Abstract
Human immunodeficiency virus type 1 (HIV-1) and other retroviruses harbor short peptide motifs in Gag that promote the release of infectious virions. These motifs, known as late assembly (L) domains, recruit a cellular budding machinery that is required for the formation of multivesicular bodies (MVBs). The primary L domain of HIV-1 maps to a PTAP motif in the p6 region of Gag and engages the MVB pathway by binding to Tsg101. Additionally, HIV-1 p6 harbors an auxiliary L domain that binds to the V domain of ALIX, another component of the MVB pathway. We now show that ALIX also binds to the nucleocapsid (NC) domain of HIV-1 Gag and that ALIX and its isolated Bro1 domain can be specifically packaged into viral particles via NC. The interaction with ALIX depended on the zinc fingers of NC, which mediate the specific packaging of genomic viral RNA, but was not disrupted by nuclease treatment. We also observed that HIV-1 zinc finger mutants were defective for particle production and exhibited a similar defect in Gag processing as a PTAP deletion mutant. The effects of the zinc finger and PTAP mutations were not additive, suggesting a functional relationship between NC and p6. However, in contrast to the PTAP deletion mutant, the double mutants could not be rescued by overexpressing ALIX, further supporting the notion that NC plays a role in virus release.
Retroviral Gag polyproteins harbor short peptide motifs that promote the detachment of assembled virions from the cell surface and from each other (16, 22, 24, 41, 49, 50, 52). These motifs, which are known as late assembly (L) domains, often remain functional even if moved to a different position within Gag and can be exchanged among unrelated viruses (39, 51). In the case of human immunodeficiency virus type 1 (HIV-1), the primary L domain consists of a conserved P(T/S)AP motif near the beginning of the C-terminal p6 domain of the Gag polyprotein (22, 24). The PTAP motif serves as a binding site for the host protein Tsg101, which is required for the release of infectious virions (21, 31, 47).
Tsg101 is a component of the endosomal sorting complex ESCRT-I, which functions in the sorting of ubiquitinated cargo into small vesicles that bud into the lumen of multivesicular bodies (MVBs) (3, 23, 27). This invagination event is topologically related to retrovirus budding and ultimately serves to deliver transmembrane proteins to lysosomal compartments for degradation. The components of this cellular budding pathway were initially identified in Saccharomyces cerevisiae, where a protein network that includes at least 18 different vacuolar protein-sorting (Vps) proteins is required for the formation of MVBs (3). The absence of any of these gene products causes the formation of an abnormal endosomal structure called the class E compartment (26). The MVB sorting pathway is conserved throughout eukaryotic evolution, and at least one homolog for each of the yeast class E Vps proteins exists in humans (48). Most of the class E Vps proteins participate in the formation of ESCRT-I or of two other high-molecular-weight complexes called ESCRT-II and ESCRT-III, which function downstream of ESCRT-I (25). MVB sorting also depends on the activity of the AAA-type ATPase VPS4, another class E Vps protein that recycles the ESCRT machinery (4). Dominant-negative versions of various ESCRT-III subunits and of VPS4 potently inhibit the functions of all known types of viral L domains (21, 30, 43, 44, 48, 53), indicating that all act through components of the MVB sorting pathway.
In addition to the PTAP motif, the p6 domain of HIV-1 Gag contains a LYPxnL motif near its C terminus that binds to ALIX/AIP1 (44), a homolog of the yeast class E Vps protein Bro1 that functions in endosomal trafficking (36). Like Tsg101, ALIX promotes viral egress, which requires the separation of the viral envelope from the plasma membrane, and is also involved in membrane fission events that are necessary for the completion of cytokinesis (10). ALIX has a modular organization, with a banana-shaped N-terminal Bro1 domain, a central domain that is shaped like a V and has therefore been termed the V domain, and a C-terminal proline-rich domain (PRD) that is thought to be unstructured (20, 29). ALIX interacts with the ESCRT-III component CHMP4 through its Bro1 domain (36), with LYPxnL-type viral L domains through its V domain (11, 33), and with Tsg101 and other endocytic proteins through its PRD (36).
The few lentiviruses that are known to lack a PTAP motif all harbor a high-affinity binding site for ALIX in the C-terminal domain of Gag (6, 44). One of these is equine infectious anemia virus, which depends on ALIX for efficient virus release (20, 30, 41, 44, 48). However, in the case of HIV-1, the ALIX binding site in p6 has only an auxiliary role in virus release that becomes more evident in a minimal Gag context (20, 44). Nevertheless, exogenous ALIX can potently rescue the profound budding defect of HIV-1 PTAP mutants, provided that the ALIX binding site in p6 remains intact (20, 46).
We previously reported that the PTAP-type L domain in HIV-1 p6 does not function autonomously but depends on other Gag regions in order to function (45). Our results suggested that the PTAP motif needs to cooperate either with another determinant in p6, which is now known to bind ALIX, or with the nucleocapsid (NC)-p1 region, which immediately precedes the p6 domain in the context of the Gag precursor (45). The NC domain contains two highly conserved CCHC motifs that coordinate zinc and are required for the selective packaging of genomic viral RNA into progeny virions (5). NC also possesses a nonspecific nucleic acid binding activity that depends on its basic residues and is crucial for Gag polymerization and assembly (13). In contrast, disruption of the two CCHC motifs had no significant effect on Gag-Gag interactions (13).
We now show that NC associates with the Bro1 domain of ALIX through its two CCHC motifs. We also show that the ability of ALIX to rescue a Tsg101 binding site mutant depends on intact CCHC motifs in NC. Disrupting both CCHC motifs led to defects in particle production and to an accumulation of the capsid (CA)-p2 and p41 Gag cleavage intermediates that is typically seen with L domain mutants (21, 22). However, the effects of disrupting the CCHC motifs and the PTAP motif in p6 were not additive, which supports the hypothesis that these motifs function in the same pathway during virus production.
MATERIALS AND METHODS
Proviral constructs.
HXBH10, the parental proviral plasmid used in this study, is a vpu-positive version of the infectious HXB2 proviral clone of HIV-1. HXBH10-gag− is unable to express Gag due to premature termination codons in the gag gene (19). The protease (PR)-negative variant HXBH10-PR−, which was used to express Pr55gag, and the HXBH10-based chimeric Gag constructs ZWT and ZWT-p6 have been described elsewhere (1). ZWT encodes an HIV-1 Gag precursor that has the NC-p1-p6 region precisely replaced by the GCN4 leucine zipper domain (1). ZWT-p6 has the HIV-1 p6 coding sequence directly fused to the 3′ end of the GCN4 sequence (1). Pr55(Y36s) and ZWT-p6(Y36s) are variants of HXBH10-PR− and ZWT-p6, respectively, that harbor the previously described Y36s mutation (22), which introduces a premature termination codon in place of Tyr-36 of p6 without altering the pol frame. The codons for NC-p1 residues 56 to 71, 52 to 71, 36 to 71, and 15 to 71 were precisely deleted from HXBH10-Y36s without affecting any other Gag coding sequences. This was achieved by introducing silent mutations into the first two p6 codons to generate a PstI restriction site. All of these deletions remove the frameshift signal required for the expression of PR and thus prevent Gag processing. In contrast, the Δ15-39, Δ15-34/C36S, Δ15-28, Δ30-31, C28,49S, and K33,34A mutations keep the frameshift signal intact and were therefore introduced into the PR-negative version of HXBH10-Y36s to prevent Gag processing. The Δ15-39, Δ15-34 C36S, and Δ15-28 mutants have two foreign codons specifying Leu-Gln inserted in place of the deleted NC codons. The C28,49S and Δ15-39 mutations were also introduced into the PR-positive proviruses HXBH10 and ΔPTAPP, a variant of HXBH10 with an in-frame deletion of codons 7 through 11 of p6 (1).
Expression vectors.
The pBJ5-based mammalian expression vectors for ALIX-HA and HA-ALIX have been described previously (44). The F676D mutant of ALIX-HA was made using the QuikChange mutagenesis strategy (Stratagene). The expression vector for HA-ALIXBro1 was obtained by inserting a PCR product encoding ALIX residues 1 to 362 with an N-terminal hemagglutinin (HA) tag between the NotI and EcoRI sites of pBJ5. The coding sequence for APOBEC3G with a C-terminal FLAG tag was amplified from BC024268 (Open Biosystems) and cloned between the HindIII and XhoI sites of pcDNA3.1(+) (Invitrogen). In all mammalian expression vectors, the start codon is preceded by a Kozak consensus sequence for efficient translation initiation.
Analysis of virus-like particles (VLP).
293T cells (3.5 × 106) or HeLa cells (8 × 105) were seeded into T80 flasks and cotransfected 24 h later with HIV-1 proviral DNA and ALIX expression vectors as indicated using a calcium phosphate precipitation technique. The total amount of transfected DNA was kept constant with carrier DNA (pBluescript). Twenty-four h (293T) or 48 h (HeLa) posttransfection, the cells were lysed in radioimmunoprecipitation assay buffer (140 mM NaCl, 8 mM Na2HPO4, 2 mM NaH2PO4, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.05% sodium dodecyl sulfate [SDS]), and the culture supernatants were clarified by low-speed centrifugation and passaged through 0.45-μm filters. Virions released into the medium were then pelleted through 20% sucrose cushions. Pelletable material and the cell lysates were analyzed by SDS-polyacrylamide gel electrophoresis (PAGE) and Western blotting as described elsewhere (1), using a rabbit anti-HIV CA serum (Advanced Biotechnologies) or the anti-HIV CA antibody 183-H12-5C (12) to detect Gag proteins. HA-tagged ALIX was detected with the anti-HA antibody HA.11 (Covance).
GST pull-down assay.
PCR fragments encoding the full-length NC (residues 1 to 55) and NC-p1 (residues 1 to 71) domains of HIV-1HXB2 Gag were inserted in frame between the EcoRI and XhoI sites of pGEX-4T-1 (GE Healthcare). Glutathione S-transferase (GST) fusion proteins were expressed in strain BL21 and immobilized on glutathione-Sepharose beads (GE Healthcare). The beads were then incubated for 3 h at 4°C with hypotonic lysates of 293T cells transiently expressing epitope-tagged ALIX or APOBEC3G, followed by extensive washing in phosphate-buffered saline. Where indicated, the beads were incubated for another 30 min at 37°C in the presence or absence of 75 U (0.75 U/μl) benzonase nuclease (EMD Biosciences) in benzonase buffer (1.2 mM MgCl2, 50 mM Tris-HCl [pH 8.0]). Bound proteins were eluted by boiling in SDS-PAGE sample buffer and resolved by SDS-PAGE. Epitope-tagged proteins were detected by Western blotting with anti-HA or anti-FLAG M2 antibody, and GST fusion proteins were visualized with colloidal Coomassie brilliant blue G-250.
RESULTS
NC-dependent incorporation of ALIX into HIV-1 virions.
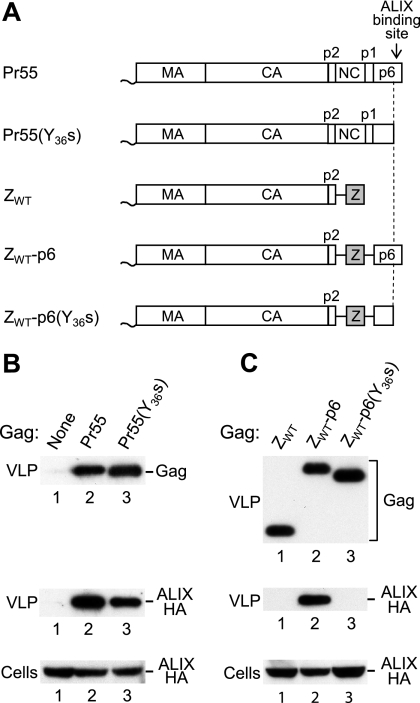
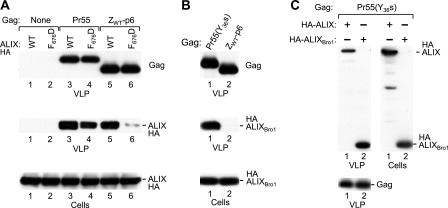
We previously examined the incorporation of ALIX into HIV-1 particles using the chimeric Gag constructs ZWT and ZWT-p6 (1, 44), which have NC-p1 replaced by the GCN4 leucine zipper domain (Fig. 1A). Because the GCN4 zipper domain rescues viral particle production by HIV-1 Gag in the absence of the NC-p1-p6 region (1), this approach allowed us to show that p6 mediates the specific incorporation of ALIX (44). Consistent with this finding, we and others have shown that both full-length ALIX and the isolated ALIX V domain bind to an LYPxnL motif near the C terminus of HIV-1 p6 (20, 29, 33, 44, 48). We therefore expected that the incorporation of ALIX into viral particles formed by authentic HIV-1 Gag would be abolished by the Y36s mutation (22), which converts the codon for Y36 of p6 into a premature termination codon and prevents the synthesis of the LYPxnL motif. To test this notion, we coexpressed HA-tagged ALIX with HIV-1 proviral constructs expressing either no Gag, the wild-type (WT) Pr55 Gag polyprotein, or Pr55(Y36s) (Fig. 1A and B). As expected, HA-ALIX was readily detected in virions produced by WT HIV-1 Gag, and the incorporation of HA-ALIX was specific, because its appearance in the particulate fraction depended on the expression of Gag (Fig. 1B, lanes 1 and 2). However, we were surprised to find that the Y36s mutation reduced the uptake of HA-ALIX into viral particles only about twofold (Fig. 1B, lane 3), indicating that in an otherwise-WT HIV-1 Gag context the LYPxnL motif contributes to the recruitment of ALIX but is not essential.
FIG. 1.
NC-mediated incorporation of ALIX into VLP. (A) Schematic illustration of the domain organizations of the HIV-1 Gag precursor (Pr55) and of mutant Gag molecules. A wavy line at the N terminus indicates the presence of a myristylation signal, and a gray box indicates the presence of a GCN4 zipper (Z) domain in place of NC-p1. (B) The ALIX binding site in p6 is not essential for the incorporation of ALIX into VLP. 293T cells were transfected with HIV-1 proviral DNA (5 μg) expressing no Gag, WT Gag, or C-terminally truncated Gag, along with 5 μg of a vector expressing ALIX-HA. VLP pellets and the cell lysates were analyzed by Western blotting to detect Gag and ALIX-HA as indicated. (C) The uptake of ALIX in the absence of the binding site in p6 depends on NC-p1. 293T cells were transfected with HIV-1 proviral DNAs (5 μg) expressing Gag molecules with a leucine zipper in place of NC-p1, along with 5 μg of a vector expressing ALIX-HA.
To determine whether the uptake of ALIX in the absence of the LYPxnL motif was mediated by the truncated p6 domain of the Y36s mutant, we introduced the Y36s mutation into ZWT-p6 (Fig. 1A). As previously reported (1, 44), the ZWT chimeric Gag molecule efficiently forms VLP that fail to incorporate coexpressed HA-ALIX (Fig. 1C, lane 1). In contrast, HA-ALIX was readily detectable in VLP produced by ZWT-p6 (Fig. 1C, lane 2), again consistent with previous results (44). Interestingly, in the context of ZWT-p6, the Y36s mutation abolished the incorporation of HA-ALIX into VLP (Fig. 1C, lane 3). Of note, the Pr55(Y36s) and ZWT-p6(Y36s) proviral constructs are precisely identical except for the region encoding NC-p1. We thus conclude that the ability of Pr55(Y36s) but not of ZWT-p6(Y36s) Gag to direct the incorporation of ALIX is dependent on NC-p1.
Role of NC zinc fingers in incorporation of ALIX.
The NC domain of HIV-1 Gag harbors two copies of a CCHC-type zinc finger motif that are involved in the specific encapsidation of genomic viral RNA into nascent viral particles. NC also exhibits a nonspecific RNA binding activity that depends on clusters of basic residues and is thought to be critical for virus assembly (8, 13). However, NC deletion mutants are not drastically defective for viral particle production as long as the N-terminal basic NC region proximal to the first CCHC motif is retained (55). Furthermore, it has been shown that particle production by a mutant that lacks nearly all of NC can be rescued by a second site mutation that inactivates PR (38). To maintain viral particle production, we therefore used a PR-deficient provirus as the parental construct to generate a series of mutants with in-frame deletions in the NC-p1 coding region. Furthermore, all NC-p1 deletions were combined with the Y36s mutation to eliminate the ALIX binding site in the p6 domain of Gag (Fig. 2A).
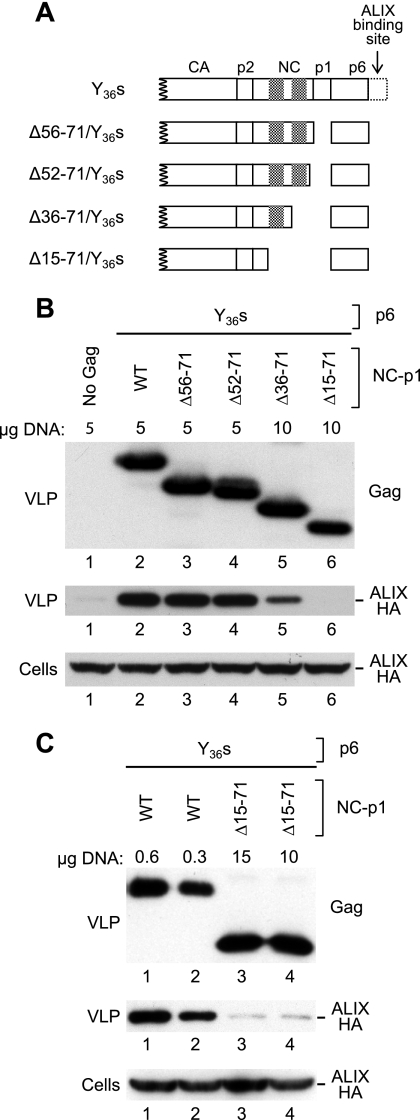
FIG. 2.
Effects of deletions in NC-p1 on the uptake of ALIX into VLP. (A) Schematic illustration of the Gag mutants examined. Cross-hatched boxes indicate positions of the zinc fingers in NC. (B) p6-independent incorporation of ALIX depends on NC but not on p1. 293T cells were transfected with the indicated amounts of mutant HIV-1 proviral DNAs together with 3 μg of a vector expressing ALIX-HA. VLP pellets and the cell lysates were analyzed by Western blotting to detect Gag and ALIX-HA as indicated. (C) Effects of a deletion that removes both zinc fingers of NC together with p1.
Since Gag multimerization is concentration dependent (40), we transfected relatively large amounts of these proviral constructs into 293T cells together with a vector expressing ALIX-HA, in an effort to override potential Gag-Gag interaction defects. As a control for Gag-independent release of ALIX in cellular vesicles, the ALIX expression vector was cotransfected with a provirus that lacked the ability to express Gag due to the presence of premature termination codons in the gag gene. As shown in Fig. 2B, deleting the region between the second CCHC motif and p6 had no effect on particle production or on the incorporation of ALIX-HA (lanes 3 and 4). In contrast, a deletion which additionally removed the second CCHC motif (Δ36-71) reduced the incorporation of ALIX-HA at least threefold (lane 5). Finally, ALIX-HA was no longer detectable in the particulate fraction if the first CCHC motif was also removed (Δ15-71) (lane 6).
In the experiment shown in Fig. 2B, the Δ15-71/Y36s mutant at 10 μg produced somewhat fewer VLP than the parental proviral construct at 5 μg. Therefore, the parental construct was titrated down to obtain equivalent levels of particle production. We found that VLP production by the Δ15-71/Y36s mutant transfected at 10 or 15 μg matched or exceeded that by the parental Y36s provirus only when less than 1 μg of the latter construct was used (Fig. 2C and data not shown). Importantly, this experiment confirmed that Δ15-71/Y36s VLP incorporate significantly less ALIX than Y36s VLP (Fig. 2C).
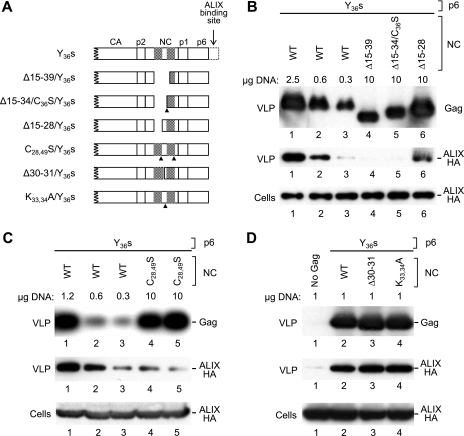
Next, we selectively targeted the CCHC motifs in NC to determine their role in the LYPxnL-independent uptake of ALIX. All NC mutants depicted in Fig. 3A harbored the Y36s mutation to prevent the incorporation of ALIX via p6 and a mutation that inactivated PR to suppress potential defects in VLP production in the absence of NC sequences. As shown in Fig. 3B, a deletion that removed the first CCHC motif and a portion of the second (Δ15-39) essentially abolished the ability of the Y36s mutant to incorporate ALIX (compare lanes 2 and 4). A similar effect was observed when a deletion that removed the first CCHC motif and a few flanking residues (Δ15-34) was combined with a point mutation that changed the second CCHC motif to SCHC (lane 5). However, the ability to incorporate ALIX was partially preserved if only the first CCHC motif was deleted and the second was kept intact (Δ15-28) (lane 6). We also note that the latter mutant produced more VLP than those which retained no intact CCHC motif (Fig. 3B). To specifically examine the role of zinc-coordinating residues in the incorporation of ALIX, we changed both CCHC motifs in NC to CCHS (Fig. 3A). As shown in Fig. 3C, VLP produced by the resulting C28,49S/Y36s mutant reproducibly incorporated less ALIX than those produced by the parental Y36s construct (compare lanes 1 and 2 to lanes 4 and 5). In contrast, mutations that targeted the region between the two CCHC motifs had no effect on the LYPxnL-independent incorporation of ALIX (Fig. 3D). Together, these results indicate that the LYPxnL-independent uptake of ALIX is mediated by the zinc fingers in NC.
FIG. 3.
Role of NC zinc fingers in uptake of ALIX into VLP. (A) Schematic illustration of the Gag mutants examined. Cross-hatched boxes indicate the positions of the zinc fingers in NC, and arrowheads indicate the positions of residues that were substituted. (B) Effects of deletions that specifically target zinc fingers. 293T cells were transfected with the indicated amounts of mutant HIV-1 proviral DNAs, together with 5 μg of a vector expressing ALIX-HA. VLP pellets and the cell lysates were analyzed by Western blotting to detect Gag and ALIX-HA as indicated. (C) Effect of disrupting both zinc fingers through point mutations. The C28,49S/Y36s mutant was examined in duplicate to verify reproducibility. (D) Effects of mutations in the region between the two zinc fingers.
ALIX physically associates with HIV-1 NC in vitro.
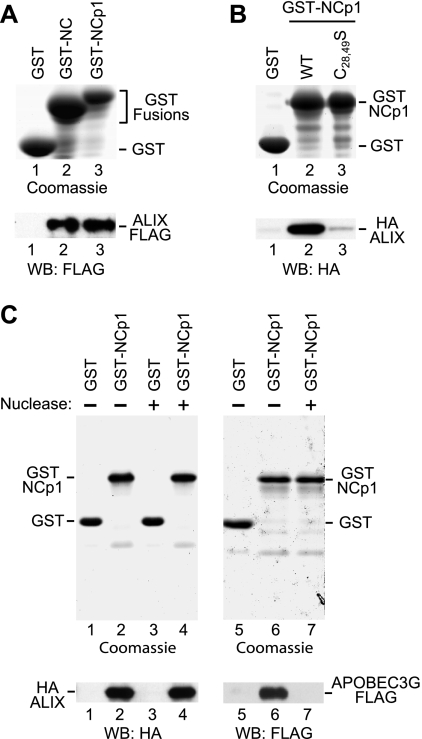
The packaging of ALIX into VLP in the absence of the LYPxnL motif in p6 suggested that ALIX may also bind to other Gag regions. To examine whether ALIX interacts with the NC-p1 region of HIV-1 Gag, GST-NC or GST-NCp1 was bound to glutathione-Sepharose beads and incubated with extracts from 293T cells expressing FLAG- or HA-tagged ALIX. Epitope-tagged ALIX was readily detected in association with both GST-NC and GST-NCp1, but not with GST alone (Fig. 4A and B). We also observed that the association of ALIX with GST-NCp1 was considerably reduced by the C28,49S mutation (Fig. 4B), consistent with the effect of this mutation on the packaging of ALIX into VLP (Fig. 3C).
FIG. 4.
In vitro interaction between ALIX and NC or NC-p1. (A) ALIX binds to GST-NC and GST-NC-p1. Glutathione-Sepharose beads decorated with bacterially expressed GST or GST fusion proteins were incubated with lysates from 293T cells expressing full-length ALIX with a C-terminal FLAG tag. GST proteins bound to the beads were detected by SDS-PAGE and Coomassie staining, and captured ALIX was detected by Western blotting with anti-FLAG. (B) A GST pull-down assay performed as for panel A, showing that the interaction between ALIX and GST-NC-p1 is impaired by the C28,49S mutation, which disrupts both CCHC motifs in NC. (C) ALIX remains bound to GST-NCp1 in the presence of nuclease, whereas the interaction between GST-NCp1 and APOBEC3G is disrupted. GST pull-down assays were performed as for panel A, followed by incubation of the beads with or without benzonase nuclease. Proteins that remained bound to the glutathione-Sepharose beads were then detected as indicated.
We previously showed that the C28,49S double mutation, which disrupts both CCHC motifs in NC, reduces the packaging of genomic viral RNA into HIV-1 virions more than 20-fold (18). We also showed that even the C28S change by itself reduced the binding of viral RNA to HIV-1 Gag 10-fold in a Northwestern blot assay (18). Since the C28,49S mutation also significantly reduced the interaction between NC-p1 and ALIX, we considered the possibility that the in vitro interaction depended on the presence of nucleic acid. To examine this possibility, HA-ALIX was pulled down with GST-NC-p1 bound to glutathione-Sepharose, followed by incubation of the beads with benzonase, a promiscuous nuclease capable of degrading all forms of RNA and DNA. This treatment had no effect on the association of HA-ALIX with GST-NC-p1 (Fig. 4C, lanes 1 to 4). As a control, we examined whether the interaction between NC-p1 and APOPEC3G was affected by benzonase. It was shown previously that the ability of Gag-GST fusion proteins to bind APOBEC3G is inhibited by RNase A (54). Similarly, we observed that benzonase completely disrupted the interaction between GST-NCp1 and APOBEC3G-FLAG under the same experimental conditions in which it had no effect on the interaction with ALIX (Fig. 4C, lanes 5 to 7). Taken together, these results indicate that the interaction between NC or NC-p1 and ALIX is not simply a consequence of unspecific bridging by nucleic acids.
NC engages the Bro1 domain of ALIX.
ALIX possesses an N-terminal Bro1 domain and a central V domain that is named for its V-like shape (20, 29). The V domain is required for the binding of ALIX to HIV-1 p6 (44), and recent structural studies revealed that Phe-676 in a conserved hydrophobic pocket of the V domain plays a central role in ALIX-L domain interactions (20, 29). Mutating Phe-676 to Asp abolished the ability of the isolated V domain to inhibit HIV-1 particle release (29) and the ability of full-length ALIX to rescue viral particle production by ΔPTAP HIV-1 (20).
Consistent with these observations, the F676D mutation nearly eliminated the packaging of ALIX into VLP formed by ZWT-p6 (Fig. 5A, compare lanes 5 and 6), which depends on the LYPxnL motif in p6 (Fig. 1C). In contrast, the same mutation had only a moderate effect on the incorporation of ALIX into particles formed by the WT Pr55 Gag polyprotein (Fig. 5A, lanes 3 and 4). Furthermore, this effect was comparable to that of removing the LYPxnL motif in p6 on the incorporation of WT ALIX (Fig. 1B). Together, these results suggested that the packaging of ALIX via NC does not depend on the presence of an intact V domain.
FIG. 5.
NC engages the Bro1 domain of ALIX. (A) Effect of disrupting the ligand binding pocket in the V domain on incorporation of ALIX into VLP. 293T cells were transfected with HIV-1 proviral DNAs (7.5 μg) expressing no Gag, WT Gag, or Gag with a leucine zipper in place of NC-p1, along with 5 μg of a vector expressing WT or mutant ALIX-HA. VLP pellets and the cell lysates were analyzed by Western blotting to detect Gag and ALIX-HA as indicated. (B) NC-dependent incorporation of the isolated Bro1 domain of ALIX. 293T cells were transfected with HIV-1 proviral DNAs (7.5 μg) expressing the indicated Gag molecules, along with 5 μg of a vector expressing HA-ALIXBro1. (C) NC-dependent incorporation of full-length HA-ALIX versus that of HA-ALIXBro1. 293T cells were transfected with HIV-1 proviral DNA (7.5 μg) expressing the Pr55(Y36s) Gag precursor, along with 5 μg of vector expressing HA-ALIX or HA-ALIXBro1.
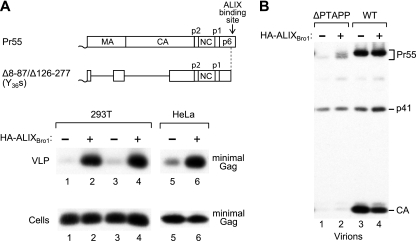
We therefore asked whether the isolated Bro1 domain of ALIX can be packaged into VLP. HA-ALIXBro1 was readily detected in VLP produced by the Y36s mutant (Fig. 5B, lane 1), which expresses a Gag polyprotein that lacks the ALIX binding site in p6 but retains the WT NC domain (Fig. 1A). Indeed, HA-ALIXBro1 was incorporated into Y36s mutant VLP at least as well as full-length ALIX (Fig. 5C). In marked contrast, HA-ALIXBro1 was not at all packaged into VLP formed by ZWT-p6 (Fig. 5B, lane 2), which lacks NC but possesses an intact p6 domain (Fig. 1A). This result was not attributable to different levels of VLP production or to differences in the expression of HA-ALIXBro1 (Fig. 5B). We thus conclude that the packaging of HA-ALIXBro1 by the Y36s mutant is specific, and we infer that NC interacts with the Bro1 domain of ALIX.
We previously described an HIV-1 mutant which encodes a minimal Gag protein called Δ8-87/Δ126-277 that lacks the globular domain of MA and the N-terminal domain of CA but produces WT levels of VLP (7). L domain function in this minimal Gag context was highly dependent on the presence of an intact ALIX binding site in p6 (44). Moreover, VLP production by ALIX binding site mutants could be rescued by overexpressing ALIX (44). This activity of ALIX depended on the presence of the PTAPP motif in p6, but not on the C-terminal PRD of ALIX (44), which is required for the rescue of ΔPTAPP HIV-1 by ALIX (20, 46). Surprisingly, ALIXΔPRD markedly increased VLP production even by a variant of Δ8-87/Δ126-277 that harbors the Y36s mutation and thus lacks the entire ALIX binding site in p6 (44). We therefore examined whether the V domain of ALIX, which binds to p6 (20, 29), is required for the rescue of VLP production in this system. In both 293T and HeLa cells, HA-ALIXBro1 increased VLP production by the Y36s version of the Δ8-87/Δ126-277 minimal Gag construct more than 10-fold without affecting Gag expression levels (Fig. 6A). As expected, HA-ALIXBro1 did not increase the release of ΔPTAPP HIV-1 (Fig. 6B, lanes 1 and 2). Furthermore, HA-ALIXBro1 did not augment virion production by WT HIV-1 (Fig. 6B, lanes 3 and 4). Since the Δ8-87/Δ126-277/Y36s minimal Gag construct retains the NC domain, these results raise the possibility that ALIX and the isolated Bro1 domain can act through NC.
FIG. 6.
Effect of HA-ALIXBro1 on particle production. (A) Rescue of VLP production by a minimal Gag construct that lacks the ALIX binding site in p6. 293T cells (lanes 1 to 4) or HeLa cells (lanes 5,6) were transfected with 1 μg (lanes 1,2), 2 μg (lanes 3,4), or 7.5 μg (lanes 5,6) of proviral DNA expressing the Y36s version of the Δ8-87/Δ126-277 minimal Gag molecule (44), along with 2 μg (lanes 1 and 2) or 5 μg (lanes 3 to 6) of empty pBJ5 or the HA-ALIXBro1 expression vector. VLP production and Gag expression levels were examined by Western blotting with anti-CA antibody 183-H12-5C. The Δ8-87/Δ126-277 minimal Gag molecule lacks the globular domain of MA and the N-terminal domain of CA, and the Y36s truncation removes the ALIX binding site in p6. (B) HA-ALIXBro1 does not increase virion production by ΔPTAPP or WT HIV-1. HeLa cells were transfected with 7.5 μg of ΔPTAPP or WT HXBH10, along with 5 μg of empty pBJ5 or the HA-ALIXBro1 expression vector, and virion production was examined as above.
NC is required for the function of ALIX in HIV-1 budding.
We and Sundquist and coworkers recently reported that the release defects of HIV-1 L domain mutants can be corrected by increasing the cellular expression levels of ALIX (20, 46). For instance, exogenous ALIX completely rescued particle production by a mutant termed ΔPTAPP, which lacks the Tsg101 binding site in p6 (46). In this ΔPTAPP rescue assay, the activity of ALIX was dependent on the presence of an intact ALIX binding site near the C terminus of p6 (20, 46).
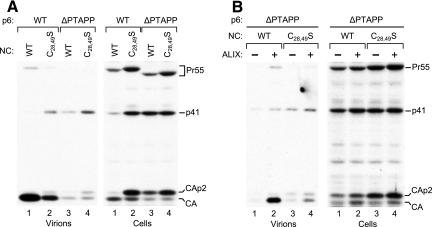
To determine whether NC also plays a role in the rescue by ALIX, we generated a variant of the ΔPTAPP mutant that harbors the C28,49S mutation in NC, which interferes with the interaction between NC and ALIX (Fig. 3 and 4). Of note, in a previous study the C28,49S mutation had no effect on Gag-Gag interactions (13). Nevertheless, the C28,49S mutation by itself reduced viral particle production in 293T cells, although this defect was relatively mild compared to that caused by the ΔPTAPP mutation (Fig. 7A). The C28,49S mutation also affected the pattern of cell-associated Gag proteins, which was altered in a manner similar to that seen with the ΔPTAPP mutant. Specifically, both mutations altered the ratio of mature CA versus the CA-p2 processing intermediate and caused an accumulation of the p41 cleavage intermediate, as is typically seen if a late assembly step is defective (21, 22). Interestingly, the C28,49S/ΔPTAPP double mutant was no more defective for particle production than the original ΔPTAPP mutant and produced a pattern of cell-associated Gag products similar to that seen with the two parental mutants (Fig. 7A). When an expression vector for ALIX was cotransfected, we observed a profound increase in viral particle production by the ΔPTAPP mutant (Fig. 7B), as previously described (46). In contrast, particle production by the C28,49S/ΔPTAPP double mutant was only marginally improved (Fig. 7B).
FIG. 7.
Effects of disrupting the two NC zinc fingers on particle production and on the ability of ALIX to rescue virus release. (A) Effects of the C28,49S mutation on particle production and Gag processing in the presence or absence of the PTAPP L domain. 293T cells were transfected with 1 μg of PR-positive HIV-1 proviral DNAs, and virion production and the pattern of cell-associated Gag products were compared by Western blotting with anti-CA serum. (B) Effect of the C28,49S mutation on the ability of ALIX to rescue ΔPTAPP HIV-1. 293T cells were transfected with 1 μg of PR-positive HIV-1 proviral DNAs and either empty pBJ5 or a version expressing HA-ALIX (2 μg).
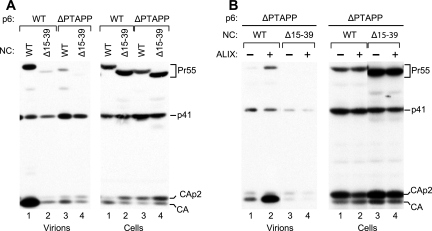
We also examined the effect of the Δ15-39 deletion in the ΔPTAPP rescue assay, since the Δ15-39 deletion appeared to completely prevent the recruitment of ALIX via NC (Fig. 3B). In the presence of WT PR, the Δ15-39 deletion in NC caused as profound a defect in viral particle production as the ΔPTAPP L domain mutation and also had a comparable effect on the processing of cell-associated Gag (Fig. 8A, lanes 2 and 3). Remarkably, when the two mutations were combined, there again was no additive effect on particle production or on Gag processing (Fig. 8A, lane 4). Moreover, particle production by the double mutant could not be rescued at all by exogenous ALIX (Fig. 8B). Taken together, these results indicate that the determinants in NC which mediate the interaction with ALIX also play a critical role in the function of ALIX in virus budding.
FIG. 8.
Effects of removing the NC zinc fingers. (A) The Δ15-39 deletion in NC, which removes all of the first zinc finger and part of the second, and the ΔPTAPP deletion in p6 have comparable effects on particle production and Gag processing, and these effects are not additive. (B) The Δ15-39/ΔPTAPP double mutant is not rescued by ALIX. These experiments were performed as described in the legend for Fig. 7.
DISCUSSION
Several studies have shown that ALIX functionally interacts with the p6 domain of HIV-1 Gag (20, 29, 33, 44, 46, 48). We now report that ALIX also interacts with the NC-p1 region, which precedes the p6 domain in the context of the unprocessed Gag polyprotein. As a consequence, the specific incorporation of ALIX into HIV-1 virions is only moderately affected by removing its binding site in p6. While we initially identified ALIX based on its p6-dependent incorporation into HIV-1 VLP, this result is explained by the fact that the Gag constructs used in our previous study had NC-p1 replaced by the GCN4 leucine zipper (44). Indeed, the present study confirms that the incorporation of ALIX into VLP becomes completely dependent on the LYPxnL motif in p6 if NC-p1 is replaced by a heterologous dimerization domain.
While the interaction with p6 is mediated by the V domain of ALIX (20, 29, 33), the association with NC was insensitive to a mutation in the ligand binding pocket of the V domain that disrupted binding to p6. Furthermore, the isolated Bro1 domain of ALIX was readily incorporated into viral particles in an NC-dependent manner, indicating that ALIX associates with NC through its N-terminal Bro1 domain. Thus, our results imply that ALIX can interact with HIV-1 Gag through both of its folded domains. The Bro1 domain of ALIX also interacts with ESCRT-III component CHMP4, and mutagenic analyses strongly suggest that this interaction is essential for the function of ALIX in virus budding (20, 46). For the interaction with NC to be functional, ALIX would thus have to be able to accommodate CHMP4 and NC simultaneously. In support of this notion, the overexpression of CHMP4B had no negative effect on the NC-dependent packaging of ALIX into HIV-1 virions (data not shown).
The two CCHC boxes in NC are thought to mediate sequence-specific interactions with the genomic viral RNA by binding to exposed guanosine bases in the packaging signal (2, 15). In the present study, we found that the CCHC boxes also mediate the packaging of ALIX into virions. In particular, substituting zinc-coordinating residues in both CCHC boxes simultaneously nearly abolished the LYPxnL-independent incorporation of ALIX and the in vitro interaction between ALIX and NC-p1. The same modification has been shown to cause a dramatic defect in genomic RNA packaging (18) and to drastically reduce the incorporation of the RNA binding protein Staufen into HIV-1 particles (32). While these parallels point to a relationship between the packaging of ALIX and of the viral genome, there is no evidence that ALIX binds nucleic acid. Moreover, we observed that the in vitro interaction between ALIX and NC-p1 was insensitive to nuclease treatment. This finding indicates that nucleic acid is not required to maintain the interaction and that specific protein-protein contacts are thus likely to be involved.
The interaction of NC with RNA plays an important role in Gag multimerization and in virus assembly, apparently because RNA is needed as a scaffold to promote Gag-Gag interactions (9, 13, 35, 55). However, it is thought that this function of NC involves its nonspecific RNA binding activity, which is primarily mediated by basic residues and does not require intact zinc fingers (8, 13, 14, 17, 37, 42, 55). Nevertheless, we have observed that the C28,49S mutation, which disrupts both zinc fingers and interferes with ALIX binding, reduces particle production in HeLa cells about 10-fold (18). This observation contrasts with the finding that the C28,49S mutation had no effect on Gag-Gag interactions in an in vitro assay and only moderately affected the nonspecific RNA binding activity of NC (13). In the present study, the C28,49S mutation also reduced particle production in 293T cells, albeit to a lesser extent than what we had previously observed in HeLa cells (18). However, a deletion which affected both zinc fingers but left the rest of NC intact reduced particle production severely. Interestingly, these zinc finger mutations had little or no effect on the residual ability of the ΔPTAPP mutant to produce viral particles, which suggests that the mutations in NC and in p6 affected the same step in virus assembly. Consistent with this possibility, the zinc finger mutations caused a similar Gag-processing defect as the ΔPTAPP mutant. These observations point to a role of HIV-1 NC in virus release, in addition to its well-documented function in virus assembly. Of note, a function of NC in virus release is also suggested by the late budding defect of certain NC mutants of the gammaretrovirus Moloney murine leukemia virus (34). Moreover, budding defects strikingly similar to those of L domain mutants have been observed upon deletion of either of the two CCHC motifs in the NC domain of an alpharetrovirus (28).
We previously presented evidence indicating that in order to promote efficient virus release, the conserved PTAPP L domain core of HIV-1 needs to cooperate either with the ALIX binding site in p6 or with NC-p1 (45). Interestingly, the release defect of PTAPP mutants can be fully rescued by overexpressing ALIX, but only if the ALIX binding site in p6 remains intact (20, 46). In the present study, we observed that this effect of ALIX also depended on the zinc fingers in NC, and specifically on determinants in NC that mediate the interaction with ALIX. One interpretation of these results is that the interactions of ALIX with NC and with p6 are both involved in its function in virus budding. However, it also remains possible that the role of ALIX in virus release strictly depends on the ability of NC to nucleate assembly and that zinc finger mutations affect this ability in a manner that has not been readily apparent in in vitro Gag multimerization assays (13). In either case, the involvement of NC in the activity of ALIX supports the notion that NC has a specific role in virus release.
Acknowledgments
We thank Suman R. Das for making the APOBEC3G-FLAG expression vector and Yoshiko Usami for making the expression vector for HA-ALIXBro1. The following reagent was obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH: HIV-1 p24 monoclonal antibody (183-H12-5C) from Bruce Chesebro and Kathy Wehrly.
This work was supported by National Institutes of Health grant AI29873.
Footnotes
Published ahead of print on 21 November 2007.
REFERENCES
- 1.Accola, M. A., B. Strack, and H. G. Gottlinger. 2000. Efficient particle production by minimal Gag constructs which retain the carboxy-terminal domain of human immunodeficiency virus type 1 capsid-p2 and a late assembly domain. J. Virol. 745395-5402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amarasinghe, G. K., R. N. De Guzman, R. B. Turner, K. J. Chancellor, Z. R. Wu, and M. F. Summers. 2000. NMR structure of the HIV-1 nucleocapsid protein bound to stem-loop SL2 of the ψ-RNA packaging signal. Implications for genome recognition. fsJ. Mol. Biol. 301491-511. [DOI] [PubMed] [Google Scholar]
- 3.Babst, M. 2005. A protein's final ESCRT. Traffic 62-9. [DOI] [PubMed] [Google Scholar]
- 4.Babst, M., B. Wendland, E. J. Estepa, and S. D. Emr. 1998. The Vps4p AAA ATPase regulates membrane association of a Vps protein complex required for normal endosome function. EMBO J. 172982-2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berkowitz, R., J. Fisher, and S. P. Goff. 1996. RNA packaging. Curr. Top. Microbiol. Immunol. 214177-218. [DOI] [PubMed] [Google Scholar]
- 6.Bibollet-Ruche, F., E. Bailes, F. Gao, X. Pourrut, K. L. Barlow, J. P. Clewley, J. M. Mwenda, D. K. Langat, G. K. Chege, H. M. McClure, E. Mpoudi-Ngole, E. Delaporte, M. Peeters, G. M. Shaw, P. M. Sharp, and B. H. Hahn. 2004. New simian immunodeficiency virus infecting De Brazza's monkeys (Cercopithecus neglectus): evidence for a Cercopithecus monkey virus clade. J. Virol. 787748-7762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Borsetti, A., A. Ohagen, and H. G. Gottlinger. 1998. The C-terminal half of the human immunodeficiency virus type 1 Gag precursor is sufficient for efficient particle assembly. J. Virol. 729313-9317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bowzard, J. B., R. P. Bennett, N. K. Krishna, S. M. Ernst, A. Rein, and J. W. Wills. 1998. Importance of basic residues in the nucleocapsid sequence for retrovirus Gag assembly and complementation rescue. J. Virol. 729034-9044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burniston, M. T., A. Cimarelli, J. Colgan, S. P. Curtis, and J. Luban. 1999. Human immunodeficiency virus type 1 Gag polyprotein multimerization requires the nucleocapsid domain and RNA and is promoted by the capsid-dimer interface and the basic region of matrix protein. J. Virol. 738527-8540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carlton, J. G., and J. Martin-Serrano. 2007. Parallels between cytokinesis and retroviral budding: a role for the ESCRT machinery. Science 3161908-1912. [DOI] [PubMed] [Google Scholar]
- 11.Chen, C., O. Vincent, J. Jin, O. A. Weisz, and R. C. Montelaro. 2005. Functions of early (AP-2) and late (AIP1/ALIX) endocytic proteins in equine infectious anemia virus budding. J. Biol. Chem. 28040474-40480. [DOI] [PubMed] [Google Scholar]
- 12.Chesebro, B., K. Wehrly, J. Nishio, and S. Perryman. 1992. Macrophage-tropic human immunodeficiency virus isolates from different patients exhibit unusual V3 envelope sequence homogeneity in comparison with T-cell-tropic isolates: definition of critical amino acids involved in cell tropism. J. Virol. 666547-6554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cimarelli, A., S. Sandin, S. Hoglund, and J. Luban. 2000. Basic residues in human immunodeficiency virus type 1 nucleocapsid promote virion assembly via interaction with RNA. J. Virol. 743046-3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dawson, L., and X. F. Yu. 1998. The role of nucleocapsid of HIV-1 in virus assembly. Virology 251141-157. [DOI] [PubMed] [Google Scholar]
- 15.De Guzman, R. N., Z. R. Wu, C. C. Stalling, L. Pappalardo, P. N. Borer, and M. F. Summers. 1998. Structure of the HIV-1 nucleocapsid protein bound to the SL3 ψ-RNA recognition element. Science 279384-388. [DOI] [PubMed] [Google Scholar]
- 16.Demirov, D. G., J. M. Orenstein, and E. O. Freed. 2002. The late domain of human immunodeficiency virus type 1 p6 promotes virus release in a cell type-dependent manner. J. Virol. 76105-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Derdowski, A., L. Ding, and P. Spearman. 2004. A novel fluorescence resonance energy transfer assay demonstrates that the human immunodeficiency virus type 1 Pr55Gag I domain mediates Gag-Gag interactions. J. Virol. 781230-1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dorfman, T., J. Luban, S. P. Goff, W. A. Haseltine, and H. G. Gottlinger. 1993. Mapping of functionally important residues of a cysteine-histidine box in the human immunodeficiency virus type 1 nucleocapsid protein. J. Virol. 676159-6169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dorfman, T., F. Mammano, W. A. Haseltine, and H. G. Gottlinger. 1994. Role of the matrix protein in the virion association of the human immunodeficiency virus type 1 envelope glycoprotein. J. Virol. 681689-1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fisher, R. D., H. Y. Chung, Q. Zhai, H. Robinson, W. I. Sundquist, and C. P. Hill. 2007. Structural and biochemical studies of ALIX/AIP1 and its role in retrovirus budding. Cell 128841-852. [DOI] [PubMed] [Google Scholar]
- 21.Garrus, J. E., U. K. von Schwedler, O. W. Pornillos, S. G. Morham, K. H. Zavitz, H. E. Wang, D. A. Wettstein, K. M. Stray, M. Cote, R. L. Rich, D. G. Myszka, and W. I. Sundquist. 2001. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell 10755-65. [DOI] [PubMed] [Google Scholar]
- 22.Gottlinger, H. G., T. Dorfman, J. G. Sodroski, and W. A. Haseltine. 1991. Effect of mutations affecting the p6 gag protein on human immunodeficiency virus particle release. Proc. Natl. Acad. Sci. USA 883195-3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gruenberg, J., and H. Stenmark. 2004. The biogenesis of multivesicular endosomes. Nat. Rev. Mol. Cell Biol. 5317-323. [DOI] [PubMed] [Google Scholar]
- 24.Huang, M., J. M. Orenstein, M. A. Martin, and E. O. Freed. 1995. p6Gag is required for particle production from full-length human immunodeficiency virus type 1 molecular clones expressing protease. J. Virol. 696810-6818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hurley, J. H., and S. D. Emr. 2006. The ESCRT complexes: structure and mechanism of a membrane-trafficking network. Annu. Rev. Biophys. Biomol. Struct. 35277-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Katzmann, D. J., M. Babst, and S. D. Emr. 2001. Ubiquitin-dependent sorting into the multivesicular body pathway requires the function of a conserved endosomal protein sorting complex, ESCRT-I. Cell 106145-155. [DOI] [PubMed] [Google Scholar]
- 27.Katzmann, D. J., G. Odorizzi, and S. D. Emr. 2002. Receptor downregulation and multivesicular-body sorting. Nat. Rev. Mol. Cell Biol. 3893-905. [DOI] [PubMed] [Google Scholar]
- 28.Lee, E. G., and M. L. Linial. 2006. Deletion of a Cys-His motif from the alpharetrovirus nucleocapsid domain reveals late domain mutant-like budding defects. Virology 347226-233. [DOI] [PubMed] [Google Scholar]
- 29.Lee, S., A. Joshi, K. Nagashima, E. O. Freed, and J. H. Hurley. 2007. Structural basis for viral late-domain binding to Alix. Nat. Struct. Mol. Biol. 14194-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martin-Serrano, J., A. Yarovoy, D. Perez-Caballero, and P. D. Bieniasz. 2003. Divergent retroviral late-budding domains recruit vacuolar protein sorting factors by using alternative adaptor proteins. Proc. Natl. Acad. Sci. USA 10012414-12419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martin-Serrano, J., T. Zang, and P. D. Bieniasz. 2001. HIV-1 and Ebola virus encode small peptide motifs that recruit Tsg101 to sites of particle assembly to facilitate egress. Nat. Med. 71313-1319. [DOI] [PubMed] [Google Scholar]
- 32.Mouland, A. J., J. Mercier, M. Luo, L. Bernier, L. DesGroseillers, and E. A. Cohen. 2000. The double-stranded RNA-binding protein Staufen is incorporated in human immunodeficiency virus type 1: evidence for a role in genomic RNA encapsidation. J. Virol. 745441-5451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Munshi, U. M., J. Kim, K. Nagashima, J. H. Hurley, and E. O. Freed. 2007. An Alix fragment potently inhibits HIV-1 budding: characterization of binding to retroviral YPXL late domains. J. Biol. Chem. 2823847-3855. [DOI] [PubMed] [Google Scholar]
- 34.Muriaux, D., S. Costes, K. Nagashima, J. Mirro, E. Cho, S. Lockett, and A. Rein. 2004. Role of murine leukemia virus nucleocapsid protein in virus assembly. J. Virol. 7812378-12385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Muriaux, D., J. Mirro, D. Harvin, and A. Rein. 2001. RNA is a structural element in retrovirus particles. Proc. Natl. Acad. Sci. USA 985246-5251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Odorizzi, G. 2006. The multiple personalities of Alix. J. Cell Sci. 1193025-3032. [DOI] [PubMed] [Google Scholar]
- 37.Ono, A., A. A. Waheed, A. Joshi, and E. O. Freed. 2005. Association of human immunodeficiency virus type 1 Gag with membrane does not require highly basic sequences in the nucleocapsid: use of a novel Gag multimerization assay. J. Virol. 7914131-14140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ott, D. E., L. V. Coren, E. N. Chertova, T. D. Gagliardi, K. Nagashima, R. C. Sowder II, D. T. Poon, and R. J. Gorelick. 2003. Elimination of protease activity restores efficient virion production to a human immunodeficiency virus type 1 nucleocapsid deletion mutant. J. Virol. 775547-5556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Parent, L. J., R. P. Bennett, R. C. Craven, T. D. Nelle, N. K. Krishna, J. B. Bowzard, C. B. Wilson, B. A. Puffer, R. C. Montelaro, and J. W. Wills. 1995. Positionally independent and exchangeable late budding functions of the Rous sarcoma virus and human immunodeficiency virus Gag proteins. J. Virol. 695455-5460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Perez-Caballero, D., T. Hatziioannou, J. Martin-Serrano, and P. D. Bieniasz. 2004. Human immunodeficiency virus type 1 matrix inhibits and confers cooperativity on Gag precursor-membrane interactions. J. Virol. 789560-9563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Puffer, B. A., L. J. Parent, J. W. Wills, and R. C. Montelaro. 1997. Equine infectious anemia virus utilizes a YXXL motif within the late assembly domain of the Gag p9 protein. J. Virol. 716541-6546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sandefur, S., R. M. Smith, V. Varthakavi, and P. Spearman. 2000. Mapping and characterization of the N-terminal I domain of human immunodeficiency virus type 1 Pr55Gag. J. Virol. 747238-7249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shim, S., L. A. Kimpler, and P. I. Hanson. 2007. Structure/function analysis of four core ESCRT-III proteins reveals common regulatory role for extreme C-terminal domain. Traffic 81068-1079. [DOI] [PubMed] [Google Scholar]
- 44.Strack, B., A. Calistri, S. Craig, E. Popova, and H. G. Gottlinger. 2003. AIP1/ALIX is a binding partner for HIV-1 p6 and EIAV p9 functioning in virus budding. Cell 114689-699. [DOI] [PubMed] [Google Scholar]
- 45.Strack, B., A. Calistri, and H. G. Gottlinger. 2002. Late assembly domain function can exhibit context dependence and involves ubiquitin residues implicated in endocytosis. J. Virol. 765472-5479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Usami, Y., S. Popov, and H. G. Gottlinger. 2007. Potent rescue of human immunodeficiency virus type 1 late domain mutants by ALIX/AIP1 depends on its CHMP4 binding site. J. Virol. 816614-6622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.VerPlank, L., F. Bouamr, T. J. LaGrassa, B. Agresta, A. Kikonyogo, J. Leis, and C. A. Carter. 2001. Tsg101, a homologue of ubiquitin-conjugating (E2) enzymes, binds the L domain in HIV type 1 Pr55(Gag). Proc. Natl. Acad. Sci. USA 987724-7729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.von Schwedler, U. K., M. Stuchell, B. Muller, D. M. Ward, H. Y. Chung, E. Morita, H. E. Wang, T. Davis, G. P. He, D. M. Cimbora, A. Scott, H. G. Krausslich, J. Kaplan, S. G. Morham, and W. I. Sundquist. 2003. The protein network of HIV budding. Cell 114701-713. [DOI] [PubMed] [Google Scholar]
- 49.Wills, J. W., C. E. Cameron, C. B. Wilson, Y. Xiang, R. P. Bennett, and J. Leis. 1994. An assembly domain of the Rous sarcoma virus Gag protein required late in budding. J. Virol. 686605-6618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yasuda, J., and E. Hunter. 1998. A proline-rich motif (PPPY) in the Gag polyprotein of Mason-Pfizer monkey virus plays a maturation-independent role in virion release. J. Virol. 724095-4103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yuan, B., S. Campbell, E. Bacharach, A. Rein, and S. P. Goff. 2000. Infectivity of Moloney murine leukemia virus defective in late assembly events is restored by late assembly domains of other retroviruses. J. Virol. 747250-7260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yuan, B., X. Li, and S. P. Goff. 1999. Mutations altering the Moloney murine leukemia virus p12 Gag protein affect virion production and early events of the virus life cycle. EMBO J. 184700-4710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zamborlini, A., Y. Usami, S. R. Radoshitzky, E. Popova, G. Palu, and H. Gottlinger. 2006. Release of autoinhibition converts ESCRT-III components into potent inhibitors of HIV-1 budding. Proc. Natl. Acad. Sci. USA 10319140-19145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zennou, V., D. Perez-Caballero, H. Gottlinger, and P. D. Bieniasz. 2004. APOBEC3G incorporation into human immunodeficiency virus type 1 particles. J. Virol. 7812058-12061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang, Y., and E. Barklis. 1997. Effects of nucleocapsid mutations on human immunodeficiency virus assembly and RNA encapsidation. J. Virol. 716765-6776. [DOI] [PMC free article] [PubMed] [Google Scholar]