Abstract
Very little is known about the role that evolutionary dynamics plays in diseases caused by mammalian DNA viruses. To address this issue in a natural host model, we compared the pathogenesis and genetics of the attenuated fibrotropic and the virulent lymphohematotropic strains of the parvovirus minute virus of mice (MVM), and of two invasive fibrotropic MVM (MVMp) variants carrying the I362S or K368R change in the VP2 major capsid protein, in the infection of severe combined immunodeficient (SCID) mice. By 14 to 18 weeks after oronasal inoculation, the I362S and K368R viruses caused lethal leukopenia characterized by tissue damage and inclusion bodies in hemopoietic organs, a pattern of disease found by 7 weeks postinfection with the lymphohematotropic MVM (MVMi) strain. The MVMp populations emerging in leukopenic mice showed consensus sequence changes in the MVMi genotype at residues G321E and A551V of VP2 in the I362S virus infections or A551V and V575A changes in the K368R virus infections, as well as a high level of genetic heterogeneity within a capsid domain at the twofold depression where these residues lay. Amino acids forming this capsid domain are important MVM tropism determinants, as exemplified by the switch in MVMi host range toward mouse fibroblasts conferred by coordinated changes of some of these residues and by the essential character of glutamate at residue 321 for maintaining MVMi tropism toward primary hemopoietic precursors. The few viruses within the spectrum of mutants from mice that maintained the respective parental 321G and 575V residues were infectious in a plaque assay, whereas the viruses with the main consensus sequences exhibited low levels of fitness in culture. Consistent with this finding, a recombinant MVMp virus carrying the consensus sequence mutations arising in the K368R virus background in mice failed to initiate infection in cell lines of different tissue origins, even though it caused rapid-course lethal leukopenia in SCID mice. The parental consensus genotype prevailed during leukopenia development, but plaque-forming viruses with the reversion of the 575A residue to valine emerged in affected organs. The disease caused by the DNA virus in mice, therefore, involves the generation of heterogeneous viral populations that may cooperatively interact for the hemopoietic syndrome. The evolutionary changes delineate a sector of the surface of the capsid that determines tropism and that surrounds the sialic acid receptor binding domain.
Evolution is a major factor in virus-host interactions and patterns of diseases (16). RNA viruses exhibit inherently high mutation rates that underlie their considerable evolutionary potential (18). The term quasispecies was introduced to describe the genetically heterogeneous populations, consisting of complex and dynamic mutant swarms, that this property engenders (15, 7). This rapid evolution is responsible for the accumulation of genetic changes in human immunodeficiency virus and poliovirus genomes and other viral genomes, particularly when the host is immunodeficient (46, 52, 31). In individuals, the interactions among dynamic virus populations of a quasispecies may determine the pathogenic features of the infection (51).
DNA viruses generally display lower rates of evolution than RNA viruses (17, 6). However, genetic heterogeneity is observed in infections of some immunodeficient hosts (24), mutation rates higher than those determined for cellular genes occur in several experimental systems, and intrinsically rapid evolution in some families of single-stranded DNA viruses has been observed previously (reviewed in reference 29). In the small single-stranded DNA viruses belonging to the Parvoviridae (50), rapid evolution may explain the diversity of adeno-associated viruses and autonomous parvoviruses circulating in primates (30, 19), the phylogenic evidence for a high mutation rate in the human B19 erythrovirus (45), the host range shift of feline parvovirus to canine hosts via transferrin receptor usage (35, 44, 21), and the isolation of mutants of minute virus of mice (MVM) under different selective pressures in vitro (38, 14, 26) or in experimental murine infections (26-28).
Tropism is a major pathogenic factor for infectious agents, and in virus evolution (5), its importance is illustrated by the diversity of circulating and emergent strains of each virus species with different tissue tropism specificities. However, few existing experimental models of viral evolution in natural hosts are amenable to an evaluation of the relevance of tropism in pathogenesis. The two host range variants of MVM, the lymphohematotropic (MVMi) and fibrotropic (MVMp) strains (8, 13), provide a useful model for the study of DNA virus tropism in disease in a mammalian host. MVM tropism is controlled predominantly by the gene encoding the 25-nm-diameter T=1 icosahedral capsid (32, 4), comprising two proteins, the major VP2 polypeptide (64 kDa) and VP1 (83 kDa), which is a minor, 142-amino-acid N-terminally extended version of VP2 (48). While MVMp and MVMi differ at only a few residues in VP2, they are reciprocally restricted for growth in each other's host cell type (49), at a point after attachment to the cell surface receptor but prior to the onset of viral gene expression and DNA replication (47). In established cell lines in vitro, two amino acid residues in MVMi, at VP2 positions 317 and 321, act coordinately as major determinants for the acquisition of fibrotropism (4) whereas the switch to lymphotropism for MVMp requires both an equivalent region of the MVMi coat protein gene and a segment of the nonstructural protein genes (11).
In mice, MVMi is pathogenic for newborns (36, 10), and in severe combined immunodeficient (SCID) adults, it causes a depletion of hemopoietic precursors in bone marrow, leading to severe leukopenia and accelerated erythropoiesis (43), which results from the ability of this virus to target committed, as well as long-term repopulating, hemopoietic stem cells (41, 42). In contrast, MVMp is apathogenic in newborn mice (22), and in oronasally inoculated adult SCID mice, infection is asymptomatic, with no clinical morbidity (39). However, in intravenous inoculations, MVMp evolves, as indicated by the single-plaque isolation of variants which carry only one of three single changes (I362S, K368R, and V325M) in the VP capsid protein (28). These variants remain fibrotropic in vitro, and their capsids show low affinity for the sialic acid component of the receptor, but unlike MVMp, they are invasive by the oronasal route and replicate in mouse tissues, eventually leading to death (28, 39).
In this report, we analyze the pathogenesis and viral genetics in SCID mice infected with two invasive MVMp variants. The development of severe hematological dysregulation correlated with the emergence of genetic heterogeneous MVMp populations showing amino acid changes at tropism determinant residues in the capsid. Most pathogenic viruses in these populations were unable to form plaques and to grow under several culture conditions.
MATERIALS AND METHODS
Cell lines and viruses.
A9 mouse fibroblasts and NB324K simian virus 40-transformed human newborn kidney, EL-4, and iD5 cells (49) were maintained in Dulbecco's modified Eagle medium (DMEM) supplemented with 5% heat-inactivated fetal calf serum (FCS; GIBCO-BRL). Murine myelomonocytic WEHI-3B D+ cells were grown in RPMI 1640 supplemented with 10% FCS. Stocks of the fibrotropic prototype strain, MVMp (13), and the virulent immunosuppressive strain, MVMi (8), and of recombinant viruses with plaque-forming capacity were prepared by large-scale transfection of the highly MVM-permissive human NB324K cell line (49) by electroporation (25) to minimize secondary mutations and were purified and filter sterilized (through a 0.22-μm-pore-size filter) as previously described (43, 39). Populations of MVMi fibrotropic forward mutants (see Fig. 3) were expanded in iD5 cells. Infectious virus titers were determined as PFU in NB324K or A9 cells by a standard assay (20, 49) or as immunofluorescence units by staining with anti-VP antibody (25). To determine titers of viruses devoid of infectivity in cell lines (see Fig. 4), recombinant and wild-type virions were extracted from transfected NB324K cells, gradient purified (25), and quantitated by slot immunoblotting (37).
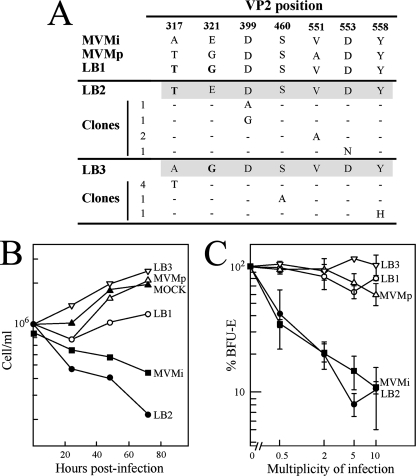
FIG. 3.
Capsid residues determining MVM tropism in culture. (A) Selection of forward mutants in A9 mouse fibroblasts from genetically independent stocks derived by transfection with infectious clones. The sequences of the assayed LB2 and LB3 mutants are shadowed, with residues changed by directed mutagenesis in bold. (B) Interaction of MVMi mutants with mouse hemopoietic cells. Nonadherent myeloid cells harvested from long-term bone marrow cultures were prestimulated for 3 days in IMDM-10% FCS-10% WEHI-3B conditioned medium as a source of interleukin-3 and infected with the indicated viruses at a multiplicity of infection of 5 PFU/cell, and the number of viable cells was scored daily. (C) Susceptibility of the BFU-E mouse hemopoietic precursors to MVMi mutants. Total C57BL or BALB/c bone marrow cells were infected at the indicated multiplicity of infection and cultured in vitro under conditions allowing the growth of this committed erythroid precursor.
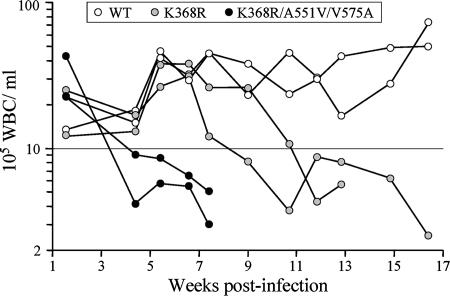
FIG. 4.
Kinetics of peripheral blood leukocytes in infected SCID mice. Two mice per each of the indicated MVMp genotypes were intravenously inoculated with 1010 gradient-purified virions/mouse, a dose equivalent to 2 × 105 PFU for the wild-type MVMp. The numbers of leukocytes (WBC) per milliliter of blood determined for each mouse over the wpi are shown. Endpoints of curves before the 17-wpi time point indicate the time of death. WT, wild type; K368R, K368R variant; K368R/A551V/V575A, RVA virus.
Mouse histology.
Eight- to 10-week-old females of the C.B-17 inbred strain of SCID mice (9) were purchased from the Jackson Laboratory (Bar Harbor, ME), infected, and monitored for levels of peripheral white blood cells (WBC) and pathological signs and survival as described previously (43). Organs freshly excised from euthanized mice were fixed by immersion in 4% paraformaldehyde for 15 min with gentle shaking and extensively washed in phosphate-buffered saline. Tissues were embedded in paraffin, sectioned at a thickness of 5 μm, and stained with hematoxylin and eosin or processed for viral antigen expression by immunohistochemistry. Staining was done with a primary MVM polyclonal antibody raised in rabbit that recognizes both capsid proteins (39), and detection was achieved with the Dako LSAB II kit with a biotinylated affinity-purified goat-anti rabbit secondary antibody and diaminobenzidine as the chromogen.
Clonogenic assays of hemopoietic progenitors.
Mouse bone marrow cells were flushed from shafts of dissected femora and tibiae in Iscove's modified DMEM (IMDM; GIBCO Laboratories, Grand Island, NY), dispersed with a 16-by-5-mm needle, and counted by trypan blue exclusion in a hemocytometer. Hemopoietic progenitors were assessed in enriched methylcellulose semisolid cultures essentially as described previously (41). The culture medium consisted of IMDM, 30% fetal bovine serum, 1% deionized bovine serum albumin (fraction V; Sigma Chemical Co., St Louis, MO), 10% WEHI-3B conditioned medium, 0.8% methylcellulose (Dow Chemical Co., Midland, MI), 2-U/ml erythropoietin (Terry Fox Laboratory, Vancouver, Canada), 10−2 mM alpha-thioglycerol, 2 × 10−3 mM l-glutamine, and 10−4 mM Na2SeO3. A total of 105 nucleated cells resuspended in 0.9 ml of medium were dispensed into a 24-well culture dish, and colonies were scored under an inverted microscope after 7 days of incubation. Progenitors from uninfected mice were assayed in parallel in the same dishes as an internal control for the cultures. Granulocyte-macrophage CFU (CFU-GM) were defined as aggregates of at least 50 cells, and erythroid burst-forming units (BFU-E) were defined as hemoglobinized bursts.
Viral genetic analysis.
MVM replicative and genomic DNA forms were isolated from mouse organ homogenates and from viral plaques by a modified Hirt procedure (2). Samples were subjected to Southern blot analysis in agarose gels as described previously (43) or to PCR amplification by using a Bio-Rad gene cycler according to published protocols. Amplified fragments were purified from agarose gels by the Concert rapid gel extraction system (GIBCO-BRL) and sequenced in a Perkin-Elmer 377 automated sequencer. The paired sets of oligonucleotides used to encompass the complete coding sequence of the MVM genome (3) have been described previously (28).
Recombinant MVM variants and clones.
Mutations were transferred into MVMp or MVMi infectious plasmids (20, 33). The MVMi-derived series of single and double mutants (LB1 to LB3) isolated from A9 cells was described previously (4, 1), as were the recombinant MVMp variants carrying the I362S and K368R in vivo-selected mutations (28). The recombinant K368R/A551V/V575A virus was constructed by PCR amplification of viral DNA isolated from the spleen of the leukopenic R6 mouse by using high-fidelity PfuI polymerase (Promega) and replacing an SspI restriction fragment (nucleotides [nt] 3233 to 4628) of the MVMp genome. Clonal analysis of viral populations in mouse tissues was addressed for PFU by direct sequencing of isolated plaques in NB324K cells (28, 26) and for molecular clones by PCR amplification from mouse tissues, with subsequent cloning into pUC19 and transformation of Escherichia coli JM109.
RESULTS AND DISCUSSION
Invasive MVMp variants elicit hemopoietic dysregulation in SCID mice.
To investigate the pathogenesis and whether the lethal disease induced by the fibrotropic MVMp I362S and K368R variants involves changes in tropism, main organs of SCID mice oronasally infected at 106 PFU/mouse were subjected to histopathological analyses at the initial stages of the infection (8 weeks postinfection [wpi]) and at the 50% survival time point determined for each virus, namely, 14 or 18 wpi for the K368R or I362S variant, respectively, and 8 wpi for MVMi. For MVMp variants, capsid expression could not be detected at 8 wpi, but at 14 to 18 wpi, positive areas of capsid staining, mainly in the capillary endothelia of main organs (e.g., livers and kidneys) and in some tissue-specific cells (e.g., occasional hepatocytes), denoted hematogenous dissemination (data not shown). However, as for MVMi, the histopathological hallmark of the MVMp variant-induced disease in SCID mice was the predominant involvement of hemopoietic organs, which contain large populations of proliferating cells potentially able to support MVM multiplication. As shown in Fig. 1A, cells with inclusion bodies staining for MVM capsid proteins were numerous in the bone marrow. In the spleen, there was also strong staining of individual cells and groups of cells often adjacent to arterioles, as well as fine granular and diffuse staining of cells throughout the organ, perhaps representing phagocytosis by resident macrophages of adjacent MVM-infected cells. These findings are consistent with dysregulated hemopoiesis in the presence of a low proportion of productively infected cells, as previously indicated by precursor analysis in MVMi infections (42, 43).
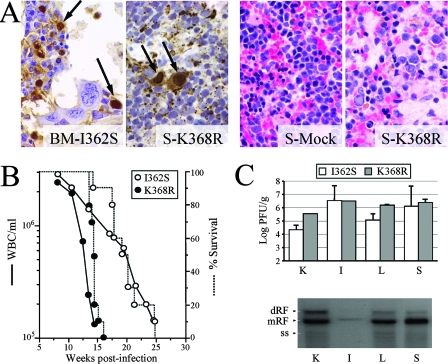
FIG. 1.
Induction of leukopenia in SCID mice by MVMp invasive variants. (A) Histopathology of MVM infection in SCID mice. (Two left panels) MVMp mutant capsid expression and intranuclear inclusion bodies (arrows) in bone marrow or spleens of infected mice (specimens were subjected to immunohistochemistry staining); (two right panels) depletion of myeloid cells and hypocellularity in the spleens of infected mice (specimens were stained with hematoxylin and eosin). BM-I362S, bone marrow from I362S virus-infected mouse; S-K368R and S-Mock, spleens from K368R virus- and mock-infected mice. (B) Survival (dotted lines) of and levels of circulating WBC (solid lines) in I362S and K368R virus-infected mice at the indicated wpi (n, 5 mice per virus from two independent experiments). Survival data were taken from reference 28. (C) Multiplication of MVMp variants in organs of leukopenic mice (n = 3) as monitored by the accumulation of infectious virus (upper panel) and of intermediates of replicative DNA (dimeric [dRF], monomeric [mRF], and single-stranded DNA [ss]) of the I362S variant (lower panel). K, kidney; L, liver; I, intestine.
The damage involving hemopoietic organs correlated with the consistent development of acute leukopenia, which became severe (<5 × 105 WBC/ml; normal range, 2 × 106 to 4 × 106 WBC/ml [43]) by 13 wpi in K368R virus infections and by 18 wpi in I362S virus infections (Fig. 1B). The development of leukopenia was accompanied by the clinical signs of hunched posture, ruffled fur, and weight loss and culminated in the death of all infected animals following the onset of severe leukopenia. The MVMp variants therefore acquired, during infection, the pathogenic features of the MVMi strain (42, 43). The delayed course of leukopenia (13 to 18 weeks) in mice inoculated with the MVMp variants compared to the 5 to 7 weeks required for MVMi to elicit severe leukopenia following oronasal inoculation with an equivalent viral dose of 106 PFU/mouse (42, 43) suggests the requirement for an adaptive process of the fibrotropic MVMp variants to target these hemopoietic cells. Titers of infectious viruses (PFU) in leukopenic animals (Fig. 1C, upper panel) were similar in the major organs examined (104 to 106 PFU/g) and, with the exception of those in the intestines, correlated well in both I362S and K368R virus infections with the levels of viral DNA replication (see the example in Fig. 1C, lower panel). Interestingly, overall PFU titers in organs did not correlate with the high numbers of MVM capsid-expressing cells found in the spleen and bone marrow (Fig. 1A and data not shown), suggesting that the bulk of viral multiplication leading to hemopoietic disease is inefficiently quantitated by this plaque assay.
Amino acid changes in MVMp capsid are associated with the development of leukopenia.
The lethal hemopoietic disease caused by the MVMp variants prompted a genetic analysis of virus isolated from major organs of leukopenic mice infected with I362S (mice S1 to S5) or K368R (mice R6 to R8) virus (Fig. 2A). With the exception of the single-nucleotide G2035A mutation of the reference MVMp genomic sequence (3), which introduces the NS1 nonstructural protein-specific S592N amino acid change in the S5 population, no genetic changes were fixed either in the NS1 or the NS2 consensus sequences present in three spleens (the spleen of mouse S4 [S4-S], S5-S, and R6-S) or in the NS1 C-terminal region (corresponding to nt 1703 to 2380) isolated from organs of independently inoculated mice (the liver of mouse S1 [S1-L], the kidneys of mouse S2 [S2-K], S3-L, and R7-S), indicating that MVMp variant nonstructural proteins do not play an evolutionary role in the development of the leukopenic syndrome. In sharp contrast, specific amino acid changes in the consensus sequence of the capsid (VP) protein were found in isolates from all leukopenic mice inoculated with either parental virus. Sequence analysis included the entire VP gene of virus from several mouse organs (S1-K, S2-S, S4-S, S5-S, R6-S, and R6-L) and, for all isolates, the region from nt 3726 to 4670, which encodes the area including the key amino acids 308 to 587 (VP2 numbering) that function in tropism (see Fig. 3), virulence (28), and nuclear targeting (25).
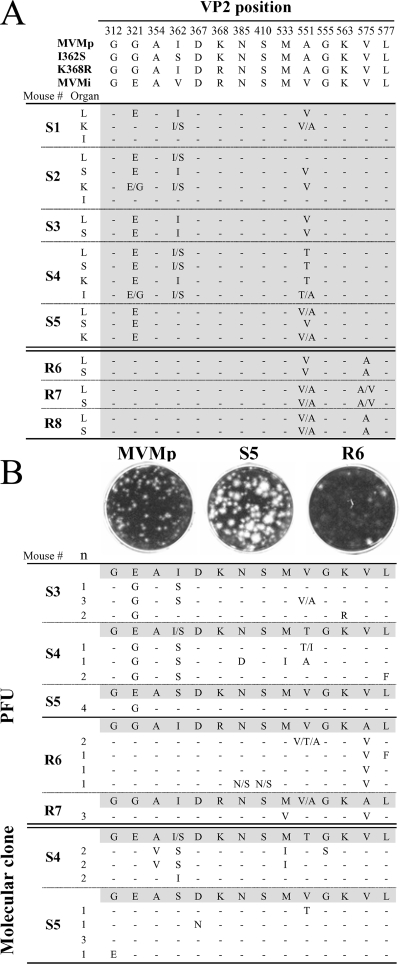
FIG. 2.
Amino acid changes corresponding to MVMp-derived genomes isolated from leukopenic mice. (A) Coding changes found in the consensus sequences of MVM populations present in organs (K, kidney; L, liver; S, spleen; I, intestine) isolated at 14 to 18 wpi from leukopenic SCID mice infected with the I362S (mice S1 to S5) and K368R (mice R6 to R8) variants. Genetic differences between the isolated populations and their respective I362S or K368R parent are outlined. (B) Clonal analysis of the MVM genome composition in spleens of leukopenic mice. The plaque heterogeneity of the S5 and R6 populations is shown above the coding changes localized in the region corresponding to nt 3726 to 4670 of the MVM genome as determined from isolated plaques (PFU) or molecular clones. Genetic changes with respect to the consensus sequence (shadowed) corresponding to viral DNA isolated from the spleens are indicated. n, number of clones with the same genotype. Single letters separated by slashes represent amino acid heterogeneity at this position in the sampled population. Dashes represent amino acids identical to those in the consensus sequence.
As shown in Fig. 2A (upper panel), the consensus VP sequence of virus from most organs (livers, spleens, and kidneys) of all of the S1 to S5 mice carried the G321E and A551V/T mutations. Both changes found in the S1 to S5 viruses were to amino acids encoded by the MVMi genotype. The emergence of the same genetic changes in MVM populations isolated from most organs, with the exception of the intestine, suggests that damaged hemopoietic organs (Fig. 1A and B) are a major source of mutants, which spread hematogenously and subsequently infect blood vessel endothelia. In the intestine, the low levels of infected cells (data not shown) and of viral DNA replication (Fig. 1C, lower panel) would explain why the consensus sequence of virus from this organ was essentially that of the parental I362S virus. In addition, viruses from some I362S virus-inoculated mice reverted partially or totally to the MVMp wild-type sequence at position 362 when the other two changes appeared (see also the discussion of clones below), a phenomenon that may reflect structural restrictions within the capsid. Likewise, specific amino acid changes in the consensus sequences of virus from the major organs were also found in all mice (R6 to R8) infected with the K368R variant (Fig. 2A, lower panel). These were A551V, as mentioned above, and V575A, a change which does not correspond to any known MVM genotype. This study demonstrates the consistent emergence, by parallel viral evolution in different mice, of a few VP amino acid changes associated with the development of leukopenia in the SCID mouse and shows that the nature of these mutations is partly specified by the virulence genotype of the parental virus, the I362S or the K368R virus. The capacity of MVM to rapidly evolve in SCID mice as described here and in our previous studies (reviewed in reference 29) can be accounted for by the remarkably high mutation frequency determined in vitro in the absence of any detectable selection force (26), together with the large-scale viral genome accumulation and the large population sizes reached in mouse organs (Fig. 1C).
MVMp evolution in mice targets tropism determinant residues of the capsid.
The significance in viral tropism of the residues changing in SCID mice was evaluated with cultured cells. A first study was focused on residues conferring fibrotropism to MVMi. Three MVMi-derived site-directed-mutagenesis mutants, LB1 to LB3, were constructed with mutations at the two residues of VP2, 317 and 321, which are coordinately required for the switch of MVMi to fibrotropism (4). The double-mutant virus LB1 had growth characteristics very similar to those of the prototype fibrotropic strain, MVMp, whereas viruses carrying only one mutation, LB2 (A317T) and LB3 (E321G), were essentially as restricted in fibroblasts as their parent, MVMi (4). Here, spontaneous fibrotropic forward mutants, selected from genetically independent stocks of each of these single mutants grown in iD5 cells, were isolated by plaque formation in A9 mouse fibroblasts. DNA sequencing of the entire VP1-VP2 gene of these MVMi host range mutants revealed in each a single additional change within VP2. The ratios of plaque titers in A9 mouse fibroblasts to those in the fully MVM-permissive NB324K human cells (49) indicated that these single host range mutants were present in independent MVMi stocks at frequencies between 1.2 × 10−3 and 7.8 × 10−5, implying, as mentioned above, a relatively high mutation rate for this virus growing in cultured cells. As shown in Fig. 3A, the A317 changed to T, reproducing the LB1 arrangement, in four out of six such mutants isolated from E321G single-mutant stocks. However, forward mutations can occur at sites other than the previously characterized 317 coordinated site, as shown by the mutations at residues 460 and 558 delineated in Fig. 3A. The other original coordinated mutation, E321G, was not recovered in any forward mutant derived from the A317T single mutant, but alternate single changes were found at residue 399, 551, or 553, suggesting that there are a significant number of possible combinations of double mutation which can confer fibrotropism upon MVMi. Significantly, of the mutations conferring the MVM tropism switch in vitro, those at 321 and 551 are among the major changes selected in leukopenic mice (Fig. 2A), and the rest, although spread apart in the primary VP2 sequence, cluster in a common domain in the three-dimensional structure of the MVM capsid (see below).
To study whether residues 317 and 321 mediating fibrotropism in the MVMi background are also involved in hematotropism, the LB mutants were assessed for their capacity to replicate in primary murine hemopoietic cultures and to kill in vitro precursors committed to the erythroid (BFU-E) and myeloid (CFU-GM) lineages. As shown in Fig. 3B, the single A317T change (LB2 mutant) did not significantly interfere with viral infection in such cultures. In contrast, the LB1 and LB3 virus mutants, which harbor the E321 residue changed to G, failed to infect hemopoietic cells productively. In agreement with this result, only the LB2 mutant and the MVMi strain suppressed the growth of the BFU-E (Fig. 3C) and of the CFU-GM (data not shown) committed precursors, underlining the importance of the glutamate residue at 321 for MVMi tropism in primary mouse hemopoietic cells. This finding is fully consistent with the selection of the G321E change in leukopenic mice infected with the I362S variant (Fig. 2A, upper panel), but this glutamate residue is dispensable for the acquisition of MVMp hematotropism in the K368R virus background (Fig. 2A, lower panel).
Heterogeneous MVM populations of limited genetic complexity arise during the development of leukopenia.
Sequence readout ambiguity at the positions of mutations in the consensus sequence of virus from the major organs (Fig. 2A) suggested that the MVM populations growing in leukopenic SCID mice were genetically heterogeneous. To estimate the complexity of the mutant spectra within these populations, two sets of MVM clones were isolated, by different procedures, from the spleens of leukopenic mice and sequenced across the region of the genome from nt 3726 to 4670. Firstly, molecular clones (n = 12), obtained by PCR cloning directly into plasmids, of MVM genomes present in the S4 and S5 spleens encoded the major amino acid changes selected in the consensus sequences of these organs (Fig. 2B, bottom panel), indicating that individual viral genomes carrying these mutations are indeed abundant in the viral populations replicating in leukopenic mice. Neutral mutations were not detected across the 968-nt region sequenced in the 12 molecular clones, but mutations determining other coding changes (e.g., A354V and V551T) appeared only in some clones and were therefore not clearly represented in the consensus sequence. Thus, the MVMp populations develop genetic heterogeneity in the capsid gene while growing in SCID mice.
Secondly, infectious viral clones (PFU) were isolated from single plaques in monolayers of NB324K human fibroblasts, a cell line highly permissive toward the MVM strains and multiple recombinants (49), and subjected to sequence analysis as described above (Fig. 2B, middle panel). Distinct plaque phenotypes were seen for isolates from S5 or R6 spleens, which formed plaques heterogeneous in size and clear or mostly diffused, respectively. Remarkably, all of the plaques isolated from S mice (n = 14) conserved the G321 residue of the parental I362S virus, instead of demonstrating selection for the G321E change found in the consensus sequence of virus from the major organs. Likewise, plaques obtained from R mice (n = 8) consistently conserved the V575 residue, as in the parental K368R virus, instead of acquiring the V575A change found in the consensus virus sequence. As in all the sequence analyses mentioned above, no neutral mutations were found in the region from nt 3726 to 4670, further supporting the action of strong selective pressures in the MVM evolutionary processes developing in SCID mice (reviewed in reference 29). Instead, other amino acid changes occurred at specific residues in some clones, with even a striking heterogeneity of DNA sequence readout corresponding to particular residues (e.g., residue 551) in a clonal, plaque-derived sequence, suggesting that these residues may be under selective pressure during plaque formation. Therefore, only the minor fraction of viruses replicating in the spleens of leukopenic mice that preserve the respective parental G321 and V575 residues, which are not represented in the consensus sequence, can form plaques on NB324K cell monolayers. In summary, this study demonstrates that late in the course of hemopoietic disease in SCID mice, MVM populations are heterogeneous but that their genetic complexity is limited to specific capsid residues and that the genotypes that predominate in the mutant spectrum carry changes that severely reduce the plaque-forming capacity.
Capsid changes selected in mice confer leukopenogenic capacity upon MVMp.
To study the role that some of the capsid changes selected in MVM populations play in the development of the hemopoietic disease, the A551V and V575A changes selected for in K368R virus-infected mice (Fig. 2A, lower panel) were engineered in the MVMp genetic background and the pathogenicity of the resulting virus carrying the three mutations K368R, A551V, and V575A (named the RVA virus) both in culture and in SCID mice was analyzed. Recombinant RVA virions were harvested and purified from a single round of transfection-initiated infection of NB324K cells to minimize the appearance of secondary mutations (see Materials and Methods), in yields indicating that these three mutations do not significantly hamper replication at postentry stages of the MVMp life cycle. However, the RVA virions were essentially unable to initiate infection, as their specific infectivity in NB324K cells was more than 100-fold lower than that of equivalent preparations of K368R virus or the MVMp wild type, further supporting the above-described restrictive effect of the V575A mutation on plaque-forming capacity. The pathogenicity of the RVA virus in SCID mice was then assayed directly by intravenously inoculating the mice with normalized amounts of purified recombinant and control wild-type MVMp virions and monitoring the level of circulating blood leukocytes and clinical signs through 17 wpi (Fig. 4). Consistent with previous findings (28, 29), MVMp did not cause hemopoietic disease by this time after infection, whereas in K368R virus-infected mice, the numbers of leukocytes had dropped significantly by 9 to 11 wpi, a few weeks earlier than those in mice inoculated intranasally (Fig. 1B). Importantly, the RVA virions caused acute and irreversible leukopenia with an onset as early as 4 wpi and led to overt clinical symptoms and death by 7 wpi. The rapid course of this disease, which resembled that caused by the naturally hematotropic MVMi strain at high intranasal doses (42, 43), demonstrates the ability of these three selected capsid mutations to confer acute leukopenogenic capacity upon MVMp.
Leukopenogenic MVMp variants show reduced fitness in culture.
To obtain further insights on the MVMp populations harvested from leukopenic mice, the fitness of these populations was studied by sequence analysis of viruses inoculated onto cells growing in vitro (Fig. 5). Consistent with the results of the clonal analysis (Fig. 2B, middle panel), the viruses of the major genotypes detected in populations from S5 and R6 leukopenic mice were significantly overgrown after 120 h in culture by viruses carrying the G321 and V575 residues characteristic of the parental strain and all of the PFU clones. Indeed, an enrichment of genomes with the change corresponding to E321G occurred immediately upon viral adsorption to NB324K cells (0 hours postinfection), indicating that glutamate at this position may interfere early in receptor recognition by the viral capsid on the NB324K cell surface while the selection for V575 enrichment may require some rounds of viral replication in culture (Fig. 5, upper panel). Importantly, both S5 and R6 populations were also restricted for growth in established cell lines of myeloid (WEHI-3B), fibroblast (A9), lymphoid (EL-4), and hybrid (iD5) origins (data not shown), which show different levels of permissiveness to MVM strains and recombinant viruses (38, 20, 2, 4, 49). Therefore, MVMp variants that are associated with leukopenia in mice show a very low level of fitness in culture. The significance of this phenomenon is unclear, though it may imply that the expression of the receptors on hemopoietic precursors targeted by virus in vivo is not maintained in cultured cells. The capacity of MVMi to form plaques (20, 49) and target committed and primitive hemopoietic precursors (41-43) may reflect an unusual property among circulating MVM strains that facilitated its early isolation (8).
FIG. 5.
Fitness of leukopenia-associated MVMp populations in culture. MVM populations harvested from the livers of leukopenic mice infected with the recombinant I362S (S5 mouse), K368R (R6 mouse), or K368R/A551V/V575A (RVA1 and RVA2 mice) viruses were inoculated onto monolayers of NB324K human fibroblasts, and low-molecular-weight DNA isolated at the indicated hours postinfection (hpi) was sequenced across the region from nt 3736 to 4687 of the MVM genome. Amino acid changes with respect to the MVM consensus sequence in the respective sampled organ (shadowed) are indicated. G/E indicates that G321 was more prevalent than E321, and vice versa for E/G.
A similar analysis of the leukopenia induced by the RVA virus (Fig. 4) was undertaken. The three engineered mutations K368R, A551V, and V575A persisted in the consensus sequences in organs of the two analyzed mice (Fig. 5, lower panel), further supporting a major role for the corresponding genotype in causing the disease. However, the viral populations had developed further genetic heterogeneity, as viruses with plaque-forming capacity could be demonstrated to be present in the major organs at titers ranging from 104 to 106 PFU/g of tissue (wet weight) (data not shown). These emerging PFU harbored the A575V reversion previously found in all plaque-forming viruses (Fig. 2B) and rapidly overgrew the RVA parental virus (Fig. 5, lower panel), which therefore exhibited a much lower level of fitness in culture, as described above for the S5 and R6 populations. The genetic and phenotypically heterogeneous MVM populations derived from the I362, K368R, or RVA virus clonal infections during leukopenia development (Fig. 2 and 5) may reflect that the targeting of different hemopoietic precursors by a variety of viral tropism variants may be required for disease development (see also below). Even some of the plaque-forming viruses conserving fibrotropism may contribute by damaging the marrow stromal fibroblasts essential for hemopoietic functions. This is an intriguing hypothesis deserving further, though technically difficult, research. Notably, a cooperative interaction between viruses within a quasispecies to cause disease in mice has been previously suggested for poliovirus infection (51).
Mapping pathogenic and tropism determinant residues in the MVM capsid structure.
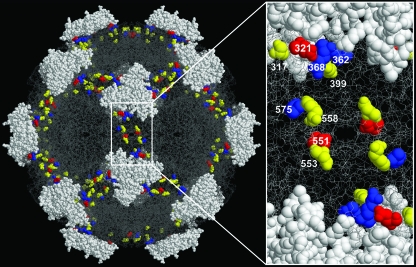
The spatial relationships between amino acid residues implicated in the tropism switches described here were determined from the three-dimensional crystal structure of the capsids of both MVM strains (1, 23). Most of these residues cluster in a domain bounded by the base of the threefold spike and a depression at the twofold axis, termed the dimple, as shown in Fig. 6. This capsid domain has also been implicated in host range determination for other parvoviruses (reviewed in references 23 and 28). Importantly, this domain contains all of the evolutionary changes selected in MVMp-infected SCID mice, including those conferring invasiveness upon the wild type (residues 362 and 368) and those in residues associated with the development of leukopenia (residues 321, 551, and 575), as residues whose side chains are exposed at the surface of the capsid (Fig. 6). Likewise, with the exception of the internal, conservative change, M533I, the rest of the changes in MVM clones isolated from mice (Fig. 3B), either as molecular clones (with changes in residues 312, 354, 367, and 555) or as plaques (with changes in residues 385, 563, and 577), are also in the same capsid region, in either surface or solvent-accessible residues (data not shown). Two of the major amino acid changes selected in vivo (those at residues 321 and 551) (Fig. 2) participate in the switch of MVMi tropism in vitro (Fig. 3), and the remainder of the forward fibrotropic residues are either on the capsid surface (Fig. 6) or likely to influence local surface conformation (1).
FIG. 6.
Residues of MVM capsid structure involved in tropism and mouse hemopoietic dysregulation. The left panel is a close-up of the solvent-accessible amino acid residues at the twofold axis of the MVM capsid involved in causing leukopenia in mice (blue), switching viral tropism in vitro (yellow), and both processes (red). The figure was prepared using the program RasMol (40) and the MVMi coordinates (1MVM) deposited in the Protein Data Bank (1). Numbers indicate residue positions in the VP2 capsid.
The entire set of pathogenic and tropism determinant residues evolving in vitro and in mice delineate an elliptical projection onto the capsid surface at the twofold axis of symmetry (Fig. 6), surrounding the binding site of sialic acid (28), a component of MVM receptors essential for infection (12). Indeed, the capsids of the I362S and K368R virulent variants of MVMp exhibited lower affinities than that of wild-type MVMp for a neuraminidase-sensitive primary receptor (39), as well as for certain glycans with terminal α2-3-linked sialic acid that bind MVMp and MVMi capsids with differential specificities (34). It is therefore conceivable that other nearby residues, such as 321, 551, and 575, associated with lethal leukopenia (Fig. 2A), contribute to the affinity for interaction of the capsid with different types of sialic acid moieties on the surfaces of hemopoietic cells that act as alternative MVM receptors. The sharply different tropism patterns within the viral populations at stages of leukopenia and among leukopenogenic viruses, such as MVMi and RVA, for cell lines and primary precursors in vitro (Fig. 2, 3, and 5) suggests that distinct sets of proliferative precursors of the complex hemopoietic repertoire are targeted by distinct viral subsets within the mutant spectrum during the development of the disease. This study may provide a first structural basis for the cooperative interactions among host range variants of dynamic populations in viral diseases.
Acknowledgments
We are indebted to T. Smailpour (MIRTH program, University of California—Irvine; NIH grant TW-00023) and Elena Merino (Centro de Biología Molecular “Severo Ochoa,” Madrid) for experimental support.
This work was supported by the European Union grant QLK3-CT-2001-01010, grant S-SAL-0185-2006 of Comunidad de Madrid, an institutional grant from Fundación Ramón Areces to the Centro de Biología Molecular “Severo Ochoa” for J.M.A., USPHS/NIH grant CA29303 to P.T., and grant SAF2005-02381 (Comisión Interministerial de Ciencia y Tecnología) to J.C.S.
Footnotes
Published ahead of print on 28 November 2007.
REFERENCES
- 1.Agbandje-McKenna, M., A. L. Llamas-Sainz, F. Wang, P. Tattersall, and M. G. Rossmann. 1998. Functional implications of the structure of the murine parvovirus, minute virus of mice. Structure 61369-1381. [DOI] [PubMed] [Google Scholar]
- 2.Antonietti, J.-P., R. Sahli, P. Beard, and B. Hirt. 1988. Characterization of the cell type-specific determinant in the genome of minute virus of mice. J. Virol. 62552-557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Astell, C. R., M. E. Gardiner, and P. Tattersall. 1986. DNA sequence of the lymphotropic variant of minute virus of mice, MVM3(i), and comparison with the DNA sequence of the fibrotropic prototype strain. J. Virol. 57656-669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ball-Goodrich, L. J., and P. Tattersall. 1992. Two amino acid substitutions within the capsid are coordinately required for acquisition of fibrotropism by the lymphotropic strain of minute virus of mice. J. Virol. 663415-3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baranowski, E., C. M. Ruiz-Jarabo, and E. Domingo. 2001. Evolution of cell recognition by viruses. Science 2921102-1105. [DOI] [PubMed] [Google Scholar]
- 6.Bernard, H. 1994. Coevolution of papillomaviruses with human populations. Trends Microbiol. 2140-143. [DOI] [PubMed] [Google Scholar]
- 7.Biebricher, C. K., and M. Eigen. 2006. What is a quasispecies? Curr. Top. Microbiol. Immunol. 2991-31. [DOI] [PubMed] [Google Scholar]
- 8.Bonnard, G. D., E. K. Manders, D. A. Campbell, R. B. Herberman, and M. J. Collins. 1976. Immunosuppressive activity of a subline of the mouse EL-4 lymphoma. J. Exp. Med. 143187-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bosma, G. C., R. P. Custer, and M. J. Bosma. 1983. A severe combined immunodeficiency mutation in the mouse. Nature 301527-530. [DOI] [PubMed] [Google Scholar]
- 10.Brownstein, D. G., A. L. Smith, R. O. Jacoby, E. A. Johnson, G. Hansen, and P. Tattersall. 1991. Pathogenesis of infection with a virulent allotropic variant of minute virus of mice and regulation by host genotype. Lab. Investig. 65357-363. [PubMed] [Google Scholar]
- 11.Colomar, M. C., B. Hirt, and P. Beard. 1998. Two segments in the genome of the immunosuppressive minute virus of mice determine the host-cell specificity, control viral DNA replication and affect viral RNA metabolism. J. Gen. Virol. 79581-586. [DOI] [PubMed] [Google Scholar]
- 12.Cotmore, S. F., and P. Tattersall. 1987. The autonomously replicating parvoviruses of vertebrates. Adv. Virus Res. 3391-173. [DOI] [PubMed] [Google Scholar]
- 13.Crawford, L. V. 1966. A minute virus of mice. Virology 29605-612. [DOI] [PubMed] [Google Scholar]
- 14.D'Abramo, A. M., Jr., A. A. Ali, F. Wang, S. F. Cotmore, and P. Tattersall. 2005. Host range mutants of minute virus of mice with a single VP2 amino acid change require additional silent mutations that regulate NS2 accumulation. Virology 340143-154. [DOI] [PubMed] [Google Scholar]
- 15.Domingo, E., and J. J. Holland. 1997. RNA virus mutations and fitness for survival. Annu. Rev. Microbiol. 51151-178. [DOI] [PubMed] [Google Scholar]
- 16.Domingo, E. 2007. Virus evolution, p. 389-421. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 5th ed. Lippincott Williams & Wilkins, Philadelphia, PA.
- 17.Drake, J. W. 1991. A constant rate of spontaneous mutations in DNA-based microbes. Proc. Natl. Acad. Sci. USA 887160-7164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Drake, J. W., and J. J. Holland. 1999. Mutation rates among RNA viruses. Proc. Natl. Acad. Sci. USA 9613910-13913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gao, G., M. R. Alvira, S. Somanathan, Y. Lu, L. H. Vandenberghe, J. J. Rux, R. Calcedo, J. Sanmiguel, Z. Abbas, and J. M. Wilson. 2003. Adeno-associated viruses undergo substantial evolution in primates during natural infections. Proc. Natl. Acad. Sci. USA 1006081-6086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gardiner, E. M., and P. Tattersall. 1988. Mapping of the fibrotropic and lymphotropic host range determinats of the parvovirus minute virus of mice. J. Virol. 622605-2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hueffer, K., J. S. L. Parker, W. Weicher, R. E. Geisesel, J.-Y. Sgro, and C. R. Parrish. 2003. The natural host range shift and subsequent evolution of canine parvovirus resulted from virus-specific binding to the canine transferrin receptor. J. Virol. 771718-1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kimsey, P. B., H. D. Engers, B. Hirt, and V. Jongeneel. 1986. Pathogenicity of fibroblast- and lymphocyte-specific variants of minute virus of mice. J. Virol. 598-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kontou, M., L. Govindasamy, H.-J. Nam, N. Bryant, A. L. Llamas-Saiz, C. Foces-Foces, E. Hernando, M.-P. Rubio, R. McKenna, J. M. Almendral, and M. Agbandje-McKenna. 2005. Structural determinants of tissue tropism and in vivo pathogenicity for the parvovirus minute virus of mice. J. Virol. 7910931-10943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lednicky, J., A. Arrington, A. R. Stewart, X. M. Dai, C. Wong, S. Jafar, M. Murphey-Corb, and J. S. Butel. 1998. Natural isolates of simian virus 40 from immunocompromised monkeys display extensive genetic heterogeneity: new implications for polyomavirus disease. J. Virol. 723980-3990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lombardo, E., J. C. Ramírez, J. García, and J. M. Almendral. 2002. Complementary roles of multiple nuclear targeting signals in the capsid proteins of the parvovirus minute virus of mice during assembly and onset of infection. J. Virol. 767049-7059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.López-Bueno, A., M. Mateu, and J. M. Almendral. 2003. High mutant frequency in populations of a DNA virus allows evasion from antibody therapy in an immunodeficient host. J. Virol. 772701-2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.López-Bueno, A., N. Valle, J. M. Gallego, J. Pérez, and J. M. Almendral. 2004. Enhanced cytoplasmic sequestration of the nuclear export receptor CRM1 by NS2 mutations developed in the host regulates parvovirus fitness. J. Virol. 787049-7059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.López-Bueno, A., M. P. Rubio, N. Bryant, R. McKenna, M. Agbandje-McKenna, and J. M. Almendral. 2006. Host-selected amino acid changes at the sialic acid binding pocket of the parvovirus capsid modulate cell binding affinity and determine virulence. J. Virol. 801563-1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.López-Bueno, A., L. P. Villarreal, and J. M. Almendral. 2006. Parvovirus variation for disease: a difference with RNA viruses? Curr. Top. Microbiol. Immunol. 299349-370. [DOI] [PubMed] [Google Scholar]
- 30.Lukashov, V., and J. Goudsmit. 2001. Evolutionary relationships among parvoviruses: virus-host coevolution among autonomous primate parvoviruses and links between adeno-associated and avian parvoviruses. J. Virol. 752729-2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martin, J., G. Dunn, R. Hull, V. Patel, and P. D. Minor. 2000. Evolution of the Sabin strain of type 3 poliovirus in an immunodeficient patient during the entire 637-day period of virus excretion. J. Virol. 743001-3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maxwell, I. H., A. L. Spitzer, F. Maxwell, and D. J. Pintel. 1995. The capsid determinant of fibrotropism for the MVMp strain of minute virus of mice functions via VP2 and not VP1. J. Virol. 695829-5832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Merchlinsky, M. J., P. J. Tattersall, J. J. Leary, S. F. Cotmore, E. M. Gardiner, and D. C. Ward. 1983. Construction of an infectious molecular clone of the autonomous parvovirus minute virus of mice. J. Virol. 47227-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nam, H. J., B. Gurda-Whitaker, W. Y. Gan, S. Ilaria, R. McKenna, P. Mehta, R. A. Alvarez, and M. Agbandje-McKenna. 2006. Identification of the sialic acid structures recognized by minute virus of mice and the role of binding affinity in virulence adaptation. J. Biol. Chem. 28125670-25677. [DOI] [PubMed] [Google Scholar]
- 35.Parrish, C. R., C. F. Aquadro, M. L. Strassheim, J. F. Evermann, J. Y. Sgro, and H. O. Mohammed. 1991. Rapid antigenic-type replacement and DNA sequence evolution of canine parvovirus. J. Virol. 656544-6552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ramírez, J. C., A. Fairén, and J. M. Almendral. 1996. Parvovirus minute virus of mice strain i multiplication and pathogenesis in the newborn mouse brain is restricted to proliferative areas and to migratory cerebellar young neurons. J. Virol. 708109-8116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reguera, J., E. Grueso, A. Carreira, C. Sanchez-Martinez, J. M. Almendral, and M. G. Mateu. 2005. Functional relevance of amino acid residues involved in interactions with ordered nucleic acid in a spherical virus. J. Biol. Chem. 28017969-17977. [DOI] [PubMed] [Google Scholar]
- 38.Ron, D., and J. Tal. 1985. Coevolution of cells and virus as a mechanism for the persistence of lymphotropic minute virus of mice in L-cells. J. Virol. 55424-430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rubio, M.-P., A. López-Bueno, and J. M. Almendral. 2005. Virulent variants emerging in mice infected by the apathogenic prototype strain of the parvovirus MVM exhibit a capsid of low avidity for a primary receptor. J. Virol. 7911280-11290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sayle, R. A., and E. J. Milner-White. 1995. RasMol: biomolecular graphics for all. Trends Biochem. Sci. 20374-376. [DOI] [PubMed] [Google Scholar]
- 41.Segovia, J. C., A. Real, J. A. Bueren, and J. M. Almendral. 1991. In vitro myelosuppressive effects of the parvovirus minute virus of mice (MVMi) on hematopoietic stem and committed progenitor cells. Blood 77980-988. [PubMed] [Google Scholar]
- 42.Segovia, J. C., G. Guenechea, J. M. Gallego, J. M. Almendral, and J. A. Bueren. 2003. Parvovirus infection suppresses long-term repopulating hematopoietic stem cells. J. Virol. 778495-8503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Segovia, J. C., J. M. Gallego, J. A. Bueren, and J. M. Almendral. 1999. Severe leukopenia and dysregulated erythropoiesis in SCID mice persistently infected by the parvovirus minute virus of mice. J. Virol. 731774-1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shackelton, L. A., C. R. Parrish, U. Truyen, and E. C. Holmes. 2005. High rate of viral evolution associated with the emergence of carnivore parvovirus. Proc. Natl. Acad. Sci. USA 102379-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shackelton, L. A., and E. C. Holmes. 2006. Phylogenetic evidence for the rapid evolution of human B19 erythrovirus. J. Virol. 803666-3669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shankarappa, R., J. B. Margolick, S. J. Gange, A. G. Rodrigo, D. Upchurch, H. Farzadegan, P. Gupta, C. R. Rinaldo, G. H. Learn, X. He, X. L. Huang, and J. I. Mullins. 1999. Consistent viral evolutionary changes associated with populations of human immunodeficiency virus type 1 infection. J. Virol. 7310489-10502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Spalholz, B. A., and P. Tattersall. 1983. Interaction of minute virus of mice with differentiated cells: strain-dependent target cell specificity is mediated by intracellular factors. J. Virol. 46937-943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tattersall, P., A. J. Shatkin, and D. C. Ward. 1977. Sequence homology between the structural polypeptides of minute virus of mice. J. Mol. Biol. 111375-394. [DOI] [PubMed] [Google Scholar]
- 49.Tattersall, P., and J. Bratton. 1983. Reciprocal productive and restrictive virus-cell interaction of immunosuppressive and prototype strains of minute virus of mice. J. Virol. 46944-955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tattersall, P. 2006. The evolution of parvovirus taxonomy, p. 5-16. In J. Kerr, S. F. Cotmore, M. E. Bloom, R. M. Linden, and C. R. Parrish (ed.), The parvoviruses. Hodder Arnold, London, United Kingdom.
- 51.Vignuzzi, M., J. K. Stone, J. J. Arnold, C. E. Cameron, and R. Andino. 2006. Quasispecies diversity determines pathogenesis through cooperative interactions in a viral population. Nature 439344-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wolinsky, S. M., B. T. M. Korber, A. U. Neumann, M. Daniels, K. J. Kunstman, A. J. Whetsell, M. R. Furtado, Y. Cao, D. D. Ho, J. T. Safrit, and R. A. Koup. 1996. Adaptative evolution of human immunodeficiency virus-type 1 during the natural course of infection. Science 272537-542. [DOI] [PubMed] [Google Scholar]