activity and the level in the early phase of infection were low but the level of activity began to rise by 2 h poststimulation. This later induction of RIG-I-mediated IFN activity may result from the poly(I:C)-induced transcriptional induction of RIG-I. These results suggest that ISGylated RIG-I protein may be nonfunctional in transmitting antiviral signals and that IFN-induced ISGylation controls IFN production through negatively regulating cellular levels of unconjugated RIG-I protein.
FIG. 3.
Overexpression of the ISG15 conjugating system negatively affects RIG-I-mediated IFN signaling. (A and B) COS-7 cells were cotransfected with the indicated expression vectors and pISRE-luc (A) or pPRDIII-I-luc (B). Cells were harvested at 48 h posttransfection and analyzed using a dual luciferase assay. The values shown are the means of results for triplicate samples, and the standard deviation (SD) is indicated. Representative results from one of three independent experiments are shown. IB, immunoblot; −, absent; +, present. (C) COS-7 cells were cotransfected with pPRDIII-I-luc and the indicated expression vectors. Cells were infected with NDV 42 h posttransfection, and luciferase activity was measured at the indicated times after infection. (D) HepG2 cells were transiently transfected with pPRDIII-I-luc and the indicated expression vectors. At 48 h posttransfection, cells were transfected with poly(I:C) for the indicated times. Total cell extracts were assayed for luciferase activity and probed with anti-RIG-I protein to examine endogenous levels of RIG-I protein. Student's t test: *, P < 0.05; **, P < 0.01.
Treatment with a proteasome inhibitor reduces levels of unconjugated RIG-I protein.
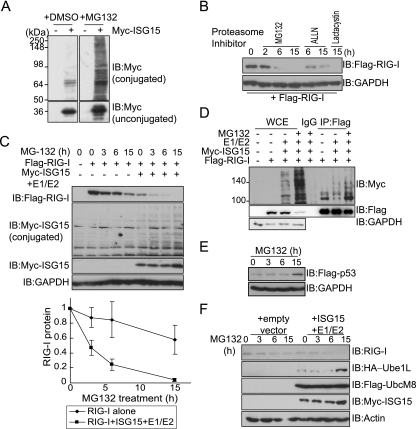
The inhibition of proteasomal activity enhances the levels of ISG15 conjugates through increased de novo conjugation (24). Therefore, we examined whether treating cells with a proteasome inhibitor also altered cellular levels of conjugated or unconjugated RIG-I protein. Treating cells with the 26S proteasome inhibitor MG132 dramatically enhanced ISGylation (Fig. 4A). We treated cells with MG132 and measured the accumulation of RIG-I protein as a function of time after drug treatment (Fig. 4B). Quite surprisingly, the inhibition of proteasomal activity was accompanied by a decrease in the levels of unconjugated RIG-I protein. Cells were exposed to two other 26S proteasome inhibitors, ALLN and Lactacystin, and similar decreases in RIG-I protein were observed. To further investigate the effect of proteasomal activity on the regulation of RIG-I, cells were treated with MG132 in the presence or absence of overexpressed components of the ISG15 conjugating system (Fig. 4C). The coexpression of the ISG15 conjugating proteins together with the inhibition of the 26S proteasome machinery induced clear reduction in the levels of unconjugated RIG-I protein, compared to those occurring after MG132 treatment alone. We next performed immunoprecipitations to examine whether the decrease in RIG-I protein was due to an increase in the ISG15 conjugation of RIG-I protein (Fig. 4D). With MG132 treatment, the ISG15 conjugation of RIG-I protein was clearly enhanced, while unconjugated RIG-I protein decreased concomitantly. MG132 worked at the concentrations used, since treatment with MG132 stabilized transfected p53 protein (Fig. 4E). Finally, we examined the effect of MG132 on endogenous RIG-I protein in HepG2 cells. With MG132 treatment, endogenous RIG-I protein levels decreased slightly, and the decrease was more extensive with the coexpression of the ISG15 conjugating system (Fig. 4F). Also, increased levels of Ube1L and ISG15 proteins were repeatedly observed in the cells treated with proteasome inhibitor. These results indicate that the inhibition of proteasome activity increases ISG15 conjugation as previously reported (23) and increases ISGylated RIG-I protein and, thus, reduces the level of unconjugated RIG-I protein.
FIG. 4.
Cellular levels of RIG-I protein decrease in response to treatment with proteasome inhibitors. (A) COS-7 cells were transfected with a Myc-ISG15 expression plasmid and treated with dimethyl sulfoxide (DMSO) or 10 μM MG132 for 15 h. Total cellular levels of ISG15-conjugated proteins (upper panels) or unconjugated free ISG15 (lower panels) were visualized by immunoblotting (IB) using anti-Myc. −, absent; +, present. (B) COS-7 cells were transfected with a Flag-RIG-I expression plasmid. At 36 h posttransfection, cells were treated with MG132 (10 μM), ALLN (10 μM), or Lactacystin (5 μM) for the indicated times, harvested, and analyzed by immmunoblotting with anti-Flag or anti-GAPDH. (C) COS-7 cells were transfected with a Flag-RIG-I expression plasmid alone or together with expression vectors for components of the ISG15 conjugating system. At 36 h posttransfection, cells were treated with 10 μM MG132 for the indicated times. Immunoblotting was performed using the indicated anti-Myc and anti-Flag antibodies. For loading control analysis, the membrane was stripped and reprobed with anti-GAPDH antibody. (Bottom panel) Cellular levels of remaining RIG-I protein following MG132 treatment. Levels of RIG-I protein were quantified by measuring the immunoblot band intensity using ImageJ (NIH) and normalized to GAPDH protein levels. Percentages of remaining RIG-I protein relative to the level of RIG-I protein at time zero (100%) were calculated. Values are averages (±SD) of results from three independent experiments. (D) COS-7 cells were transfected with various expression vectors, as indicated. Total cell extracts were subjected to immunoprecipitation (IP) with anti-Flag or with mouse IgG, followed by immunoblotting with anti-Myc, anti-Flag, or anti-GAPDH. WCE, whole-cell extracts. (E) HepG2 cells were transfected with Flag-p53 expression plasmid. After 36 h, cells were treated with 10 μM MG132 for the indicated times. Total cell extracts were harvested at the end of the incubations, and immunoblotting was performed using anti-Flag antibody. (F) HepG2 cells were transfected with the indicated plasmids and, at 36 h posttransfection, were treated with 10 μM MG132 for the indicated times. Total cell extracts were harvested at the end of the incubations, and immunoblotting was performed using the indicated antibodies.
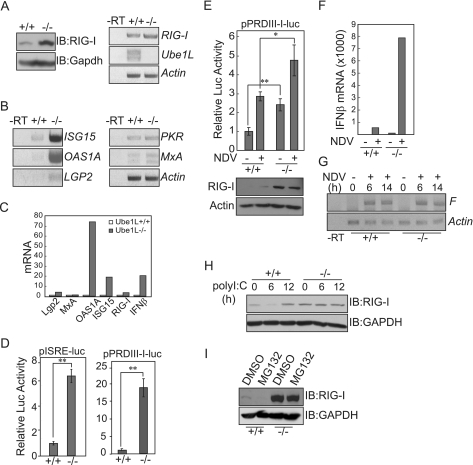
The RIG-I-mediated antiviral signaling response is enhanced in Ube1L-deficient MEF cells.
If ISG15 conjugation is the key event regulating levels of RIG-I protein, then the basal levels of RIG-I protein should be higher in cells deficient in ISG15 conjugating components. Therefore, we compared basal levels of RIG-I protein and transcripts in Ube1L−/− and wild-type MEF cells (Fig. 5A). Levels of RIG-I protein in Ube1L-deficient cells were much higher than those in wild-type MEFs. However, RIG-I transcript levels were not enhanced as dramatically as the protein levels in Ube1L−/− cells, suggesting that the higher RIG-I protein levels in Ube1L-deficient cells were due to posttranslational control. Since Ube1L−/− cells expressed a higher level of RIG-I, we hypothesized that RIG-I-mediated antiviral signaling may be intensified in cells deficient in ISG15 conjugating components. We therefore measured basal levels of transcripts of ISGs, such as ISG15, OAS1A, MxA, PKR, and Lgp2 genes, in Ube1L−/− MEF cells (Fig. 5B and C). Basal levels of ISG15, OAS1A, and Lgp2 transcripts were higher in Ube1L-deficient cells than in wild-type MEFs. In addition, Ube1L-deficient cells exhibited constitutively higher IFN signaling-related transcription factor activity, as assayed by pISRE and pPRDIII-I reporter activity (Fig. 5D). When cells were infected with NDV, both basal and virus-induced levels of IFN promoter activity in Ube1L−/− MEF cells were significantly increased compared to those in wild-type cells (Fig. 5E). More importantly, Ube1L−/− MEF cells produced higher levels of basal and NDV-induced IFN-β transcripts (Fig. 5F). However, the actual levels of NDV replication were not affected dramatically, as judged by NDV F gene-specific RT-PCR analysis of the cells infected with NDV for 6 and 14 h (Fig. 5G). In Ube1L+/+ MEF cells, levels of unconjugated RIG-I protein initially decreased and then increased upon poly(I:C) stimulation, as previously observed in HEK293 and HepG2 cells (Fig. 5H). However, levels of RIG-I protein in Ube1L-deficient cells were not altered after poly(I:C) stimulation, confirming that the downregulation of RIG-I protein observed during poly(I:C) treatment was due primarily to ISGylation activity. We also treated cells with the proteasome inhibitor MG132, which enhances the ISGylation of RIG-I protein (Fig. 5I). Again, unconjugated RIG-I protein was reduced to undetectable levels in wild-type cells but no reduction was observed in cells deficient in ISG15 conjugation system components.
FIG. 5.
The RIG-I-mediated antiviral signaling response is enhanced in UbelL-deficient MEFs. (A) Ube1L+/+ and Ube1L−/− MEF cells were analyzed for RIG-I expression. (Left panel) Immunoblot (IB) assay with anti-RIG-I protein and anti-GAPDH. (Right panel) RT-PCR analysis to measure RIG-I, UbelL, and β-actin gene transcripts. −RT, without reverse transcriptase. (B and C) Ube1L+/+ and Ube1L−/− MEF cells were analyzed for IFN-stimulated gene expression. Total RNA was extracted from MEF cells, and levels of ISG15, OAS1A, MxA, PKR, and Lgp2 gene transcripts were measured by conventional (B) or real-time (C) RT-PCR. The degree of induction (n-fold) of each transcript compared to the level in wild-type cells is shown. (D) Ube1L+/+ and Ube1L−/− MEF cells were analyzed for basal IRF activity. MEFs were transfected with the pISRE-luc or pPRDIII-I-luc reporter plasmid, and total extracts were assayed for luciferase activity. Representative results from one of three independent experiments are shown. Error bars indicate SDs for the three experiments. (E) Ube1L+/+ and Ube1L−/− MEF cells were analyzed for virus-induced IFN promoter activity. At 45 h posttransfection, MEFs were infected with NDV for 3 h and total extracts were assayed for IFN promoter activity. Error bars indicate SDs for results from three experiments. Student's t test: *, P < 0.05; **, P < 0.01. (F) Ube1L+/+ and Ube1L−/− MEF cells were analyzed for IFN-β transcripts. MEFs were infected with NDV for 3 h, and IFN-β transcripts were quantified by real-time RT-PCR. (G) Ube1L+/+ and Ube1L−/− MEF cells were analyzed for the levels of NDV replication. MEFs were infected with NDV for 6 or 14 h, and transcripts of the F gene were analyzed by RT-PCR. (H) MEFs were transfected with poly(I:C) for the indicated times, and total extracts were immunoassayed for endogenous RIG-I protein. (I) Ube1L+/+ and Ube1L−/− MEF cells were treated with 10 μM MG132 for 12 h, and levels of endogenous RIG-I protein were analyzed by immunoblotting. DMSO, dimethyl sulfoxide.
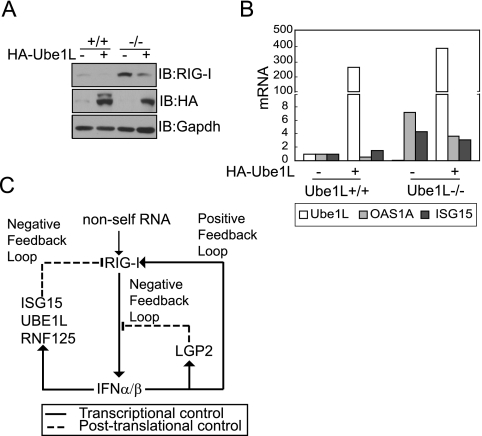
Both basal and LPS-induced levels of IRF7 and ISG15 transcripts in Ube1L−/− and Ube1L+/+ macrophages were similar (20). Since we observed that basal levels of some ISG proteins were enhanced in Ube1L−/− MEFs, we examined whether the cells were stressed under the culture conditions we used, possibly leading to artifactual results. We transfected MEF cells with a Ube1L expression plasmid and examined whether the Ube1L−/− phenotype could be abolished. In the presence of Ube1L overexpression, basal levels of RIG-I protein in Ube1L−/− MEF cells were clearly reduced (Fig. 6A). Furthermore, basal levels of ISG15 and OAS1A gene transcripts in HA-Ube1L-overexpressing Ube1L−/− MEF cells were decreased (Fig. 6B), confirming that the enhanced levels of RIG-I protein in Ube1L-deficient cells were due to the lack of ISGylation.
FIG. 6.
Overexpression of Ube1L restored the wild-type phenotype to Ube1L−/− MEFs. (A) Ube1L+/+ and Ube1L−/− MEF cells were transfected with an HA-Ube1L expression plasmid. After 48 h, total extracts were immunoassayed for endogenous RIG-I protein and HA-Ube1L expression. The experiment was repeated more than three times, with similar results. IB, immunoblot; −, absent; +, present. (B) Ube1L+/+ and Ube1L−/− MEF cells were transfected with an HA-Ube1L expression plasmid. After 48 h, levels of ISG15, OAS1A, and Ube1L gene transcripts were quantified by real-time RT-PCR. The degree of induction (n-fold) compared to the level in untreated wild-type MEFs is shown. (C) Proposed RIG-I signaling pathway regulated by multiple positive and negative feedback loops.
DISCUSSION
In the present study, we demonstrated that IFN-inducible ISG15 conjugation of RIG-I protein negatively regulates RIG-I-mediated signaling. Intracellular viral signals are initially recognized by RIG-I protein and induce the synthesis of type I IFN, which activates the transcription of RIG-I, LGP2, and components of the ISG15 conjugation system. Newly synthesized RIG-I protein binds to intracellular viral RNAs, if present, to further upregulate IFN synthesis and, thus, forms a positive feedback loop for the propagation of antiviral signals (42). Concomitantly, IFN activates the transcription of LGP2, the product of which binds dsRNAs but fails to signal downstream and, hence, forms a negative feedback loop regulating RIG-I-mediated signals (32). In addition to these regulatory circuits, we have identified a novel negative feedback loop that controls cellular levels of RIG-I protein. IFN induces ISG15 conjugation to RIG-I protein and lowers cellular levels of unconjugated RIG-I protein and, thus, negatively regulates RIG-I-mediated antiviral signaling. Although the negative regulation of RIG-I by IFN-induced ISGylation is a novel finding, it is not the only posttranslational feedback control regulating RIG-I protein levels. Recently, studies have shown that RIG-I protein can be ubiquitin conjugated by at least two different E3 ligases, TRIM25 and RNF125 (4, 9). While the ubiquitination of RIG-I protein at Lys 172 by TRIM25 is required to mediate antiviral signaling responses, ubiquitination by RNF125 controls the proteasomal degradation of RIG-I protein, thereby negatively regulating RIG-I-mediated signaling pathways. The expression of RNF125 is enhanced 24 to 36 h after poly(I:C) stimulation, providing another negative feedback loop for IFN production by controlling cellular levels of RIG-I protein. The presence of multiple positive and negative feedback loops regulating RIG-I indicates that maintaining optimal levels of RIG-I protein is critical to balance defense reactions and hypersensitivity in antiviral responses (Fig. 6C). Because IFN stimulates the transcription of components that are mutually inhibitory, the outcome of the viral infection may be determined by competition between negative and positive regulators whose activity depends on the basal levels of proteins and transcripts, the kinetics of IFN-inducible transcription and protein modifications, and the intracellular levels of viral components over the time course of the infection.
When we treated cells with MG132, we observed an increase in total ISG15 conjugates, as previously reported (24). ISG15 conjugation to RIG-I protein increased, and therefore, the unconjugated form of RIG-I protein decreased. This result is somewhat controversial, as RIG-I protein is degraded by the overexpression of the ubiquitin E3 ligase RNF125 (4). Based on the observation that RIG-I was not stabilized by treatment with MG132 alone, we propose that proteasome-dependent degradation of RIG-I protein occurs only after RNF125-mediated ubiquitination, which requires intracellular poly(I:C) and/or IFN signals. We initially reported that levels of RIG-I protein did not correlate with levels of RIG-I transcripts over time in response to intracellular poly(I:C) (Fig. 1). Given the negative control by poly(I:C) and IFN-induced ISGylation and ubiquitination (by RNF125), this result should be reinterpreted as a dynamic modulation of levels of unconjugated RIG-I protein by posttranslational modification during antiviral responses. Free or unconjugated RIG-I protein may be functional and is presumably a target for TRIM25-mediated ubiquitination. In the experimental scheme we used, it was not possible to quantify the exact amount of total RIG-I protein (the sum of unconjugated and conjugated forms) over the time course of the viral infection. Similarly, we do not yet know the molecular fate of the ISGylated RIG-I protein. Since RNF125-mediated ubiquitination degrades RIG-I, it will be of interest to determine whether ISGylated RIG-I protein can be ubiquitinated by RNF125. When we examined the effects of UBP43 on ISG15-mediated RIG-I protein control, decreased levels of unconjugated RIG-I protein were not restored to former levels by the overexpression of UBP43 (Fig. 2). We have hypothesized that ISGylated RIG-I protein becomes subject to an irreversible biochemical process, such as proteolysis or proteasomeal degradation, and hence, the overexpression of UBP43 was unable to alter levels of RIG-I protein. Alternatively, overexpressed UBP43 functions as a negative regulator of IFN signals, and therefore, levels of RIG-I protein were unchanged. Lastly, it is possible that our proposed mechanism is due to indirect effects of ISGylation, which require as-yet-unidentified proteins to control the stability of RIG-I.
We observed that the basal levels of RIG-I and ISG proteins were higher in Ube1L-deficient MEF cells (Fig. 5). Together with enhanced IFN-stimulated response element and IFN promoter activity, these data indicate that Ube1L-deficient MEF cells have elevated levels of antiviral components in the absence of viral infection. Since there is no negative regulation by ISGylation in Ube1L-deficient MEF cells, RIG-I-mediated antiviral responses should be enhanced upon viral infection. Based on these observations, we hypothesized that Ube1L-deficient mice would exhibit hyperresponses to viral infections, due to unregulated RIG-I protein accumulation and the consequent hyperproduction of antiviral molecules. In contrast, however, Kim et al. reported that Ube1L-deficient mice pretreated with IFN-β exhibit normal responses to LCMV and VSV (20). In addition, basal levels of ISG15 were unchanged in Ube1L-deficient macrophages. There are several explanations for these discrepancies. First, RIG-I is essential for IFN induction in response to intracellular viral infection in fibroblasts and conventional dendritic cells, but not in macrophages (15). The reason we observed a dramatic effect in Ube1L-deficient MEFs, but not in Ube1L-deficient macrophages, may be this cell-specific contribution of RIG-I during the antiviral response. Secondly, RIG-I protein senses specific types of RNA viruses, such as NDV, Sendai virus, and influenza virus, to mediate antiviral responses (16). Therefore, antiviral signaling in LCMV infection may be RIG-I independent. It is noteworthy that ISG15 knockout mice exhibited normal responses to LCMV and VSV infections but showed hyperresponsiveness to influenza, herpes, and Sindbis viral infections (23, 30). Finally, virus- or IFN-induced transcriptional induction of RIG-I may overcome the negative regulatory effect of ISGylation.
In summary, our findings reveal a novel regulatory mechanism of IFN production through the negative control of RIG-I-mediated antiviral responses by IFN-induced ISGylation. A complete understanding of this regulation will require elucidating the relationship between the ubiquitination and the ISGylation of RIG-I protein and the functional interactions of these processes during antiviral responses.
Acknowledgments
We thank K. I. Kim for valuable discussions and T. Fujita, D.-E. Zhang, and C. H. Chung for plasmids and Ube1L−/− MEFs.
This work was supported by grants from the Korean Health 21 R&D Project of the Ministry of Health and Welfare (0412-BM01-716-0001) and the POSCO Research Fund.
Footnotes
Published ahead of print on 5 December 2007.
REFERENCES
- 1.Akira, S., and K. Takeda. 2004. Toll-like receptor signalling. Nat. Rev. Immunol. 4499-511. [DOI] [PubMed] [Google Scholar]
- 2.Akira, S., S. Uematsu, and O. Takeuchi. 2006. Pathogen recognition and innate immunity. Cell 124783-801. [DOI] [PubMed] [Google Scholar]
- 3.Andrejeva, J., K. S. Childs, D. F. Young, T. S. Carlos, N. Stock, S. Goodbourn, and R. E. Randall. 2004. The V proteins of paramyxoviruses bind the IFN-inducible RNA helicase, mda-5, and inhibit its activation of the IFN-beta promoter. Proc. Natl. Acad. Sci. USA 10117264-17269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arimoto, K., H. Takahashi, T. Hishiki, H. Konishi, T. Fujita, and K. Shimotohno. 2007. Negative regulation of the RIG-I signaling by the ubiquitin ligase RNF125. Proc. Natl. Acad. Sci. USA 1047500-7505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blomstrom, D. C., D. Fahey, R. Kutny, B. D. Korant, and E. Knight, Jr. 1986. Molecular characterization of the interferon-induced 15-kDa protein. Molecular cloning and nucleotide and amino acid sequence. J. Biol. Chem. 2618811-8816. [PubMed] [Google Scholar]
- 6.Dastur, A., S. Beaudenon, M. Kelley, R. M. Krug, and J. M. Huibregtse. 2006. Herc5, an interferon-induced HECT E3 enzyme, is required for conjugation of ISG15 in human cells. J. Biol. Chem. 2814334-4338. [DOI] [PubMed] [Google Scholar]
- 7.Farrell, P. J., R. J. Broeze, and P. Lengyel. 1979. Accumulation of an mRNA and protein in interferon-treated Ehrlich ascites tumour cells. Nature 279523-525. [DOI] [PubMed] [Google Scholar]
- 8.Foy, E., K. Li, R. Sumpter, Jr., Y. M. Loo, C. L. Johnson, C. Wang, P. M. Fish, M. Yoneyama, T. Fujita, S. M. Lemon, and M. Gale, Jr. 2005. Control of antiviral defenses through hepatitis C virus disruption of retinoic acid-inducible gene-I signaling. Proc. Natl. Acad. Sci. USA 1022986-2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gack, M. U., Y. C. Shin, C. H. Joo, T. Urano, C. Liang, L. Sun, O. Takeuchi, S. Akira, Z. Chen, S. Inoue, and J. U. Jung. 2007. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature 446916-920. [DOI] [PubMed] [Google Scholar]
- 10.Haas, A. L., P. Ahrens, P. M. Bright, and H. Ankel. 1987. Interferon induces a 15-kilodalton protein exhibiting marked homology to ubiquitin. J. Biol. Chem. 26211315-11323. [PubMed] [Google Scholar]
- 11.Heil, F., H. Hemmi, H. Hochrein, F. Ampenberger, C. Kirschning, S. Akira, G. Lipford, H. Wagner, and S. Bauer. 2004. Species-specific recognition of single-stranded RNA via Toll-like receptor 7 and 8. Science 3031526-1529. [DOI] [PubMed] [Google Scholar]
- 12.Hemmi, H., T. Kaisho, O. Takeuchi, S. Sato, H. Sanjo, K. Hoshino, T. Horiuchi, H. Tomizawa, K. Takeda, and S. Akira. 2002. Small anti-viral compounds activate immune cells via the TLR7 MyD88-dependent signaling pathway. Nat. Immunol. 3196-200. [DOI] [PubMed] [Google Scholar]
- 13.Hornung, V., J. Ellegast, S. Kim, K. Brzozka, A. Jung, H. Kato, H. Poeck, S. Akira, K. K. Conzelmann, M. Schlee, S. Endres, and G. Hartmann. 2006. 5′-Triphosphate RNA is the ligand for RIG-I. Science 314994-997. [DOI] [PubMed] [Google Scholar]
- 14.Imaizumi, T., S. Aratani, T. Nakajima, M. Carlson, T. Matsumiya, K. Tanji, K. Ookawa, H. Yoshida, S. Tsuchida, T. M. McIntyre, S. M. Prescott, G. A. Zimmerman, and K. Satoh. 2002. Retinoic acid-inducible gene-I is induced in endothelial cells by LPS and regulates expression of COX-2. Biochem. Biophys. Res. Commun. 292274-279. [DOI] [PubMed] [Google Scholar]
- 15.Kato, H., S. Sato, M. Yoneyama, M. Yamamoto, S. Uematsu, K. Matsui, T. Tsujimura, K. Takeda, T. Fujita, O. Takeuchi, and S. Akira. 2005. Cell type-specific involvement of RIG-I in antiviral response. Immunity 2319-28. [DOI] [PubMed] [Google Scholar]
- 16.Kato, H., O. Takeuchi, S. Sato, M. Yoneyama, M. Yamamoto, K. Matsui, S. Uematsu, A. Jung, T. Kawai, K. J. Ishii, O. Yamaguchi, K. Otsu, T. Tsujimura, C. S. Koh, C. Reis e Sousa, Y. Matsuura, T. Fujita, and S. Akira. 2006. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature 441101-105. [DOI] [PubMed] [Google Scholar]
- 17.Kawai, T., and S. Akira. 2006. Innate immune recognition of viral infection. Nat. Immunol. 7131-137. [DOI] [PubMed] [Google Scholar]
- 18.Kawai, T., K. Takahashi, S. Sato, C. Coban, H. Kumar, H. Kato, K. J. Ishii, O. Takeuchi, and S. Akira. 2005. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat. Immunol. 6981-988. [DOI] [PubMed] [Google Scholar]
- 19.Kim, K. I., N. V. Giannakopoulos, H. W. Virgin, and D. E. Zhang. 2004. Interferon-inducible ubiquitin E2, Ubc8, is a conjugating enzyme for protein ISGylation. Mol. Cell. Biol. 249592-9600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim, K. I., M. Yan, O. Malakhova, J. K. Luo, M. F. Shen, W. Zou, J. C. de la Torre, and D. E. Zhang. 2006. Ube1L and protein ISGylation are not essential for alpha/beta interferon signaling. Mol. Cell. Biol. 26472-479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee, Y. J., H. W. Sung, J. G. Choi, J. H. Kim, and C. S. Song. 2004. Molecular epidemiology of Newcastle disease viruses isolated in South Korea using sequencing of the fusion protein cleavage site region and phylogenetic relationships. Avian Pathol. 33482-491. [DOI] [PubMed] [Google Scholar]
- 22.Lenschow, D. J., N. V. Giannakopoulos, L. J. Gunn, C. Johnston, A. K. O'Guin, R. E. Schmidt, B. Levine, and H. W. Virgin IV. 2005. Identification of interferon-stimulated gene 15 as an antiviral molecule during Sindbis virus infection in vivo. J. Virol. 7913974-13983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lenschow, D. J., C. Lai, N. Frias-Staheli, N. V. Giannakopoulos, A. Lutz, T. Wolff, A. Osiak, B. Levine, R. E. Schmidt, A. Garcia-Sastre, D. A. Leib, A. Pekosz, K. P. Knobeloch, I. Horak, and H. W. Virgin IV. 2007. IFN-stimulated gene 15 functions as a critical antiviral molecule against influenza, herpes, and Sindbis viruses. Proc. Natl. Acad. Sci. USA 1041371-1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu, M., X. L. Li, and B. A. Hassel. 2003. Proteasomes modulate conjugation to the ubiquitin-like protein, ISG15. J. Biol. Chem. 2781594-1602. [DOI] [PubMed] [Google Scholar]
- 25.Loeb, K. R., and A. L. Haas. 1992. The interferon-inducible 15-kDa ubiquitin homolog conjugates to intracellular proteins. J. Biol. Chem. 2677806-7813. [PubMed] [Google Scholar]
- 26.Malakhov, M. P., K. I. Kim, O. A. Malakhova, B. S. Jacobs, E. C. Borden, and D. E. Zhang. 2003. High-throughput immunoblotting. Ubiquitin-like protein ISG15 modifies key regulators of signal transduction. J. Biol. Chem. 27816608-16613. [DOI] [PubMed] [Google Scholar]
- 27.Malakhov, M. P., O. A. Malakhova, K. I. Kim, K. J. Ritchie, and D. E. Zhang. 2002. UBP43 (USP18) specifically removes ISG15 from conjugated proteins. J. Biol. Chem. 2779976-9981. [DOI] [PubMed] [Google Scholar]
- 28.Meylan, E., J. Curran, K. Hofmann, D. Moradpour, M. Binder, R. Bartenschlager, and J. Tschopp. 2005. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature 4371167-1172. [DOI] [PubMed] [Google Scholar]
- 29.Mibayashi, M., L. Martinez-Sobrido, Y. M. Loo, W. B. Cardenas, M. Gale, Jr., and A. Garcia-Sastre. 2007. Inhibition of retinoic acid-inducible gene I-mediated induction of beta interferon by the NS1 protein of influenza A virus. J. Virol. 81514-524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Osiak, A., O. Utermohlen, S. Niendorf, I. Horak, and K. P. Knobeloch. 2005. ISG15, an interferon-stimulated ubiquitin-like protein, is not essential for STAT1 signaling and responses against vesicular stomatitis and lymphocytic choriomeningitis virus. Mol. Cell. Biol. 256338-6345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pichlmair, A., O. Schulz, C. P. Tan, T. I. Naslund, P. Liljestrom, F. Weber, and C. Reis e Sousa. 2006. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science 314997-1001. [DOI] [PubMed] [Google Scholar]
- 32.Rothenfusser, S., N. Goutagny, G. DiPerna, M. Gong, B. G. Monks, A. Schoenemeyer, M. Yamamoto, S. Akira, and K. A. Fitzgerald. 2005. The RNA helicase Lgp2 inhibits TLR-independent sensing of viral replication by retinoic acid-inducible gene-I. J. Immunol. 1755260-5268. [DOI] [PubMed] [Google Scholar]
- 33.Saito, T., R. Hirai, Y. M. Loo, D. Owen, C. L. Johnson, S. C. Sinha, S. Akira, T. Fujita, and M. Gale, Jr. 2007. Regulation of innate antiviral defenses through a shared repressor domain in RIG-I and LGP2. Proc. Natl. Acad. Sci. USA 104582-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schroder, M., and A. G. Bowie. 2005. TLR3 in antiviral immunity: key player or bystander? Trends Immunol. 26462-468. [DOI] [PubMed] [Google Scholar]
- 35.Seth, R. B., L. Sun, C. K. Ea, and Z. J. Chen. 2005. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell 122669-682. [DOI] [PubMed] [Google Scholar]
- 36.Soares, P. B., C. Demetrio, L. Sanfilippo, A. H. Kawanoto, L. Brentano, and E. L. Durigon. 2005. Standardization of a duplex RT-PCR for the detection of influenza A and Newcastle disease viruses in migratory birds. J. Virol. Methods 123125-130. [DOI] [PubMed] [Google Scholar]
- 37.Stark, G. R., I. M. Kerr, B. R. Williams, R. H. Silverman, and R. D. Schreiber. 1998. How cells respond to interferons. Annu. Rev. Biochem. 67227-264. [DOI] [PubMed] [Google Scholar]
- 38.Sumpter, R., Jr., Y. M. Loo, E. Foy, K. Li, M. Yoneyama, T. Fujita, S. M. Lemon, and M. Gale, Jr. 2005. Regulating intracellular antiviral defense and permissiveness to hepatitis C virus RNA replication through a cellular RNA helicase, RIG-I. J. Virol. 792689-2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wong, J. J., Y. F. Pung, N. S. Sze, and K. C. Chin. 2006. HERC5 is an IFN-induced HECT-type E3 protein ligase that mediates type I IFN-induced ISGylation of protein targets. Proc. Natl. Acad. Sci. USA 10310735-10740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xu, L. G., Y. Y. Wang, K. J. Han, L. Y. Li, Z. Zhai, and H. B. Shu. 2005. VISA is an adapter protein required for virus-triggered IFN-beta signaling. Mol. Cell 19727-740. [DOI] [PubMed] [Google Scholar]
- 41.Yoneyama, M., M. Kikuchi, K. Matsumoto, T. Imaizumi, M. Miyagishi, K. Taira, E. Foy, Y. M. Loo, M. Gale, Jr., S. Akira, S. Yonehara, A. Kato, and T. Fujita. 2005. Shared and unique functions of the DExD/H-box helicases RIG-I, MDA5, and LGP2 in antiviral innate immunity. J. Immunol. 1752851-2858. [DOI] [PubMed] [Google Scholar]
- 42.Yoneyama, M., M. Kikuchi, T. Natsukawa, N. Shinobu, T. Imaizumi, M. Miyagishi, K. Taira, S. Akira, and T. Fujita. 2004. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat. Immunol. 5730-737. [DOI] [PubMed] [Google Scholar]
- 43.Zhao, C., C. Denison, J. M. Huibregtse, S. Gygi, and R. M. Krug. 2005. Human ISG15 conjugation targets both IFN-induced and constitutively expressed proteins functioning in diverse cellular pathways. Proc. Natl. Acad. Sci. USA 10210200-10205. [DOI] [PMC free article] [PubMed] [Google Scholar]