Abstract
A key function of the Nef protein of immunodeficiency viruses is the downregulation of the T-cell and macrophage coreceptor, CD4, from the surfaces of infected cells. CD4 downregulation depends on a conserved (D/E)XXXL(L/I)-type dileucine motif in the C-terminal, flexible loop of Nef, which mediates binding to the clathrin adaptor complexes AP-1, AP-2, and AP-3. We now report the identification of a consensus (D/E)D motif within this loop as a second, conserved determinant of interaction of Nef with AP-2, though not with AP-1 and AP-3. Mutations in this diacidic motif abrogate both AP-2 binding and CD4 downregulation. We also show that a dileucine motif from tyrosinase, both in its native context and in the context of Nef, can bind to AP-2 independently of a diacidic motif. These results thus identify a novel type of AP-2 interaction determinant, support the notion that AP-2 is the key clathrin adaptor for the downregulation of CD4 by Nef, and reveal a previously unrecognized diversity among dileucine sorting signals.
The Nef protein encoded by the primate lentiviruses, human immunodeficiency virus type 1 (HIV-1), HIV-2, and simian immunodeficiency virus (SIV), is critical for progression from infection to the disease, AIDS. Nef is an accessory factor that is produced early after infection and regulates various signaling and trafficking pathways in the host cells, T lymphocytes and macrophages/monocytes (reviewed in references 3, 36, and 47). Perhaps the best-characterized function of Nef is the downregulation of CD4, a transmembrane protein expressed on the surfaces of the host cells. CD4 serves as a coreceptor for both class II molecules of the major histocompatibility complex (MHC-II molecules) on antigen-presenting cells and the Env surface glycoprotein of the primate lentiviruses. Nef-induced CD4 downregulation involves removal of the receptor from the cell surface and its subsequent targeting for degradation in lysosomes (1, 14, 45). This has been proposed to accomplish the dual purpose of prevention of superinfection and enhancement of virus release (31, 35).
Nef has been postulated to alter CD4 trafficking by linking the cytosolic tail of CD4 to three clathrin-associated, heterotetrameric adaptor protein (AP) complexes, AP-1, AP-2, and AP-3, which mediate protein-sorting events at the trans-Golgi network, plasma membrane, and endosomes, respectively (44, 46). Indeed, Nef has been shown to interact with the CD4 cytosolic tail (22, 23, 50) and with the three AP complexes (6, 8, 10, 12, 17, 27, 28, 33). However, recent work using RNA interference has demonstrated that Nef-induced CD4 downregulation is dependent mainly on AP-2, not on AP-1 or AP-3 (8, 29, 48). In addition, Nef has been localized to plasma membrane clathrin-coated pits that contain AP-2 (7, 13, 18). These findings are consistent with the proposal that Nef downregulates CD4 mainly by inducing its rapid endocytosis from the cell surface by a clathrin/AP-2-dependent mechanism (1, 8, 13, 17, 18, 29).
Nef contains a highly mobile loop comprising residues 154 to 180, which is inserted between the last two strands of a β-sheet (20, 32) (see Fig. 1A) (unless otherwise indicated, sequences and residue numbers correspond to the NL4-3 variant of HIV-1). Both the downregulation of CD4 and the interaction with AP-2 are critically dependent on a dileucine-containing sequence (ENTSLL [residues 160 to 165]; referred to as the “dileucine motif”) within this loop (6, 8, 11, 12, 17, 28). This sequence fits the general consensus motif (D/E)XXXL(L/I) (where X is any amino acid) for dileucine-based sorting signals involved in the binding of numerous cellular proteins to AP complexes, thereby enabling their clathrin-dependent sorting (4). CD4 downregulation also depends on two other sets of residues within the Nef C-terminal flexible loop: a diacidic pair, DD (residues 174 and 175) (see Fig. 1A), and the charged residues ERE (residues 177 to 179) (2). Mutation of these residues abrogates the ability of Nef to downregulate CD4 to the same extent as mutation of the dileucine motif. Moreover, a mutation of D174 to K was found in an HIV-1 strain isolated from a long-term nonprogressor (that is, an HIV-1-infected patient who did not develop AIDS for 10 years or longer) (42), underscoring the functional importance of the diacidic motif. Unlike the ENTSLL sequence, however, neither the DD nor the ERE sequence has been shown to interact with AP-2 or other AP complexes. Instead, the DD sequence has been implicated in binding to several other host cell proteins: the c-Raf1 protein kinase (25), the Eed Polycomb group protein (56), and the V1H subunit of the vacuolar ATPase (37). The latter protein has also been shown to bind to the μ2 subunit of AP-2, thus leading to the proposal that V1H acts as a bridge between Nef and AP-2 (16). The recent development of assays that detect robust binding of Nef to AP-2 (8, 12) allowed us to assess whether DD, ERE, or other residues within the loop play a direct or indirect role in this interaction.
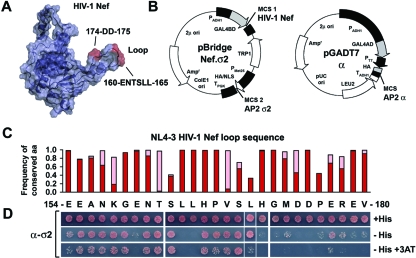
FIG. 1.
Identification of residues within the HIV-1 Nef C-terminal flexible loop that contribute to AP-2 binding. (A) Ribbon diagram and surface rendering of the 3-dimensional structure of HIV-1 Nef. Determinants for CD4 downregulation and AP-2 binding within the C-terminal flexible loop are highlighted in red. This model was generated with PyMOL (www.pymol.org) by using coordinates from accession codes 1QA5 (15) and 2NEF (21), with additional residues of the loop region modeled arbitrarily. The structure, sequences, and residue numbers correspond to the NL4-3 variant of HIV-1 Nef. (B) Plasmids used for Y3H experiments. Nef was expressed as a GAL4BD fusion protein from the pBridge vector along with the AP-2 σ2 subunit. AP-2 α (αC isoform) was expressed as a GAL4AD fusion protein from the pGADT7 vector. (C) Comparison of the loop sequence (residues 154 to 180) of NL4-3 Nef (GenBank accession number U26942) to an alignment of 1,290 HIV-1 and SIVcpz Nef sequences found on the Los Alamos HIV sequence database (www.hiv.lanl.gov). The NL4-3 residues were scored for the frequency of identity and similarity at each position within the alignment. Similarity was defined by a log odds score of ≥0 on the BLOSUM62 substitution matrix (24). The bar graph indicates the frequency of occurrence of each NL4-3 residue in the aligned sequences (red bars) as well as the frequency of similar residues at each position (pink bars). (D) Y3H analysis of binding of full-length NL4-3 HIV-1 Nef loop mutants to the AP-2 α-σ2 hemicomplex. Each colony represents an alanine substitution mutant at the corresponding residue of the Nef loop coexpressed with α and σ2 by cotransformation of HF7c yeast with the plasmids diagramed in panel B. The third position (residue 156) is a naturally occurring alanine in the wild-type sequence. Growth in the absence of histidine (−His) or in the combined absence of histidine and presence of 3 mM 3-aminotriazole (+3AT) is indicative of interactions at two levels of stringency. Immunoblot analysis showed that all the fusion proteins were expressed at similar levels in the transformed yeast cells (data not shown).
We report that the DD sequence is absolutely required for direct binding of Nef to AP-2. This requirement is specific to AP-2, since the DD sequence is dispensable for binding to AP-1 and AP-3. A number of other residues within the flexible loop, including residue E179 of the ERE sequence, appear to make additional contributions to AP-2 binding. These findings provide a simpler, more straightforward explanation for the requirement of the DD sequence in CD4 downregulation, and they further support the preponderant role of AP-2-dependent endocytosis in Nef-induced CD4 downregulation. In addition, they identify a novel determinant for interaction of a cargo protein with AP-2. Finally, swapping of dileucine signals between Nef and the cellular protein tyrosinase reveals important differences in the requirements of (D/E)XXXL(L/I)-type signals for binding to the AP complexes.
MATERIALS AND METHODS
Recombinant DNA constructs.
The NL4-3 Nef cDNA that was used to make all Nef constructs was a gift from Sundararajan Venkatesan, NIAID, NIH. The pNL4-3 Nef.IRES.GFP, pBridge.Nef.σ1, pBridge.Nef.σ2, and pBridge.Nef.σ3 vectors have been described previously (8, 28). All mutations within these vectors were created by site-directed mutagenesis using the QuikChange II kit (Stratagene, Cedar Creek, TX). Typically, the mutation was introduced into the pBridgeNef.σ2 plasmid, and then a fragment containing the Nef open reading frame was subcloned into the EcoRI-SalI restriction sites of pBridge.Nef.σ3. The same fragments were additionally subcloned into pIRES2.GFP (Clontech, Mountain View, CA) and pCI.neo (Promega, Madison, WI) to make the relevant vectors (pNL4-3 Nef.IRES.GFP and pCI.Nef variants). Because σ1 has an internal EcoRI restriction site, the σ2 open reading frame was excised from the mutated pBridge.Nef.σ2 plasmids using the NotI and BglII enzymes and was replaced with the σ1 open reading frame to make the different permutations of pBridge.Nef.σ1. The pGADT7.γ1, pGADT7.αC, and pGADT7.δ vectors have also been described previously (8, 28). pCMV.CD4 (5) was kindly provided by Klaus Strebel (NIAID, NIH). All open reading frames were verified by nucleotide sequence analysis.
Antibodies and other reagents.
Mouse monoclonal antibodies to human CD4, either unconjugated or conjugated with allophycocyanin (APC), were purchased from Caltag (Burlingame, CA). The Alexa Fluor 594-conjugated secondary antibody to mouse immunoglobulin G was purchased from Invitrogen (Carlsbad, CA). Horseradish peroxidase-conjugated secondary antibodies were purchased from GE Healthcare (Piscataway, NJ). HIV-1 Nef antiserum (51) was obtained from the NIH AIDS Research and Reference Reagent Program.
Cell culture and transfection.
HeLa cells (American Type Culture Collection, Manassas, VA) were grown in Dulbecco's modified Eagle medium supplemented with 100 U/ml penicillin, 0.1 μg/ml streptomycin, 2 mM l-glutamine, and 10% (vol/vol) fetal bovine serum. Cells were transiently transfected with the indicated plasmids by using Lipofectamine 2000 (Invitrogen) according to the instructions provided by the manufacturer.
FACS analysis.
HeLa cells were stained with the appropriate antibodies conjugated to APC and were prepared for fluorescence-activated cell sorter (FACS) analysis as previously described (8). Untransfected HeLa cells were used as a negative control for antibody labeling. Green fluorescent protein (GFP) fluorescence expressed from pIRES2.GFP or its derivatives was used to identify and gate around transfected cells. The amount of cell-associated APC fluorescence in GFP-positive cells was measured with a FACSCalibur flow cytometer and analyzed using CellQuest software (Becton Dickinson, Franklin Lakes, NJ).
Endocytosis assays.
HeLa cells were cotransfected with pCMV.CD4 (5) and pNL4-3 Nef.IRES.GFP (or Nef mutants) on 10-cm-diameter tissue culture dishes at ∼70% confluence. The following day, the plates were washed with phosphate-buffered saline (PBS), and cells were harvested after a brief incubation in PBS plus 2 mM EDTA to promote detachment. The harvested cells were washed twice with ice-cold PBS and then incubated on ice for 30 min in binding buffer (OptiMEM [Invitrogen] supplemented with 2% [wt/vol] bovine serum albumin) with anti-CD4 (1:100 dilution). An aliquot of cells was incubated without antibody as a control. After incubation, the cells were washed twice with ice-cold PBS, resuspended with prewarmed binding buffer, and incubated at 37°C. An aliquot of cells that remained on ice was reserved for the 0-min time point. At intervals, aliquots of the cells were transferred from the reaction tubes to ice-cold PBS. At the end of the time course, the samples were subjected to two additional washes in ice-cold PBS and were then resuspended in PBS plus 1% (wt/vol) bovine serum albumin and 0.1% (wt/vol) NaN3 containing a 1:100 dilution of an APC-conjugated secondary antibody. After additional washes, the samples were analyzed for CD4 surface levels by FACS.
Immunofluorescence confocal microscopy.
HeLa cells were seeded on glass coverslips to ∼25% density and were transfected the following day with plasmids pCMV.CD4 and pCI.Nef at a 4:5 ratio. All subsequent steps were performed at room temperature. One day after transfection, cells were fixed for 15 min with 4% (wt/vol) paraformaldehyde in PBS, permeabilized for 5 min with 0.2% (wt/vol) Triton X-100 in PBS, and incubated for 1 h in blocking buffer (PBS plus 4% fetal bovine serum). This was followed by incubation with blocking buffer plus a monoclonal antibody to CD4 (1:100 dilution). After several washes with PBS, cells were incubated with an Alexa Fluor 594-conjugated secondary antibody, washed again, and mounted on slides. Cells were imaged on a Zeiss LSM510 laser scanning confocal microscope (Carl Zeiss, Thornwood, NY) with a 63× Plan Apochromat 1.4-numerical-aperture objective by using the 543-nm line of a He-Ne laser. Emission was collected over the range of 560 to 660 nm with appropriate filter sets.
Immunoblotting.
Cells were lysed for 20 min on ice by using 1% (vol/vol) NP-40 in PBS supplemented with a protease inhibitor cocktail (Roche Diagnostics GmbH, Mannheim, Germany). Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), transfer to nitrocellulose membranes, and detection with antibodies were performed as described previously (8).
Y3H analysis.
Yeast three-hybrid (Y3H) assays were performed as described in previous publications (8, 28). Briefly, wild-type and mutant versions of NL4-3 Nef were expressed as GAL4 DNA-binding domain (GAL4BD) fusion proteins in the pBridge vector (Clontech, Mountain View, CA) along with rat σ1A, σ2, or σ3A. For other experiments, wild-type and mutant versions of the mouse tyrosinase cytosolic tail were expressed as GAL4BD fusion proteins in pBridge vectors containing the AP σ subunits. The large AP subunits (mouse γ1, rat αC, and human δ) were expressed as GAL4 activation domain (GAL4AD) fusion proteins from the pGADT7 vector (Clontech). Saccharomyces cerevisiae strain HF7c was transformed with pairs of pBridge and pGADT7 vectors using the standard lithium acetate procedure, and transformants were selected on dropout agar plates lacking Leu, Trp, and Met. After several days, colonies were transferred to three sets of dropout agar plates: those lacking Leu, Trp, and Met; those lacking His, Leu, Trp, and Met; and those lacking His, Leu, Trp, and Met and supplemented with 3 mM 3-aminotriazole. Colony growth on the selective plates (i.e., those lacking His) was checked 3 to 4 days later. Each Y3H experiment was performed a minimum of three times.
Recombinant protein expression and purification and GST pulldown experiments.
NL4-3 Nef was expressed as an N-terminal hexahistidine-tagged fusion protein in Escherichia coli BL21(DE3) as described previously (8). The Nef mutant in which the D residues at positions 174 and 175 had been changed to A (the DD174,175AA mutant) was generated using the QuikChange II kit (Stratagene). AP-2 core, comprising residues 1 to 621 from rat αC (α trunk), 1 to 591 from rat β2 (β2 trunk), 1 to 141 from mouse μ2 (μ2 N-terminal domain), and 1 to 143 from rat σ2 (full-length σ2), was expressed in E. coli Rosetta 2 (DE3) cells (Novagen, San Diego, CA) (8). The amino acid sequences of the mouse and rat AP-2 core components are 98 to 100% identical to those of their human orthologs. In addition, the structural requirements for HIV-1 Nef to downregulate human CD4 are conserved from Drosophila melanogaster to humans (8), justifying the use of a heterologous system. The α trunk was produced as a C-terminal glutathione S-transferase (GST) fusion and the β2 trunk as an N-terminal hexahistidine fusion protein to facilitate a two-step purification strategy (8). The methodology for pulldown experiments has been described previously (8). Briefly, the GST-AP-2 core and the GST-ɛ ear domain (residues 849 to 1137) of AP4 were immobilized onto glutathione-Sepharose. Hexahistidine-tagged wild-type or DD174,175AA Nef (final concentration, 100 μM) was incubated with the immobilized GST fusion proteins for 30 min at 4°C. The beads were subsequently centrifuged at 1,000 × g, washed twice with Tris-buffered saline (10 mM Tris-HCl [pH 7.4]-150 mM NaCl), supplemented with 0.1% (wt/vol) Triton X-100, resuspended in SDS sample buffer, boiled, and resolved on a 4 to 12% NuPAGE gel (Invitrogen) using morpholineethanesulfonic acid running buffer. To analyze the salt sensitivity of the interactions, identical aliquots of GST-AP-2 core were incubated with hexahistidine-tagged wild-type Nef (final concentration, 100 μM) in the presence of varying amounts of NaCl from 0.1 M to 1 M. The beads were prepared for analysis as described above.
Surface plasmon resonance (SPR).
The binding of untagged AP-2 core to hexahistidine-tagged wild-type and mutant Nef proteins was measured with a BIAcore T100 instrument (BIAcore AB, Uppsala, Sweden) at 25°C with a flow rate of 20 μl min−1. All binding experiments were performed in HBS (10 mM sodium HEPES [pH 7.4]-150 mM NaCl). A CM5 chip was activated using N-hydroxysuccinimide-1-ethyl-3(3-dimethylaminopropyl) carbodiimide (1:1) at a flow rate of 5 μl min−1 for 400 s. The GST-ɛ ear domain of AP4 and hexahistidine-tagged wild-type or DD174,175AA Nef (30 μg ml−1) in 10 mM acetate buffer (pH 5.0) were passed over individual flow cells at a flow rate of 20 μl min−1 using HBS at 25°C with a response level of 10,000 response units. The binding of the recombinant untagged AP-2 core to wild-type and mutant Nef was measured simultaneously by passing AP-2 core over consecutive flow cells with association and dissociation times of 120 s and 400 s, respectively. Between subsequent injections of AP-2 core proteins, surfaces were regenerated with an injection of HBS supplemented with 500 mM NaCl for 15 s at 100 μl min−1. Sensorgram response unit (RU) values were subtracted from that of a control sensor surface with the immobilized GST-ɛ ear domain of AP4. Steady-state binding data of AP-2 for Nef were fitted using BIAevaluation software (BIAcore) with globally floating KD (equilibrium dissociation constant), Rmax, and refractive index values.
RESULTS
Identification of a diacidic motif that is required for interaction of HIV-1 Nef with the AP-2 α-σ2 hemicomplex.
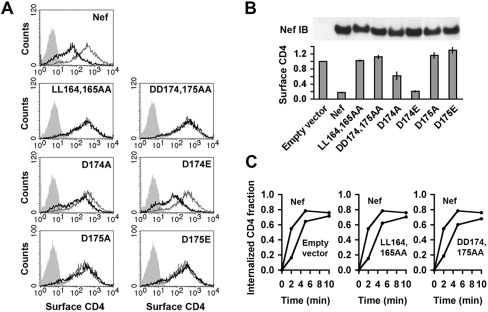
The dileucine motif (ENTSLL in NL4-3; EXXXLL consensus for all variants) contained within the 27-residue C-terminal loop of Nef (residues 154 to 180) (Fig. 1A) is highly conserved among all strains of HIV and SIV (34, 43). Conservation of this motif is consistent with its critical roles in CD4 downregulation and AP-2 binding (6, 8, 11, 12, 17, 28). An analysis of 1,290 Nef sequences obtained from the Los Alamos HIV sequence database showed that conservation is not limited to the dileucine motif but extends to most residues within the loop (Fig. 1C). To determine whether these other residues are also important for interaction with AP-2, we generated a series of alanine substitution mutants at 26 of the 27 loop residues in the context of full-length Nef (residue 156 is a naturally occurring alanine) and tested for binding to AP-2 subunits by using aY3H assay (vectors are shown in Fig. 1B) (8). Of the four subunits of AP-2 (i.e., α, β2, μ2, and σ2), an assembly of two, α-σ2, has previously been shown to be required for interaction with Nef (8, 12), thus necessitating the use of the Y3H system. As expected from previous work (8, 12), mutation of either L164 or L165 of the dileucine motif completely abrogated binding to α-σ2 (Fig. 1D). Mutation of the E160 residue, which is part of the consensus EXXXLL motif, caused a partial loss of binding (Fig. 1D), as previously shown (8). Strikingly, we found that mutation of several residues at the C-terminal half of the loop also caused defects in α-σ2 binding, with various degrees of severity (Fig. 1D). The strongest defects were observed for mutants with alterations of the acidic residues D174 and D175, which exhibited no binding to α-σ2 (Fig. 1D). Mutation of several other residues in this region, including H171, G172, M173, P176, R178, E179, and V180, caused partial binding defects (Fig. 1D). The charged residues in this region have previously been implicated in CD4 downregulation (2, 26) and localization of Nef to clathrin-coated pits (18). In particular, the DD174,175AA and ERE177-179AAA mutants have been shown to be null for CD4 downregulation (2, 18). Our findings thus suggested that the functional requirement for these residues was due to their roles in mediating the interaction with AP-2. Because mutation of D174 and D175 caused the most severe defects, subsequent studies focused on these two residues (referred to below as the “diacidic motif”).
The diacidic motif is required for direct binding of HIV-1 Nef to AP-2.
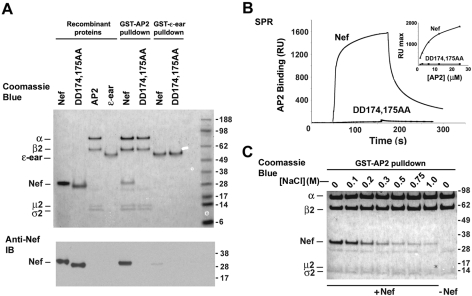
To ascertain if the diacidic motif was required for direct interaction of Nef with AP-2, we performed in vitro binding experiments using recombinant proteins produced in bacteria. Analyses were done using an AP-2 core complex devoid of hinge and ear domains and of the μ2 C-terminal domain. We showed previously that this recombinant AP-2 core construct is capable of interacting with Nef in a manner dependent on the dileucine motif (8). Pulldown assays showed that the GST-tagged AP-2 core complex bound wild-type Nef but not DD174,175AA mutant Nef, as detected by SDS-PAGE followed by Coomassie blue staining (Fig. 2A, top) and immunoblot analysis (Fig. 2A, bottom). Confirming the specificity of these interactions, we observed negligible binding of both wild-type and mutant Nef to the ear domain (residues 849 to 1137) of the AP4 ɛ subunit (44, 46) fused to GST (Fig. 2A). These results were corroborated by SPR spectroscopy, which showed binding of untagged AP-2 core to wild-type Nef but not to DD174,175AA mutant Nef immobilized on biosensor chips (Fig. 2B). The affinity of the AP-2 core for wild-type Nef calculated from these experiments was 6 ± 1 μM (n = 3). These experiments thus demonstrated that the diacidic motif is required for direct interaction of Nef with AP-2.
FIG. 2.
In vitro analyses of Nef-AP-2 interaction determinants. (A) Purified recombinant proteins used in this experiment include hexahistidine-tagged wild-type and DD174,175AA mutant Nef, AP-2 core tagged with GST at the C terminus of α and hexahistidine at the N terminus of β2, and the ear domain of AP4 ɛ tagged at its N terminus with GST. Binding of Nef proteins to GST-tagged AP-2 core and ɛ-ear was analyzed by GST pulldown and SDS-PAGE, followed by Coomassie blue staining (top) or immunoblotting (IB) with an antibody to Nef (bottom). The input recombinant proteins were run alongside for comparison (first four lanes). Wild-type hexahistidine-tagged Nef is visible as a ∼27-kDa band in the fifth lane. This experiment is representative of three experiments with similar results. (B) GST-ɛ-ear (control) or wild-type or DD174,175AA mutant hexahistidine-tagged Nef was covalently attached to the surface of a CM5 sensor chip. Subsequently, the surfaces were probed for binding of untagged AP-2 core by SPR. Shown are control-subtracted sensorgrams of individual injections of AP-2 core (25 μM) over surfaces containing wild-type or DD174,175AA mutant hexahistidine-tagged Nef. (Inset) Plot of varying concentrations (0.5 μM to 25 μM) of AP-2 core. (C) The interaction of hexahistidine-tagged Nef with GST-tagged AP-2 core is sensitive to ionic strength. GST-tagged AP-2 core was incubated with hexahistidine-tagged Nef in the presence of increasing NaCl concentrations (0.1 M to 1 M) during the pulldown experiment. The positions of the recombinant proteins (for simplicity, tags are not indicated) are shown on the left and those of molecular mass markers (in kilodaltons) on the right of the gel.
Binding of HIV-1 Nef to AP-2 is dependent on electrostatic interactions.
The requirement of the diacidic motif, as well as other charged residues, for Nef binding to AP-2 suggested that electrostatic interactions might be important contributors to the overall binding affinity. If this is the case, binding should be sensitive to high concentrations of salt. To test this prediction, we used the GST pulldown assay to examine the binding of wild-type Nef to the GST-tagged AP-2 core in the presence of increasing NaCl concentrations (Fig. 2C). We observed that this binding was distinctly salt sensitive; dramatic losses of binding were observed at NaCl concentrations in excess of physiological levels (150 mM). This indicated that electrostatic interactions contribute to the formation of the Nef-AP-2 complex.
The diacidic motif fits a (D/E)D consensus and is not required for interaction with the AP-1 γ-σ1 and AP-3 δ-σ3 hemicomplexes.
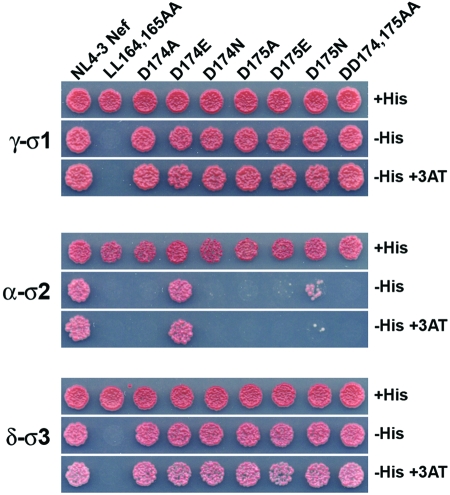
To further characterize the specific requirements for each residue of the diacidic motif, we generated a series of Nef constructs with single alterations of aspartate to alanine, glutamate, or asparagine, and we assessed the effect of each mutation on binding to the AP-2 α-σ2 hemicomplex and the homologous AP-1 γ-σ1 and AP-3 δ-σ3 hemicomplexes by using the Y3H system. As previously shown (8, 12, 28), mutation of L164 and L165 to alanine abolished binding to the three hemicomplexes (Fig. 3). Single or double mutation of D174 and D175 to alanine abolished binding to α-σ2 (Fig. 3). The isoelectric D174E mutation, on the other hand, had no effect on binding to α-σ2 (Fig. 3). This is not surprising, since this position is either D or E in nearly all Nef sequences (46.0% D, 52.2% E among all HIV-1 Nef variants) (Fig. 1C). In contrast, D175E displayed severely reduced binding to α-σ2 (Fig. 3). This is in accordance with the almost exclusive occurrence of D at this position (98.9% D in all HIV-1 variants) (Fig. 1C). Additionally, we observed that the isosteric D174N and D175N substitutions resulted in elimination and reduction of binding to α-σ2, respectively (Fig. 3). Thus, the diacidic motif can be generally defined as (D/E)D, with N being a weak substitute at the second position. Remarkably, none of the mutations in the diacidic motif had any effect on the interaction of Nef with γ-σ1 or δ-σ3 (Fig. 3). Therefore, the interaction of Nef with AP-2 depends on both the dileucine and diacidic motifs, whereas interaction with AP-1 and AP-3 is exclusively dependent on the dileucine motif. This strongly suggests that the conservation of the diacidic motif is closely tied to a specific requirement of Nef for binding to AP-2.
FIG. 3.
Mutational analysis of Nef binding to the AP-1 γ-σ1, AP-2 α-σ2, and AP-3 δ-σ3 hemicomplexes. Interactions were analyzed as described in the legend to Fig. 1 and in Materials and Methods.
Correlation with the requirements of the diacidic motif for CD4 downregulation and enhanced internalization.
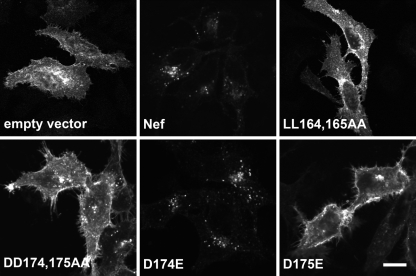
We next assessed the activities of the various Nef mutants for CD4 downregulation in transfected HeLa cells by immunofluorescence microscopy (Fig. 4) and FACS analysis (Fig. 5). In agreement with previous work (2, 26), DD174,175AA Nef, like LL164,165AA Nef, failed to downregulate CD4 in both assays (Fig. 4 and 5A and B). Single mutations of D174 or D175 to alanine also largely prevented the ability of Nef to downregulate CD4 (Fig. 5A and B). Remarkably, mutation of D174 to glutamate had no effect on CD4 downregulation, whereas mutation of D175 to glutamate abolished downregulation in both assays (Fig. 4 and Fig. 5A and B). Immunoblot analysis showed that all constructs were expressed at similar levels (Fig. 5B), indicating that the failure of some mutants to downregulate CD4 was not due to a lack of expression. These effects on CD4 downregulation mirrored the effects on α-σ2 binding observed in the Y3H experiments (Fig. 3), which we take to be further evidence for a causal relationship between the two processes. The consensus for the diacidic motif that emerges from the binding and functional analyses is thus (D/E)D.
FIG. 4.
Immunofluorescence microscopy analysis of CD4 downregulation by wild-type and mutant Nef proteins. HeLa cells were cotransfected with two different plasmids: pCMV.CD4 and wild-type or mutant versions of pCI.Nef. Cells were fixed, permeabilized, and stained with mouse anti-CD4, followed by Alexa Fluor 594-conjugated anti-mouse immunoglobulin G. The distribution of CD4 was imaged by confocal microscopy. Bar, 10 μm.
FIG. 5.
FACS analysis of CD4 downregulation by wild-type and mutant Nef proteins. HeLa cells were cotransfected with pCMV.CD4 and either wild-type or mutant versions of pNL4-3 Nef.IRES.GFP, which is designed to express Nef and GFP as separate proteins from the same bicistronic transcript. For the FACS analyses, GFP fluorescence was used as a marker to identify cells that had been transfected with a Nef expression plasmid. (A) FACS histograms of CD4 surface levels in cells transfected with the plasmids mentioned above. For each condition, the relative amount of CD4 at the plasma membrane is shown for cells transfected with the pIRES2.GFP empty vector (no Nef expression) (thin lines) and for cells transfected with wild-type or mutant Nef (bold lines). The fluorescence profile of cells left untransfected (no CD4 expression) (solid gray shading) is also shown. (B) (Top) Immunoblot (IB) analysis of the expression of wild-type and mutant versions of Nef by using an antiserum to HIV-1 Nef. Cells transfected with the empty vector pIRES2.GFP (no Nef expression) were used as a control. (Bottom) Quantification of the results in panel A. Bars represent geometric means ± standard errors of the means from three independent experiments. Levels of CD4 on the surfaces of Nef-expressing cells are compared to those on the surfaces of cells transfected with the pIRES2.GFP empty vector (taken as 1). (C) FACS analysis of CD4 endocytosis in cells expressing wild-type or mutant versions of Nef. Data for cells that were transfected with the empty vector and do not express Nef are included as a control.
Mutation of D174 and D175 to alanine had no effect on the ability of Nef to downregulate MHC-I molecules (data not shown), in accordance with data in a previous report (52) and with the fact that MHC-I downregulation occurs by a mechanism distinct from that of CD4 (19, 30, 38, 41, 47).
Although both decreased transport to the plasma membrane and enhanced endocytosis have been proposed to account for CD4 downregulation by Nef (1, 8, 13, 17, 18, 29, 40, 49), the latter mechanism is most consistent with the role of AP-2 in this process. To explore this correlation in more detail, we measured the rates of endocytosis of CD4 upon expression of wild-type or mutant forms of Nef (Fig. 5C). As expected, mutation of the dileucine motif (LL164,165AA mutant) abolished the ability of Nef to accelerate the endocytosis of CD4 in HeLa cells (Fig. 5C). Importantly, mutation of the diacidic motif (DD174,175AA mutant) had the same inhibitory effect (Fig. 5C). Thus, both the dileucine and diacidic motifs of Nef are required for the enhanced endocytosis that leads to the downregulation of CD4.
Distinct requirement of different dileucine motifs for the contribution of diacidic motifs.
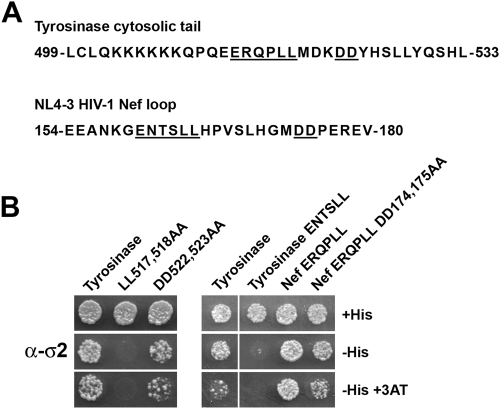
We sought to extend these studies by assessing whether the (D/E)XXXL(L/I)-type dileucine motif from the cytosolic tail of the cellular protein tyrosinase exhibited a similar requirement for downstream acidic residues. The mouse tyrosinase tail (residues 499 to 533) has been previously shown, using the Y3H system, to bind to the AP-1 γ-σ1 and AP-3 δ-σ3 hemicomplexes (54). Interestingly, the tyrosinase dileucine motif (ERQPLL; residues 513 to 518) is followed by a DD sequence (residues 522 and 523) (Fig. 6A). We tested the wild-type tyrosinase tail as well as the LL517,518AA and DD522,523AA mutants for their abilities to bind to α-σ2. As shown in Fig. 6B, tyrosinase bound to α-σ2 in a manner that was completely dependent on the dileucine motif but only slightly dependent on the diacidic motif. What, then, might be the cause of the absolute requirement for the diacidic sequence in order for Nef to bind AP-2? We reasoned that the dileucine motif of Nef might be weaker than that of tyrosinase, necessitating the additional contribution of the diacidic motif for detectable binding. To test this hypothesis, we generated tyrosinase tail constructs in which the ERQPLL sequence was replaced by the Nef ENTSLL sequence, and vice versa. We observed that, in contrast to the wild-type tyrosinase tail, tyrosinase with the ENTSLL sequence could not bind to AP-2 (Fig. 6B). Importantly, Nef with ERQPLL bound to AP-2 in a manner that was largely independent of the diacidic motif (Fig. 6B). Since swapping of the dileucine signals involved replacement of only the intervening NTS and RQP residues [i.e., the X positions in the (D/E)XXXL(L/I) consensus], these findings indicate that these residues are important determinants of interaction. These experiments thus highlighted functional differences among (D/E)XXXL(L/I)-type signals, some of which require additional determinants for binding to AP-2.
FIG. 6.
Y3H analysis reveals qualitative differences among dileucine motifs. (A) Sequences of the mouse tyrosinase cytosolic tail and the NL4-3 HIV-1 Nef flexible loop. The dileucine and diacidic motifs are underlined. Amino acid numbers are given to the left and right of the sequences. (B) Y3H analysis of the interaction of the α-σ2 hemicomplex with wild-type and mutant versions of the tyrosinase cytosolic tail and with mutant versions of full-length Nef. The Y3H experiments were performed as described in the legend to Fig. 1 and in Materials and Methods.
DISCUSSION
We have presented evidence for the existence of a novel, conserved diacidic motif [DD174-175 in NL4-3; (D/E)D in general] within the C-terminal flexible loop of all Nef variants, which cooperates with the previously characterized dileucine motif (i.e., ENTSLL160-165 in NL4-3; EXXXLL in general) to bind to AP-2. The diacidic motif is located 9 residues C-terminal to the dileucine motif and appears to be the key element of a longer (∼11-residue) determinant that contains other conserved acidic residues. Both the diacidic and dileucine motifs are required for the binding of Nef to AP-2 in Y3H, GST pulldown, and SPR assays. This requirement parallels that for CD4 downregulation, which also depends on the integrity of both motifs (2; also this work). This provides strong additional support for the proposed role of AP-2 in Nef-induced CD4 downregulation.
The dileucine and diacidic motifs are likely to make direct, simultaneous contact with two sites on the surface of AP-2. The Nef dileucine motif belongs to the larger class of dileucine-based sorting signals defined by the (D/E)XXXL(L/I) consensus motif that mediate the transport of transmembrane proteins in post-Golgi trafficking pathways (4). Sequences conforming to this motif bind to AP-1, AP-2, and AP-3 on what is presumably a conserved pocket on the surfaces of the γ-σ1, α-σ2, and δ-σ3 hemicomplexes, respectively (8, 12, 28). The diacidic motif described here is also required for binding to the α-σ2 hemicomplex, though not to the homologous γ-σ1 and δ-σ3 hemicomplexes, indicating that the binding site for this motif is specific to AP-2. The properties of the two binding sites appear quite distinct, since the key residues of their ligands are bulky hydrophobic and acidic residues, respectively. Consistent with an important contribution of electrostatic forces to the overall strength of interaction, Nef-AP-2 binding in vitro is inhibited by high salt concentrations. It should be noted that our Y3H interaction studies were performed using the rat AP-2 α and σ2 sequences. However, human and rat AP-2 α proteins share a high degree of sequence similarity, particularly within the trunk domain (98.8% identity and 99.7% similarity among the first 600 residues), which has been shown to mediate interactions with Nef (8, 28) (Fig. 2). The human and rat AP-2 σ2 sequences are 100% identical. We cannot exclude the possibility that minor differences among α subunits may affect their affinities for Nef, and we are currently performing mutagenesis studies on AP-2 to address this issue.
Among the residues downstream of DD174-175, mutation of E179 to alanine caused the greatest loss in AP-2 binding (Fig. 1). Despite the fact that all of the Nef-GAL4BD fusion proteins used in this study were expressed at similar levels in yeast (data not shown), immunoblot analysis of Nef E179A expression in HeLa cells revealed decreased protein levels in HeLa cells relative to those for the wild type (data not shown), a finding similar to that reported previously for the ERE177-179AAA triple mutant (2). Thus, this mutation appears to destabilize Nef, preventing us from concluding that E179 is directly involved in CD4 downregulation. It is notable that among the C-terminal loop residues, only mutations within the dileucine and diacidic sites produced completely null AP-2-binding and CD4 downregulation phenotypes (Fig. 1). Other sites within the loop that produced only a partial loss of binding, such as E160 (28), resulted in minor changes in CD4 downregulation (data not shown). These data indicate the central importance of the dileucine and diacidic motifs.
Other dileucine-based sorting signals that fit the (D/E)XXXL(L/I) consensus motif, such as those in tyrosinase (ERQPLL) (Fig. 6) and Limp2 (ERAPLI) (data not shown), are capable of binding AP-2 independently of any acidic motifs. How might we explain the different properties of these signals vis-à-vis the Nef dileucine motif? We think that the tyrosinase and Limp2 signals are intrinsically strong ligands for the dileucine-binding site on AP-2, obviating the need for secondary determinants. In contrast, both the dileucine and diacidic motifs in Nef appear to be inherently weak, necessitating the presence of both for binding to AP-2. Indeed, substitution of the Nef ENTSLL sequence for the ERQPLL sequence in the tyrosinase cytosolic tail abrogates binding to AP-2 (Fig. 6B). Conversely, substitution of ERQPLL for ENTSLL in Nef confers diacidic-motif-independent binding to AP-2 (Fig. 6B). Remarkably, the Nef ENTSLL signal mediates strong, diacidic-motif-independent binding to AP-1 and AP-3, highlighting the different preferences of these AP complexes for particular dileucine motifs. These findings demonstrate the importance of the X residues within (D/E)XXXL(L/I) signals as key determinants of binding affinity and specificity.
The functional differences between the Nef and tyrosinase dileucine motifs beg the question as to why such a weak AP-2-binding motif is so highly conserved among Nef proteins from different HIV and SIV isolates. One possibility is that bivalent binding imparts a particular orientation to Nef and/or AP-2 that may be required for function. Another possibility is that the specific nature of the Nef dileucine motif is conserved for purposes other than just AP-2 binding and CD4 downregulation. Some evidence already exists for this. Mutations of the acidic residue of the Nef dileucine motif (E160) and, to a lesser extent, of N161, T162, and S163, have deleterious effects on Nef-dependent upregulation of DC-SIGN and the MHC-II-associated invariant chain (9), despite the fact that these residues play little if any role in CD4 downregulation (28).
The distinct requirement of the diacidic motif for binding to AP-2 lends further support to the involvement of this complex in CD4 downregulation and to enhanced endocytosis as the mechanism for CD4 downregulation. Since the diacidic motif is also required for Nef to induce downregulation of CD28 and CD8αβ (53), we expect that downregulation of these proteins also will be found to be AP-2 dependent and mediated by enhanced endocytosis. In contrast, neither the diacidic motif nor AP-2 is required for MHC-I downregulation by Nef (38, 48, 52; also data not shown). Instead, this downregulation depends on a predicted N-terminal α helix (41), a separate acidic cluster (residues 62 to 65), a polyproline motif in Nef (19), and the AP-1 complex (38, 48). Thus, Nef has evolved different means of interfering with the surface expression of various host cell proteins.
The conserved diacidic motif present in all Nef variants represents a third type of determinant of interaction with AP-2, in addition to (D/E)XXXL(L/I) dileucine-based signals and YXXØ tyrosine-based signals (where Ø is a bulky hydrophobic residue) (4). This multiplicity of binding determinants is reminiscent of that of the endoplasmic-reticulum-associated COPII coat, which is also capable of interacting with diverse signals through different interaction surfaces (39). The Nef diacidic motif differs from most dileucine- and tyrosine-based signals, however, in that it does not seem capable of binding AP-2 without the contribution of the dileucine motif. It remains to be determined whether acidic motifs from other proteins also mediate interaction with AP-2. A diacidic pair in the cytosolic tail of tyrosinase does not contribute significantly to AP-2 binding mediated by a typical dileucine-based signal (Fig. 6). However, the cytosolic tails of many transmembrane proteins have acidic clusters that function as sorting signals (4). One in particular, found on the cytosolic tail of furin, mediates endocytosis in addition to trans-Golgi network localization (55) and is therefore a good candidate for interaction with AP-2. Alternatively, the interaction of a diacidic motif with AP-2 might be peculiar to Nef. This would make this interaction an “Achilles' heel,” which could be targeted for pharmacologic prevention of the pathogenic effects of Nef.
Acknowledgments
We thank B. Beach, N. Tsai, and X. Zhu for technical assistance and R. Mattera and E. Dell'Angelica for critical readings of the manuscript.
This work was supported by the intramural programs of NICHD and NIDDK and by the NIH Intramural AIDS Targeted Antiviral Program (IATAP). W.J.S. is the recipient of NICHD Career Transition Award K22 HD54602. R.C. was supported by the NIH-Cambridge and Gates-Cambridge graduate scholarships.
Footnotes
Published ahead of print on 21 November 2007.
REFERENCES
- 1.Aiken, C., J. Konner, N. R. Landau, M. E. Lenburg, and D. Trono. 1994. Nef induces CD4 endocytosis: requirement for a critical dileucine motif in the membrane-proximal CD4 cytoplasmic domain. Cell 76853-864. [DOI] [PubMed] [Google Scholar]
- 2.Aiken, C., L. Krause, Y. L. Chen, and D. Trono. 1996. Mutational analysis of HIV-1 Nef: identification of two mutants that are temperature-sensitive for CD4 downregulation. Virology 217293-300. [DOI] [PubMed] [Google Scholar]
- 3.Arora, V. K., B. L. Fredericksen, and J. V. Garcia. 2002. Nef: agent of cell subversion. Microbes Infect. 4189-199. [DOI] [PubMed] [Google Scholar]
- 4.Bonifacino, J. S., and L. M. Traub. 2003. Signals for sorting of transmembrane proteins to endosomes and lysosomes. Annu. Rev. Biochem. 72395-447. [DOI] [PubMed] [Google Scholar]
- 5.Bour, S., U. Schubert, and K. Strebel. 1995. The human immunodeficiency virus type 1 Vpu protein specifically binds to the cytoplasmic domain of CD4: implications for the mechanism of degradation. J. Virol. 691510-1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bresnahan, P. A., W. Yonemoto, S. Ferrell, D. Williams-Herman, R. Geleziunas, and W. C. Greene. 1998. A dileucine motif in HIV-1 Nef acts as an internalization signal for CD4 downregulation and binds the AP-1 clathrin adaptor. Curr. Biol. 81235-1238. [DOI] [PubMed] [Google Scholar]
- 7.Burtey, A., J. Z. Rappoport, J. Bouchet, S. Basmaciogullari, J. Guatelli, S. M. Simon, S. Benichou, and A. Benmerah. 2007. Dynamic interaction of HIV-1 Nef with the clathrin-mediated endocytic pathway at the plasma membrane. Traffic 861-76. [DOI] [PubMed] [Google Scholar]
- 8.Chaudhuri, R., O. W. Lindwasser, W. J. Smith, J. H. Hurley, and J. S. Bonifacino. 2007. Downregulation of CD4 by human immunodeficiency virus type 1 Nef is dependent on clathrin and involves direct interaction of Nef with the AP2 clathrin adaptor. J. Virol. 813877-3890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coleman, S. H., R. Madrid, N. Van Damme, R. S. Mitchell, J. Bouchet, C. Servant, S. Pillai, S. Benichou, and J. C. Guatelli. 2006. Modulation of cellular protein trafficking by human immunodeficiency virus type 1 Nef: role of the acidic residue in the ExxxLL motif. J. Virol. 801837-1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coleman, S. H., N. Van Damme, J. R. Day, C. M. Noviello, D. Hitchin, R. Madrid, S. Benichou, and J. C. Guatelli. 2005. Leucine-specific, functional interactions between human immunodeficiency virus type 1 Nef and adaptor protein complexes. J. Virol. 792066-2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Craig, H. M., M. W. Pandori, and J. C. Guatelli. 1998. Interaction of HIV-1 Nef with the cellular dileucine-based sorting pathway is required for CD4 down-regulation and optimal viral infectivity. Proc. Natl. Acad. Sci. USA 9511229-11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Doray, B., I. Lee, J. Knisely, G. Bu, and S. Kornfeld. 2007. The γ/σ1 and α/σ2 hemicomplexes of clathrin adaptors AP-1 and AP-2 harbor the dileucine recognition site. Mol. Biol. Cell 181887-1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Foti, M., A. Mangasarian, V. Piguet, D. P. Lew, K. H. Krause, D. Trono, and J. L. Carpentier. 1997. Nef-mediated clathrin-coated pit formation. J. Cell Biol. 13937-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garcia, J. V., and A. D. Miller. 1991. Serine phosphorylation-independent downregulation of cell-surface CD4 by nef. Nature 350508-511. [DOI] [PubMed] [Google Scholar]
- 15.Geyer, M., C. E. Munte, J. Schorr, R. Kellner, and H. R. Kalbitzer. 1999. Structure of the anchor-domain of myristoylated and non-myristoylated HIV-1 Nef protein. J. Mol. Biol. 289123-138. [DOI] [PubMed] [Google Scholar]
- 16.Geyer, M., H. Yu, R. Mandic, T. Linnemann, Y. H. Zheng, O. T. Fackler, and B. M. Peterlin. 2002. Subunit H of the V-ATPase binds to the medium chain of adaptor protein complex 2 and connects Nef to the endocytic machinery. J. Biol. Chem. 27728521-28529. [DOI] [PubMed] [Google Scholar]
- 17.Greenberg, M., L. DeTulleo, I. Rapoport, J. Skowronski, and T. Kirchhausen. 1998. A dileucine motif in HIV-1 Nef is essential for sorting into clathrin-coated pits and for downregulation of CD4. Curr. Biol. 81239-1242. [DOI] [PubMed] [Google Scholar]
- 18.Greenberg, M. E., S. Bronson, M. Lock, M. Neumann, G. N. Pavlakis, and J. Skowronski. 1997. Co-localization of HIV-1 Nef with the AP-2 adaptor protein complex correlates with Nef-induced CD4 down-regulation. EMBO J. 166964-6976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Greenberg, M. E., A. J. Iafrate, and J. Skowronski. 1998. The SH3 domain-binding surface and an acidic motif in HIV-1 Nef regulate trafficking of class I MHC complexes. EMBO J. 172777-2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grzesiek, S., A. Bax, G. M. Clore, A. M. Gronenborn, J. S. Hu, J. Kaufman, I. Palmer, S. J. Stahl, and P. T. Wingfield. 1996. The solution structure of HIV-1 Nef reveals an unexpected fold and permits delineation of the binding surface for the SH3 domain of Hck tyrosine protein kinase. Nat. Struct. Biol. 3340-345. [DOI] [PubMed] [Google Scholar]
- 21.Grzesiek, S., A. Bax, J. S. Hu, J. Kaufman, I. Palmer, S. J. Stahl, N. Tjandra, and P. T. Wingfield. 1997. Refined solution structure and backbone dynamics of HIV-1 Nef. Protein Sci. 61248-1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grzesiek, S., S. J. Stahl, P. T. Wingfield, and A. Bax. 1996. The CD4 determinant for downregulation by HIV-1 Nef directly binds to Nef. Mapping of the Nef binding surface by NMR. Biochemistry 3510256-10261. [DOI] [PubMed] [Google Scholar]
- 23.Harris, M. P., and J. C. Neil. 1994. Myristoylation-dependent binding of HIV-1 Nef to CD4. J. Mol. Biol. 241136-142. [DOI] [PubMed] [Google Scholar]
- 24.Henikoff, S., and J. G. Henikoff. 1992. Amino acid substitution matrices from protein blocks. Proc. Natl. Acad. Sci. USA 8910915-10919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hodge, D. R., K. J. Dunn, G. K. Pei, M. K. Chakrabarty, G. Heidecker, J. A. Lautenberger, and K. P. Samuel. 1998. Binding of c-Raf1 kinase to a conserved acidic sequence within the carboxyl-terminal region of the HIV-1 Nef protein. J. Biol. Chem. 27315727-15733. [DOI] [PubMed] [Google Scholar]
- 26.Iafrate, A. J., S. Bronson, and J. Skowronski. 1997. Separable functions of Nef disrupt two aspects of T cell receptor machinery: CD4 expression and CD3 signaling. EMBO J. 16673-684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Janvier, K., H. Craig, D. Hitchin, R. Madrid, N. Sol-Foulon, L. Renault, J. Cherfils, D. Cassel, S. Benichou, and J. Guatelli. 2003. HIV-1 Nef stabilizes the association of adaptor protein complexes with membranes. J. Biol. Chem. 2788725-8732. [DOI] [PubMed] [Google Scholar]
- 28.Janvier, K., Y. Kato, M. Boehm, J. R. Rose, J. A. Martina, B. Y. Kim, S. Venkatesan, and J. S. Bonifacino. 2003. Recognition of dileucine-based sorting signals from HIV-1 Nef and LIMP-II by the AP-1 γ-σ1 and AP-3 δ-σ3 hemicomplexes. J. Cell Biol. 1631281-1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jin, Y. J., C. Y. Cai, X. Zhang, H. T. Zhang, J. A. Hirst, and S. J. Burakoff. 2005. HIV Nef-mediated CD4 down-regulation is adaptor protein complex 2 dependent. J. Immunol. 1753157-3164. [DOI] [PubMed] [Google Scholar]
- 30.Kasper, M. R., J. F. Roeth, M. Williams, T. M. Filzen, R. I. Fleis, and K. L. Collins. 2005. HIV-1 Nef disrupts antigen presentation early in the secretory pathway. J. Biol. Chem. 28012840-12848. [DOI] [PubMed] [Google Scholar]
- 31.Lama, J. 2003. The physiological relevance of CD4 receptor down-modulation during HIV infection. Curr. HIV Res. 1167-184. [DOI] [PubMed] [Google Scholar]
- 32.Lee, C. H., K. Saksela, U. A. Mirza, B. T. Chait, and J. Kuriyan. 1996. Crystal structure of the conserved core of HIV-1 Nef complexed with a Src family SH3 domain. Cell 85931-942. [DOI] [PubMed] [Google Scholar]
- 33.Le Gall, S., L. Erdtmann, S. Benichou, C. Berlioz-Torrent, L. Liu, R. Benarous, J. M. Heard, and O. Schwartz. 1998. Nef interacts with the mu subunit of clathrin adaptor complexes and reveals a cryptic sorting signal in MHC I molecules. Immunity 8483-495. [DOI] [PubMed] [Google Scholar]
- 34.Leitner, T., B. Foley, B. Hahn, P. Marx, F. McCutchan, J. Mellors, S. Wolinsky, and B. Korber (ed.). 2005. HIV sequence compendium 2005. Theoretical Biology and Biophysics Group, Los Alamos National Laboratory, Los Alamos, NM.
- 35.Levesque, K., A. Finzi, J. Binette, and E. A. Cohen. 2004. Role of CD4 receptor down-regulation during HIV-1 infection. Curr. HIV Res. 251-59. [DOI] [PubMed] [Google Scholar]
- 36.Lindwasser, O. W., R. Chaudhuri, and J. S. Bonifacino. 2007. Mechanisms of CD4 downregulation by the Nef and Vpu proteins of primate immunodeficiency viruses. Curr. Mol. Med. 7171-184. [DOI] [PubMed] [Google Scholar]
- 37.Lu, X., H. Yu, S. H. Liu, F. M. Brodsky, and B. M. Peterlin. 1998. Interactions between HIV1 Nef and vacuolar ATPase facilitate the internalization of CD4. Immunity 8647-656. [DOI] [PubMed] [Google Scholar]
- 38.Lubben, N. B., D. A. Sahlender, A. M. Motley, P. J. Lehner, P. Benaroch, and M. S. Robinson. 2007. HIV-1 Nef-induced down-regulation of MHC class I requires AP-1 and clathrin but not PACS-1 and is impeded by AP-2. Mol. Biol. Cell 183351-3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mancias, J. D., and J. Goldberg. 2007. The transport signal on Sec22 for packaging into COPII-coated vesicles is a conformational epitope. Mol. Cell 26403-414. [DOI] [PubMed] [Google Scholar]
- 40.Mangasarian, A., M. Foti, C. Aiken, D. Chin, J. L. Carpentier, and D. Trono. 1997. The HIV-1 Nef protein acts as a connector with sorting pathways in the Golgi and at the plasma membrane. Immunity 667-77. [DOI] [PubMed] [Google Scholar]
- 41.Mangasarian, A., V. Piguet, J. K. Wang, Y. L. Chen, and D. Trono. 1999. Nef-induced CD4 and major histocompatibility complex class I (MHC-I) down-regulation are governed by distinct determinants: N-terminal alpha helix and proline repeat of Nef selectively regulate MHC-I trafficking. J. Virol. 731964-1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mariani, R., F. Kirchhoff, T. C. Greenough, J. L. Sullivan, R. C. Desrosiers, and J. Skowronski. 1996. High frequency of defective nef alleles in a long-term survivor with nonprogressive human immunodeficiency virus type 1 infection. J. Virol. 707752-7764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.O'Neill, E., L. S. Kuo, J. F. Krisko, D. R. Tomchick, J. V. Garcia, and J. L. Foster. 2006. Dynamic evolution of the human immunodeficiency virus type 1 pathogenic factor, Nef. J. Virol. 801311-1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Owen, D. J., B. M. Collins, and P. R. Evans. 2004. Adaptors for clathrin coats: structure and function. Annu. Rev. Cell Dev. Biol. 20153-191. [DOI] [PubMed] [Google Scholar]
- 45.Rhee, S. S., and J. W. Marsh. 1994. Human immunodeficiency virus type 1 Nef-induced down-modulation of CD4 is due to rapid internalization and degradation of surface CD4. J. Virol. 685156-5163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Robinson, M. S. 2004. Adaptable adaptors for coated vesicles. Trends Cell Biol. 14167-174. [DOI] [PubMed] [Google Scholar]
- 47.Roeth, J. F., and K. L. Collins. 2006. Human immunodeficiency virus type 1 Nef: adapting to intracellular trafficking pathways. Microbiol. Mol. Biol. Rev. 70548-563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Roeth, J. F., M. Williams, M. R. Kasper, T. M. Filzen, and K. L. Collins. 2004. HIV-1 Nef disrupts MHC-I trafficking by recruiting AP-1 to the MHC-I cytoplasmic tail. J. Cell Biol. 167903-913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rose, J. J., K. Janvier, S. Chandrasekhar, R. P. Sekaly, J. S. Bonifacino, and S. Venkatesan. 2005. CD4 down-regulation by HIV-1 and simian immunodeficiency virus (SIV) Nef proteins involves both internalization and intracellular retention mechanisms. J. Biol. Chem. 2807413-7426. [DOI] [PubMed] [Google Scholar]
- 50.Rossi, F., A. Gallina, and G. Milanesi. 1996. Nef-CD4 physical interaction sensed with the yeast two-hybrid system. Virology 217397-403. [DOI] [PubMed] [Google Scholar]
- 51.Shugars, D. C., M. S. Smith, D. H. Glueck, P. V. Nantermet, F. Seillier-Moiseiwitsch, and R. Swanstrom. 1993. Analysis of human immunodeficiency virus type 1 nef gene sequences present in vivo. J. Virol. 674639-4650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stoddart, C. A., R. Geleziunas, S. Ferrell, V. Linquist-Stepps, M. E. Moreno, C. Bare, W. Xu, W. Yonemoto, P. A. Bresnahan, J. M. McCune, and W. C. Greene. 2003. Human immunodeficiency virus type 1 Nef-mediated downregulation of CD4 correlates with Nef enhancement of viral pathogenesis. J. Virol. 772124-2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stove, V., I. Van de Walle, E. Naessens, E. Coene, C. Stove, J. Plum, and B. Verhasselt. 2005. Human immunodeficiency virus Nef induces rapid internalization of the T-cell coreceptor CD8αβ. J. Virol. 7911422-11433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Theos, A. C., D. Tenza, J. A. Martina, I. Hurbain, A. A. Peden, E. V. Sviderskaya, A. Stewart, M. S. Robinson, D. C. Bennett, D. F. Cutler, J. S. Bonifacino, M. S. Marks, and G. Raposo. 2005. Functions of adaptor protein (AP)-3 and AP-1 in tyrosinase sorting from endosomes to melanosomes. Mol. Biol. Cell 165356-5372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Voorhees, P., E. Deignan, E. van Donselaar, J. Humphrey, M. S. Marks, P. J. Peters, and J. S. Bonifacino. 1995. An acidic sequence within the cytoplasmic domain of furin functions as a determinant of trans-Golgi network localization and internalization from the cell surface. EMBO J. 144961-4975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Witte, V., B. Laffert, O. Rosorius, P. Lischka, K. Blume, G. Galler, A. Stilper, D. Willbold, P. D'Aloja, M. Sixt, J. Kolanus, M. Ott, W. Kolanus, G. Schuler, and A. S. Baur. 2004. HIV-1 Nef mimics an integrin receptor signal that recruits the polycomb group protein Eed to the plasma membrane. Mol. Cell 13179-190. [DOI] [PubMed] [Google Scholar]