Abstract
The protein encoded by the UL14 gene of herpes simplex virus type 1 (HSV-1) and HSV-2 is expressed late in infection and is a minor component of the virion tegument. An UL14-deficient HSV-1 mutant (UL14D) forms small plaques and exhibits an extended growth cycle at low multiplicities of infection (MOI) compared to wild-type virus. Although UL14 is likely to be involved in the process of viral maturation and egress, its precise role in viral replication is still enigmatic. In this study, we found that immediate-early viral mRNA expression was decreased in UL14D-infected cells. Transient coexpression of UL14 and VP16 in the absence of infection stimulated the nuclear accumulation of both proteins. We intended to visualize the fate of VP16 released from the infected virion and constructed UL14-null (14D-VP16G) and rescued (14R-VP16G) viruses that expressed a VP16-green fluorescent protein (GFP) fusion protein. Synchronous high-multiplicity infection of the viruses was performed at 4°C in the absence of de novo protein synthesis. We found that the presence of UL14 in the virion had an enhancing effect on the nuclear accumulation of VP16-GFP. The lack of UL14 did not significantly alter virus internalization but affected incoming capsid transport to the nuclear pore. These observations suggested that UL14 (i) enhanced VP16 nuclear localization at the immediately early phase, thus indirectly regulating the expression of immediate-early genes, and (ii) was associated with efficient nuclear targeting of capsids. The tegument protein UL14 could be part of the machinery that regulates HSV-1 replication.
Herpes simplex virus type 1 (HSV-1) is a large-DNA, enveloped virus with icosahedral symmetry. The herpes simplex virion has four components: an electron-dense core containing the double-stranded DNA genome, the icosahedral capsid, the tegument, and an outer envelope containing glycoprotein (31). The envelope contains at least 8 of 11 different glycoproteins: gB, gC, gD, gE, gG, gH, gI, gJ, gK, gL, and gM (31). The tegument is an amorphous protein layer that contains about 20 virus-encoded proteins, including VP1/2 (UL36), VP11/12 (UL46), VP13/14 (UL47), VP16 (UL48), VP22 (UL49), ICP0, ICP4, and the virion host shutoff protein (UL41) plus the products of genes US2, US3, US10, US11, UL11, UL13, UL14, UL16, UL17, UL21, UL37, UL51, and UL56 (12, 25, 42). The tegument contains important regulatory proteins that are released into the newly infected cell following fusion of the viral envelope with the host cell plasma membrane, but its structure is largely unknown. UL36 is present in the deepest layers of the tegument attached to the vertices of the capsid (45) and interacts with the capsid protein VP5 (24). The use of cryoelectron tomography showed that the tegument frequently forms an asymmetrical cap between the capsid and the outer envelope (9). Tegument proteins that are present in high amounts (1,000 to 2,000 copies per virion), such as UL46, UL47, UL48, and UL49, are likely to play a structural role (11, 44) in contrast to the lower-copy-number ICP0 and ICP4 (100 to 150 copies per virion), which probably play regulatory roles (42).
UL14 is a 32-kDa protein that is expressed late in infection, after viral DNA synthesis (4, 36). There are no more than a few dozen molecules of UL14 per HSV-1 virion, and some of the UL14 protein is phosphorylated (4). UL14 is conserved in the alpha-, beta-, and gammaherpesviruses, and the coding region of UL14 in HSV-1 overlaps that of UL13. The most conserved residues are located in the nonoverlapping region, and the overlapping region seems to encode a variable-length C-terminal domain that is poorly conserved.
An HSV-1 UL14 mutant virus exhibited an extended growth cycle at low multiplicities of infection (MOI) and appeared to be compromised in the release of virus particles from the infected cell. In mice injected intracranially, the 50% lethal dose of the mutant was reduced more than 30,000-fold. Furthermore, recovery of the UL14D mutant from latently infected sacral ganglia of mice injected peripherally was significantly less than that of wild-type virus, suggesting a marked defect in the establishment of, or in reactivation from, latent infection (4).
The HSV-2 UL14 protein enhanced the nuclear localization of the packaging protein UL33 and capsid protein VP26 in a transient expression system (38), though neither UL14 nor UL33 nor VP26 possesses a consensus nuclear localization signal. The translocation of VP26 was largely dependent on the N-terminal half of UL14, which contains a sequence similar to the peptide-binding domain of the human heat shock protein Hsp70. In Vero and HEp2 cells expressing HSV-2 UL14, the activity of coexpressed luciferase was greatly enhanced, suggesting that UL14's presence upregulated the folding of luciferase in the absence of other viral proteins (39). In addition, HSV-2 UL14 also suppresses apoptosis in expressing cells (40). The phenotype of the HSV-1 UL14 mutant virus suggests that UL14 protein is important for successful virus growth, especially at low MOI.
VP16 is an abundant 65-kDa virion phosphoprotein that is synthesized late in infection and subsequently packaged into virions (21-23, 27). VP16 acts during the earliest stages of infection in concert with HCF-1 and Oct-1 to induce transcription of the viral immediate-early (IE) genes, thereby facilitating the onset of lytic gene expression (reviewed in references 30 and 31). The highly acidic C-terminal domain of VP16 has been shown to recruit host RNA polymerase II and associated initiation components (3, 8, 32). Recombinant viruses which are defective in this activity of VP16 show reduced levels of IE transcription and significantly impaired replication (1, 33, 41).
In this study we further characterized the properties of UL14-deficient mutants of HSV-1. Here we show that UL14 plays a significant role in the nuclear localization of VP16 at the IE phase of infection, thus regulating IE gene expression.
MATERIALS AND METHODS
Cells.
The immortalized African green monkey kidney cell line Vero, the human larynx carcinoma cell line HEp-2, the human osteosarcoma cell line U2OS (a gift from Y. Nishida), and rabbit skin cells (a gift from B. Roizman) were used. Cells were propagated in Dulbecco's modified Eagle's medium supplemented with 5% calf serum (Vero), 10% fetal bovine serum (HEp-2, U2OS), or 5% fetal bovine serum (rabbit skin cells) plus 100 units/ml penicillin, 100 μg/ml streptomycin, and 2 mM glutamine at 37°C in 5% CO2.
Viruses.
HSV-1 wild-type strain 17syn+, UL14 mutant virus UL14D, and rescued virus UL14R were kindly provided by C. Cunningham. HSV-1 strain MP44 (a syncytium-type 17syn+ VP16-green fluorescent protein [GFP] virus) was a gift from P. O'Hare. Viral stocks were prepared in Vero or rabbit skin cells and titrated on Vero cell monolayers. Growth curves were obtained as previously reported (4).
Electron microscopy.
Cells were grown to monolayers on sapphire disks coated with carbon and infected with wild-type or UL14D virus. At 16 to 24 h postinfection, the cells were washed in mammalian Ringer solution, fixed in half Karnovsky's fixative (2% glutaraldehyde, 2% paraformaldehyde in 30 mM HEPES buffer), and dehydrated with an ascending series of ethanol (up to 100%). Dehydrated samples were embedded in Lowicryl K4M. Embedded samples were glued onto Epon stages with the glass side facing up. After the Epon was trimmed to expose the glass surface, the sample was emerged into liquid nitrogen for 2 s in order to facilitate the dissociation of the glass disk from the underlying carbon. Ultrathin sections were cut with a diamond knife and collected onto Formvar-coated copper grids and were double stained with 5% uranyl acetate followed by Reynold's lead citrate solution. Sections were examined with a JEOL JEM-1200EX transmission electron microscope.
RNA extraction.
HEp-2 cells were plated at a density of 5 × 105 cells the day before infection. Wild-type or UL14D virus was infected at an MOI of 3 PFU/cell. At 2 h postinfection, the cells were washed extensively in cold phosphate-buffered saline (PBS), and the cells were harvested with a sterile cell scraper. Total RNA was extracted using a QIAamp RNeasy Mini kit (Qiagen) according to the manufacturer's instructions. The RNAs were digested with an RNase-free DNase set (Qiagen) for 15 min at room temperature. cDNAs were generated from 15 μg of total RNA by use of Superscript II reverse transcriptase (Invitrogen) according to the manufacturer's instructions.
Real-time PCR analysis.
Total mRNA was quantified by using primers for the β2 microglobulin human housekeeping gene (predeveloped TaqMan assay reagents; Applied Biosystems), and the HSV-1 transcripts ICP0 and ICP4 (5) were as follows: ICP4 forward primer (5′-GCAGCAGTACGCCCTGA-3′), ICP4 reverse primer (5′-TTCTGGAGCCACCCCATG-3′), and probe [5′-(6-carboxyfluorescein)CACGCGGCTGCTGCTGTACA(minor groove binder)-3′]; ICP0 forward primer (5′-CACCACGGACGAGGATGAC-3′), ICP0 reverse primer (5′-CGGCGCCTCTGCGT-3′), and probe [5′-(6-carboxyfluorescein)ACGACGCAGACTACG (minor groove binder)-3′].
Plasmids.
For the transient expression of HSV-1 UL14 and VP16, the coding sequences were cloned into the pcDNA3.1(+) vector (Invitrogen). Primers UL14F (CGGGATCCATGGACCGAGAT) and UL14R (CGGAATTCGCGGCTCATTCG) and VP16F (CGGGATCCATGGACCTCTTG and VP16R (CGGAATTCCCTACCCACCGT) were used to generate PCR products for UL14 and VP16, respectively. UL14 and VP16 DNAs were both cloned into the BamHI, EcoRI sites to generate pcUL14 and pcVP16. Transfections were carried out using Lipofectamine 2000 (Invitrogen) as recommended by the manufacturer.
Construction of VP16-GFP viruses.
Two strains, 14D-VP16G and 14R-VP16G, were constructed in this study. Viral DNA from HSV-1 MP44 was purified as a template to amplify the VP16-GFP flanked by approximately 200 bp of the native 5′ promoter-leader sequence and 3′ untranslated sequence by PCR. The primers used were VP16GF (CCCGAATTCGAATCTACACGACAG) and VP16GR (CGTCTAGAGGGTTGCTTAAATGC). The PCR product was inserted into the EcoRI, XbaI sites of pBluescript II KS to generate the plasmid pBS-VP16G. Purified plasmid DNA was linearized and cotransfected by the DEAE-dextran method (13) onto rabbit skin cells with the purified genome of HSV-1 UL14D. UL14D lacks the KpnI site at nucleotide 28624 within UL14 (4). GFP-positive plaques were purified by three sequences of plaque purification. One hundred-percent-purity stocks were designated 14D-VP16G. For the construction of a revertant virus, 14R-VP16G, the purified genome of 14D-VP16G was cotransfected with a 2.6-kbp fragment containing the HSV-1 wild-type UL14 gene. This fragment was amplified by PCR using the HSV-1 17syn+ DNA as a template with primers Rev-UL14F (CGTCTAGACGATCTTGATCC) and Rev-UL14R (GGGTCGACGTACACGCTAAC) and inserted into the XbaI, SalI sites of pBluescript II KS. Reverted viruses that contained the KpnI site were detected by amplification of the UL14 gene by PCR, followed by restriction digestion with KpnI. Three sequences of plaque purification were performed to obtain reverted virus stocks designated 14R-VP16G.
Purification of viral DNA.
Vero cells (2 × 108) were infected with viruses at an MOI of 0.01. When complete cytopathic effect was observed, infected cells and media were centrifuged, and the cell pellet was resuspended in 150 mM NaCl, 10 mM Tris (pH 7.4), 1.5 mM MgCl2, 0.1% NP-40. After centrifugation, the supernatant was treated with 0.2% sodium dodecyl sulfate (SDS), 0.5 mM EDTA, 50 mM β-mercaptoethanol to lyse virus particles. Protein was extracted twice by phenol-chloroform extraction. DNA was precipitated with ethanol and resuspended in water. After treatment with RNase A, the suspension was loaded on a potassium acetate gradient (5% to 20% w/vol) made in TE buffer (10 mM Tris [pH 8.0], 5 mM EDTA) and centrifuged at 27,000 rpm for 16 h at 20°C. DNA was concentrated by ethanol precipitation.
Southern blotting of virus DNA.
DNA was digested with either KpnI or EcoRV and fractionated by electrophoresis in 0.5% gels. Following transfer to Hybond-N membranes, the blots were probed by ECL direct nucleic acid labeling and detection systems (Amersham Biosciences). The probes for UL14 and VP16 were the inserts for pcUL14 and pcVP16, respectively. Prehybridization and hybridization washing were performed by standard methods.
Virion purification.
For virus purification, cells were infected with HSV-1 wild type (17syn+) or UL14D, UL14R, MP44, 14D-VP16G, or 14R-VP16G at a low MOI and incubated until a complete cytopathic effect developed. Extracellular medium was harvested and cellular debris was removed by low-speed centrifugation. For reduction of volume, the virus-containing supernatant was centrifuged for 1 h at 22,000 rpm in a TST-28 rotor (Hitachi). The pellet was resuspended in TBSal (200 mM NaCl, 2.6 mM KCl, 10 mM Tris-HCl [pH 7.5], 20 mM MgCl2, 1.8 mM CaCl2) (14), layered onto a discontinuous sucrose gradient (30, 40, and 50% sucrose), and centrifuged for 2 h at 20,000 rpm in a TST-28 rotor. Virions accumulated at the boundary between 40 and 50% sucrose and were harvested by aspiration, pelleted, and resuspended in TBSal.
Synchronous infection.
High-MOI synchronous infection was carried out with VP16-GFP viruses in the presence of 100 μM cycloheximide (Sigma) or in some cases also in that of 10 μg/ml actinomycin D (Sigma) in all stages of the experiment. First, the virus inoculant (MOI, 40 to 200 PFU/cell of cell-associated virus) was adsorbed onto cell monolayers at 4°C for 2 h with continuous rocking. After this period, cells were washed three times in fresh medium and either fixed immediately in 4% paraformaldehyde in PBS or fixed at intervals after incubation at 37°C in a CO2 incubator.
Analysis of virus internalization.
To assay for virus internalization, viruses were bound to cells as described above. The cells were washed to remove unbound virus and shifted to 37°C for various lengths of time. The cells were washed with cold sterile PBS and incubated with PBS containing 2 mg/ml proteinase K (Sigma) at 4°C. After 1 h, the cells were washed briefly in PBS and fixed immediately.
Immunofluorescence.
Cells grown on coverslips were washed in PBS and fixed for 10 min in 4% paraformaldehyde in PBS at room temperature. For GFP visualization, the slips were mounted directly onto glass slides with Shandon PermaFluor (Thermo). For indirect immunofluorescence, fixed cells were permeabilized in 0.1% or 1% Triton X-100 in PBS for 5 min at room temperature. The coverslips were inverted onto droplets (20 μl) of blocking buffer (4% goat serum, 1% bovine serum albumin in PBS-Tween [0.05%]) on a clean Parafilm sheet for 30 min at room temperature. Primary and secondary antibodies (Alexa Fluor; Molecular Probes) were diluted in blocking buffer and reacted for 30 min at room temperature. For examination of HCF-1, infected HEp-2 cells were fixed/permeabilized in cold methanol for 10 min and subjected to indirect immunofluorescence. Samples were examined under a Zeiss LSM510 confocal immunofluorescence microscope. Quantification of fluorescence intensity was performed using the LSM510 version 3.2 software.
Immunoprecipitation.
HEp-2 cells grown in 60-mm culture dishes were infected with either 14D- or 14R-VP16G virus at an MOI of 5 PFU/cell. At 15 hours postinfection (h.p.i.), cells were harvested and VP16-GFP was immunoprecipitated with anti-GFP polyclonal antibody. The procedure for immunoprecipitation has been described previously (16). Protein A-agarose (Roche) was used for precipitation.
Antibodies.
Anti-UL14, anti-UL42, and anti-VP16 antibodies were described previously. Anti-HCF-1 antibody was kindly provided by P. O'Hare. Anti-ICP0 monoclonal antibody (MAb) and anti-ICP4 MAb were purchased from Virusys, and anti-HSV1+HSV2 VP5 MAb was from Abcam. Anti-β actin MAb and anti-GFP polyclonal antibody was purchased from Medical & Biological Laboratories (MBL). Normal rabbit serum was obtained from DAKO.
RESULTS
Growth kinetics of HSV-1 UL14D.
Cunningham et al. previously reported that when RS and HFL cells were infected with the UL14 mutant virus UL14D at a high MOI, the growth kinetics were slightly delayed compared to wild-type and revertant UL14R virus, and the final yield of UL14D virus was seven- to eightfold less. When infected at a low MOI, the maximum yield of UL14D was three- to fourfold less than seen for the wild type and took about three times longer to attain. UL14D released roughly one-fifth of the proportion of virus released by the wild type and UL14R, indicating that UL14D virions are inefficiently packaged and released from infected cells (4).
We reexamined the growth kinetics of UL14D virus in HEp-2 cells. Cells were infected with wild-type or UL14D virus at an MOI of 5 PFU/cell, and virus infectivity was determined by titration at appropriate times after infection. There was a significant delay in viral growth in UL14D-infected cells at early times postinfection (up to 9 h), and the maximum yield of total UL14D virus in HEp-2 cells was five- to sixfold less than that for the wild type (Fig. 1A). UL14R growth was similar to that of the wild-type virus, so for all subsequent experiments we compared UL14D only to the wild type. The particle-to-PFU (or copy-to-PFU) ratios of the strains were not significantly different (4). We next reexamined wild-type- and UL14D-infected HEp-2 cells by conventional Epon embedding and electron microscopy (not shown). Compared to the wild type, UL14D virus showed significantly low extracellular virions both at 16 and 24 h postinfection, and many particles in the perinuclear region were nonenveloped or in the process of becoming enveloped or de-enveloped. These observations were identical to those reported previously (4) and suggested that the mutant virus had a defect in egress.
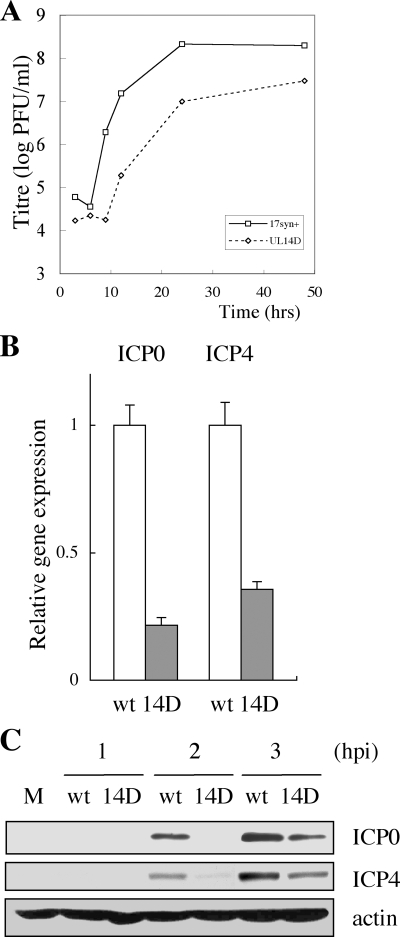
FIG. 1.
(A) A representative one-step growth curve of HSV-1 wild-type (17syn+) and UL14D viruses in HEp-2 cells. Cells were infected at an MOI of 5 PFU/cell and harvested at 3, 6, 9, 12, 24, and 36 h postinfection. Cells were scraped into the medium, sonicated, and titrated on Vero cell monolayers. (B) Quantitative real-time PCR analysis of HEp-2 cells infected with wild-type (wt) and UL14D viruses. Cells were infected with the viruses at an MOI of 3 PFU/cell, and total RNA was harvested at 2 h postinfection. After DNase treatment and cDNA synthesis by reverse transcriptase treatment, the gene expression of ICP0 and ICP4 was normalized by β2 microglobulin and quantified in triplicate by real-time PCR. The results are representative of multiple experiments and show that the gene expressions of ICP0 (P = 0.024; Student's t test) and ICP4 are lower in UL14D-infected cells than in wild-type-infected cells. (C) Expression of IE protein ICP0 and ICP4 in HEp-2 cells infected with wild-type and UL14D viruses at an MOI of 5 PFU/cell. Cells were infected with the viruses and harvested at 1, 2, and 3 h postinfection. Whole-cell lysates were prepared and subjected to SDS-PAGE and Western blotting, followed by detection with anti-ICP0, anti-ICP4, and β-actin MAbs. M, mock-infected cells.
Expression of IE genes decreases in UL14D-infected cells.
The growth curve of the UL14D virus at a high-multiplicity infection exhibits a characteristic delay at early times postinfection in a number of different cell lines (4). We hypothesized that this phenotype may be the result of an unknown defect in the onset of lytic infection. To determine if the initial delay in the growth of UL14D was due to a defect in IE gene expression, we examined the expression of ICP0 and ICP4 in infected cells. HEp-2 cells were synchronously infected at an MOI of 3 PFU/cell with UL14D or wild-type virus. The mRNA levels were quantified by real-time reverse transcription-PCR. At 2 h postinfection, ICP0 mRNA was nearly fivefold less (P = 0.024; Student's t test), and ICP4 mRNA was threefold less in UL14D-infected cells than in wild-type-infected cells (Fig. 1B). This suggested that the UL14D virus had a defect in the expression of IE proteins. Therefore, we next examined protein synthesis levels of ICP0 and ICP4 in UL14D-infected cells. HEp-2 cells were infected with wild-type and UL14D viruses at an MOI of 3 PFU/cell, and whole-cell lysates were harvested at 1, 2, and 3 h.p.i. and subjected to SDS-polyacrylamide gel electrophoresis (PAGE) and Western blotting (Fig. 1C). The expression of ICP0 and ICP4 proteins was indeed delayed in UL14D-infected cells, showing that IE protein expression was affected in UL14D-infected cells.
UL14 protein enhances the nuclear accumulation of VP16.
Because of the finding that UL14D-infected cells exhibited lower IE gene expression, we were interested in the dynamics of virion-supplied VP16. This was because VP16, also called α-TIF (trans-inducing factor), is a strong inducer of viral IE gene expression in the newly infected cell (37). On the other hand, one of the functions of the HSV-2 UL14 protein in uninfected cells was to translocate viral proteins such as UL33 and VP26 into the nucleus upon coexpression (38). It was possible that the nuclear accumulation of these proteins was an artifact of overexpression but also suggested that UL14 had intrinsic abilities to alter the compartmentalization of other viral proteins. Therefore, we hypothesized that UL14 may be associated in the dynamics of VP16, which is a major constituent of the tegument.
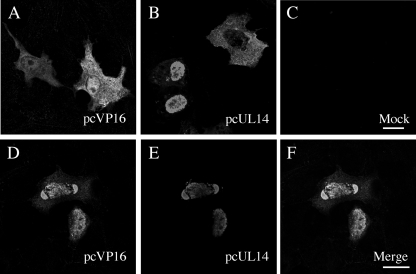
In order to pursue this hypothesis, we first used a transient expression system to determine if the presence of UL14 protein affected VP16's cellular localization in the absence of infection. In cells expressing VP16 only, the protein was diffusely spread throughout the cytoplasm and nucleus (Fig. 2A) as shown by others (15), reflecting the fact that VP16 does not have a consensus nuclear localization signal. Under single expression, UL14 was both nuclear and cytoplasmic (Fig. 2B) as reported previously (36). Interestingly, upon coexpression, the two viral proteins both showed significant nuclear colocalization and accumulation (Fig. 2D to F). This observation suggested that UL14 protein and VP16 mutually enhanced their nuclear localizations.
FIG. 2.
Intracellular localization of VP16 in transfected cells. HEp-2 cells were transfected with plasmids expressing HSV-1 VP16 (pcVP16) and HSV-1 UL14 (pcUL14). Cells were transfected with pcVP16 (A) or pcUL14 (B) alone, mock transfected (C), or cotransfected with pcVP16 and pcUL14 (D to F). Twenty four hours after transfection, cells were fixed, permeabilized, and subsequently reacted with anti-VP16 rabbit polyclonal antibody (A and D) and anti-UL14 mouse polyclonal antibody (B and E) and appropriate secondary antibodies. When coexpression took place with UL14, the diffused localization of VP16 (A) became significantly nuclear and colocalization of the proteins was observed (D to F). Panels A to C are of the same magnifications as panels D to F, respectively. Bars in panels C and F, 10 μm.
In HSV-1 wild-type-infected cells, UL14 was reported to localize to 10 to 40 punctate foci in each infected cell nucleus (4) that form adjacent to or in the proximity of replication compartments (data not shown). The significance of these foci is unknown. However, this pattern was not observed in transiently expressing cells, indicating that viral proteins other than VP16 are required for the recruitment of UL14 to such foci.
Construction of VP16-GFP viruses to visualize VP16 in virions.
Given the fact that the coexpression of UL14 and VP16 enhanced the nuclear accumulation of VP16, we further set out to examine whether virion-supplied VP16 conformed to this occurrence. However, it was difficult to analyze VP16 localization by indirect immunofluorescence due to the reduced accessibility of viral antigens in fixed cells and difficulties in eliminating nonspecific fluorescence (data not shown).
La Boissiere et al. reported that VP16-GFP fusion proteins are incorporated normally into HSV virions and that the activation domain of VP16 is functional in chimeric proteins (17). Thus, we decided to construct VP16-GFP viruses in order to visualize the fate of the input protein with greater sensitivity. The recombinant virus was designed to express VP16-GFP (VP16 with a GFP at the C terminus) instead of VP16. VP16-GFP DNA was made by PCR amplification of the relative sequence from the purified DNA of strain HSV-1 MP44 (a syncytium-forming clone of strain HSV-1 v44) as described in Materials and Methods.
After the purification of GFP-expressing plaques, the incorporation of VP16-GFP into the virus genome was confirmed by probing EcoRV digests with VP16 (17). The EcoRV I fragment increased in size from 7.4 kb (UL14D) to approximately 8.2 kb in both 14D- and 14R-VP16-GFP viruses (Fig. 3A). In addition, the incorporation of the KpnI site (which is located at nucleotide 28624 of the HSV-1 DNA sequence within UL14) in the reverted virus was confirmed by the detection of KpnI fragments E (10.8 kb) and O (3.8 kb) with a UL14 probe (Fig. 3A).
FIG. 3.
(A) Southern bolt analysis of VP16-GFP-expressing viruses constructed in this study. Purified viral DNAs of UL14D, 14D-VP16G, and 14R-VP16G were digested with EcoRV (lanes 1 to 3). Similarly, 14D-VP16G and 14R-VP16G DNA was digested with KpnI (lanes 4 and 5), followed by Southern blotting and detection of VP16 (lanes 1 to 3) and UL14 (lanes 4 and 5) with an appropriate probe. The incorporation of VP16-GFP into the genome corresponds to an increase from 7.4 kb (lane 1) to 8.2 kb (lanes 2 and 3) of the EcoRV I fragment. The KpnI site at nucleotide 28624 of the HSV-1 genome is within the UL14 gene. In 14D-VP16G, the probe detected a 14.6-kb band (lane 4). In 14R-VP16G, the probe detected two bands which correspond to KpnI E (10.8 kb) and O (3.8 kb) fragments (lane 5). (B) Expression of viral proteins in 14R-VP16G- and 14D-VP16G-infected cells. HEp-2 cells were infected at an MOI of 3 PFU/cell, harvested at intervals, and subjected to SDS-PAGE followed by Western blotting for detection of VP5, UL14, and β actin. VP16-GFP (95 kDa) was detected both by anti-VP16 and anti-GFP antibodies on separate membranes. M, mock-infected cells. (C) Growth kinetics of 14R-VP16G and 14D-VP16G viruses. HEp-2 cells were infected at an MOI of 3 PFU/cell, harvested at intervals, and titrated on Vero cell monolayers. (D) Similar amounts of VP16-GFP are present in cell-associated preparations of 14R-VP16G and 14D-VP16G viruses. A sequential threefold dilution of cell-associated virus stocks starting at an MOI of 1 PFU/cell was submitted to SDS-PAGE and Western blotting, and examined for VP16-GFP with anti-GFP antibody. (E) Similar amounts of VP16-GFP are incorporated into the 14R- and 14D-VP16G virions. Extracellular virions were submitted to twofold dilution, SDS-PAGE, and Western blotting and detected with anti-VP5 MAb and anti-GFP antibody. Scanned images were submitted to densitometry analysis using Adobe Photoshop 7.0, and the intensity of the bands for VP16-GFP was normalized to that of VP5. The ratios of VP16GFP to VP5 were similar between the two strains.
To examine the synthesis of VP16-GFP fusion proteins, HEp-2 cells were infected with 14D-VP16G and 14R-VP16G at an MOI of 3 PFU/cell. The time course of VP5 and VP16-GFP accumulation was slightly slower in 14D-VP16G-infected cells (Fig. 3B). We next examined the growth kinetics of the two viruses by infecting HEp-2 cells at an MOI of 3 PFU/cell. The growth patterns of 14R- and 14D-VP16G (Fig. 3C) were comparable to those of the non-GFP HSV-1 wild-type 17syn+ and UL14D (Fig. 1A). 14R- and 14D-VP16G reached maximum yields of 2.4 × 108 and 2.4 × 107 PFU/ml, respectively, suggesting that these viruses were fully capable of productive growth.
Analysis of VP16-GFP input during synchronous infection.
There are 1,500 to 2,000 molecules of VP16 incorporated into each HSV virion (17). We thought that at a sufficiently high MOI we would be able to visualize the fate of VP16-GFP released from the tegument into the cytoplasm following the internalization of 14D- or 14R-VP16G virus. In a study by La Boissiere et al., however, the examination of live cells after infection with a GFP-VP16 virus (MOI of 10 PFU/cell) in the presence of cycloheximide did not lead to any firm evidence for the fate of input VP16 (17). This suggested that a much higher MOI was required for visualization and also that it would be better to synchronize the viral internalization process.
Cell-associated 14D- and 14R-VP16G viruses were concentrated to achieve an MOI of over 40 PFU/cell. As our strategy for examining VP16 dynamics depended solely on GFP fluorescence, the virus preparations were subjected to threefold serial dilutions and SDS-PAGE followed by Western blotting to compare VP16-GFP levels. When we normalized the virus preparations by MOI, the amounts of VP16-GFP were similar in 14R- and 14D-VP16G preparations (Fig. 3D). In addition, densitometry analysis of VP16-GFP incorporated into extracellular 14R- and 14D-VP16G virions showed that these too were similar (Fig. 3E). This indicated that the virus preparations were suitable for an examination of input VP16-GFP.
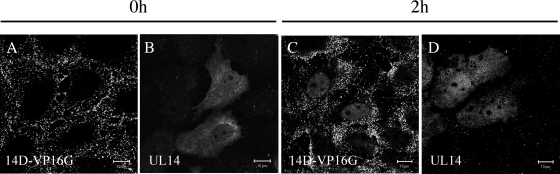
Cell monolayers (HEp-2, Vero, U2OS) were synchronously infected with 14D-VP16G or 14R-VP16G at an MOI of 40 PFU/cell and fixed in paraformaldehyde after the 2-h adsorption period at 4°C, and also at 1, 2, and 4 h after a 37°C incubation period. To inhibit de novo protein synthesis, cycloheximide (100 μM) was included throughout the experiment (Fig. 4J). The cells were visualized by confocal fluorescent microscopy and analyzed by z-axis sectioning (0.25-μm intervals, 30 to 40 slices per cell). Results shown in Fig. 4A to H are pictures from U2OS-infected cells.
FIG. 4.
Examination of input VP16-GFP in newly infected cells shows that the nuclear accumulation of VP16-GFP is enhanced by the presence of UL14 protein. (A to H) U2OS cells were inoculated with 14R- and 14D-VP16G viruses at an MOI of 40 PFU/cell in the presence of 100 μM cycloheximide. Adsorption was performed at 4°C with continuous rocking of the culture dishes. After 2 h of virus binding, the cells were washed three times in fresh medium containing cycloheximide and either fixed in 4% paraformaldehyde in PBS or cultured in the presence of cycloheximide at 37°C. Cells were fixed at 1, 2 and 4 h after shifting up to 37°C and analyzed by confocal immunofluorescence microscopy. Samples were analyzed by z-axis sectioning, and a single representative plane is shown. The nuclear accumulation of VP16-GFP was apparent in 14R-VP16G-infected cells at 1 h postinfection. In contrast, 14D-VP16G-infected cells accumulated VP16-GFP with slower kinetics and a lower peak nuclear fluorescence. Identical results were obtained using HSV-1 MP44 in place of 14R-VP16G (not shown). Bars, 10 μm. (I) Quantification of the nuclear fluorescence intensity of VP16-GFP. The nuclear fluorescence intensity of the cells shown in the photographs in panels A to H was analyzed using LSM version 3.2 software. The average absolute value and standard deviation is shown. (J) Cycloheximide sufficiently inhibits de novo protein synthesis in cells synchronously infected with 14R-VP16G at an MOI of 40 PFU/cell. Prior to infection, cells were chilled at 4°C for 15 min. Vero cells were infected with the virus in the presence (lanes 1 to 4) or absence (lanes 5 to 7) of 100 μM cycloheximide. Viruses were bound to the cell surface for 2 h at 4°C. Cells were either washed in cold PBS and harvested (lanes 2 and 5) or washed extensively (three times) in cold fresh medium in the presence (lanes 1 to 4) or absence (lanes 5 to 7) of cycloheximide, followed by incubation at 37°C in fresh medium in the presence (lanes 1 and 2) or absence (lanes 3 to 7) of cycloheximide. Cells were harvested after 2 h (lanes 3 and 6) or 4 h (lanes 1, 4, and 7) of incubation. M, mock-infected cells. Whole-cell lysates were separated by SDS-PAGE and Western blotting and detected with anti-UL42, anti-GFP, and anti-tubulin antibodies. The early protein UL42 was undetectable in cells infected in the presence of cycloheximide throughout the experiment (lane 1). Input VP16-GFP was readily detectable (lanes 2 and 5), and there was no marked change (due to de novo synthesis) in the levels of VP16-GFP (a late protein) in lanes 1 to 6.
Immediately after the adsorption period, VP16-GFP was scattered throughout the surfaces of cells infected with either 14D-VP16G or 14R-VP16G (Fig. 4A and E and data not shown). Strikingly, after 1 h at 37°C, nuclear GFP fluorescence was observed in cells infected with the revertant virus (Fig. 4B), whereas in cells infected with 14D-VP16G, GFP fluorescence was only faintly visible in the nucleus (Fig. 4F). After 2 h (Fig. 4C) and 4 h (Fig. 4D) at 37°C, 14R-VP16G-infected cells exhibited rich, clearer nuclear fluorescence while cytoplasmic VP16 diminished in some cells. In contrast, nuclear GFP fluorescence in 14D-VP16G-infected cells increased only slightly throughout this period (Fig. 4G and H). The same phenomenon as seen for 14R-VP16G-infected cells was observed after synchronous high-multiplicity infection of the parental strain HSV-1 MP44 (data not shown). In addition, these observations were reproducible in HEp-2 (not shown) and Vero cells.
The photographs of fixed cells harboring VP16-GFP showed clearly that there was a defect in the nuclear accumulation of VP16-GFP in cells infected with the UL14-deficient virus. We further quantified the intensity of VP16-GFP fluorescence in the nucleus in the pictures shown in Fig. 4A to H. The nuclear VP16-GFP fluorescence was considerably stronger in 14R-VP16G-infected cells than in 14D-VP16G-infected cells (Fig. 4I). However, though VP16-GFP transport was greatly reduced in cells infected with 14D-VP16G, it was not completely blocked. This is slightly similar to the patterns of VP16 localization observable in Fig. 2A and D.
The addition of cycloheximide in the culture media would block not only viral but also cellular protein synthesis. There was a possibility that the novel production of cellular proteins such as molecular chaperones was required for VP16-GFP to enter the nucleus with efficiency and that the synthesis block was detrimental. However, the localization of VP16-GFP in cells synchronously infected without cycloheximide showed essentially similar results until 2 h after a shift up to 37°C (not shown). This ruled out the possibility that the defect in VP16-GFP transport in 14D-VP16G-infected cells was a consequence of the protein synthesis block. Therefore, the presence of UL14 was required for the efficient nuclear transport of VP16-GFP rather than the synthesis of cellular proteins. We conclude that in 14D-VP16G-infected cells, the nuclear accumulation of VP16-GFP was considerably reduced due to a lack of UL14.
Finally, to make certain that the above observations truly reflected the dynamics of input VP16-GFP and not newly synthesized VP16-GFP, the effect of cycloheximide was examined in detail. Vero cells were synchronously infected with 14R-VP16G at an MOI of 40 PFU/cell. 14R-VP16G was bound either in the presence (Fig. 4J, lanes 1 to 4) or in the absence (lanes 5 to 7) of 100 μM cycloheximide for 2 h at 4°C. After this period, cells were washed extensively with fresh medium in the presence (lanes 1 to 4) or the absence (lanes 5 to 7) of cycloheximide then either harvested directly (lanes 2 and 5) or further incubated at 37°C for 2 h (lanes 3 and 6) or 4 h (lanes 1, 4, and 7) in the presence (lanes 1 and 2) or the absence (lanes 3 to 7) of cycloheximide. At the end of the incubation period, cells were harvested and submitted to SDS-PAGE and Western blotting. The blot was detected for UL42 protein (an early protein not incorporated into virions) to assay for de novo protein synthesis and also for VP16-GFP. The results clearly showed that cycloheximide sufficiently inhibited viral protein synthesis (compare lane 1 to lanes 4 and 7). Under cycloheximide, no UL42 protein synthesis was detected at 4 h after shifting up to 37°C. Furthermore, inclusion of the drug only during virus binding delayed the synthesis of UL42 (compare lanes 4 and 7). Also, input VP16-GFP was readily detectable (lanes 2 and 5), but no marked increase was detected in its expression. Considering that VP16 is a late-expressed protein, it was unlikely that the de novo synthesis of VP16-GFP occurred in the presence of 100 μM cycloheximide. Therefore, the pictures in Fig. 4 represent only the dynamics of virion-supplied VP16-GFP.
Transient expression of UL14 increases nuclear accumulation of VP16-GFP.
We next examined if the exogenous expression of HSV-1 UL14 protein could complement the nuclear import of VP16-GFP in 14D-VP16G-infected cells. Vero cells were transfected with plasmid pcUL14. Twenty-four hours later, the cells were synchronously infected with 14D-VP16G at an MOI of 150 PFU/cell in the presence of cycloheximide. Cells were fixed 2 h after upshift to 37°C, permeabilized, and examined for VP16-GFP and UL14 protein by indirect immunofluorescence. To our interest, a significant population of cells that expressed moderate UL14 protein levels showed distinguishable VP16-GFP nuclear localization, in contrast to most nonexpressing cells (Fig. 5C and D). This suggested that the transient expression of UL14 protein could partially enhance the nuclear accumulation of VP16-GFP in 14D-VP16G-infected cells.
FIG. 5.
Transient expression of UL14 protein increases VP16-GFP nuclear localization in 14D-VP16G-infected cells. Vero cells were transfected with 2 μg of pcUL14 plasmid DNA and 24 h later were synchronously infected with 14D-VP16G at an MOI of 150 PFU/cell in the presence of cycloheximide. Cells were fixed, permeabilized, and examined for UL14 protein by indirect immunofluorescence with anti-UL14 polyclonal antibody. The localization of VP16-GFP and UL14 protein immediately after adsorption (A and B) and 2 h after shifting up to 37°C (C and D). Enhanced nuclear VP16-GFP fluorescence was observed in cells expressing moderate levels of UL14 protein (C). Bars, 10 μm.
Comparison of 14D- and 14R-VP16G internalizations.
We did not discriminate between intracellular and extracellular VP16-GFP fluorescence in previous experiments (Fig. 4). To examine whether there was a defect in the binding or penetration of 14D-VP16G virus, we performed a virus internalization assay (34). Vero cells were synchronously infected with the viruses at an MOI of 50 PFU/cell in the presence of cycloheximide. After adsorption for 2 h at 4°C, cells were shifted to 37°C for various times to allow for virus penetration. Immediately after adsorption or after incubation at 37°C, cells were treated with 2 mg/ml proteinase K at 4°C in order to digest noninternalized virus and subsequently the GFP fluorescence. The cells were then fixed and examined for VP16-GFP.
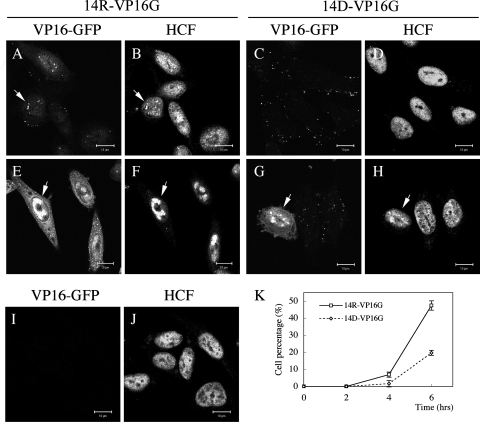
Immediately after the adsorption period (untreated with proteinase K), VP16-GFP fluorescence intensities were similar in 14D- and 14R-VP16G-infected cells (Fig. 6A and E). Cells with bound 14D-VP16G showed slightly stronger fluorescence, which coincided with a slightly higher copy number/PFU ratio for 14D-VP16G than for 14R-VP16G (data not shown). Cells treated with proteinase K immediately after adsorption showed very weak fluorescence, indicating that most of the particles were located extracellularly (Fig. 6B and F). After 30 min at 37°C, internalized VP16-GFP was resistant to the treatment and was localized mainly to cytoplasmic regions in both 14D- and 14R-VP16G-infected cells (Fig. 6C and G). There was no recognizable difference in GFP fluorescence between the viruses. At 60 min, 14R-VP16G-infected cells exhibited nuclear VP16-GFP fluorescence. In contrast, VP16-GFP was mainly cytoplasmic in 14D-VP16G-infected cells (Fig. 6D and H), similar to the result shown in Fig. 4. A detailed analysis of the infected cell population confirmed that the total GFP fluorescence intensities were similar (to slightly stronger in 14D-VP16G-infected cells) in both viruses. This suggested that the two viruses did not have any significant discrepancy in their levels of virus internalization. This experiment underlines that VP16-GFP had a defect in nuclear transport after infection with the 14D-VP16G virus. VP16-GFP may have a problem in cytoplasmic dispersal after internalization, but this could not be looked at by the methods used in this study. These experiments showed that UL14 played an important role in the correct nuclear targeting of VP16.
FIG. 6.
Internalization of VP16-GFP in 14R-VP16G- and 14D-VP16G-infected cells. Vero cells were synchronously infected with the two strains at an MOI of 50 PFU/cell in the presence of cycloheximide. Immediately after adsorption, cells were fixed to visualize bound virus (A and E) or treated with 2 mg/ml proteinase K for 1 h either immediately (B and F) or following 30 min (C and G) or 1 h (D and H) of incubation at 37°C. Treated cells were fixed thereafter to examine internalized VP16-GFP. 14R-VP16G and 14D-VP16G did not exhibit a marked difference in internalization or in total GFP fluorescence. 14R-VP16G-infected cells exhibited VP16-GFP nuclear localization (arrows in panel D), whereas 14D-VP16G-infected cells did not (H). This suggested that UL14 protein was important for the dispersal and/or nuclear targeting of VP16-GFP. Bars, 10 μm.
Delayed nuclear accumulation of capsids in 14D-VP16G-infected cells.
IE gene induction is influenced by at least two factors. One is the transinducing activity of VP16, and the other is the efficiency of retrograde capsid transport. In the next experiment, we analyzed the transport of infecting capsid in 14R- and 14D-VP16G-infected cells. Identical samples from the previous experiment (Fig. 4) were utilized for indirect immunofluorescence analysis of the major capsid protein VP5. As expected, 14R-VP16G-infected cells showed marked accumulation of capsids at the rim of the nucleus (Fig. 7B). In contrast, in 14D-VP16G-infected cells, capsids were found at the nuclear rim to a much lesser extent (Fig. 7F). This suggested that UL14-deficient virus also had a defect in the transport of incoming capsids, the reason for which is unknown. This defect could not be rescued by exogenous expression of the UL14 protein (data not shown). Thus, the lower expression kinetics of IE genes in UL14-deficient virus-infected cells was influenced by two factors: the defect in VP16 compartmentalization and the nuclear targeting of capsids. However, considering that the reduced expression of IE genes was observed for cells infected at a high MOI and that the transinducing activity of VP16 is extremely strong, we estimated that the phenotype of the UL14-deficient viruses were impacted more by the nuclear transport defect of VP16.
FIG. 7.
Capsids do not accumulate efficiently in the nuclei of 14D-VP16G-infected cells. U2OS cells synchronously infected with 14R-VP16G or 14D-VP16G at an MOI of 40 PFU/cell in the presence of cycloheximide were fixed at 2 h after shifting up to 37°C and permeabilized, followed by detection by indirect immunofluorescence with anti-VP5 MAb. 14R-VP16G-infected cells exhibited nuclear VP16-GFP, and capsids were found lining the nucleus-cytoplasm boundary (A to D). Neither VP16-GFP nor capsids assembled as efficiently in 14D-VP16G-infected cells (E to H). Bars, 10 μm.
Analysis of HCF-1 in 14D- and 14R-VP16G-infected cells.
VP16 induces the formation of a transcriptional regulatory complex with Oct-1 and the cell proliferation factor HCF-1 in order to activate viral IE gene expression (19). Therefore, we examined the localization of HCF-1 in 14D- and 14R-VP16G-infected cells. HEp-2 cells were infected with the viruses at an MOI of 10 PFU/cell, and the localization of HCF-1 was examined at various times postinfection (Fig. 8). In mock-infected cells, HCF-1 localized to the nucleus with the exclusion of nucleoli (Fig. 8J). As infection progressed, we observed the reorganization of HCF-1 in the nucleus as represented by speckled foci followed by honeycomb-like structures (16). Bright foci of HCF-1 colocalizing with VP16-GFP were clearly seen in 14R-VP16G-infected cells at 4 h.p.i. (Fig. 8A and B), and larger globular structures were readily observed at 6 h.p.i. (Fig. 8E and F). In 14D-VP16G-infected cells, the colocalization of HCF-1 with VP16-GFP remained intact but the appearance of such foci was delayed (Fig. 8G and H). The number of cells that displayed reorganization of HCF-1 increased only at later times of infection (not shown). Quantification of the percentage of cells harboring prominent signs of HCF-1 reorganization (such as bright foci and globular structures) showed a delay in the accumulation of such cells in 14D-VP16G-infected cells (Fig. 8K).
FIG. 8.
Formation of HCF-1 foci is delayed in 14D-VP16G-infected cells. HCF-1 localization was analyzed in 14R-VP16G (A, B, E, and F)- and 14D-VP16G (C, D, G, and H)-infected HEp-2 cells. Cells were either mock infected (I and J) or infected with the viruses at an MOI of 10 PFU/cell (A to H). Cells were fixed in cold methanol prior to infection (I and J) or at 2 h.p.i. (not shown), 4 h.p.i. (A, B, C, and D), or 6 h.p.i. (E, F, G, and H). In uninfected cells, HCF-1 localized in the nucleus, mainly excluding the nucleoli, with some cells exhibiting subtle foci in the nucleus regardless of infection (not shown). Speckled foci of VP16-GFP and HCF-1 (arrows in A and B) could be readily observed at 4 h.p.i. in 14R-VP16G-infected cells. As reported previously (16), the foci gradually grew into larger globular structures (arrows in E and F). In 14D-VP16G-infected cells, the majority of focus-positive cells had not formed globular structures at 4 or 6 h.p.i. (arrows in G and H). (K) The percentage of infected cells presenting clear intranuclear foci of HCF-1 and VP16-GFP (such as those shown by arrows in panels A and B) was quantified for a total of 200 cells from three or four randomly chosen fields. The vertical lines in the plot show standard deviations.
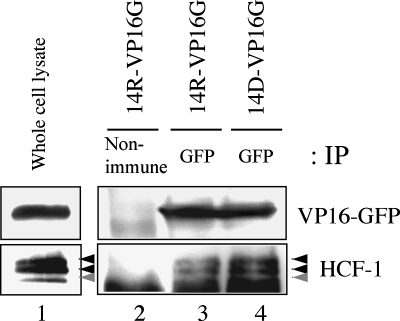
Having shown that VP16-GFP colocalized with HCF-1, we next examined whether these two still interacted. In view of the fact that viral growth was not affected by the incorporation of VP16-GFP, it was plausible that they still formed a complex. We confirmed this assumption by the coprecipitation of HCF-1 and VP16-GFP in both 14R- and 14D-VP16G-infected cells (Fig. 9). Thus, the fusion of GFP to VP16 did not disrupt the formation of the complex. This showed that though the lack of UL14 affected the nuclear transport of VP16-GFP during infection, it did not affect the interaction between VP16-GFP and HCF-1. This also agreed with the report that the complex can form in transfected cells in the absence of all other viral proteins (15). It is worth mentioning that a slightly larger amount of HCF-1 coprecipitated repeatedly with VP16-GFP in 14D-VP16G-infected cells (Fig. 9, lane 4). The relevance of this result is unclear. However, cells infected with an UL14-deficient virus exhibited noticeable alterations in cell (especially nuclear) morphology at late times postinfection compared to wild-type virus-infected cells (not shown). It may be that a discrepancy in the cellular environment due to the lack of UL14 could have affected the results of the coimmunoprecipitation.
FIG. 9.
VP16-GFP is capable of binding with HCF-1 in both 14R- and 14D-VP16G-infected cells. HEp-2 cells were infected with the viruses at an MOI of 5 PFU/cell. At 15 h.p.i., cell lysates were immunoprecipitated (IP) with either nonimmune rabbit serum (lane 2) as a control or anti-GFP polyclonal antibody (lanes 3 and 4). The samples were separated by SDS-PAGE, submitted to Western blotting, and examined for GFP or HCF-1 (lane 1; whole-cell lysate). For HCF-1, two major bands (black arrowheads) and one minor band (gray arrowhead) were usually detected.
Finally, we demonstrated that an important role of the tegument protein UL14 in newly infected cells was to increase the nuclear localization of the major tegument VP16. We hypothesize that in the absence of UL14, IE transcription was affected due to a reduced supply of the transinducing factor. Under high-multiplicity infection, the slower migration of capsids to the nuclear pore is probably a secondary factor when considering this phenotype. However, under low-multiplicity infection, this factor could affect the speed of cell-to-cell spread of the UL14-deficient virus.
DISCUSSION
After HSV enters the cytoplasm of an infected cell, VP16 molecules that are released are said to bind directly to the cellular cofactor HCF-1, promoting subsequent interaction of the VP16-HCF-1 complex with the POU DNA-binding domain of Oct-1 and selective assembly onto HSV IE promoters (15, 37). Oct-1 has been reported to be critical but not essential for IE gene expression using Oct-1 deficient mouse embryonic fibroblasts (29). HCF-1 is the key factor for HSV IE gene expression, which is strictly required for VP16-mediated transcriptional induction such that the depletion of HCF-1 significantly impacted the basal level of IE expression mediated by VP16-dependent transcriptional induction (28).
In infected cells, UL14 is most likely to play a role in the sophistication of HSV replication, especially in the late phase. However, its role as a tegument protein in the infecting virion remained unknown. UL14 protein is a phosphoprotein of 32 kDa, and only a few dozen molecules of UL14 are said to be incorporated per virion (4). Our previous studies showed that UL14 of HSV-2 exerts tolerance to heat-induced stress and to chemically induced apoptosis. It is also capable of localizing the capsid protein VP26 and the packaging protein UL33 into the nucleus of coexpressing cells. In addition, a 15-amino-acid stretch in HSV UL14 (amino acids 60 to 74) share 67% similarity to a sequence of the peptide-binding domain of human Hsp70 (39). The foremost objective of this study was to identify a more specific role for UL14 in the viral infection process.
Analysis using reverse transcription real-time PCR and Western blotting demonstrated that the accumulation of ICP0 and ICP4 mRNAs and proteins were delayed in UL14D-infected cells. ICP0 stimulates the expression of all temporal classes of HSV-1 genes and many heterologous genes in transfection reporter assays (reviewed in reference 7) and is required for the expression of IE genes when the C-terminal activation domain of VP16 is deleted, and ICP0 can partially substitute for the VP16 activation function (26). In addition, ICP0 confers a significant growth advantage for the virus, especially at a low MOI; plays an important role for HSV reactivation; and may be essential for infectious virus production during the reactivation of latent HSV in vivo (2, 10, 20, 35, 43).
We hypothesized that reduced IE gene expression at early times postinfection was due to an alteration in the function of input VP16. Analysis of the dynamics of VP16-GFP in newly infected cells showed that the nuclear accumulation of VP16-GFP was decreased and/or delayed in 14D-VP16G-infected cells. These differences in VP16 localization in 14D-VP16G-infected nuclei were due to defective dispersal and/or transport of VP16. It was crucial that newly synthesized proteins were nonexistent, so synchronous-infection assays were carried out with higher concentrations of cycloheximide or with a combination of actinomycin D, cycloheximide, and methionine-free medium. These experiments produced identical results (data not shown), supporting the hypothesis that the tegument UL14 protein participated in the nuclear targeting of VP16.
As there are so many proteins that comprise the tegument, we cannot dismiss the possibility that there was a deficiency in the functions of other tegument proteins in UL14-deficient virus-infected cells. However, we have not pursued this hypothesis. A recent report indicated that VP22, one of the most abundant tegument proteins, is important for viral spread during HSV-1 infection at low MOI and for the accumulation of extracellular virus to wild-type levels (6). This study is another example suggesting that tegument proteins are influential on many aspects of HSV-1 growth. Though our study suggests an attractive role for the UL14 protein, we feel that much more examination is needed to elucidate its position in the replication cycle of the virus.
Observations by others indicate that HCF-1 itself is a limiting factor in VP16 nuclear import and acts as a chaperone for the nuclear entry of VP16 by direct interaction (15). Our study indicated that UL14 protein was not a required element for VP16-GFP to interact with HCF-1. As stated by others (16), the nature of interactions during infection can differ from that in the replication phase. One cannot simply take the results from the replication phase and apply those to the infection phase. In addition, in order to detect a protein supplied by the virion, hypersensitive measures or, alternatively, unnatural man-made conditions, such as ultrahigh-multiplicity infection, are required. Again, more experiments or a completely new design of experiment may be required to obtain optimal results.
The transport of incoming capsids to the nuclear pore was impaired though not blocked in 14D-VP16G-infected cells. This alone may decrease the expression of IE genes due to a shortage of template DNA. However, we think that this phenotype may be more influential when the virus is infected at a low MOI. At a low MOI, the UL14D virus forms small plaques compared to the wild type and exhibits an extended growth cycle (4). When the slower migration of capsids is combined with a VP16 transport defect, it is likely that the virus will display such a phenotype, not to mention the functional loss of the late protein UL14. As the retrograde transport of capsids is still not completely understood, this observation itself presents a demanding issue. Transient expression of UL14 was unable to rescue capsid transport (data not shown); thus, the unavailability of UL14 in the replication cycle could be disrupting effective tegumentation or important tegument-tegument interactions that are supposed to function in the capsid transport process.
In this study, we suggested that UL14 has an important purpose as a virion component. Given that VP16 is present in vast excess in the infecting virion compared to UL14, questions arise as to how UL14 is actually functioning. However, the molecular basis for this remains unsolved. The present study raises challenging questions and also identifies future areas for research.
Acknowledgments
We are grateful to Charles Cunningham and Andrew Davison for the HSV-1 strains 17syn+, UL14D, and UL14R and also for providing useful technical information. We thank Bernard Roizman for the RS cell line, Peter O'Hare for HSV-1 MP44 and anti-HCF-1 antibody, and Yoshihiro Nishida for the U2OS cell line. We thank Yoshikazu Fujita (Division for Medical Research Engineering, Nagoya University) for support with electron microscopy and Kazuko Nagamoto and Eiko Iwata for expert technical assistance.
This work was supported by Grants-in-Aid for Scientific Research on Priority Areas from the Ministry of Education, Culture, Sport, Science and Technology of Japan (18073007 and 16017240).
Footnotes
Published ahead of print on 21 November 2007.
REFERENCES
- 1.Ace, C. I., T. A. McKee, J. M. Ryan, J. M. Cameron, and C. M. Preston. 1989. Construction and characterization of a herpes simplex virus type 1 mutant unable to transinduce immediate-early gene expression. J. Virol. 632260-2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cai, W. Z., and P. A. Schaffer. 1989. Herpes simplex virus type 1 ICP0 plays a critical role in the de novo synthesis of infectious virus following transfection of viral DNA. J. Virol. 634579-4589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cousens, D. J., R. Greaves, C. R. Goding, and P. O'Hare. 1989. The C-terminal 79 amino acids of the herpes simplex virus regulatory protein, Vmw65, efficiently activate transcription in yeast and mammalian cells in chimeric DNA-binding proteins. EMBO J. 82337-2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cunningham, C., A. J. Davison, A. R. MacLean, N. S. Taus, and J. D. Baines. 2000. Herpes simplex virus type 1 gene UL14: phenotype of a null mutant and identification of the encoded protein. J. Virol. 7433-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Decman, V., P. R. Kinchington, S. A. Harvey, and R. L. Hendricks. 2005. Gamma interferon can block herpes simplex virus type 1 reactivation from latency, even in the presence of late gene expression. J. Virol. 7910339-10347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duffy, C., J. H. Lavail, A. N. Tauscher, E. G. Wills, J. A. Blaho, and J. D. Baines. 2006. Characterization of a UL49-null mutant: VP22 of herpes simplex virus type 1 facilitates viral spread in cultured cells and the mouse cornea. J. Virol. 808664-8675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Everett, R. D. 2000. ICP0, a regulator of herpes simplex virus during lytic and latent infection. Bioessays 22761-770. [DOI] [PubMed] [Google Scholar]
- 8.Greaves, R., and P. O'Hare. 1989. Separation of requirements for protein-DNA complex assembly from those for functional activity in the herpes simplex virus regulatory protein Vmw65. J. Virol. 631641-1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grunewald, K., P. Desai, D. C. Winkler, J. B. Heymann, D. M. Belnap, W. Baumeister, and A. C. Steven. 2003. Three-dimensional structure of herpes simplex virus from cryo-electron tomography. Science 3021396-1398. [DOI] [PubMed] [Google Scholar]
- 10.Harris, R. A., R. D. Everett, X. X. Zhu, S. Silverstein, and C. M. Preston. 1989. Herpes simplex virus type 1 immediate-early protein Vmw110 reactivates latent herpes simplex virus type 2 in an in vitro latency system. J. Virol. 633513-3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heine, J. W., R. W. Honess, E. Cassai, and B. Roizman. 1974. Proteins specified by herpes simplex virus. XII. The virion polypeptides of type 1 strains. J. Virol. 14640-651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Homa, F. L., and J. C. Brown. 1997. Capsid assembly and DNA packaging in herpes simplex virus. Rev. Med. Virol. 7107-122. [DOI] [PubMed] [Google Scholar]
- 13.Kawaguchi, Y., M. Tanaka, A. Yokoymama, G. Matsuda, K. Kato, H. Kagawa, K. Hirai, and B. Roizman. 2001. Herpes simplex virus 1 alpha regulatory protein ICP0 functionally interacts with cellular transcription factor BMAL1. Proc. Natl. Acad. Sci. USA 981877-1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kopp, M., B. G. Klupp, H. Granzow, W. Fuchs, and T. C. Mettenleiter. 2002. Identification and characterization of the pseudorabies virus tegument proteins UL46 and UL47: role for UL47 in virion morphogenesis in the cytoplasm. J. Virol. 768820-8833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.La Boissiere, S., T. Hughes, and P. O'Hare. 1999. HCF-1-dependent nuclear import of VP16. EMBO J. 18480-489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.La Boissiere, S., and P. O'Hare. 2000. Analysis of HCF-1, the cellular cofactor of VP16, in herpes simplex virus-infected cells. J. Virol. 7499-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.La Boissiere, S., A. Izeta, S. Malcomber, and P. O'Hare. 2004. Compartmentalization of VP16 in cells infected with recombinant herpes simplex virus expressing VP16-green fluorescent protein fusion proteins. J. Virol. 788002-8014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reference deleted.
- 19.Lee, S., and W. Herr. 2001. Stabilization but not the transcriptional activity of herpes simplex virus VP16-induced complexes is evolutionarily conserved among HCF-1 family members. J. Virol. 7512402-12411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leib, D. A., C. L. Bogard, M. Kosz-Vnenchak, K. A. Hicks, D. M. Coen, D. M. Knipe, and P. A. Schaffer. 1989. A deletion mutant of the latency-associated transcript of herpes simplex virus type 1 reactivates from the latent state with reduced frequency. J. Virol. 632893-2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lemaster, S., and B. Roizman. 1980. Herpes simplex virus phosphoproteins. II. Characterization of the virion protein kinase and of the polypeptides phosphorylated in the virion. J. Virol. 35798-811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marsden, H. S., N. D. Stow, V. G. Preston, M. C. Timbury, and N. M. Wilkie. 1978. Physical mapping of herpes simplex virus-induced polypeptides. J. Virol. 28624-642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McLean, G., F. Rixon, N. Langeland, L. Haarr, and H. Marsden. 1990. Identification and characterization of the virion protein products of herpes simplex virus type 1 gene UL47. J. Gen. Virol. 712953-2960. [DOI] [PubMed] [Google Scholar]
- 24.McNabb, D. S., and R. J. Courtney. 1992. Characterization of the large tegument protein (ICP1/2) of herpes simplex virus type 1. Virology 190221-232. [DOI] [PubMed] [Google Scholar]
- 25.Mettenleiter, T. C. 2002. Herpesvirus assembly and egress. J. Virol. 761537-1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mossman, K. L., and J. R. Smiley. 1999. Truncation of the C-terminal acidic transcriptional activation domain of herpes simplex virus VP16 renders expression of the immediate-early genes almost entirely dependent on ICP0. J. Virol. 739726-9733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Naldinho-Souto, R., H. Browne, and T. Minson. 2006. Herpes simplex virus tegument protein VP16 is a component of primary enveloped virions. J. Virol. 802582-2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Narayanan, A., M. L. Nogueira, W. T. Ruyechan, and T. M. Kristie. 2005. Combinatorial transcription of herpes simplex virus and varicella zoster virus immediate early genes is strictly determined by the cellular coactivator HCF-1. J. Biol. Chem. 2801369-1375. [DOI] [PubMed] [Google Scholar]
- 29.Nogueira, M. L., V. E. Wang, D. Tantin, P. A. Sharp, and T. M. Kristie. 2004. Herpes simplex virus infections are arrested in Oct-1-deficient cells. Proc. Natl. Acad. Sci. USA 1011473-1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.O'Hare, P. 1993. The virion transactivator of herpes simplex virus. Semin. Virol. 4145-155. [Google Scholar]
- 31.Roizman, B., and D. M. Knipe. 2001. Herpes simplex viruses and their replication, p. 2399-2460. In D. M. Knipe, P. M. Howley, et al. (ed.), Fields virology, 4th ed. Lippincott, Williams and Wilkins, Philadelphia, PA.
- 32.Sadowski, I., J. Ma, S. Triezenberg, and M. Ptashne. 1988. GAL4-VP16 is an unusually potent transcriptional activator. Nature 335563-564. [DOI] [PubMed] [Google Scholar]
- 33.Smiley, J. R., and J. Duncan. 1997. Truncation of the C-terminal acidic transcriptional activation domain of herpes simplex virus VP16 produces a phenotype similar to that of the in1814 linker insertion mutation. J. Virol. 716191-6193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sodeik, B., M. W. Ebersold, and A. Helenius. 1997. Microtubule-mediated transport of incoming herpes simplex virus 1 capsids to the nucleus. J. Cell Biol. 1361007-1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thompson, R. L., and N. M. Sawtell. 2006. Evidence that the herpes simplex virus type 1 ICP0 protein does not initiate reactivation from latency in vivo. J. Virol. 8010919-10930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wada, K., F. Goshima, H. Takakuwa, H. Yamada, T. Daikoku, and Y. Nishiyama. 1999. Identification and characterization of the UL14 gene product of herpes simplex virus type 2. J. Gen. Virol. 802423-2431. [DOI] [PubMed] [Google Scholar]
- 37.Wysocka, J., and W. Herr. 2003. The herpes simplex virus VP16-induced complex: the makings of a regulatory switch. Trends Biochem. Sci. 28294-304. [DOI] [PubMed] [Google Scholar]
- 38.Yamauchi, Y., K. Wada, F. Goshima, H. Takakuwa, T. Daikoku, M. Yamada, and Y. Nishiyama. 2001. The UL14 protein of herpes simplex virus type 2 translocates the minor capsid protein VP26 and the DNA cleavage and packaging UL33 protein into the nucleus of coexpressing cells. J. Gen. Virol. 82321-330. [DOI] [PubMed] [Google Scholar]
- 39.Yamauchi, Y., K. Wada, F. Goshima, T. Daikoku, K. Ohtsuka, and Y. Nishiyama. 2002. Herpes simplex virus type 2 UL14 gene product has heat shock protein (HSP)-like functions. J. Cell Sci. 1152517-2527. [DOI] [PubMed] [Google Scholar]
- 40.Yamauchi, Y., T. Daikoku, F. Goshima, and Y. Nishiyama. 2003. Herpes simplex virus UL14 protein blocks apoptosis. Microbiol. Immunol. 47685-689. [DOI] [PubMed] [Google Scholar]
- 41.Yang, W. C., G. V. Devi-Rao, P. Ghazal, E. K. Wagner, and S. J. Triezenberg. 2002. General and specific alterations in programming of global viral gene expression during infection by VP16 activation-deficient mutants of herpes simplex virus type 1. J. Virol. 7612758-12774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yao, F., and P. A. Schaffer. 1994. Physical interaction between the herpes simplex virus type 1 immediate-early regulatory proteins ICP0 and ICP4. J. Virol. 688158-8168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yao, F., and P. A. Schaffer. 1995. An activity specified by the osteosarcoma line U2OS can substitute functionally for ICP0, a major regulatory protein of herpes simplex virus type 1. J. Virol. 696249-6258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang, Y., and J. L. McKnight. 1993. Herpes simplex virus type 1 UL46 and UL47 deletion mutants lack VP11 and VP12 or VP13 and VP14, respectively, and exhibit altered viral thymidine kinase expression. J. Virol. 671482-1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhou, Z. H., D. H. Chen, J. Jakana, F. J. Rixon, and W. Chiu. 1999. Visualization of tegument-capsid interactions and DNA in intact herpes simplex virus type 1 virions. J. Virol. 733210-3218. [DOI] [PMC free article] [PubMed] [Google Scholar]