Abstract
Two similar, large double-stranded DNA viruses, Feldmannia species virus 158 (FsV-158) and FsV-178, replicate only in the unilocular reproductive cells (sporangia) of a brown filamentous alga in the genus Feldmannia. Virus particles are not present in vegetative cells but they are produced in the sporangia formed on vegetative filaments that have been transferred newly into culture. Thus, we proposed that these viruses exist in the vegetative cells in a latent form (R. G. Ivey, E. C. Henry, A. M. Lee, L. Klepper, S. K. Krueger, and R. H. Meints, Virology 220:267-273, 1996). In this article we present evidence that the two FsV genomes are integrated into the host genome during vegetative growth. The FsV genome integration sites were identified by cloning the regions where the FsV genome is linked to the host DNA. FsV-158 and FsV-178 are integrated into two distinct locations in the algal genome. In contrast, the integration sites in the two viral genomes are identical. Notably, the integration sites in the host and viruses contain GC and CG dinucleotide sequences, respectively, from which the GC sequences are recovered at both host-virus junctions. The splice sites in the two FsV genomes are predicted to form a stem-loop structure with the CG dinucleotide in the loop portion.
The discovery of large double-stranded DNA (dsDNA) viruses over the past several decades has extended the upper limit of the size range of virus genomes severalfold. These viruses infect widely disparate organisms ranging from simple filamentous brown algae, like Feldmannia and Ectocarpus (Feldmannia species virus [FsV] and Ectocarpus siliculosus virus [EsV], respectively; ∼ 160 to 320 kbp) (7, 14), and single-celled green algae, an internal symbiont of Paramecium (Paramecium bursaria chlorella virus type 1; ∼330 kbp) (18), to the amoeba Acanthameoba, which may be infected by the recently discovered 1.2-Mbp mimivirus (3, 16). This virus has the largest virus genome described to date and contains as many as 911 open reading frames (ORFs).
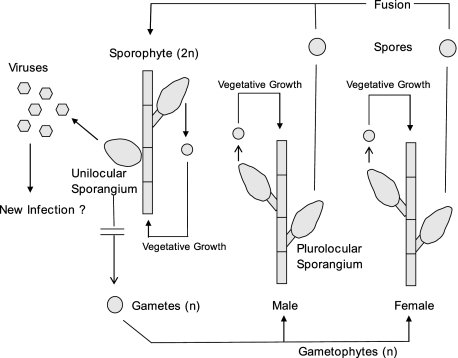
Previously, we reported that cultured isolates of a taxonomically undesignated brown algal species in the genus Feldmannia produce a large dsDNA virus, FsV, only within reproductive cells (sporangia) (7). Algae in the genus Feldmannia typically grow in marine habitats in long filaments (∼10 cm). They reproduce using a complex “alternation of generations” life cycle consisting of vegetative and/or a sexually reproductive diploid sporophytes and a vegetative haploid gametophyte stage (Fig. 1). Although it has been reported that both diploid sporophyte and haploid gametophyte are isomorphic stages, the gametophytes contain only plurilocular (multicelled) sporangia as reproductive structures, and the sporophytes contain two forms, similar plurilocular sporangia and unique, unilocular (single-celled) sporangia wherein meiosis occurs, producing haploid gametes.
FIG. 1.
Life cycle cartoon of Feldmannia sp. The alga has both a haploid and diploid form. Vegetative growth and germination of spores allow for complete survival in the absence of sexual reproduction. In this isolate, the sporophyte is not produced because viruses are produced in the unilocular sporangia rather than gametes. n, number of genome copies.
Both gametophyte and sporophyte algae grow vigorously in culture when started from either a microfilament or individual cells. In healthy sporophytes the unicellular, unilocular sporangia produce an estimated 1 million haploid gametes through meiosis. However, in the Feldmannia sp. sporophytic isolate described here, the unilocular sporangium does not produce gametes but instead produces up to 5 million virus particles per cell (7). Apparently, in nature viable gametes are produced occasionally from a virus-infected sporophyte that can develop normally to give a gametophyte generation. Sporophyte cultures started from single vegetative cells or spores resulted in mature plants, which develop unilocular sporangia that produce virus. An uninfected form of this alga has not been isolated from nature; therefore, infection experiments cannot be conducted with FsV.
There is no evidence of virus replication in the vegetative cells or plurilocular reproductive cells of either gametophyte or sporophyte plants. However, Southern blot analysis of the gametophyte DNA probed with a virus-specific probe indicated that virus DNA was present in the host (12). In this form the viruses are stably transmitted during meiosis and vegetative growth and expressed only in cells normally programmed for meiotic, gamete production.
The icosahedral capsid of FsV is 150 nm in diameter and encloses a dsDNA genome (7). In laboratory-grown algal isolates, viruses with two genome size-classes of 158 and 178 kbp are produced (designated FsV-158 and FsV-178), whose ratio is dependent on the temperature used to grow the host (8). The two viral genomes with circular restriction maps are very similar but differ in the number of small, ∼170-bp repeats (8, 11). Both virus genomes are stable in the host after 7 years of monthly serial passage (8). Genes encoding DNA polymerase, protein kinases, integrases, transposases, and other enzymes have been identified in the FsV genome (9, 12, 13, 15). A comparative phylogenetic analysis of the FsV DNA polymerase with other viral DNA polymerases indicates that FsV belongs to a family of algal viruses named Phycodnaviridae (13). Villarreal and DeFilippis (20) have suggested that the FsV DNA polymerase may be the closest relative to the progenitor of all DNA polymerase delta molecules.
In the current article we identify the FsV genomic integration sites in the host algal DNA and clone the host-virus genome integration sites.
MATERIALS AND METHODS
Algal cultures.
Unialgal cultures were initiated from single vegetative cells of the filamentous brown alga, Feldmannia sp. (Fig. 1). Algal culture and virus DNA purification were carried out as previously described (7, 11).
Southern hybridization.
Electrophoresed DNA was depurinated in 0.25 N HCl, denatured in 1.5 M NaCl-0.5 N NaOH, transferred by capillary action onto a nylon membrane (Hybond-NTM; Amersham) overnight, and baked for 2 h at 80°C. Probes were prepared by radiolabeling 25 to 50 ng of gel-purified DNA with 50 μCi of [γ-32P]dCTP (3,000 Ci/mM) using a random primer extension kit (United States Biochemicals, Cleveland, OH). Southern blots of FsV DNA were prehybridized for 1 h in hybridization buffer (50% formamide, 7% sodium dodecyl sulfate, 0.12 M Na-phosphate buffer, pH 8.2) and then hybridized at 42°C for 6 h in 15 ml of the same buffer containing 1.5 × 107 to 3 × 107 dpm of labeled probe. Hybridized membranes were washed twice for 20 min in 0.1× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) and 0.1% sodium dodecyl sulfate and autoradiographed at −80°C with intensifying screens.
Cosmid library construction.
Total genomic DNA was isolated from Feldmannia gametophyte culture FsP-G4A. Genomic DNA was partially digested with Sau3AI, treated with calf intestinal alkaline phosphatase, and ligated into the BamHI site of Supercos 1 (Stratagene, La Jolla, CA). The ligated DNA was packaged using Gigapack II packaging extract (Stratagene) and transfected into Escherichia coli XL1-Blue MR cells. The library was screened by colony hybridization (17) with radiolabeled DNA from previously mapped virus fragments. Cosmid DNA was isolated by alkaline lysis DNA minipreps (17). Restriction fragment order was determined by analysis of overlapping clones and by comparison to previously mapped virus clones (8).
Sequencing.
The subfragments of the DNA inserts in cosmids were cloned into the pGEM-3Zf(+) vector (Promega) in the E. coli host, DH5α (17). Template DNA for automated sequencing was prepared using QIAprep miniprep kits (Qiagen, Santa Clarita, CA). Automated DNA sequencing was performed by the Central Services Laboratory, Center for Gene Research and Biotechnology, Oregon State University, Corvallis, OR. Assembly and analysis of DNA sequence data were performed by using the GCG program package (Program manual for the Wisconsin package, version 8, Genetics Computer Group, Madison, WI).
PCR amplification of the unoccupied host integration site.
PCR primers P35HL (5′-ACTGGTACCAGTATTACAATATTCGTATCATAGGTC-3′) and X35HR (5′-ACTCTGCAGGGTGCCATGATGAGTGTTGTGCATACA-3′) were designed to target each of the host DNA regions flanking the integration sites based on the sequence of clones P3.5-7A and X3.5-4, respectively (Fig. 2g and h), and PCR was conducted using total sporophyte DNA as a template. Template DNA (1, 10, and 100 ng of total sporophyte DNA) was mixed individually with 20 pM of the oligonucleotide primers in 1× Taq polymerase buffer with 0.2 mM concentrations of each deoxynucleoside triphosphate and 1.5 mM MgCl2. The reaction mixture was heated at 94°C for 4 min to denature the template, and 2.5 units of Taq polymerase (Gibco/BRL, Gaithersburg, MD) was added. For the first PCR, a 4-min incubation at 94°C was followed by 30 cycles of 94°C for 1 min, 65°C for 1 min, and 72°C for 1 min. After the reaction was completed, 15 μl of the PCR samples was analyzed by electrophoresis. The appropriate PCR products were gel purified and cloned.
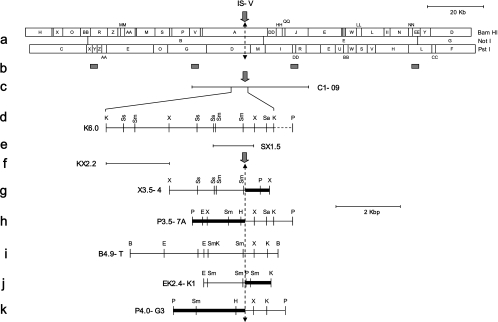
FIG. 2.
Map of the integration site (IS-V and IS-A) containing clones. Location of cosmid clone C1-09 and regions used as probes for G4A library screening are depicted below the linear format of the full-length restriction map of FsV-158 (8). The dashed vertical line represents the junction regions for integration. Restriction maps of the various subclones containing integration sites are below the full-length restriction map, with their names to the left of each map. Thick lines represent host DNA. Restriction sites are as follows: B, BamHI; E, EcoRI; H, HindIII; K, KpnI; P, PstI; Sa, SalI; Sm, SmaI; Ss, SstI; X, XbaI. Refer to the text for the details of the clones.
RESULTS
Viral, algal, and algal/viral map sites.
Fig. 2 represents map elements helpful for understanding the results presented here. These elements include the following: a linearized BamHI, NotI, and PstI map of the normally circular FsV-158 genome (Fig. 2a); the sites on the FsV-158 map of four ∼400-bp cloned fragments used as probes (b); cosmid C1-09, one of five cosmids previously prepared that cover FsV-158 (c); K6.0, a 6-kbp KpnI subclone of C1-09 (d); SX1.5, an SstI-XhoI subclone of K6.0 (e); and KX2.2, a KpnI-XhoI subclone of K6.0 (f). The sequence in the virus genome (see also Fig. 4A and B) which opens (Fig. 2, solid arrows) to permit integration was designated the integration site-virus (IS-V). For nomenclature symmetry the algal chromosome site into which virus insertion occurs was designated the integration site-alga (IS-A). The left and right borders of the opened algal site were also determined. The junctions formed upon viral integration (see also Fig. 4B) are included in X3.5-4, an XhoI subclone containing algal-viral junction right (A-VJR) from FsV-158 integration (Fig. 2g); P3.5-7A, a PstI subclone containing algal-viral junction left (A-VJL) of FsV-158 (h); B4.9-T, a BamHI subclone from viral cosmid C1-41 covering the IS-V region of FsV-178 (i); EK2.4-K1, an EcoRI-KpnI subclone containing A-VJR of the FsV-178 integration (j); and P4.0-G3, the PstI subclone containing A-VJL of the FsV-178 integration (k). The bold lines in Fig. 2g, h, j, and k represent areas of the algal chromosome while the thin lines are those of the virus. The 20-kbp marker refers to the map in Fig. 2a only; all other maps should reference the 2-kpb marker.
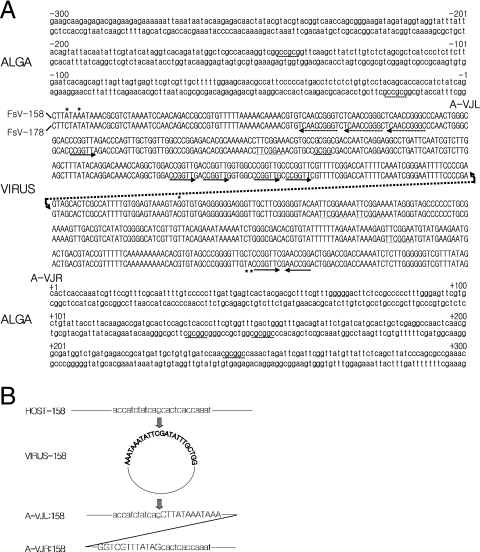
FIG. 4.
(A) Sequence of the host and FsV-158 and FsV-178 around the site of integration. The sequences are numbered from −300 to +300, with zero representing the IS-A. The regions containing virus DNA sequence are represented in uppercase letters while host DNA sequence is in lowercase. The repeated sequences, described in the text, are underlined. The right arrow marks the virus sequence CCGGTT while the left arrow marks the reverse complement, AACCGG. An asterisk marks bases which vary between virus sequences. The virus integration sites and unoccupied host sites are aligned, and the FsV-158 is represented by the upper line of the couplet, and FsV-178 is represented by the lower. The conserved GC dinucleotide present in the viral genome and host integration sites is shown resulting in junctions A-VJL and A-VJR. (B). Detailed sequence representation in the close vicinity of IS-V and IS-A and the junctions created upon integration. Uppercase and lowercase letters are as described for panel A.
Integration site fragment identification.
A cosmid library was prepared from the plant line, G4A, a Feldmannia gametophyte culture which does not produce unilocular sporangia or virus (Fig. 1). Colonies that hybridized with a pool of four short, approximately 400-bp gel-isolated, radiolabeled fragments prepared from purified virus DNA were selected. The probe sites chosen were spaced approximately every 40 kbp on the FsV-158 genome (Fig. 2b) and were taken from conserved regions based on map data from FsV-158 and FsV-178.
This library should contain three insert types: (i) those demonstrating solely algal DNA, (ii) those with internal DNA from the integrated virus, and (iii) those with both algal and virus DNA. The last group should contain the algal chromosome-viral junctions A-VJL or A-VJR. Over 50 cosmids that hybridized with these viral probes were selected. Restriction fragment patterns for each clone were compared with virus DNA patterns on gels, and each viral insert was mapped against previously published maps prepared for FsV-158 and FsV-178 (8). Most of the inserts produced maps identical to those already known for purified virus. These inserts were presumed to arise from viral DNA internal to the integration junction sites.
There were, in addition, distinctly aberrant insert types that hybridized with one specific probe, the PstI fragment DD isolated from C1-09 (Fig. 2b). These inserts gave different patterns compared to gels of purified virus DNA. They also failed to reproduce established FsV restriction fragment maps.
Southern blots from these aberrant cosmids were positive when probed with the entire viral probe, C1-09 (data not shown). This result suggested that C1-09 contains the viral sequence IS-V, which must open to produce right and left ends of a linearized form for integration into the algal chromosome. Furthermore, it suggested that these aberrant cosmids contained in addition the junctions A-VJL and A-VJR created upon viral integration.
Isolation of IS-V, A-VJL, and A-VJR.
Based on the viral integration hypothesis, we proposed that small subfragments containing IS-V from C1-09 should disappear from gel patterns of the G4A library and be replaced by newly created fragments containing A-VJL and A-VJR. This linkage of viral and algal sequences resulted in size shifts of two restriction fragments in the G4A library. The fragments P3.5-7A and P4.0-G3 (Fig. 2h and k) were gel isolated and determined to fulfill this criterion. Through mapping and sequence analysis, they were shown to contain A-VJLs of distinct FsV-158 and FsV-178 DNA integrations.
Two subclones, a 6.0-kbp KpnI-fragment (Fig. 2d, K6.0) from C1-09 and a 4.9-kbp BamHI-fragment (Fig. 2i, B4.9-T) from C1-41 covering the same region of FsV-158 and -178, respectively, were isolated from viral cosmid libraries (8). SX1.5, a subclone from K6.0, was identified by positive hybridization to P3.5-7A and P4.0-G3 and shown to possibly contain IS-V (Fig. 2e). A second subclone, KX2.2 (Fig. 2f) from K6.0, known to reside outside of the sequence of IS-V, was isolated. Aberrant cosmids from the G4A library which hybridized with KX2.2 should contain the other junctions of both FsV-158 and -178 integrations. From these cosmids two independent A-VJR-containing subclones, X3.5-4 and EK2.4-K1, were obtained (Fig. 2g and j).
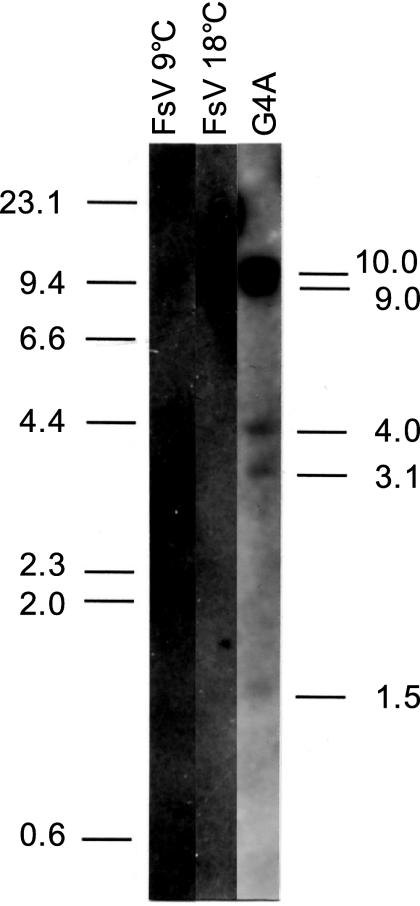
Genomic DNA of FsV, prepared from host cells cultured at 9°C and 18°C, and total G4A genomic DNA were digested with EcoRI and PstI, and Southern blots were probed with SX1.5 (Fig. 2e). Viral genomic DNA prepared from host cells cultured at 9°C, in which the large-size FsV-178 is predominant (8), showed a strong band of the expected size of 2.5 kbp and a faint band of about 10 kb from the FsV-158 (Fig. 3, lane 1). The band pattern was reversed in the viral genomic DNA prepared from 18°C in which the FsV-158 is dominant (Fig. 3, lane 2). The SX1.5 probe hybridized to five DNA fragments (Fig. 3, lane 3). The sizes of four such DNA fragments, 9.0, 4.0, 3.1, and 1.5 kbp, corresponded to the sizes of each DNA fragment containing the algal-virus junctions in the G4A library (data not shown). Unexpectedly, a 10-kbp DNA fragment containing the IS-V of the intact virus also hybridized to SX1.5 (Fig. 3, lane 3).
FIG. 3.
Autoradiograph of gametophyte DNA (G4A) and FsV DNA (from either 9°C or 18°C cultures) digested with both EcoRI and PstI and probed with SX1.5-K6, a subclone of K6.0 (−1064 to +334), containing the IS-V site. Positions of the size markers (in kbp) are indicated on the left. The sizes of the individual hybridization fragments are labeled on the right. The 10.0-kbp fragment corresponds to intact FsV-158, 9.0 corresponds to X3.5-4, 4.0 corresponds to P4.0-G3, 3.1 corresponds to P3.5-7A, and 1.5 corresponds to EK2.4-K1. The 2.5 kbp FsV-178 fragment is absent in the G4A lane.
Sequencing of integration sites.
Four DNA fragments containing A-VJLs and A-VJRs of the FsV-158 and FsV-178 integrations were sequenced in both directions. The IS-Vs from both viral forms were sequenced as well. Alignment of the nucleotide sequence data indicates that the IS-Vs from both FsV-158 and FsV-178 which open to permit integration are virtually identical (Fig. 4A). Two nucleotide differences occur at positions 4 and 7 of the left IS-V, and three occur at positions −170, −53, and −54 of right IS-V (Fig. 4A, asterisks).
However, the two sites in the algal chromosome where the two virus genomes integrated were entirely unique (Fig. 4A). A CG dinucleotide pair defines the integration cut site, which occurs in the IS-Vs of both FsV-158 and FsV-178. A complementary GC dinucleotide is found in the IS-A where either FsV-158 or FsV-178 integrates. Two GC sequences are reformed upon insertion at both the A-VJL and A-VJR of both the FsV-158 and FsV-178 integrations (Fig. 4B).
Based on restriction maps and sequence data, the virus portions of P3.5-7A and P4.0-G3 (Fig. 2, thin lines on maps) are similar to K6.0 from the 158-kbp genome and B4.9-T from the 178-kbp genome, respectively. These were designated A-VJL for both virus forms. Furthermore, in the virus DNA-containing regions of the A-VJR clones, X3.5-4 is similar to K6.0 (158-kbp virus genome) while EK2.4-K1 is more similar to B4.9-T (178-kbp virus genome). However, there was some variation at the nucleotide sequence level between sequences from purified viruses and their integrated counterparts. The region of homology between K6.0 and P3.5-7A differs by only a single base pair between the integration site and the KpnI site 1,020 bp distant. Greater sequence variation exists between B4.9-T and P4.0-G3; there are two restriction site polymorphisms and 13 base differences between the two sequences. The sequence between the SstI site and the integration site (1,064 bp) is identical between the K6.0 and X3.5-4 clones while the sequence of EK2.4-K1 differs at 29 positions relative to B4.9-T.
Fine structure at the integration site.
The IS-V region contains short repeated sequences on both sides of the cut site (Fig. 4A). The sequence TTCGGAA occurs three times in the right border region and once to the left within 200 bp of the IS-V (Fig. 4A, underlined). The sequence CCGGTT occurs five times to the left border of IS-V and once to the right (Fig. 4A, rightward arrows). There, an inverse complement AACCGG is found once to the right in close proximity and three times to the left within 300 bp of IS-V (leftward arrows). At AACCGG sites in the left border, the repeats are contained within the larger repeat, TCAACCGGG (dotted leftward arrows). The 5-bp sequence CGCGG occurs in different regions of both algal genome border regions within 300 bp of where FsV-158 and FsV-178 integrate and is included in the longer GCCGCG sequence in the left of the integration site and the reverse CGCGGC to the right (dotted underlines). The significance or utility of these small repeats has not been established.
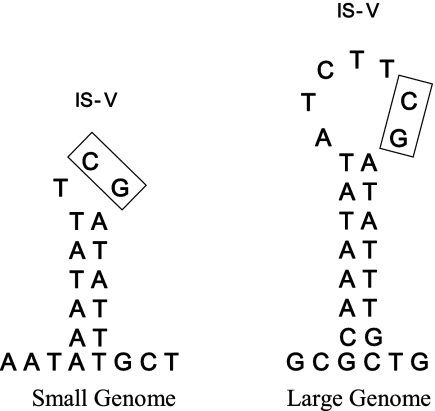
The IS-Vs of both viral genomes contain short inverted repeats that are capable of forming stem-loop structures often associated with the recombination sites (Fig. 5). The IS-V sequence from FsV-158 is capable of forming a 6-bp stem with a 3-bp loop while the sequence from the IS-V of the 178-kbp genome can form an 8-bp stem with a 7-bp loop (Fig. 5). In both cases the CG dinucleotide occurs in the loop structure as a part of a common TTCGATATTTGCT sequence. There are no regions of homology between the virus and algal sequences on either side of the junction sites beyond these small sequences. Stem-loops cannot be drawn based on sequences from the algal IS-A borders.
FIG. 5.
Stem-loop structures are found at the integration site of the small (158 kbp) and large (178 kbp) genomes of FsV. The boxes indicate the conserved CG dinucleotides. The conserved CG dinucleotide is present in both viral genomes, the host integration sites, and the resulting junctions A-VJL and A-VJR.
The sequence data were used to query the sequence databases using the BLAST program (1) to identify ORFs encoding proteins with known functions near IS-V and IS-A. The only significant finding was a 495-bp ORF located in reverse orientation 175 bp to the left of IS-V. This ORF contains a RING zinc finger motif (9).
Isolation of unoccupied integration sites.
Primers based on algal DNA regions flanking the A-VJL and A-VJR were used for PCR analysis of total G4A DNA to determine if an unoccupied site might exist. A DNA fragment of 587 bp was amplified (data not shown). This fragment is the expected size based on sequence data that assumes the virus is not integrated. The alignment of the PCR product sequence and algal regions of the A-VJL and A-VJR showed that the unoccupied IS-A would form the A-VJL and A-VJR upon the integration of the FsV genome (Fig. 4A).
DISCUSSION
FsV is produced only within unilocular sporangia that release millions of spores in healthy plants. The persistent infection of host algal culture in the absence of demonstrable infection suggested that the FsV genome either integrates into the host genome or occurs as an intact intracellular virus episome (8).
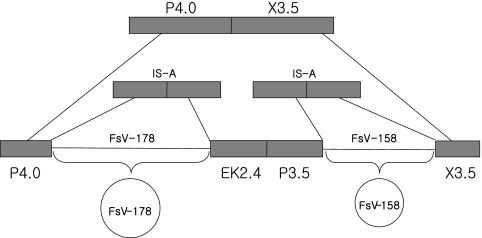
In the present study, two virus integration sites for a large dsDNA FsV genome were identified in the gametophyte G4A line. DNA sequence alignment clearly demonstrates that integration occurred at two sequence-specific host locations, likely on the same linkage group as explained in Fig. 6. In contrast, the viral site (IS-V) that opens for preintegration linearization of the normally circular viral genome is identical for the two viral forms. All four host-viral junctions were identified and sequenced. A host site that lacked virus DNA was also identified.
FIG. 6.
A model for the generation of the unoccupied host integration sites. Two different virus genomes are integrated into the same host chromosome (middle). The host-virus junctions and virus genomes are labeled. The virus genome excises at sites P3.5 and X3.5 to generate a functional virus capable of replication and packaging while restoring the host sites P3.5/X3.5 (top). An alternative excision generates two FsV genomes spliced together and unable to replicate. This would bring junctions P4.0 and X3.5 into juxtaposition.
In addition, a 10-kbp virus DNA fragment that spanned the IS-V of FsV-158 was identified (Fig. 3, lane 3). Two possible explanations for this observation are that another IS-V site exists which we failed to identify or that intact virus DNA is present in the vegetative cells. This would be the case if there were an infection or an excision. Calculations based on Southern hybridization signals indicate that if the virus genome is maintained as an intact form, each virus genome is present at only one or two copies per cell. This could be the case if integration and excision were in progress.
Based on restriction site analysis and sequence data, we suggest that clones P3.5-7A and X3.5-4 contain the borders of a single IS-A site for FsV-158. Similarly, P4.0-G3 and EK2.4-K1 define a second host location for FsV-178 (Fig. 6). Using PCR primers designed from P3.5-7A and X3.5-4, we were able to demonstrate an unoccupied IS-A from the host. We did not identify an unoccupied host site for FsV-178 using P4.0-G3 and EK2.4-K1 primers.
Unexpectedly, we amplified a fragment using primers derived from P4.0-G3 (FsV-158) and X3.5-4 (FsV-178). A possible explanation for the P4.0-G3/X3.5-4 PCR product is depicted in Fig. 6. It suggests that the two IS-A sites occur in tandem, possibly with a region of intervening host DNA. The most parsimonious explanation is that while a single excision of FsV-158 led to a proper host site, FsV-178 does not excise alone, hence, our inability to amplify such a site. However, if both viruses excise simultaneously, then the intervening algal DNA will be lost, and a hybrid site described above would be created.
The two independent IS-A sites from P3.5-7A/X3.5-4 and P4.0-G3/EK2.4-K1 show little sequence identity other than a GC motif at the site of integration and GCCGCG to the left of the integration site and its reverse complement, CGCGGC, to the right.
Two characteristics of phaeoviruses, found in FsV as well as EsV, are their long latency period and apparent lysogeny (2, 4, 18, 19). The nucleotide sequence analysis of the EsV type 1 genome (5) identified a gene with significant similarities to the carboxy-terminal, catalytic domain of an integrase. There was no report on the integration mechanism of this virus. Based on recently completed sequencing and annotation of the FsV-158 genome, an ORF with 29% amino acid sequence identity to the putative integrase gene of EsV was identified; the expression and role of this ORF in virus replication need to be elucidated.
Sequence analysis of the IS-V and IS-A, the unoccupied site, and the resulting alga-virus junctions identified a common GC dinucleotide motif (Fig. 4B). The well-studied bacteriophage λ integration into the genome of its host, E. coli, shows substantial sequence homology between the integrating virus and target host DNA. The recombination sites of the phage and the bacterial host, called attP and attB, respectively, contain a common “core” 15-bp sequence (10). The core sequence is cleaved in a staggered fashion, with seven overhanging nucleotides at the beginning of recombination, and recovered at each end of the prophage genome by recombination. This process is different from retrovirus integration, where staggered cleavage of the target DNA results in duplication of host target DNA at both ends of the provirus genome while two nucleotides at each end of the long terminal repeats are lost (6). Although the GC dinucleotides in the FsV and its host genome are recovered at the A-VJLs and A-VJRs after integration, no duplication of host target sequence occurs (Fig. 3). It is not clear whether the GC dinucleotides are cleaved in a staggered fashion or in the middle, but recovery of this sequence at both junctions without any loss or duplication of the virus and host genome is uncommon.
It has been suggested that a secondary structure formed by a palindromic sequence may influence integration efficiency by affecting the A-philicity, DNA bendability, and protein-induced deformability of the DNA (21). Also, AT-rich sequence can generate DNA curvature, which makes this region accessible for recombination. No palindromic or AT-rich sequence was found in the Feldmannia genome near the integration site of the two FsV genomes. AT-rich stem-loop structures can be produced around the IS-V by positioning the GC dinucleotides in the loop area (Fig. 5). Further analyses on the function of these stem-loop structures and repeated sequences around the IS-V, including possible involvement in the interaction with viral or host factors, will help explain the mechanism of this unique integration process. The present report is the first description of the integration of an algal virus genome into its host genome, but details of the integration and excision processes and viral and host factors involved in these processes need to be further characterized.
Footnotes
Published ahead of print on 21 November 2007.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215403-410. [DOI] [PubMed] [Google Scholar]
- 2.Bräutigam, M., M. Klein, R. Knippers, and D. G. Műller. 1995. Inheritance and meiotic elimination of a virus genome in the host Ectocarpus siliculosus (Phaeophyceae) J. Phycol. 31823-827. [Google Scholar]
- 3.Claverie, J. M., H. Ogata, S. Audic, C. Abergel, K. Suhre, and P. E. Fournier. 2006. Mimivirus and the emerging concept of giant virus. Virus Res. 117133-144. [DOI] [PubMed] [Google Scholar]
- 4.Delaroque, N., I. Maier, R. Knippers, and D. G. Műller. 1999. Persistent virus integration into the genome of its algal host, Ectocarpus siliculosus (Phaeophyceae). J. Gen. Virol. 801367-1370. [DOI] [PubMed] [Google Scholar]
- 5.Delaroque, N., D. G. Mûller, G. Bothe, T. Pohl, R. Knippers, and W. Boland. 2001. The complete DNA sequence of the Ectocarpus siliculosus virus EsV-1 genome. Virology 287112-132. [DOI] [PubMed] [Google Scholar]
- 6.Engelman, A., K. Mizuuchi, and R. Craigie. 1991. HIV-1 DNA integration: mechanism of viral DNA cleavage and DNA strand transfer. Cell 671211-1221. [DOI] [PubMed] [Google Scholar]
- 7.Henry, E. C., and R. H. Meints. 1992. A persistent virus infection in Feldmannia (Phaeophyceae). J. Phycol. 28517-528. [Google Scholar]
- 8.Ivey, R. G., E. C. Henry, A. M. Lee, L. Klepper, S. K. Krueger, and R. H. Meints. 1996. A Feldmannia algal virus has two genome size-classes. Virology 220267-273. [DOI] [PubMed] [Google Scholar]
- 9.Krueger, S. K., R. G. Ivey, E. C. Henry, and R. H. Meints. 1996. A brown algal virus genome contains a “RING” zinc finger motif. Virology 219301-303. [DOI] [PubMed] [Google Scholar]
- 10.Landy, A. 1989. Dynamic, structural, and regulatory aspects of λ site-specific recombination. Annu. Rev. Biochem. 58913-949. [DOI] [PubMed] [Google Scholar]
- 11.Lee, A. M., R. G. Ivey, E. C. Henry, and R. H. Meints. 1995. Characterization of a repetitive element in a brown algal virus. Virology 21474-480. [DOI] [PubMed] [Google Scholar]
- 12.Lee, A. M., R. G. Ivey, and R. H. Meints. 1998. Repetitive DNA insertion in a protein kinase ORF of a latent FSV (Feldmannia sp. virus) genome. Virology 24835-45. [DOI] [PubMed] [Google Scholar]
- 13.Lee, A. M., R. G. Ivey, and R. H. Meints. 1998. The polymerase gene of a brown algal viruses: structure and phylogeny. J. Phycol. 34608-615. [Google Scholar]
- 14.Mûller, D. G. 1991. Mendelian segregation of a virus genome during host meiosis in marine brown alga Ecotocarpus siliculosus. J. Plant Physiol. 137739-743. [Google Scholar]
- 15.Park, Y., G. D. Kim, and T. J. Choi. 2007. Molecular cloning and characterization of the DNA adenine methyltransferase gene in Feldmannia sp. virus. Virus Genes 34177-183. [DOI] [PubMed] [Google Scholar]
- 16.Raoult, D., S. Audic, C. Robert, C. Abergel, P. Renesto, H. Ogata, B. La Scola, M. Suzan, and J. M. Claverie. 2004. The 1.2-megabase genome sequence of mimivirus. Science 3061344-1350. [DOI] [PubMed] [Google Scholar]
- 17.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 18.Van Etten, J. L., and R. H. Meints. 1999. Giant viruses infecting algae. Annu. Rev. Microbiol. 53447-494. [DOI] [PubMed] [Google Scholar]
- 19.Van Etten, J. L., M. V. Graves, D. G. Mûller, W. Boland, and N. Delaroque. 2002. Phycodnaviridae-large DNA algal viruses. Arch. Virology 1471479-1516. [DOI] [PubMed] [Google Scholar]
- 20.Villarreal, L. P., and V. R. DeFilippis. 2000. A hypothesis for DNA viruses as the origin of eukaryotic replication proteins. J. Gen. Virol. 747079-7084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu, X., Y. Li, B. Crise, S. M. Burgess, and D. J. Munroe. 2005. Weak palindromic consensus sequences are a common feature found at the integration target sites of many retroviruses. J. Virol. 795211-5214. [DOI] [PMC free article] [PubMed] [Google Scholar]