Regulated protein degradation is a process that controls many cellular functions, including cell cycle progression, checkpoint activation, and apoptosis induction, and has also been implicated in development, cancer, and neurodegenerative diseases (45, 47, 48). Most of the regulated protein destruction is accomplished by the ubiquitin proteasome system (UPS). Proteins that are destined for degradation are first marked by chains of polyubiquitin that are appended to the epsilon amino groups of lysine residues in the target protein. Polyubiquitin chains target the substrate for degradation in the 26S proteasome.
Ubiquitin is a small protein that is highly conserved in eukaryotes. Covalent attachment of ubiquitin to the target protein requires the sequential activities of three enzymes (reviewed in references 41 and 45). In the first step, a ubiquitin-activating enzyme (E1) forms a thioester bond with ubiquitin. The ubiquitin molecule is then transferred to a ubiquitin-conjugating enzyme (E2). The E2 enzyme then interacts with a ubiquitin ligase (E3), which acts as a bridge to bring E2 and the target protein in the vicinity of each other. The ubiquitin chain is then transferred to a lysine residue on the target protein. The specificity of this process is ultimately determined by the E3 ligase complex. There are three types of E3 ligase complexes (for a schematic representation, see Fig. 1A): HECT ubiquitin ligases, single RING finger ubiquitin ligases, and multisubunit RING finger ubiquitin ligases, referred to as CRUL (Cullin RING ubiquitin ligases) (Fig. 1A shows a Cullin 4 [Cul4]-damaged-DNA-specific binding protein 1 [DDB1]-based CRUL). CRUL ligases are also referred to as SCF (Skp1/Cul1/F-box)-type E3 ligases.
FIG. 1.
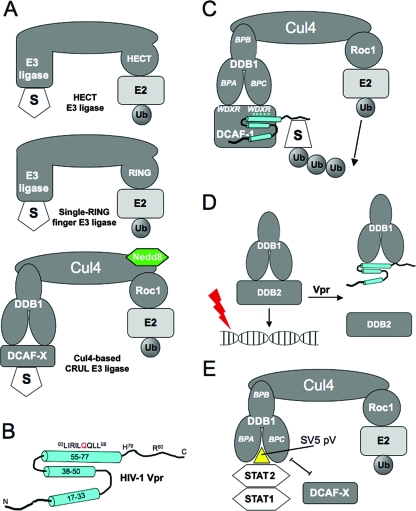
E3 ubiquitin ligases and their interactions with viral proteins. (A) The three main types of E3 ligases. (B) The nuclear magnetic resonance structure of HIV-1 Vpr. Cylinders represent α-helices, and lines are unstructured regions. (C) The Vpr-UPS model predicts the interaction of Vpr with DCAF1 and a putative degradation substrate, “S.” BPA, BPB, and BPC denote the three beta-propeller domains of DDB1. Le Rouzic et al. (34) found that the WDXR motifs of DCAF1 are required for the interaction with Vpr. For simplicity, we have represented Vpr interacting with the WDXR motif on BPC; there is no evidence to support which of the two WDXR motifs is required for Vpr interaction. (D) The Vpr-UV-DDB model predicts that the interaction of Vpr with DDB1 displaces DDB2 and prevents the DDB1-DDB2 complex, also known as UV-DDB, from recognizing damaged DNA. (E) SV5 protein V (pV) interacts with DDB1 and displaces DCAF, creating a new interface that recruits STAT2 as an adaptor, which then recruits STAT1 for ubiquitination. Ub, ubiquitin.
Viruses can manipulate many aspects of the biology of the infected cell, including the UPS. Manipulation of the UPS by a viral protein was first demonstrated in 1993 in a landmark study by Scheffner and colleagues (52), in which they reported that the E6 proteins from human papillomavirus (HPV) types 16 and 18 induced the polyubiquitination and degradation of the tumor suppressor p53. Degradation of p53 allows HPV-infected cells to circumvent cell cycle arrest and induction of apoptosis, effects that arise from viral infection, thereby providing a permissive cellular environment where HPV can efficiently replicate. Together with the ability of HPV E7 to block the function of the retinoblastoma protein (Rb), E6-mediated destruction of p53 is a key activity responsible for HPV's tumorigenic capacity (20).
Since the discovery of HPV E6's function, it has become increasingly apparent that manipulation of the UPS is a shared strategy used by unrelated viruses (Table 1). Recent reports have demonstrated that the paramyxovirus V proteins, hepatitis B virus protein X, and the human immunodeficiency virus type 1 (HIV-1) Vpu and Vif proteins are all able to manipulate E3 ubiquitin ligase complexes (24, 36, 40, 43, 55, 56, 64).
TABLE 1.
Examples of viral proteins that manipulate the UPS
| Virus | Viral protein | UPS interactions | Target | Functional effects | Reference |
|---|---|---|---|---|---|
| HPV | E6 | E6AP (HECT type) | p53 | Dysregulates cell cycle | 52 |
| Paramyxovirus SV5 | V | Cul4-DDB1 | STAT1, STAT2 | Overcomes type I interferon responses | 24 |
| Hepatitis B virus | HBX | Cul4-DDB1 | Unknown | Affects apoptosis, DNA repair, and cell cycle and enhances viral replication | 36, 56 |
| Vpu | Cul1-Skp1-β-TrCP1 | CD4 | Enhances viral release, inhibits IκB degradation | 8, 40 | |
| HIV-1 | Vif | Cul5-EloginB-C | APOBEC3G/F | Overcomes restriction | 43, 55, 64 |
| Vpr | DDB1-DCAF1 | Unknown | Induces ATR-dependent cell cycle arrest in G2 | 12, 25, 34, 53, 62 |
The most recent addition to this list of viral proteins is HIV-1 Vpr (11, 25, 34, 53, 54, 62). This review will focus on the newly identified interaction of Vpr with the UPS and the role it plays in the biology of HIV-1 Vpr.
Vpr INDUCES REPLICATION STRESS
HIV-1 vpr encodes a 96-amino-acid, 14-kDa protein. Research from a number of laboratories in the last decade has shown that Vpr performs multiple functions, including the induction of cell cycle arrest in the G2 phase, transactivation of the viral promoter, nuclear import of preintegration complexes in macrophages, induction of apoptosis, and enhancement of the fidelity of reverse transcription (reviewed in references 2 and 35). The molecular structure of Vpr has been resolved by using nuclear magnetic resonance (44) and shown to consist of a core with three interacting α-helices flanked by flexible, unstructured N- and C-terminal domains (for a schematic representation, see Fig. 1B).
Vpr induces G2 arrest by activating the ataxia telagiectasia-mutated and Rad3-related protein (ATR). ATR is a sensor of replication stress. Replication stress is a cellular condition that results from the stalling of replication forks as a consequence of various cellular insults, such as deoxyribonucleotide depletion, topoisomerase inhibition, and UV light-induced DNA damage (reviewed in reference 42). Through its serine/threonine kinase activity, ATR, in response to replication stress, phosphorylates a number of well-known protein targets and also leads to the formation of DNA-damaged nuclear foci. The physiological roles of this response are to halt DNA replication and to recruit DNA repair machinery. Expression of Vpr, either alone or in the context of full-length HIV-1, results in phosphorylation of the following ATR targets: histone 2A variant X (H2A-X), the replication protein A 32-kDa subunit (RPA32), checkpoint kinase 1 (Chk1), breast cancer-associated protein 1 (BRCA1), and p53-binding protein 1 (53BP1) (33, 38, 51, 68).
There is strong selection in favor of the HIV-1 vpr gene in vivo (16). In a vaccine study conducted with chimpanzees (14), two animals were challenged with a virus stock derived from HIVIIIB, which encodes a truncated, nonfunctional Vpr protein. A retrospective analysis of the vpr gene sequence in these animals’ virus populations revealed restoration of the truncated open reading frame in both chimpanzees (16). In addition, in an accidental infection of a laboratory worker with a stock of HIVIIIB, which also contained the above inactivating mutation (50, 61), the vpr gene reverted to full-length within 2 years (16). In further support of a role for Vpr in viral pathogenesis, an abnormal accumulation of infected cells in G2 has recently been demonstrated in cells from HIV-1-infected patients (69).
Vpr-induced G2 arrest has two downstream effects with important consequences for both the infected cell and the virus. First, the transcriptional activity of the viral promoter is increased in G2/M compared to that of cells in G1/S (16). Accordingly, production of viral particles is also enhanced during the G2 phase (16). It has also been suggested that accumulation of infected cells in G2 may favor selective translation of viral products, owing to the presence of an internal ribosome entry site in the HIV-1 genome (9).
The second consequence of G2 arrest is the subsequent onset of apoptosis in the infected cells. Induction of apoptosis by Vpr is linked to induction of G2 arrest, as both effects depend on the presence and function of ATR (1).
Vpr INTERACTS WITH A CRUL E3 UBIQUITIN LIGASE CONTAINING CUL4A, DDB1, AND DCAF1
The most intriguing question regarding the biology of Vpr is how it induces ATR activation leading to induction of G2 arrest. The answer to this question has remained a mystery until very recently. Plausible mechanisms that have been proposed include the ability of Vpr to bind directly to DNA (12, 66), its potential ability to bind to members of the ATR complex (68), activation of Wee1 kinase (65), the ability of Vpr to bind to Cdc25 (15), and the ability of Vpr to influence DNA repair (30). Ironically, a most-revealing clue to Vpr's mechanism of action was obtained in 1994, when a novel cellular protein of unknown function initially named Vpr-interacting protein (RIP) and later renamed Vpr-binding protein (VprBP) was isolated as a coprecipitation partner of Vpr (67).
The function of RIP/VprBP remained enigmatic until 2006, when several groups identified a family of proteins that were associated with the damaged-DNA-specific binding protein 1 (DDB1), a Cul4 adaptor (3, 19, 22, 28). This novel family of proteins, which includes VprBP, acts as the substrate specificity modules in a Cul4- and DDB1-based E3 ubiquitin ligase complex (3, 19, 22, 28). VprBP was, accordingly, renamed DDB1- and Cul4A-associated factor 1 (DCAF1).
Through coimmunoprecipitation experiments, several groups have very recently confirmed the physical association of Vpr with DCAF1 and extended this notion to show that Vpr is capable of binding a larger complex consisting of Cul4A, DDB1, and DCAF1 (11, 25, 34, 53, 62). In addition, several lines of evidence from the above groups strongly suggest that activation of the G2 checkpoint by Vpr is induced via binding and, possibly, activation of the Cul4A-DDB1-DCAF1 ligase.
DDB1 IS A Cul4 ADAPTOR
DDB1 is the only known Cul4 adaptor, and it links Cul4 to a number of possible substrate specificity subunits, which constitute a family of WD repeat proteins collectively referred to as DCAFs. The ubiquitination targets for several DCAFs have been identified. For example, CDT2 (DCAF2) recruits the origin of replication licensing factor CDT1 (21, 26) to prevent rereplication of DNA. Cockayne syndrome protein A (CSA; another WD repeat protein that associates with DDB1; not updated to the “DCAF” nomenclature) targets Cockayne syndrome B (CSB) for destruction as part of the recovery phase of transcription-coupled DNA repair (18). The DDB2/xeroderma pigmentosum complementation group E protein (XPE) is another DCAF that interacts with DDB1-Cul4A to promote degradation of XPC (57) and the histones 3 and 4 (59) as part of the response to DNA damage. Cul4A- and Cul4B-containing E3 ligases are also responsible for destruction of the cyclin-dependent kinase inhibitor p27 and cyclin E, respectively (23). A recent report has also found MDM2 and p53 to be degraded by Cul4-DDB1 complexes (5). Thus, the general roles of Cul4-DDB1 E3 ligases appear to consistently involve genome stability, DNA replication, and cell cycle checkpoint control. Cul4-based E3 ligases have also been implicated in the formation of heterochromatin via recruitment of methyl transferases to the chromatin (22, 27). It is not clear at the moment whether the ubiquitin transferase activity of the Cul4 complex is required for its role in heterochromatin formation.
DCAF1 LINKS Vpr TO DDB1 AND IS REQUIRED FOR Vpr-INDUCED G2 ARREST
Five laboratories recently detected the association of Vpr with a Cul4A-containing E3 ligase complex. While most of the evidence came from the observation that Vpr coimmunoprecipitated with DCAF1 and/or DDB1 (11, 25, 34, 53, 62), dissection of the interactions through mutational analysis of both DCAF1 and Vpr revealed that DCAF1 acts to bridge Vpr to DDB1 and the larger E3 ligase complex. Le Rouzic et al., in an effort to ablate the DDB1-interacting domains in DCAF1 (these are the WDXR repeats, which are conserved in most DCAFs) by mutagenesis, demonstrated that the WDXR motifs are required for binding not only to DDB1 but also to Vpr (34). Furthermore, overexpression of a DCAF1 fragment encompassing the WDXR repeats increased Vpr-mediated G2 arrest, indicating that the ability of DCAF1 to bind—and, possibly, bridge—DDB1 and Vpr is central to Vpr function (34). A small caveat to the above mutagenesis experiments is that the WDXR motifs could be essential elements of the protein structure of DCAF1 (and not necessarily the binding regions for DDB1 and Vpr), and therefore, their disruption precludes binding to both DDB1 and Vpr in a nonspecific fashion.
Experiments performed by DeHart et al., Wen et al., and Hrecka et al., revealed that depletion of DCAF1 by RNA interference eliminated the ability of Vpr to coprecipitate with DDB1 (11, 25, 62) and indicated that DCAF1 bridges Vpr onto DDB1-Cul4A.
Based on the above evidence, a model has emerged in which Vpr binds to a Cul4-DDB1-DCAF1 E3 ligase to trigger polyubiquitination and subsequent degradation of a putative cellular protein (Fig. 1 C), resulting in activation of the G2 checkpoint (11, 25, 34, 62). We will refer to the previous model as the Vpr-UPS model (Fig. 1C). An alternative model, which is based on the ability of DDB1 to function in detection of DNA damage as part of the UV-DDB complex, has been proposed (53). We will refer to this model, discussed below, as the Vpr-UV-DDB model (Fig. 1D).
The Vpr-UPS model predicts that Vpr simultaneously interacts with two cellular proteins: DCAF1 and a putative ubiquitination substrate (Fig. 1C). Thus, Vpr would be using two different interfaces, which could be independently mutated to generate two types of mutants: those that disrupt DCAF1 binding and those that disrupt substrate binding. While both types of Vpr mutants would be predicted to be inactive, those that retain the ability to bind to DCAF1 but are unable to recruit substrates should act as dominant-negative (DN) proteins.
The above predictions were confirmed as follows. The domain of Vpr that binds to DCAF1 was mapped to the leucine-rich (LR) motif 60LIRILQQLL68 within the third α-helix of HIV-189.6 Vpr (67). The first type of mutant (disrupting DCAF1 interaction) is exemplified by the Q65R substitution described by Le Rouzic et al. (34) (Q65 is shown in red in Fig. 1B). Consistent with the hypothesis that the DCAF1-Vpr interaction is required for Vpr function, Vpr(Q65R) failed to induce G2 arrest (34). Truncation of the last 18 residues of Vpr [Vpr(1-78)] or replacement of arginine at position 80 by alanine [Vpr(R80A)] resulted in proteins with intact binding to DCAF1 but unable to induce G2 arrest (11, 34). Coexpression of either Vpr(1-78) or Vpr(R80A) with wild-type Vpr resulted in a DN effect by the mutant on the induction of G2 arrest by wild-type Vpr (11, 34). Mutation of the LR domain (Q65R) in the context of Vpr(R80A) ablated the DN character of Vpr(R80A), indicating that the DN character of Vpr(R80A) depended on its ability to bind DCAF1. Together, these experiments indicate (i) that binding of Vpr to DCAF1 is necessary but not sufficient for induction of G2 arrest and (ii) that the carboxy-terminal domain of Vpr is likely required for the recruitment of a cellular protein whose ubiquitination and degradation leads to G2 arrest.
A FUNCTIONAL UPS IS REQUIRED FOR Vpr FUNCTION
The interaction of Vpr with an E3 ubiquitin ligase could result in two theoretical outcomes: inhibition or activation of the enzymatic activity. In the first case, one would predict that RNA interference-mediated depletion of DCAF1 would mimic the activity of Vpr. On the other hand, if Vpr promoted activation of the E3 ligase, then depletion of DCAF1 should counteract the effect of Vpr. The evidence overwhelmingly (albeit indirectly) points in the second direction, as depletion of DCAF1 uniformly restores a normal cell cycle profile in the presence of Vpr (11, 25, 34, 62).
DeHart et al. provide two additional lines of evidence to support that a functional UPS is required for Vpr function (11). First, incubation with epoxomicin, a pharmacologic inhibitor of the proteasome, overcomes the ability of Vpr to induce G2 arrest. Secondly, overexpression of a DN ubiquitin mutant, Ub(K48R), which blocks polyubiquitination (43), restores a normal cell cycle profile in the presence of Vpr.
The amino-terminal domain of Cul4A is responsible for binding to the adaptor, DDB1, whereas the C terminus interacts with the ring of Cul1 (ROC1) and E2. The C-terminal domain of Cul4A also contains the site for neddylation, where a ubiquitin-like molecule, Nedd8, needs to be covalently linked in order for CRUL ligases to become active. Thus, truncation of the C-terminal domain of Cul4A should lead to a DN mutant that is unable to recruit E2 or to be neddylated, as was previously demonstrated for a similar mutant in Cul1 (63). Wen et al. constructed a DN Cul4A and observed that its overexpression relieved Vpr-induced G2 arrest (62). This observation further supports the requirement for an active Cul4A-based E3 ubiquitin ligase in Vpr-induced G2 arrest. It is noteworthy that overexpression of a DN-Cul1 construct also alleviated G2 arrest to a similar degree (62). Although this result may appear to undermine the specificity of DN Cullin reagents, one must bear in mind that activation of the G2 checkpoint has been shown to require, at a downstream step, degradation of the Cdc25A phosphatase via a Cul1-Skp1-β-TRCP E3 ligase (10, 29). Degradation of Cdc25A through this pathway requires phosphorylation of a phosphodegron domain that is a known target of Chk1 kinase. Since Chk1 is a target of ATR upon activation by Vpr, this seems like a plausible explanation for the involvement of Cul1 in Vpr-induced G2 arrest. The relative roles of Cul1 and Cul4 (whether redundant or sequential) in the signaling of Vpr-induced G2 arrest will obviously require further clarification.
Vpr AND OTHER VIRAL MANIPULATORS OF THE UPS
The details of the interaction of Vpr with the E3 complex are important in the context of similar interactions of viral proteins with DDB1-Cul4A. DDB1 contains three beta propeller domains, A, B, and C (BPA, BPB, and BPC) (Fig. 1E). Proteins V and X from simian virus 5 (SV5), a paramyxovirus, and HBV, respectively, interact directly with DDB1 by positioning themselves in the cleft that lies between BPA and BPC. The interaction of proteins V and X with DDB1 is mutually exclusive with the DCAF-DDB1 interaction, at least for the several DCAFs tested so far (3, 37). However, proteins V and X lack WDXR motifs (3). In this fashion, protein V recruits STAT2, which is then used as an adaptor to recruit STAT1. STAT1, a protein that is not normally turned over via proteasome-dependent proteolysis, is then polyubiquitinated in a DCAF-independent manner and targeted for degradation (24). The target for HBV protein X remains unknown.
In contrast to proteins V and X, Vpr appears to use the WD repeat motifs to interact with DCAF1, and importantly, Vpr binding to DCAF1 does not exclude DCAF1 from the E3 ligase complex. Thus, Vpr behaves like Vpu, which does not directly bind to Skp1 (the Cul1 adaptor that is equivalent to DDB1 in Cul4) but instead uses the β-TRCP subunit. Interestingly, in addition to promoting degradation of CD4, Vpu acts as an inhibitor of β-TRCP-mediated ubiquitination of IκB, a natural substrate for β-TRCP (8).
Since DCAF1 is part of the Vpr-associated E3 ligase, it is formally possible that the substrate being recruited to the E3 complex by Vpr is a natural substrate for DCAF1. If this is the case, Vpr represents a departure from its precedents, wherein the viral proteins recruit noncognate targets to the E3 ligase (Table 1). Unfortunately, since the identity of the protein(s) being polyubiquitinated under the influence of Vpr is still unknown, one can only speculate on this point. The observation that DCAF1 depletion relieves low-dose aphidicolin (like Vpr, an inducer of replication stress)-mediated G2 arrest (11) lends support to the idea that Vpr may enhance degradation of a normal DCAF1 substrate. How could Vpr enhance the E3 ligase activity toward a normal DCAF1 substrate? The results published by Hrecka et al. suggest that Vpr may induce activation of the neddylation of Cul4A-DDB1 (25) and therefore raise the possibility that Vpr regulates the activity, and perhaps not the specificity, of the complex.
DDB1 ALSO PARTICIPATES IN NER
In addition to its role as a Cul4 adaptor, DDB1 plays a role in nucleotide excision repair (NER). DDB1, in concert with DDB2/XPE, forms the UV-damaged-DNA-binding protein complex (UV-DDB) (32). In this complex, DDB2 has intrinsic DNA-binding ability. Upon UV-induced DNA damage, DDB1 translocates to the nucleus, where it is tethered to the damaged DNA via a direct interaction with DDB2, initiating repair of the damaged DNA by recruiting NER factors (32).
Schrofelbauer et al. reported that Vpr, by directly binding to DDB1, inhibited the DDB1-DDB2 interaction (53). As a consequence of this interference, Vpr impaired the ability of DDB1 to translocate to the nucleus in response to UV irradiation, and Vpr-expressing cells had a decreased ability to repair DNA following UV irradiation. Taken together, these data suggest that Vpr inhibits repair of DNA damage by blocking the interaction between DDB1 and DDB2 (Vpr-UV-DDB model) (Fig. 1D). Damaged DNA accumulates in the presence of Vpr, and this damage is ultimately the trigger of the G2 checkpoint.
This is an attractive model of how Vpr may act and, in principle, is not mutually exclusive with the Vpr-UPS model. However, a close look at the evidence raises some potential contradictions between the two models. First, the function of the DDB1-DDB2 complex in recognizing DNA damage does not require the presence of DCAF1, whereas Vpr-induced G2 arrest does. Second, the binding of Vpr to DDB1 is not direct and is instead mediated by DCAF1. Third, certain carboxy-terminal mutants of Vpr, such as Vpr(1-78) and Vpr(R80A), efficiently coimmunoprecipitate with DDB1 but are unable to induce G2 arrest, suggesting that binding to DDB1-DCAF1 is not sufficient for checkpoint activation.
Xeroderma pigmentosum complementation group E cells lack DDB2/XPE function due to mutations in DDB2 but appear to express normal DDB1. DeHart et al. examined the role of DDB2/XPE by introducing Vpr in XPE cells (11). Expression of Vpr arrested XPE cells in G2 in a manner that was indistinguishable from that of the control fibroblasts. This finding is also inconsistent with the idea that Vpr activates the G2 checkpoint by disrupting UV-DDB activity.
It is tempting to conclude from the above findings that the actions of Vpr on an E3 ubiquitin ligase and on the UV-DDB complex represent independent effects of Vpr and that perhaps the disruption of UV-DDB is yet another way in which Vpr exerts cytopathicity in the infected cell.
DOES Vpr MODIFY THE INTRINSIC UBIQUITIN LIGASE ACTIVITY OF CUL4A-DDB1-DCAF1?
The activity of CRUL ligases is regulated via neddylation (Fig. 1A). Neddylation results in enhanced recruitment of E2 to the Cullin complex and increases E3 ligase activity (60). The COP1 signalosome, which associates with Cullins, deneddylates the complex, rendering it inactive by allowing interaction with CAND1 (60). CAND1 is a repressor of CRUL ligases, because it triggers dissociation of the substrate receptor subunits. Hrecka et al. examined the neddylation status of Cul4A in the presence of Vpr and found that overexpression of Vpr and DCAF1 led to increased neddylation of Cul4A (25). While the activity of Cul4A-DDB1-DCAF1 could not be measured against its normal target(s) simply because none are known, Hrecka et al. used autoubiquitination of Cul4A as a surrogate marker. They found that in the presence Vpr, Cul4A was extensively polyubiquitinated (25).
DEGRADATION TARGET(S) OF Vpr AND DCAF1
As mentioned above, natural substrates for DCAF1 are not known, nor is the one targeted by Vpr in order to induce G2 arrest. Schrofelbauer observed that Vpr induced proteasomal degradation of uracil N glycosylase (UNG), an effect also mediated by a Cul4-DDB1 E3 ligase (53, 54). Wen et al., in contrast, observed that UDG was constitutively (i.e., in a manner that was independent of Vpr) targeted by a Cul4A-DDB1 ligase and also reported that DCAF1 knockdown did not affect UNG stability, whether in the presence of Vpr or in its absence (62). Both groups concurred that degradation of UNG is independent of Vpr-induced G2 arrest (53, 62). The relevance of UNG degradation to HIV-1 replication has recently been put in question (31). Therefore, understanding whether and how UPS-mediated degradation of UNG may play a role in HIV-1 biology will require further investigation.
Several substrates have been identified for DCAFs other than DCAF1. Of relevance here, Cdt1, an important regulator of replication origin licensing, is subjected to proteasomal degradation following polyubiquitination by the Cul4A-DDB1 E3 ligase (4, 46). Enforced inhibition of DDB1 can lead to the stabilization of Cdt1, resulting in endoreduplication and activation of ATR and the G2 checkpoint (39). Endoreduplication, in fact, was found to occur in the presence of Vpr, leading to the formation of cells containing 8N chromosomes (6). DeHart et al. tested the hypothesis that Vpr, by redirecting the specificity of the E3 ligase to a particular substrate, might in turn hinder ubiquitination of Cdt1 (11). Cdt1 steady-state levels, however, did not change in the presence or absence of Vpr, and therefore, it was concluded that Vpr does not activate the G2 checkpoint via inhibition of the Cul4A-DDB1 E3 ligase towards degradation of Cdt1.
CONSERVATION OF DCAF1 BINDING IN SIVmac/HIV-2 Vpx
Vpr is structurally and functionally conserved in five of the primate lentiviral lineages, including HIV-1/SIVcpz, HIV-2/SIVmac/SIVsm, SIVagm, SIVsyk, and SIVmnd (13, 49, 58). The HIV-2/SIVmac/SIVsm group carries two highly related genes termed vpr and vpx. Vpr and Vpx share significant sequence identity with HIV-1 Vpr. Two functions ascribed to HIV-1 Vpr have segregated in HIV-2/SIVmac/SIVsm such that HIV-2/SIVmac/SIVsm Vpr induces G2 arrest, whereas Vpx participates in nuclear transport of preintegration complexes in nondividing cells (13, 49). In view of this, one would predict that HIV-2/SIVmac/SIVsm Vpr proteins would be able to bind to DCAF1, while the Vpx counterparts would not. In line with this expectation, it was shown that SIVmac and HIV-2 Vpr interacted with DCAF1 (34, 62). Surprisingly, however, Le Rouzic et al. found that the SIVmac and HIV-2 Vpx proteins, which are unable to manipulate the cell cycle, also bind to DCAF1, using a LR motif that is conserved with that of HIV-1 and SIVmac Vpr (34).
Thus, for SIVmac, both Vpr and Vpx are able to bind DCAF1, but only Vpr has an effect on the cell cycle. This raises the question of whether or not the ability of Vpx to interact with DCAF1 is necessary for Vpx to promote infection of nondividing cells (7, 13, 17). And if so, is this function exerted through manipulation of the corresponding E3 ubiquitin ligase? Clearly, the recent findings about Vpr and the UPS have generated many new questions that should be the subject of intense investigation in the near future.
It should be pointed out that Wen et al., in contrast, found that HIV-2 Vpx failed to bind to DCAF1 (62). It appears that this discrepancy could be explained by the different HIV-2 isolates used for the cloning of the vpx gene, HIV-2ROD (62) and HIV-2GH-1 (34), although further studies will be required to clarify this issue (E. Le Rouzic and C. de Noronha, personal communication).
CONCLUDING REMARKS
Vpr is the third accessory protein from HIV-1 to be identified as a manipulator of E3 ubiquitin ligases. And, pending further confirmation, the same could apply to lentiviruses in the HIV-2/SIVmac/SIVsm group (Vpr, Vif, and Vpx). It is striking that Vpr, Vpu, and Vif (and, by extension, HPV E6, SV5 protein V, and HBV protein X), without evidence for common ancestry, have evolved to use the same strategy for very different purposes. Vpu uses Cul1-Skp1-β-TRCP to degrade CD4 and allow optimal virus production. Vif uses Cul5-ElonginB-C to degrade APOBEC3G and F and overcome cytidine deamination leading to hypermutation. And now we know that Vpr manipulates Cul4-DDB1-DCAF1 to induce G2 arrest and apoptosis.
It is also intriguing how the degradation of a putative cellular factor instigated by Vpr leads to replication stress and G2 arrest. Although documented instances exist for how the UPS controls cell cycle progression and checkpoint activation, how Vpr connects both sets of machinery appears to be different, and therefore its mechanism represents a missing link.
Outside of a moderate transactivation effect associated with the G2 phase, it is not yet clear how HIV-1 profits from manipulation of the UPS. We anticipate that the discovery of the ubiquitination target(s) for Vpr will not only present us with the missing link mentioned above but may also reveal previously unsuspected ways in which lentiviral Vpr manipulates the biology of host cells.
ADDENDUM IN PROOF
During preparation of this manuscript, two additional reports were published that confirm the activity of Vpr in recruiting and activating a Cul4-DDBl-DCAF1 E3 ubiquitin ligase, resulting in G2 arrest (J. P. Belzile, G. Duisit, N. Rougeau, J. Mercier, A. Finzi, and E. A. Cohen, PLoS Pathog. 3:e85, 2007, and L. Tan, E. Ehrlich, and X. F. Yu, J. Virol. 81:10822-10830, 2007).
Acknowledgments
We are grateful to Catherine Transy, Florence Margottin-Goguet, and Erwann Le Rouzic (Institut Cochin, Paris, France); Carlos de Noronha (Albany Medical College, Albany, NY); and Erik Zimmerman (University of Utah, Salt Lake City, UT) for enlightening discussions and valuable help with preparation of the manuscript.
This work was supported by NIH research grant AI49057 to V.P.
Footnotes
Published ahead of print on 12 September 2007.
REFERENCES
- 1.Andersen, J. L., J. L. Dehart, E. S. Zimmerman, O. Ardon, B. Kim, G. Jacquot, S. Benichou, and V. Planelles. 2006. HIV-1 Vpr-induced apoptosis is cell cycle dependent and requires Bax but not ANT. PLoS Pathog. 2e127. doi: 10.1371/journal.ppat.0020127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andersen, J. L., and V. Planelles. 2005. The role of Vpr in HIV-1 pathogenesis. Curr. HIV Res. 343-51. [DOI] [PubMed] [Google Scholar]
- 3.Angers, S., T. Li, X. Yi, M. J. MacCoss, R. T. Moon, and N. Zheng. 2006. Molecular architecture and assembly of the DDB1-CUL4A ubiquitin ligase machinery. Nature 443590-593. [DOI] [PubMed] [Google Scholar]
- 4.Arias, E. E., and J. C. Walter. 2006. PCNA functions as a molecular platform to trigger Cdt1 destruction and prevent re-replication. Nat. Cell Biol. 884-90. [DOI] [PubMed] [Google Scholar]
- 5.Banks, D., M. Wu, L. A. Higa, N. Gavrilova, J. Quan, T. Ye, R. Kobayashi, H. Sun, and H. Zhang. 2006. L2DTL/CDT2 and PCNA interact with p53 and regulate p53 polyubiquitination and protein stability through MDM2 and CUL4A/DDB1 complexes. Cell Cycle 51719-1729. [DOI] [PubMed] [Google Scholar]
- 6.Bartz, S. R., M. E. Rogel, and M. Emerman. 1996. Human immunodeficiency virus type 1 cell cycle control: Vpr is cytostatic and mediates G2 accumulation by a mechanism which differs from DNA damage checkpoint control. J. Virol. 702324-2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Belshan, M., L. A. Mahnke, and L. Ratner. 2006. Conserved amino acids of the human immunodeficiency virus type 2 Vpx nuclear localization signal are critical for nuclear targeting of the viral preintegration complex in non-dividing cells. Virology 346118-126. [DOI] [PubMed] [Google Scholar]
- 8.Bour, S., C. Perrin, H. Akari, and K. Strebel. 2001. The human immunodeficiency virus type 1 Vpu protein inhibits NF-κB activation by interfering with βTrCP-mediated degradation of IκB. J. Biol. Chem. 27615920-15928. [DOI] [PubMed] [Google Scholar]
- 9.Brasey, A., M. Lopez-Lastra, T. Ohlmann, N. Beerens, B. Berkhout, J.-L. Darlix, and N. Sonenberg. 2003. The leader of human immunodeficiency virus type 1 genomic RNA harbors an internal ribosome entry segment that is active during the G2/M phase of the cell cycle. J. Virol. 773939-3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Busino, L., M. Donzelli, M. Chiesa, D. Guardavaccaro, D. Ganoth, N. V. Dorrello, A. Hershko, M. Pagano, and G. F. Draetta. 2003. Degradation of Cdc25A by β-TrCP during S phase and in response to DNA damage. Nature 42687-91. [DOI] [PubMed] [Google Scholar]
- 11.DeHart, J. L., E. S. Zimmerman, O. Ardon, C. M. R. Monteiro-Filho, E. R. Argañaraz, and V. Planelles. 2007. HIV-1 Vpr activates the G2 checkpoint through manipulation of the ubiquitin proteasome system. Virol. J. 457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Rocquigny, H., A. Caneparo, T. Delaunay, J. Bischerour, J.-F. Mouscadet, and B. P. Roques. 2000. Interactions of the C-terminus of viral protein R with nucleic acids are modulated by its N-terminus. Eur. J. Biochem. 2673654-3660. [DOI] [PubMed] [Google Scholar]
- 13.Fletcher, T. M., III, B. Brichacek, N. Sharova, M. A. Newman, G. Stivahtis, P. M. Sharp, M. Emerman, B. H. Hahn, and M. Stevenson. 1996. Nuclear import and cell cycle arrest functions of the HIV-1 Vpr protein are encoded by two separate genes in HIV-2/SIVSM. EMBO J. 156155-6165. [PMC free article] [PubMed] [Google Scholar]
- 14.Fultz, P. N., P. Nara, F. Barre-Sinoussi, A. Chaput, M. L. Greenberg, E. Muchmore, M. P. Kieny, and M. Girard. 1992. Vaccine protection of chimpanzees against challenge with HIV-1-infected peripheral blood mononuclear cells. Science 2561687-1690. [DOI] [PubMed] [Google Scholar]
- 15.Goh, W. C., N. Manel, and M. Emerman. 2004. The human immunodeficiency virus Vpr protein binds Cdc25C: implications for G2 arrest. Virology 318337-349. [DOI] [PubMed] [Google Scholar]
- 16.Goh, W. C., M. E. Rogel, C. M. Kinsey, S. F. Michael, P. N. Fultz, M. A. Nowak, B. H. Hahn, and M. Emerman. 1998. HIV-1 Vpr increases viral expression by manipulation of the cell cycle: a mechanism for selection of Vpr in vivo. Nat. Med. 465-71. [DOI] [PubMed] [Google Scholar]
- 17.Goujon, C., L. Rivière, L. Jarrosson-Wuilleme, J. Bernaud, D. Rigal, J.-L. Darlix, and A. Cimarelli. 2007. SIVSM/HIV-2 Vpx proteins promote retroviral escape from a proteasome-dependent restriction pathway present in human dendritic cells. Retrovirology 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Groisman, R., I. Kuraoka, O. Chevallier, N. Gaye, T. Magnaldo, K. Tanaka, A. F. Kisselev, A. Harel-Bellan, and Y. Nakatani. 2006. CSA-dependent degradation of CSB by the ubiquitin-proteasome pathway establishes a link between complementation factors of the Cockayne syndrome. Genes Dev. 201429-1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.He, Y. J., C. M. McCall, J. Hu, Y. Zeng, and Y. Xiong. 2006. DDB1 functions as a linker to recruit receptor WD40 proteins to CUL4-ROC1 ubiquitin ligases. Genes Dev. 202949-2954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hebner, C. M., and L. A. Laimins. 2006. Human papillomaviruses: basic mechanisms of pathogenesis and oncogenicity. Rev. Med. Virol. 1683-97. [DOI] [PubMed] [Google Scholar]
- 21.Higa, L. A., I. S. Mihaylov, D. P. Banks, J. Zheng, and H. Zhang. 2003. Radiation-mediated proteolysis of CDT1 by CUL4-ROC1 and CSN complexes constitutes a new checkpoint. Nat. Cell Biol. 51008-1015. [DOI] [PubMed] [Google Scholar]
- 22.Higa, L. A., M. Wu, T. Ye, R. Kobayashi, H. Sun, and H. Zhang. 2006. CUL4-DDB1 ubiquitin ligase interacts with multiple WD40-repeat proteins and regulates histone methylation. Nat. Cell Biol. 81277-1283. [DOI] [PubMed] [Google Scholar]
- 23.Higa, L. A., X. Yang, J. Zheng, D. Banks, M. Wu, P. Ghosh, H. Sun, and H. Zhang. 2006. Involvement of CUL4 ubiquitin E3 ligases in regulating CDK inhibitors Dacapo/p27Kip1 and cyclin E degradation. Cell Cycle 571-77. [DOI] [PubMed] [Google Scholar]
- 24.Horvath, C. M. 2004. Weapons of STAT destruction. Interferon evasion by paramyxovirus V protein. Eur. J. Biochem. 2714621-4628. [DOI] [PubMed] [Google Scholar]
- 25.Hrecka, K., M. Gierszewska, S. Srivastava, L. Kozaczkiewicz, S. K. Swanson, L. Florens, M. P. Washburn, and J. Skowronski. 2007. Lentiviral Vpr usurps Cul4-DDB1[VprBP] E3 ubiquitin ligase to modulate cell cycle. Proc. Natl. Acad. Sci. USA 10411778-11783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hu, J., C. M. McCall, T. Ohta, and Y. Xiong. 2004. Targeted ubiquitination of CDT1 by the DDB1-CUL4A-ROC1 ligase in response to DNA damage. Nat. Cell Biol. 61003-1009. [DOI] [PubMed] [Google Scholar]
- 27.Jia, S., R. Kobayashi, and S. I. Grewal. 2005. Ubiquitin ligase component Cul4 associates with Clr4 histone methyltransferase to assemble heterochromatin. Nat. Cell Biol. 71007-1013. [DOI] [PubMed] [Google Scholar]
- 28.Jin, J., E. E. Arias, J. Chen, J. W. Harper, and J. C. Walter. 2006. A family of diverse Cul4-Ddb1-interacting proteins includes Cdt2, which is required for S phase destruction of the replication factor Cdt1. Mol. Cell 23709-721. [DOI] [PubMed] [Google Scholar]
- 29.Jin, J., T. Shirogane, L. Xu, G. Nalepa, J. Qin, S. J. Elledge, and J. W. Harper. 2003. SCFβ-TRCP links Chk1 signaling to degradation of the Cdc25A protein phosphatase. Genes Dev. 173062-3074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jowett, J. B., Y. M. Xie, and I. S. Chen. 1999. The presence of human immunodeficiency virus type 1 Vpr correlates with a decrease in the frequency of mutations in a plasmid shuttle vector. J. Virol. 737132-7137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kaiser, S. M., and M. Emerman. 2006. Uracil DNA glycosylase is dispensable for human immunodeficiency virus type 1 replication and does not contribute to the antiviral effects of the cytidine deaminase Apobec3G. J. Virol. 80875-882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kulaksiz, G., J. T. Reardon, and A. Sancar. 2005. Xeroderma pigmentosum complementation group E protein (XPE/DDB2): purification of various complexes of XPE and analyses of their damaged DNA binding and putative DNA repair properties. Mol. Cell. Biol. 259784-9792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lai, M., E. S. Zimmerman, V. Planelles, and J. Chen. 2005. Activation of the ATR pathway by human immunodeficiency virus type 1 Vpr involves its direct binding to chromatin in vivo. J. Virol. 7915443-15451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Le Rouzic, E., N. Belaïdouni, E. Estrabaud, M. Morel, J.-C. Rain, C. Transy, and F. Margottin-Goguet. 2007. HIV1 Vpr arrests the cell cycle by recruiting DCAF1/VprBP, a receptor of the Cul4-DDB1 ubiquitin ligase. Cell Cycle 6182-188. [DOI] [PubMed] [Google Scholar]
- 35.Le Rouzic, E., and S. Benichou. 2005. The Vpr protein from HIV-1: distinct roles along the viral life cycle. Retrovirology 211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leupin, O., S. Bontron, C. Schaeffer, and M. Strubin. 2005. Hepatitis B virus X protein stimulates viral genome replication via a DDB1-dependent pathway distinct from that leading to cell death. J. Virol. 794238-4245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Leupin, O., S. Bontron, and M. Strubin. 2003. Hepatitis B virus X protein and simian virus 5 V protein exhibit similar UV-DDB1 binding properties to mediate distinct activities. J. Virol. 776274-6283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li, G., R. T. Elder, K. Qin, H. U. Park, D. Liang, and R. Y. Zhao. 2007. PP2A dependent and independent pathways for ATR phosphorylation of Chk1. J. Biol. Chem. 2827287-7298. [DOI] [PubMed] [Google Scholar]
- 39.Lovejoy, C. A., K. Lock, A. Yenamandra, and D. Cortez. 2006. DDB1 maintains genome integrity through regulation of Cdt1. Mol. Cell. Biol. 267977-7990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Margottin, F., S. P. Bour, H. Durand, L. Selig, S. Benichou, V. Richard, D. Thomas, K. Strebel, and R. Benarous. 1998. A novel human WD protein, h-βTrCp, that interacts with HIV-1 Vpu connects CD4 to the ER degradation pathway through an F-box motif. Mol. Cell 1565-574. [DOI] [PubMed] [Google Scholar]
- 41.McCall, C. M., J. Hu, and Y. Xiong. 2005. Recruiting substrates to cullin 4-dependent ubiquitin ligases by DDB1. Cell Cycle 427-29. [DOI] [PubMed] [Google Scholar]
- 42.McGowan, C. H., and P. Russell. 2004. The DNA damage response: sensing and signaling. Curr. Opin. Cell Biol. 16629-633. [DOI] [PubMed] [Google Scholar]
- 43.Mehle, A., B. Strack, P. Ancuta, C. Zhang, M. McPike, and D. Gabuzda. 2004. Vif overcomes the innate antiviral activity of APOBEC3G by promoting its degradation in the ubiquitin-proteasome pathway. J. Biol. Chem. 2797792-7798. [DOI] [PubMed] [Google Scholar]
- 44.Morellet, N., S. Bouaziz, P. Petitjean, and B. P. Roques. 2003. NMR structure of the HIV-1 regulatory protein VPR. J. Mol. Biol. 327215-227. [DOI] [PubMed] [Google Scholar]
- 45.Nalepa, G., M. Rolfe, and J. W. Harper. 2006. Drug discovery in the ubiquitin-proteasome system. Nat. Rev. Drug Discov. 5596-613. [DOI] [PubMed] [Google Scholar]
- 46.Nishitani, H., and Z. Lygerou. 2002. Control of DNA replication licensing in a cell cycle. Genes Cells 7523-534. [DOI] [PubMed] [Google Scholar]
- 47.O'Connell, B. C., and J. W. Harper. 2007. Ubiquitin proteasome system (UPS): what can chromatin do for you? Curr. Opin. Cell Biol. 19206-214. [DOI] [PubMed] [Google Scholar]
- 48.Petroski, M. D., and R. J. Deshaies. 2005. Function and regulation of cullin-RING ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 69-20. [DOI] [PubMed] [Google Scholar]
- 49.Planelles, V., J. B. Jowett, Q. X. Li, Y. Xie, B. Hahn, and I. S. Chen. 1996. Vpr-induced cell cycle arrest is conserved among primate lentiviruses. J. Virol. 702516-2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Reitz, M. S., Jr., L. Hall, M. Robert-Guroff, J. Lautenberger, B. M. Hahn, G. M. Shaw, L. I. Kong, S. H. Weiss, D. Waters, R. C. Gallo, et al. 1994. Viral variability and serum antibody response in a laboratory worker infected with HIV type 1 (HTLV type IIIB). AIDS Res. Hum. Retrovir. 101143-1155. [DOI] [PubMed] [Google Scholar]
- 51.Roshal, M., B. Kim, Y. Zhu, P. Nghiem, and V. Planelles. 2003. Activation of the ATR-mediated DNA damage response by the HIV-1 viral protein R. J. Biol. Chem. 27825879-25886. [DOI] [PubMed] [Google Scholar]
- 52.Scheffner, M., J. M. Huibregtse, R. D. Vierstra, and P. M. Howley. 1993. The HPV-16 E6 and E6-AP complex functions as a ubiquitin-protein ligase in the ubiquitination of p53. Cell 75495-505. [DOI] [PubMed] [Google Scholar]
- 53.Schröfelbauer, B., Y. Hakata, and N. R. Landau. 2007. HIV-1 Vpr function is mediated by interaction with the damage-specific DNA-binding protein DDB1. Proc. Natl. Acad. Sci. USA 1044130-4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schröfelbauer, B., Q. Yu, S. G. Zeitlin, and N. R. Landau. 2005. Human immunodeficiency virus type 1 Vpr induces the degradation of the UNG and SMUG uracil-DNA glycosylases. J. Virol. 7910978-10987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sheehy, A. M., N. C. Gaddis, and M. H. Malim. 2003. The antiretroviral enzyme APOBEC3G is degraded by the proteasome in response to HIV-1 Vif. Nat. Med. 91404-1407. [DOI] [PubMed] [Google Scholar]
- 56.Sitterlin, D., F. Bergametti, and C. Transy. 2000. UVDDB p127-binding modulates activities and intracellular distribution of hepatitis B virus X protein. Oncogene 194417-4426. [DOI] [PubMed] [Google Scholar]
- 57.Sugasawa, K., Y. Okuda, M. Saijo, R. Nishi, N. Matsuda, G. Chu, T. Mori, S. Iwai, K. Tanaka, K. Tanaka, and F. Hanaoka. 2005. UV-induced ubiquitylation of XPC protein mediated by UV-DDB-ubiquitin ligase complex. Cell 121387-400. [DOI] [PubMed] [Google Scholar]
- 58.Tristem, M., A. Purvis, and D. L. Quicke. 1998. Complex evolutionary history of primate lentiviral vpr genes. Virology 240232-237. [DOI] [PubMed] [Google Scholar]
- 59.Wang, H., L. Zhai, J. Xu, H. Y. Joo, S. Jackson, H. Erdjument-Bromage, P. Tempst, Y. Xiong, and Y. Zhang. 2006. Histone H3 and H4 ubiquitylation by the CUL4-DDB-ROC1 ubiquitin ligase facilitates cellular response to DNA damage. Mol. Cell 22383-394. [DOI] [PubMed] [Google Scholar]
- 60.Wei, N., and X. W. Deng. 2003. The COP9 signalosome. Annu. Rev. Cell Dev. Biol. 19261-286. [DOI] [PubMed] [Google Scholar]
- 61.Weiss, S. H., J. J. Goedert, S. Gartner, M. Popovic, D. Waters, P. Markham, F. di Marzo Veronese, M. H. Gail, W. E. Barkley, J. Gibbons, et al. 1988. Risk of human immunodeficiency virus (HIV-1) infection among laboratory workers. Science 23968-71. [DOI] [PubMed] [Google Scholar]
- 62.Wen, X., K. M. Duus, T. D. Friedrich, and C. M. de Noronha. 2007. The HIV1 protein Vpr acts to promote G2 cell cycle arrest by engaging a DDB1 and Cullin4A-containing ubiquitin ligase complex using VprBP/DCAF1 as an adaptor. J. Biol. Chem. 28227046-27057. [DOI] [PubMed] [Google Scholar]
- 63.Wu, K., A. Chen, and Z. Q. Pan. 2000. Conjugation of Nedd8 to CUL1 enhances the ability of the ROC1-CUL1 complex to promote ubiquitin polymerization. J. Biol. Chem. 27532317-32324. [DOI] [PubMed] [Google Scholar]
- 64.Yu, X., Y. Yu, B. Liu, K. Luo, W. Kong, P. Mao, and X. F. Yu. 2003. Induction of APOBEC3G ubiquitination and degradation by an HIV-1 Vif-Cul5-SCF complex. Science 3021056-1060. [DOI] [PubMed] [Google Scholar]
- 65.Yuan, H., M. Kamata, Y.-M. Xie, and I. S. Y. Chen. 2004. Increased levels of Wee-1 kinase in G2 are necessary for Vpr- and gamma irradiation-induced G2 arrest. J. Virol. 788183-8190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang, S., D. Pointer, G. Singer, Y. Feng, K. Park, and L. J. Zhao. 1998. Direct binding to nucleic acids by Vpr of human immunodeficiency virus type 1. Gene 212157-166. [DOI] [PubMed] [Google Scholar]
- 67.Zhao, L. J., S. Mukherjee, and O. Narayan. 1994. Biochemical mechanism of HIV-I Vpr function. Specific interaction with a cellular protein. J. Biol. Chem. 26915577-15582. [PubMed] [Google Scholar]
- 68.Zimmerman, E. S., J. Chen, J. L. Andersen, O. Ardon, J. L. DeHart, J. Blackett, S. K. Choudhary, D. Camerini, P. Nghiem, and V. Planelles. 2004. Human immunodeficiency virus type 1 Vpr-mediated G2 arrest requires Rad17 and Hus1 and induces nuclear BRCA1 and γ-H2AX focus formation. Mol. Cell. Biol. 249286-9294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zimmerman, E. S., M. P. Sherman, J. L. Blackett, J. A. Neidleman, C. Kreis, P. Mundt, S. A. Williams, M. Warmerdam, J. Kahn, F. M. Hecht, R. M. Grant, C. M. de Noronha, A. S. Weyrich, W. C. Greene, and V. Planelles. 2006. Human immunodeficiency virus type 1 Vpr induces DNA replication stress in vitro and in vivo. J. Virol. 8010407-10418. [DOI] [PMC free article] [PubMed] [Google Scholar]