Abstract
The equine lentivirus receptor 1 (ELR1), a member of the tumor necrosis factor receptor (TNFR) protein family, has been identified as a functional receptor for equine infectious anemia virus (EIAV). Toward defining the functional interactions between the EIAV SU protein (gp90) and its ELR1 receptor, we mapped the gp90 binding domain of ELR1 by a combination of binding and functional assays using the EIAV SU gp90 protein and various chimeric receptor proteins derived from exchanges between the functional ELR1 and the nonbinding homolog, mouse herpesvirus entry mediator (murine HveA). Complementary exchanges of the respective cysteine-rich domains (CRD) between the ELR1 and murine HveA proteins revealed CRD1 as the predominant determinant of functional gp90 binding to ELR1 and also to a chimeric murine HveA protein expressed on the surface of transfected Cf2Th cells. Mutations of individual amino acids in the CRD1 segment of ELR1 and murine HveA indicated the Leu70 in CRD1 as essential for functional binding of EIAV gp90 and for virus infection of transduced Cf2Th cells. The specificity of the EIAV SU binding domain identified for the ELR1 receptor is fundamentally identical to that reported previously for functional binding of feline immunodeficiency virus SU to its coreceptor CD134, another TNFR protein. These results indicate unexpected common features of the specific mechanisms by which diverse lentiviruses can employ TNFR proteins as functional receptors.
Equine infectious anemia virus (EIAV), a member of the lentivirus subfamily, establishes a persistent infection in horses and causes a uniquely dynamic lentiviral disease characterized by recurrent waves of viremia and disease cycles with clinical signs that include fever, diarrhea, edema, lethargy, anemia, thrombocytopenia, and occasional encephalitis or ataxia (24). EIAV disease in horses is apparently related to an exclusive infection of monocytes and macrophages, making EIA a relevant model for studying lentiviral pathogenicity from macrophage infections without the complications of lymphocyte infections associated with the immunodeficiency lentiviruses. Toward defining EIAV-macrophage interactions, we recently identified a tumor necrosis factor receptor (TNFR) family protein, designated equine lentivirus receptor 1 (ELR1), as a functional cellular receptor for both primary and cell-adapted strains of EIAV (35). This finding of a single functional receptor protein for EIAV is in distinct contrast to the common use of dual coreceptors reported for immunodeficiency lentiviruses (human, simian, and feline immunodeficiency viruses) (3, 12, 13, 30, 33) but similar to the single receptor usage reported for simple retroviruses, such as murine or avian leukemia viruses (1). Of particular interest is the fact that feline immunodeficiency virus (FIV) also uses a TNFR protein, CD134, as a coreceptor with CD4 for infection of target cells (12, 30). The single TNFR protein receptor target for EIAV combined with its relatively simple genetic composition is consistent with our previous suggestion that EIAV may reflect a retrovirus at the interface between the simple oncornaviruses and complex lentiviruses. The use of a single receptor by EIAV also raises a number of interesting questions about the specificity of the interactions between ELR1 and the SU protein (gp90) of EIAV that activate fusion and mediate infection of target cells.
Based on the highly conserved structure of TNFR proteins, the ELR1 is predicted to be a type I transmembrane protein, with its ectodomain containing the characteristic four cysteine-rich domains designated CRD1 to CRD4 (26). The solved crystal structures of various TNFR proteins reveal a highly conserved three-dimensional rod-like ectodomain structure to which ligand binding induces conformational alterations that activate diverse signal transduction pathways leading to cell proliferation or death (20, 31). Among the family of ELR1 proteins, the most homologous protein sequence to ELR1 is the herpesvirus entry mediator proteins expressed on the surface of human (HveA, also known as HVEM and TNFR-14) (25) and mouse (mHveA) cells (27, 32). In an earlier study, Elder and colleagues took advantage of the structural similarities of feline and human CD134 to map the critical determinants in CD134 that mediate functional binding to the SU protein of FIV (11).
The goal of the current study was to identify amino acid residues in ELR1 that are critical for EIAV gp90 binding, as a part of ongoing studies that utilize EIAV as a model to define the mechanisms of retrovirus entry into target cells. Following the exemplary mapping studies described by Elder and colleagues, we employed domain exchanges and single amino acid substitutions between ELR1 and murine HveA to identify the specific cysteine-rich domain(s) critical for functional interactions with EIAV gp90. Chimeric receptor proteins were evaluated both for their ability to bind EIAV gp90 in a cell-cell binding assay and their ability to support virus infection in transduced target cells. The results of these studies for the first time reveal the binding sites on ELR1 and highlight a remarkable conservation of structure and function among TNFR proteins that serve as receptors for different viruses.
MATERIALS AND METHODS
ELR1 mutagenesis.
Murine HveA was cloned by reverse transcriptase PCR (RT-PCR) from mouse thymus cells using primers based on the published murine HveA sequence (27, 32). ELR1 and murine HveA domain exchange mutants or point mutations were generated using an overlapping PCR strategy (19). Briefly, 5′ and 3′ halves of the appropriate receptor genes were amplified in separate PCRs. The internal primers were designed to overlap the position selected for mutation. The agarose gel-purified PCR products were then used to amplify the full-length receptor gene using two outer primers. The full-length PCR products were then digested using EcoRI and BamHI and inserted into a pFB-Neo-LacZ vector (Stratagene, La Jolla, CA) with the same digestion sites. As outlined below in Fig. 2, all chimeric constructs contained the ELR1 leader sequence, transmembrane domain, and cytoplasmic domain with a hemagglutinin (HA) tag. For the point mutation constructs based on murine HveA, all receptor gene sequences were from wild-type murine HveA.
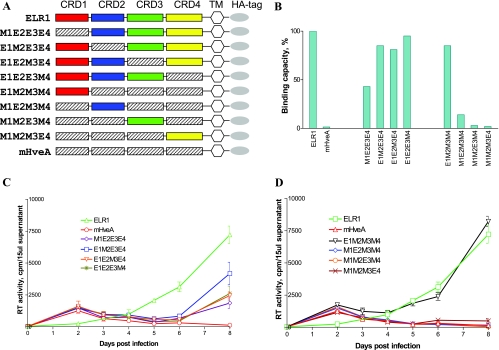
FIG. 2.
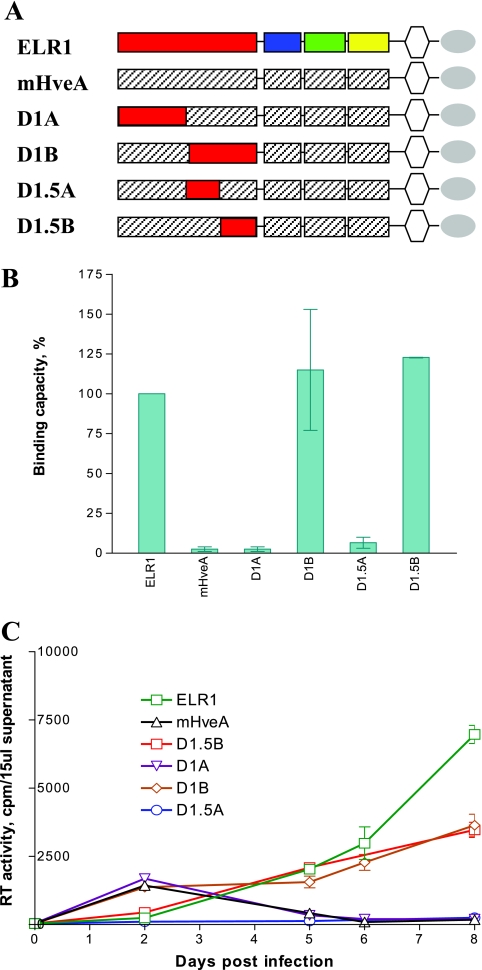
The EIAV gp90 binding domain of ELR1 maps to CRD1. The roles of individual CRD segments of ELR1 in gp90 binding and virus infection were evaluated by functional assay of chimeric receptor proteins. (A) Schematic representation of chimeric constructs with domain exchanges between gp90 binding ELR1 and nonbinding murine HveA. Color-coded CRD are from ELR1; shaded CRD segments are from murine HveA. All chimeras contain an ELR1 leader sequence, transmembrane domain, and cytoplasmic domain (TM) followed by an HA tag. The construct nomenclature indicates the source of the specific CRD as the equine (E) ELR1 or murine (M) HveA protein. (B) EIAV gp90 binding capacity of each of the chimeric receptor constructs expressed on the surface of transduced CHO cells in a cell-cell binding assay, as described in Materials and Methods. (C) EIAV replication in Cf2Th cells transduced with the indicated chimeric receptor constructs with individual ELR1 CRD segments substituted into the murine HveA protein in a gain-of-function assay. (D) EIAV replication in Cf2Th cells transduced with the indicated chimeric receptor constructs with individual murine HveA CRD segments substituted into the ELR1 protein in a loss-of-function assay. Virus replication levels in panels C and D were quantified by assays of the production of RT activity in culture supernatants.
Virus strains and cells.
The cell-adapted pathogenic EIAVUK molecular clone was used in this study (10). Equine dermal (ED), fetal equine kidney (FEK), CHO, and Cf2Th (ATCC CRL-1430) cells were grown in minimal essential medium plus 10% fetal bovine serum and 1% penicillin-streptomycin. For the selection of a stable cell line expressing ELR1 or chimeric receptors, transduced Cf2Th cells were grown in standard medium plus 800 μg/ml G418. Selection medium was refreshed every 3 days.
Anti-ELR1 polyclonal sera.
The pEctD2 construct was developed to express the histidine peptide-tagged ectodomain of the ELR1 protein. For construction of the pEctD2 expression vector, the ELR1 gene ectodomain sequence was amplified by PCR. The PCR products were subcloned into the pQE-80L plasmid (Qiagen, Valencia, CA) by using the BamHI and HindIII restriction enzyme sites. A six-histidine peptide was fused to the 5′ end of the protein for purification purposes. Protein expression and purification were carried out as described previously (15). Briefly, the pEctD2-transformed Escherichia coli cells were precultured in 5 ml of Luria broth medium supplemented with 2 g of glucose/liter and 100 μg of ampicillin/ml overnight at 37°C with vigorous shaking. The overnight culture was used to inoculate 100 ml of Luria broth medium. The expression of ELR1 protein was induced by addition of 1 mM isopropyl-β-d-thiogalactopyranoside to the log-phase culture. The bacterial pellets were harvested at 4 h postinduction by centrifugation at 6,000 × g for 10 min. The cells were then lysed with a bacterial protein extraction reagent (Pierce, Rockford, IL), filtered through 0.45-μm-pore-size filters, and applied to a column equilibrated with the binding buffer according to the manufacturer's directions (Amersham, Piscataway, NJ). The column was then washed with the elution buffer containing 10 mM Na2HPO4, 10 mM NaH2PO4, 0.5 M NaCl, and 10 mM imidazole (pH 7.4) at a flow rate of 5 ml/min to elute the bound ELR1 protein. To examine the quality of the purified ELR1 protein, column fractions were collected, assayed for protein content by measuring absorbance at 280 nm, and then analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. The eluted ELR1 protein was dialyzed against phosphate-buffered saline (PBS) overnight. Anti-ELR1 antibody was produced commercially by immunizing New Zealand White rabbits with the purified ELR1 protein (Washington Biotechnology, Columbia, MD).
Mutant ELR1-transduced Cf2Th cells.
All mutant receptor proteins were inserted into the pFB-Neo-LacZ vector, and vesicular stomatitis virus-pseudotyped retroviruses expressing the receptor and its mutant derivatives were made as described previously (35). EIAVUK nonpermissive Cf2Th cells were seeded in T25 flasks 1 day before retrovirus infection. Cells were infected with receptor recombinant retrovirus at a multiplicity of infection of 0.1. At 24 h postinfection, 800 μg/ml G418 was added to the culture medium. After 2 weeks of selection, G418-resistant cells were expanded and used for assays of receptor expression and EIAV gp90 binding.
Construction of expression vectors for GFP-labeled EIAV gp90.
The EIAV gp90 gene used for these studies was a codon-optimized version of the parental pathogenic proviral molecular clone EIAVUK described in detail by Cook et al. (10). The green fluorescent protein (GFP) gene was from the pGFP-N3 vector (BD Biosciences Clontech, Mountain View, CA). The EIAV gp90-GFP fusion genes were amplified by overlapping PCR (19) and then cloned into a p2CI vector derived from PCR2.1 by insertion of a cytomegalovirus promoter, PCR-amplified IRES-neomycin-resistant sequences from the pFB-Neo-LacZ vector (Stratagene, La Jolla, CA), and poly(A) signal sequences from pcDNA3.1 (Invitrogen, Carlsbad, CA). Overlapping PCR was performed as described above for the ELR1 mutagenesis. The p2CI-GFP plasmid containing only the GFP gene was also constructed as a control for the expression of GFP.
Production of CHO cells expressing EIAV gp90-GFP.
Plasmid DNA expressing either EIAV gp90-GFP or GFP was transfected into CHO cells using Fugene 6 (Roche, Indianapolis, IN) transfection reagent, followed by G418 selection with 400 μg/μl G418 in growth medium. Single G418-resistant colonies were subcultured individually in six-well plates. Colonies were analyzed by flow cytometry for GFP expression, and the colony displaying the highest level of GFP expression was expanded and maintained under G418 selection for subsequent studies of gp90 binding. Selected transduced cells were shown to display consistent surface expression of EIAV gp90-GFP through repeated passages with about 90% of cells positive for staining with gp90-specific monoclonal antibodies (data not shown).
Flow cytometry analysis.
The surface expression of ELR1 or EIAV gp90-GFP in transduced cells was evaluated by flow cytometry. Briefly, 5 × 105 cells were harvested from flasks using 10 mM EDTA-PBS and then fixed in 1% paraformaldehyde for 1 h at 4°C. The cells were then washed three times with wash buffer (PBS containing 5% heat-inactivated fetal bovine serum) and then incubated with 2 μl rabbit anti-ELR1 polyclonal antibody or 1 μl anti-gp90 monoclonal antibody in 100 μl wash buffer for 1 h at 4°C. The cells were then washed three times with wash buffer and incubated with 1 μl secondary conjugate in 100 μl wash buffer for 1 h at 4°C. Following three washes with wash buffer, antibody labeling of ELR1 and gp90 labeling were analyzed by flow cytometry using a BD FACSCalibur (BD Biosciences, San Jose, CA). HA tag staining was performed following the protocol described previously (35).
Cell-cell binding assay.
The cell-cell binding protocols used to assay the level of binding between various ELR1 constructs and EIAV gp90, individually expressed on surfaces of different cells, are based on procedures previously described for the study of binding between the S protein of severe acute respiratory coronavirus and its cellular receptor (8). For the cell-cell binding assay, Cf2th cells transduced with ELR1 or its mutant derivatives were used as the target monolayer, and CHO cells transduced with the EIAV gp90-GFP served as the ligand cells. Approximately 2 × 105 receptor-transduced Cf2Th cells expressing individual ELR1 protein constructs were seeded in each well of six-well plates (Becton Dickinson Labware, Bedford, MA). Cells were cultured at 37°C for 2 days and then transferred to 4°C for 30 min. Immediately prior to use, the transduced gp90-GFP CHO or GFP CHO cell monolayers were detached by adding 10 mM EDTA-PBS and washed three times with ice-cold minimal essential medium plus 10% fetal bovine serum. About 3 × 106 transduced CHO cells (ligand) were then added to each well of receptor-transduced Cf2Th cells (target) at 4°C and incubated for 1.5 h. Unbound CHO cells were then removed by washing three times with PBS and one time with PBS containing 0.5 M NaCl. Plates were then moved to room temperature, and the attached cells were removed by pipetting using wide-bore tips with 0.5 ml PBS in each well. Resuspended cells were subjected to HA tag staining and flow cytometry analysis to measure bound Cf2Th cells.
Virus replication and RT assay.
The extracellular RT activity in cell-free culture medium was measured to determine the levels of virus production, as described previously (18). Reported RT levels represent the averages of duplicate samples.
Modeling of ELR1.
Homology modeling was conducted using MODELLER 8.0 (www.salilab.org/modeler/modeler.html) on a Macintosh platform. Sequence alignments for input into MODELLER (28) were generated by CLUSTALW (www.ebi.ac.uk/clustalw). The refined structure of HveA at 2.65-Å resolution (Protein Data Bank ID 1JMA) was used as a template. The amino acid sequences of murine HveA and ELR1 (NCBI PID AAY21177 and AAI04055, respectively) were obtained from the National Center for Biotechnology Information (www.ncbi.nlm.nih.gov/entrez). Only CRD1, a modular folding unit within the full-length receptor, was used in modeling studies. Single point mutations to murine HveA and ELR1 corresponding to position 70 in ELR1 were also modeled. In all cases, the homology domain with the lowest MODELLER objective function score was selected and visualized using VMD 1.86 (14).
RESULTS
Validation of cell-cell binding assay.
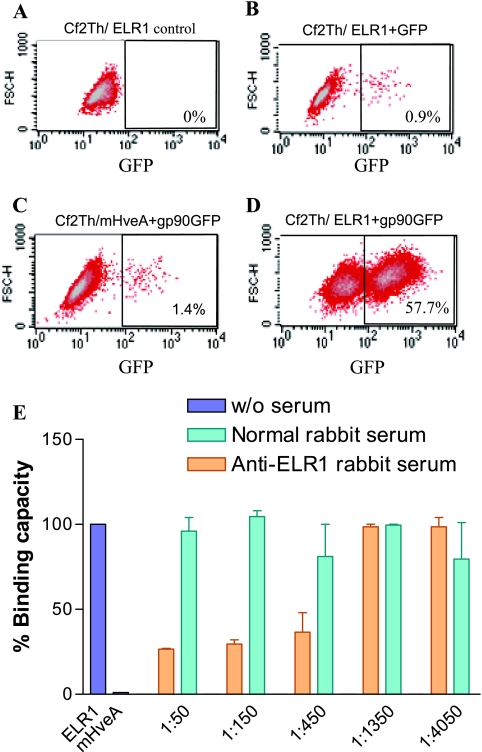
The data in Fig. 1 demonstrate the specificity of the cell-cell binding assay for measuring ELR1 binding with EIAV gp90. In the cell-cell binding assay, only the CHO cells expressing gp90GFP bound to the target Cf2Th/ELR1 cells in flow cytometry (Fig. 1D). In contrast, there was no significant binding of the CHO gp90-GFP cells to Cf2Th cells expressing the nonfunctional mouse herpes simplex virus receptor mHveA (Fig. 1C), and CHO cells expressing only GFP also failed to significantly bind the target Cf2Th/ELR1 cells (Fig. 1B). The specificity of the binding observed between the ligand CHO gp90-GFP cells and the target Cf2Th/ELR1 cells was further tested by determining the ability of a reference polyclonal anti-ELR1 antibody to block the cell-cell binding under the standard assay conditions. As summarized in Fig. 1E, the reference polyclonal immune serum to ELR1 inhibited the cell-cell binding, and the observed inhibition was directly related to the dilution of the serum added to the binding assay. In distinct contrast, normal rabbit serum failed to reduce the level of cell-cell binding. Thus, these data indicate that the conditions employed for the cell-cell binding assays specifically measure interactions mediated by EIAV gp90 and ELR1 expressed on the cell surfaces.
FIG. 1.
Validation of the cell-cell binding assay. Shown are representative flow cytometry analysis data that summarize the specificity of the binding of target Cf2Th/ELR1 to the ligand CHO cells expressing gp90-GFP under standard assay conditions. (A) Cf2Th/ELR1 cells incubated with nontransduced CHO cells. (B) Cf2Th/ELR1 cells incubated with transduced CHO cells expressing only GFP. (C) Cf2Th cells expressing nonfunctional mHveA receptor incubated with CHO cells expressing gp90-GFP. (D) Cf2Th/ELR1 cells incubated with CHO cells expressing gp90-GFP. (E) The specificity of the cell-cell binding assay was demonstrated by blocking with ELR1-specific rabbit serum antibodies. Ligand CHO cells expressing gp90-GFP were mixed with target Cf2Th/ELR1 cells and incubated in the presence of medium alone or medium containing the indicated dilutions of normal or immune rabbit serum.
EIAV gp90 binds to the C terminus of CRD1 of ELR1.
Based on a comparison of TNFR family protein sequences using CLUSTALW, ELR1 is most closely related to the herpes simplex virus entry mediator HveA and it mouse version, murine HveA (data not shown). A closer comparison revealed that ELR1 and murine HveA contain predicted ectodomains and analogous CRD segments, as described by Naismith and Sprang (26). In preliminary studies, murine HveA expressed on the surface of transduced Cf2Th cells did not bind EIAV gp90 or support infection (data not shown). The lack of EIAV-specific receptor function is in contrast to the gp90 binding and productive virus infection mediated by ELR1 expressed on the surface of transduced Cf2Th cells. Thus, we pursued an initial mapping of the functional binding domains of ELR1 by two complementary domain exchange strategies. In the first strategy (loss of function), we substituted selected CRD segments from murine HveA into ELR1 and measured the chimeric receptor proteins for gp90 binding and for their ability to mediate infection of Cf2Th cells transduced with the respective chimeric receptor. In the second strategy (gain of function), we substituted selected CRD segments from ELR1 into the murine HveA backbone and assayed the product chimeric receptors for their ability to bind gp90 and to mediate infection of transduced Cf2Th cells. The panel of chimeric receptor constructs is summarized in Fig. 2.
Prior to assaying the functional properties of the various chimeric receptors, the total expression of each receptor construct in transduced Cf2Th cells was evaluated by flow cytometry using the HA tag contained in each receptor protein. These assays indicated that 85 to 95% of the transduced cells for each receptor construct were positive for HA, indicating similar levels of expression for each receptor protein (data not shown). Surface expression of the chimeric receptors was confirmed by flow cytometry of the various transduced cell lines labeled with the rabbit immune serum to ELR1 (data not shown). While qualitatively confirming the surface expression of each receptor construct, these assays could not provide a quantitative comparison of surface expression levels, as the amount of potential antibody-reactive ELR1 sequences varied markedly among the constructs. However, for those point mutation constructs based on ELR1, we observed similar levels of surface expression compared to wild-type ELR1.
We next assayed each cell surface-expressed receptor construct in the cell-cell binding assay for its capacity to bind EIAV gp90 expressed on the surface of transduced CHO cells (Fig. 2). Figure 2B summarizes the loss-of-function experiments in which selected CRD segments of murine HveA were substituted into the ELR1 protein backbone. Cell-cell binding assays indicate about 57% binding for the parental ELR1 and less than 1% binding for the murine HveA, demonstrating the specificity of the assay procedures. In our analysis, wild-type ELR1 binding capacity was set as 100%, and the binding capacities of the various mutant receptor constructs were calculated as a proportion of ELR1 binding. Substitution of the CRD1 from murine HveA into ELR1 (M1E2E3E4) reduced the binding capacity by about 50% compared to the binding level observed with parental ELR1. Individual substitutions of the murine HveA CRD2, CRD3, or CRD4 into ELR1 resulted in 15%, 19%, and 5% loss of binding capacity, respectively, compared to parental ELR1.
The results of the cell-cell binding assays for the murine HveA receptor proteins containing individual CRD segments from ELR1 are also summarized in Fig. 2B. These data demonstrate that the chimeric receptor containing the CRD1 from ELR1 (E1M2M3M4) gained 85% of the binding capacity compared to ELR1. In contrast, the chimeric receptors with substitutions of CRD2, CRD3, or CRD4 from ELR1 into the murine HveA backbone recovered only 14%, 3%, and 2% of ELR1 binding capacity, respectively. Taken together, the loss of binding function and gain of binding function assays both indicate a predominant role for CRD1 in gp90 binding, with only a relatively minor influence of the other three CRD segments.
Having established the gp90 binding properties of the chimeric receptor constructs, we next assayed the functional consequences by measuring the ability of each chimeric construct to support EIAV infection and replication in transduced Cf2Th cells. Figure 2C summarizes the replication properties of EIAV in the panel of Cf2Th cells stably transduced with the various ELR1 constructs containing a CRD substitution from murine HveA. Figure 2D summarizes the replication data for the complementary set of substitution mutations into the murine HveA protein backbone. In the assays of the murine CRD substitutions into ELR1 that measure loss of function, virus replication levels in the cells transduced with the M1E2E3E4 chimeric receptor were reduced by about 60% compared to virus replication in ELR1-transduced cells. Virus replication levels produced by the remaining chimeric receptor proteins with substitutions in CRD2, CRD3, or CRD4 were reduced to various intermediate levels between those observed with M1E2E3E4 and ELR1 (Fig. 2C). In the assays of the equine CRD substitutions into murine HveA that measured gain of function, virus replication in the E1M2M3M4-tranduced cells was equal to that observed in the ELR1-transduced cells; no other substitution in this panel of receptor constructs was able to support EIAV replication (Fig. 2D). Therefore, these data clearly demonstrate that domain exchanges between ELR1 and mHveA did not alter the overall structure of the receptors as binding ligands. Importantly, the binding chimeric receptors also support virus entry and subsequently virus replication in transduced cells, at various levels between ELR1 and mHveA. The virus replication assays further highlight the role of the CRD1 domain in receptor function, with the remarkable observation that substitution of this domain into the murine HveA protein produced a fully functional receptor for EIAV.
To further define the critical sequences in the ELR1 CRD1 that mediate receptor function, we next employed the same domain exchange strategy, but with smaller segments of the equine CRD1 substituted into the murine HveA protein backbone (Fig. 3A). The constructs were then assayed for gp90 binding (Fig. 3B) and virus replication (Fig. 3C) in transduced Cf2Th cells. As summarized in Fig. 3B, only receptor constructs containing the C-terminal sequences of the ELR1 CRD1 segment gained gp90 binding and EIAV infection functions; the chimeric receptor (D1A) containing the N-terminal sequences of the ELR1 CRD1 failed to bind gp90 or to support virus replication. Specifically, the D1B and D1.5B mutants both displayed gp90 binding levels equal to ELR1 (Fig. 3B). The D1B and D1.5B receptors also supported EIAV replication as effectively as ELR1, while the D1.5A chimeric receptor failed to support virus replication. Taken together, these data indicated that the gp90 binding and EIAV infection functions were associated with the C-terminal half of the CRD1 of ELR1.
FIG. 3.
The C terminus of ELR1 CRD1 is critical for EIAV gp90 binding. Fragments of ELR1 CRD1 were substituted into murine HveA, and the chimeric receptors were tested for gp90 binding and virus infection functions. (A) Schematic representation of the panel of receptor constructs containing segments of the ELR1 CRD1 substituted into the murine HveA protein. (B) Relative EIAV gp90 binding capacity of each chimeric receptor compared to ELR1 in the cell-cell binding assay. The gp90 binding levels observed for each chimeric receptor protein are expressed as a percentage of the binding observed with ELR1. (C) EIAV replication in Cf2Th cells transduced with the indicated CRD1 chimeric receptor constructs, as described for Fig. 2.
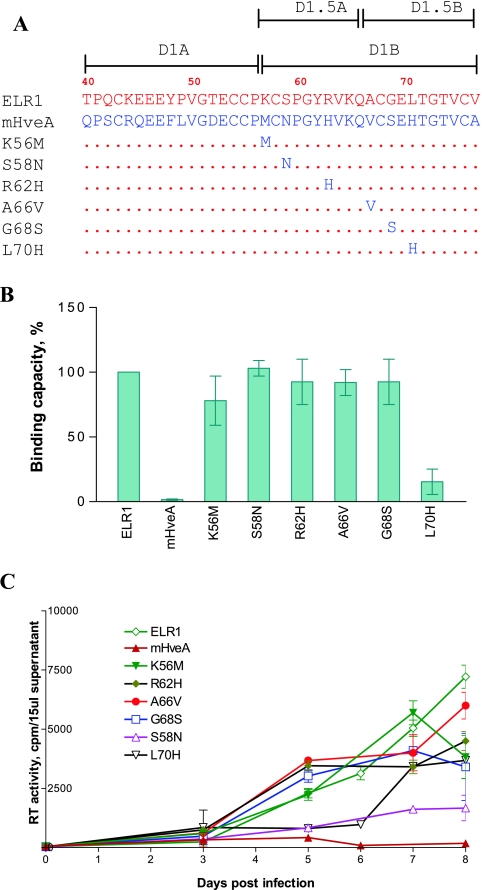
Mapping of CRD1 residues critical for receptor-envelope interactions.
Even though ELR1 and murine HveA differ markedly in their EIAV gp90 binding capacity, the critical sequences identified in the D1B subdomain of the CRD1 of these respective receptor proteins differ by only a few amino acids (Fig. 4A). To further define the specific residues in CRD1 that mediate gp90 binding, a panel of receptor mutations containing single amino acid substitutions were constructed, each converting the equine amino acid to its corresponding murine amino acid (Fig. 4A). To eliminate the influence of other CRD sequences in ELR1 on functional properties, we introduced these point mutations into the backbone of the fully functional E1M2M3M4 chimeric receptor that contained only the CRD1 sequences from ELR1 substituted into the murine HveA protein.
FIG. 4.
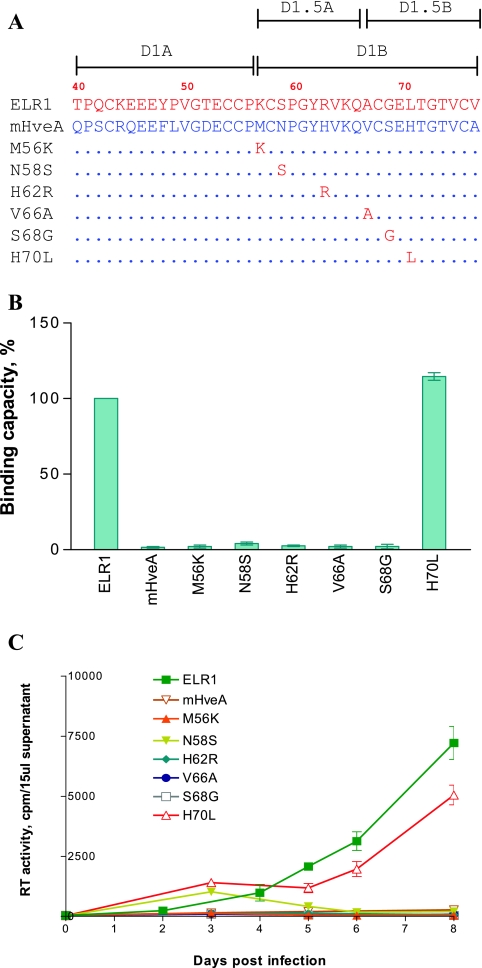
Mapping of CRD1 residues critical for ELR1 function by loss-of-function analyses of point mutations converting ELR1 to murine HveA amino acids. (A) Schematic representation of the series of point mutations constructed by substituting the indicated amino acids of ELR1 CRD1 with the corresponding amino acid from the murine HveA CRD1 sequence. All point mutations were made in the E1M2M3M4 chimeric receptor to isolate the equine CRD1 contributions to function. The names and C-terminal CRD1 sequences of the various receptor mutants derived from E1M2M3M4 are shown. Amino acids of ELR1 and mHveA are in red and blue letters, respectively. Numbers indicate positions of amino acids in the predicted protein sequence of full-length ELR1. Dots in the mutant receptor sequences indicate identical amino acids to ELR1. (B) EIAV gp90 binding capacity of each receptor mutant expressed as a percentage of ELR1 binding in the cell-cell binding assay. (C) EIAV replication in Cf2Th cells transduced by the indicated chimeric receptor protein constructs, as described for Fig. 2.
Following the same procedure as used for the CRD exchange mutants, the various point mutations in the context of stably transduced Cf2Th cells were compared for their abilities to bind EIAV gp90 and to support virus replication. As shown in Fig. 4B, CRD1 mutations of K56M caused a 20% decrease in gp90 binding activity compared to ELR1, while the S58N, R62H, A66V, and G68S mutations displayed gp90 binding levels similar to ELR1. In contrast, the mutant L70H displayed an 80% reduction in gp90 binding capacity. Interestingly, all of the CRD1 point mutants were able to support EIAV replication in transduced Cf2Th cells (Fig. 4C), although the observed levels of virus replication differed among the individual mutants. Interestingly, the virus replication levels produced with each mutant were not tightly correlated to the gp90 binding levels observed with the various point mutations. For example, the R62H, A66V, and G68S mutants that displayed wild-type gp90 levels supported virus replication as well as the ELR1 protein. Similarly, the L70H mutant, which displayed a greatly reduced gp90 binding capacity, mediated virus replication levels that were about 10-fold less than the replication levels observed with ELR1. However, the S58N mutant displayed a wild-type gp90 binding capacity but produced virus replication levels that were also about 10-fold lower than ELR1. While the mechanistic explanations for the lack of correlation between gp90 binding and virus replication capacities remain to be defined, the current results do emphasize the importance of the ELR1 L70 as a determinant for gp90 binding and suggest that S58 may play an important role in virus replication following the initial binding events.
Conversion of murine HveA to a functional EIAV receptor by a single point mutation.
In order to further explore the roles of the individual variant amino acids of CRD1 of ELR1 and murine HveA, a series of point mutations were made in the murine HveA backbone to convert the residue to its corresponding amino acid in ELR1 (Fig. 5A). The receptor mutants were tested for their gp90 binding capacities and abilities to support virus replication. As shown in Fig. 5B, mutations in the murine HveA protein at position 56, 58, 62, 66, or 68 failed to bind gp90. In contrast, mutant H70L receptor protein displayed gp90 binding levels that were equivalent to the ELR1 protein. Consistent with the observed gp90 binding capacity of the murine HveA point mutations, only the H70L receptor protein was able to support virus replication in transduced Cf2Th cells; the remaining point mutants were defective for virus replication (Fig. 5C). This ability of a single amino acid mutation (mH70L) to transform a nonfunctional receptor into a functional receptor further confirmed the important role of Leu70 in the receptor binding site for EIAV gp90.
FIG. 5.
Mapping of CRD1 residues critical for ELR1 function by gain-of-function analyses of point mutations converting murine HveA to equine ELR1 amino acids. (A) Schematic representation of the series of point mutations constructed by substituting the indicated amino acids of murine HveA CRD1 with the corresponding amino acid from the murine ELR1 CRD1 sequence. The names and C-terminal CRD1 sequences of the various receptor mutants derived from murine HveA are shown. Amino acids of ELR1 and mHveA are in red and blue letters, respectively. Dots in the mutant receptor sequences indicate identical amino acids to ELR1. (B) EIAV gp90 binding capacity of each receptor mutant expressed as a percentage of ELR1 binding in the cell-cell binding assay. (C) EIAV replication in Cf2Th cells transduced by the indicated chimeric receptor protein constructs, as described for Fig. 2.
Requirement of a small hydrophobic amino acid at ELR1 residue 70 for functional binding.
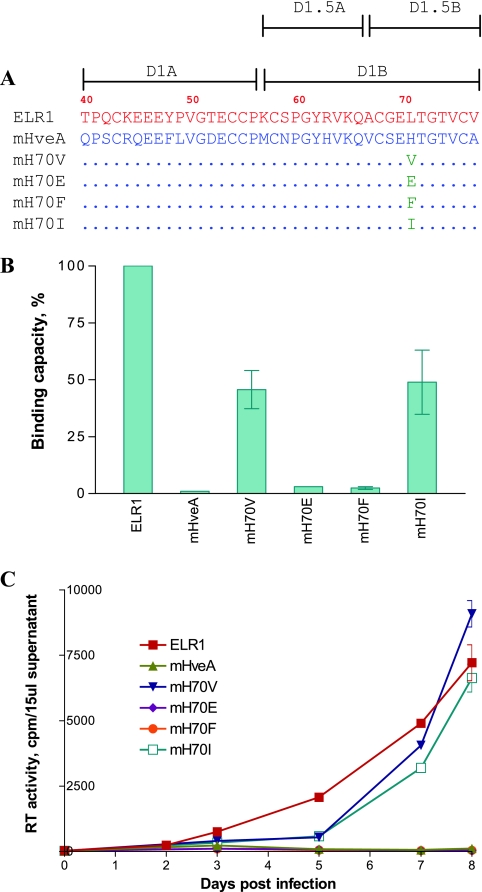
The previous experiments indicated Leu70 as critical for functional binding to EIAV gp90. To explore the nature of the side chain at this position that is necessary for receptor function, we mutated H70 in murine HveA to a series of selected amino acids with different chemical properties and then assayed the gp90 binding and virus replication properties of the mutated receptors. As summarized in Fig. 6, substitution of the murine HveA H70 with relatively small hydrophobic amino acids, valine or isoleucine, produced receptor proteins that displayed gp90 binding levels that were about 50% of the activity observed for ELR1 (Fig. 6B). However, the receptors with these two point mutations were both functional for supporting EIAV replication in the transduced Cf2Th cells (Fig. 6C). In contrast to this gain of function with the valine and isoleucine substitutions, the H70F and the H70E receptors both failed to bind gp90 (Fig. 6B) or to support EIAV replication (Fig. 6C).
FIG. 6.
Structural requirements for residue 70 of CRD1 for binding and infection functions. Residue 70 of the murine HveA receptor construct was mutated individually to the indicated amino acids, and the product mutant receptor protein was assayed for binding and infection functions. (A) Schematic representation (C-terminal CRD1 sequences) of the individual point mutations introduced into residue 70 of the receptor protein, including valine, isoleucine, phenylalanine, and glutamate. Amino acids of ELR1 and mHveA are in red and blue letters, respectively. Dots in mutant receptors indicate identical amino acids to mHveA. (B) EIAV gp90 binding capacity of each receptor mutant expressed as a percentage of ELR1 binding in the cell-cell binding assay. (C) EIAV replication in Cf2Th cells transduced by the indicated chimeric receptor protein constructs, as described for Fig. 2.
Modeling of ELR1.
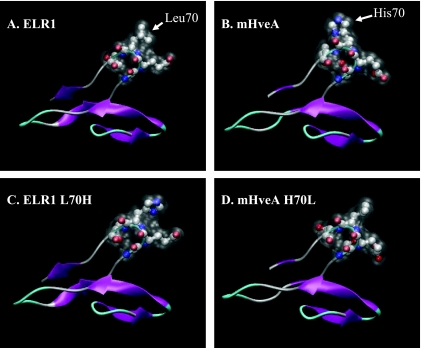
Given the critical role of L70 of ELR1 for gp90 binding and the support of viral replication, the CRD1 domain of the receptor protein was examined by homology modeling. For comparative evaluation, the same domain of murine HveA was also modeled using the crystal structure of the corresponding domain of HveA as a template. The highly conserved sequences of ELR1 and murine HveA show 89% and 64% identity in CRD1, respectively, to HveA. Since mutations at position 70 of ELR1 are critical for activity, the inactivating mutant L70H ELR1 was also modeled, as was the analogous mutation in murine HveA (H70L), which fully supported receptor binding. Given the high sequence conservation, it is unsurprising that this domain in all models retained nearly identical overall structures to that of the CRD1 of HveA. All of the modeled structures retained similar backbone coordinates, with root mean square deviations differing less than 0.41 Å from the template, suggesting that the differing activities of the various constructs are not due to any change in the global fold of the domain. Closer examination of the β-turn containing L70 of ELR1 (Fig. 7A) shows that the surface-accessible volume of this loop is primarily defined by the acidic Glu at position 69 and the hydrophobic Leu at position 70. In all four models (Fig. 7A to D), the Glu is conserved. The most striking differences between the four model structures are the alterations in polarity and surface profile attributable to the Leu/His at position 70.
FIG. 7.
Schematic representation of ELR1, mHveA, and point mutation models. In all figures, the β-turn containing residues 68 to 71 is shown as a CPK model, with overlay of accessible surface area shown as white translucence. In all models, the Leu (A and D) or His (B and C) at position 70 projects outward at the top right of the figure. (A) ELR1; (B) murine HveA; (C) His mutant of ELR1; (D) Leu mutant of mHveA.
DISCUSSION
In the current study we have used domain exchange and point mutation strategies between the ELR1 protein with a high binding capacity for EIAV gp90 and the murine HveA protein, which lacks substantial gp90 binding capacity, despite a high degree of homology between the two TNFR family proteins. The results of these studies, employing both loss-of-function and gain-of-function assays, clearly identify the C-terminal half of the CRD1 segment of ELR1 as the critical domain mediating gp90 binding and Leu70 as the critical residue within the CRD1 necessary for functional binding of ELR1 to gp90. In addition, the data presented here demonstrate that the CRD2, CRD3, and CRD4 sequences in ELR1, while not essential for gp90 binding, can influence the level of gp90 binding, presumably through allosteric effects on overall receptor protein conformation and interactions with the gp90 protein.
The current observations suggest that the relatively small number of variant amino acid residues between the functional ELR1 and the nonfunctional murine HveA proteins, including L70, may actually represent contact points with the EIAV gp90, rather than producing marked differences in the conformation of the respective receptor proteins. In this regard, the homology modeling of CRD1 (Fig. 7) indicates remarkably similar conformations for the respective CRD1 segments of the ELR1 and murine HveA receptor proteins. The observation that no single point mutation in ELR1 completely abrogated gp90 binding suggests further that receptor binding to the viral envelope protein likely involves multiple contact points with residues within and outside of the CRD1 sequences. Thus, these data are consistent with other reports that multiple amino acid point mutations are required to completely eliminate receptor activity for FIV (11), avian sarcoma-leukosis virus (16, 17), human immunodeficiency virus (23), and murine leukemia virus (21).
A remarkable observation in these studies was that the single substitution of H70 in murine HveA to a Leu residue transformed the murine protein into a functional receptor for EIAV infection that was equivalent to ELR1. This finding allowed us to more closely examine the structural requirements for this critical residue to provide a functional receptor phenotype to the murine HveA protein. Our evaluation of His70 substitutions by various amino acids with different chemical properties revealed a strong influence of amino acid structure on gp90 binding properties, consistent with the concept of this residue providing a critical contact point with the viral envelope protein. Thus, substitutions of the His70 residue with amino acids containing aromatic side chains or charged side chains failed to produce gp90 binding, while substitutions with amino acids with hydrophobic alkyl side chains resulted in gp90 binding and functioned as a receptor for EIAV infection. The molecular modeling studies of the CRD1 domain indicate that the hydrophobic Leu in the β-turn projects outward, dominating the surface profile of this putative binding determinant. We interpret that the properties of this side chain are critical for binding to the receptor binding groove of the EIAV envelope. The results suggested that not only is hydrophobic important, but also the size and shape of the side chain for the amino acid at residue position 70 are critical for optimal envelope-receptor binding.
TNFR family molecules have been identified as cellular receptors for several viruses, such as herpes simplex virus, avian sarcoma-leukosis viruses, feline immunodeficiency virus and, most recently, equine infectious anemia virus (2, 6, 22, 35). It is intriguing that two lentiviruses, EIAV and FIV, both use receptor proteins belonging to the TNFR family. In the case of FIV-CD134 interactions, amino acids critical for envelope gp120 binding have been mapped to Asp60 and Asp62 of the CD134 protein (11). Comparison of the predicted three-dimensional structures for CD134 and ELR1 reveals that the CD134 Asp60 is at the same position as the Leu70 of ELR1. In addition, crystallographic studies of the binding interactions between herpes simplex virus (HSV) gD protein and its HveA receptor (7) have demonstrated a critical role for the C-terminal segment of CRD1 of the HveA protein for binding to the viral gD envelope protein (9, 34). These similarities between EIAV, FIV, and HSV envelope-receptor interactions appear to reveal common themes in the use of TNFR family proteins as virus receptors and also suggest common structural motifs in the envelope proteins of these diverse viruses that are utilized for receptor binding. In the cases of FIV and HSV, the natural ligand binding sites of the receptor proteins are distinct from their respective viral binding sites (11, 29, 34). The ligand specificity of ELR1 and its relationship to the EIAV gp90 binding site remain to be determined.
The use of various immune regulatory proteins by diverse lentiviruses for infection of target cells suggests a common required function, beyond the induction of viral fusogenicity, that is achieved by the interaction of the viral envelope proteins with these cell surface receptor molecules. We hypothesize that lentiviruses have evolved to utilize TNFR proteins and other immune regulatory proteins as receptors to achieve an activation of target cells, macrophages, and/or lymphocytes that results in enhanced levels of gene expression and virus replication in nondividing cellular targets. In this regard, it is interesting that visna virus binding and infection of target primary cells has been associated with a membrane serine/threonine kinase complex and a sequential activation of mitogen-activated protein kinase pathways that is required for visna virus replication; specific inhibitors of the mitogen-activated protein kinase pathway were shown to completely block visa virus replication (4, 5). Thus, more detailed studies of early downstream events resulting from lentiviral envelope binding to specific receptors may reveal critical cellular processes that can be evaluated as potential targets for antiviral drug development.
Acknowledgments
This research was supported by NIH grants 5R01 CA49296 from the National Cancer Institute and 9R56 AI073261 from the National Institutes of Allergy and Infectious Diseases.
We thank Tim Sturgeon and Jodi Craigo for titration of virus stocks and Berthony Deslouches for assistance with the cloning of the mouse herpesvirus mediator gene. We thank Shannon Barnes for excellent editorial review of the manuscript.
Footnotes
Published ahead of print on 21 November 2007.
REFERENCES
- 1.Adkins, H. B., J. Brojatsch, J. Naughton, M. M. Rolls, J. M. Pesola, and J. A. Young. 1997. Identification of a cellular receptor for subgroup E avian leukosis virus. Proc. Natl. Acad. Sci. USA 9411617-11622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adkins, H. B., J. Brojatsch, and J. A. Young. 2000. Identification and characterization of a shared TNFR-related receptor for subgroup B, D, and E avian leukosis viruses reveal cysteine residues required specifically for subgroup E viral entry. J. Virol. 743572-3578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alkhatib, G., C. Combadiere, C. C. Broder, Y. Feng, P. E. Kennedy, P. M. Murphy, and E. A. Berger. 1996. CC CKR5: a RANTES, MIP-1α, MIP-1β receptor as a fusion cofactor for macrophage-tropic HIV-1. Science 2721955-1958. [DOI] [PubMed] [Google Scholar]
- 4.Barber, S. A., L. Bruett, and J. E. Clements. 2000. Involvement of a membrane-associated serine/threonine kinase complex in cellular binding of visna virus. Virology 274321-330. [DOI] [PubMed] [Google Scholar]
- 5.Barber, S. A., L. Bruett, B. R. Douglass, D. S. Herbst, M. C. Zink, and J. E. Clements. 2002. Visna virus-induced activation of MAPK is required for virus replication and correlates with virus-induced neuropathology. J. Virol. 76817-828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brojatsch, J., J. Naughton, M. M. Rolls, K. Zingler, and J. A. Young. 1996. CAR1, a TNFR-related protein, is a cellular receptor for cytopathic avian leukosis-sarcoma viruses and mediates apoptosis. Cell 87845-855. [DOI] [PubMed] [Google Scholar]
- 7.Carfi, A., S. H. Willis, J. C. Whitbeck, C. Krummenacher, G. H. Cohen, R. J. Eisenberg, and D. C. Wiley. 2001. Herpes simplex virus glycoprotein D bound to the human receptor HveA. Mol. Cell 8169-179. [DOI] [PubMed] [Google Scholar]
- 8.Chou, C. F., S. Shen, Y. J. Tan, B. C. Fielding, T. H. Tan, J. Fu, Q. Xu, S. G. Lim, and W. Hong. 2005. A novel cell-based binding assay system reconstituting interaction between SARS-CoV S protein and its cellular receptor. J. Virol. Methods 12341-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Connolly, S. A., D. J. Landsburg, A. Carfi, D. C. Wiley, R. J. Eisenberg, and G. H. Cohen. 2002. Structure-based analysis of the herpes simplex virus glycoprotein D binding site present on herpesvirus entry mediator HveA (HVEM). J. Virol. 7610894-10904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cook, R. F., C. Leroux, S. J. Cook, S. L. Berger, D. L. Lichtenstein, N. N. Ghabrial, R. C. Montelaro, and C. J. Issel. 1998. Development and characterization of an in vivo pathogenic molecular clone of equine infectious anemia virus. J. Virol. 721383-1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Parseval, A., U. Chatterji, G. Morris, P. Sun, A. J. Olson, and J. H. Elder. 2004. Structural mapping of CD134 residues critical for interaction with feline immunodeficiency virus. Nat. Struct. Mol. Biol. 1260-66. [DOI] [PubMed] [Google Scholar]
- 12.de Parseval, A., U. Chatterji, P. Sun, and J. H. Elder. 2004. Feline immunodeficiency virus targets activated CD4+ T cells by using CD134 as a binding receptor. Proc. Natl. Acad. Sci. USA 10113044-13049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feng, Y., C. C. Broder, P. E. Kennedy, and E. A. Berger. 1996. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science 272872-877. [DOI] [PubMed] [Google Scholar]
- 14.Humphrey, W., A. Dalke, and K. Schulten. 1996. VMD: visual molecular dynamics. J. Mol. Graph. 1433-38. [DOI] [PubMed] [Google Scholar]
- 15.Jin, S., C. J. Issel, and R. C. Montelaro. 2004. Serological method using recombinant S2 protein to differentiate equine infectious anemia virus (EIAV)-infected and EIAV-vaccinated horses. Clin. Diagn. Lab. Immunol. 111120-1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Klucking, S., and J. A. Young. 2004. Amino acid residues Tyr-67, Asn-72, and Asp-73 of the TVB receptor are important for subgroup E avian sarcoma and leukosis virus interaction. Virology 318371-380. [DOI] [PubMed] [Google Scholar]
- 17.Knauss, D. J., and J. A. Young. 2002. A fifteen-amino-acid TVB peptide serves as a minimal soluble receptor for subgroup B avian leukosis and sarcoma viruses. J. Virol. 765404-5410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lichtenstein, D. L., K. E. Rushlow, R. F. Cook, M. L. Raabe, C. J. Swardson, G. J. Kociba, C. J. Issel, and R. C. Montelaro. 1995. Replication in vitro and in vivo of an equine infectious anemia virus mutant deficient in dUTPase activity. J. Virol. 692881-2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu, Q., E. C. Thorland, J. A. Heit, and S. S. Sommer. 1997. Overlapping PCR for bidirectional PCR amplification of specific alleles: a rapid one-tube method for simultaneously differentiating homozygotes and heterozygotes. Genome Res. 7389-398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Locksley, R. M., N. Killeen, and M. J. Lenardo. 2001. The TNF and TNF receptor superfamilies: integrating mammalian biology. Cell 104487-501. [DOI] [PubMed] [Google Scholar]
- 21.Lundorf, M. D., F. S. Pedersen, B. O'Hara, and L. Pedersen. 1999. Amphotropic murine leukemia virus entry is determined by specific combinations of residues from receptor loops 2 and 4. J. Virol. 733169-3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marsters, S. A., T. M. Ayres, M. Skubatch, C. L. Gray, M. Rothe, and A. Ashkenazi. 1997. Herpesvirus entry mediator, a member of the tumor necrosis factor receptor (TNFR) family, interacts with members of the TNFR-associated factor family and activates the transcription factors NF-κB and AP-1. J. Biol. Chem. 27214029-14032. [DOI] [PubMed] [Google Scholar]
- 23.Moebius, U., L. K. Clayton, S. Abraham, S. C. Harrison, and E. L. Reinherz. 1992. The human immunodeficiency virus gp120 binding site on CD4: delineation by quantitative equilibrium and kinetic binding studies of mutants in conjunction with a high-resolution CD4 atomic structure. J. Exp. Med. 176507-517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Montelaro, R. C., J. M. Ball, and K. E. Rushlow. 1993. Equine retroviruses, p. 257-360. In J. Levy (ed.), The Retroviridae. Plenum Press, New York, NY.
- 25.Montgomery, R. I., M. S. Warner, B. J. Lum, and P. G. Spear. 1996. Herpes simplex virus-1 entry into cells mediated by a novel member of the TNF/NGF receptor family. Cell 87427-436. [DOI] [PubMed] [Google Scholar]
- 26.Naismith, J. H., and S. R. Sprang. 1998. Modularity in the TNF-receptor family. Trends Biochem. Sci. 2374-79. [DOI] [PubMed] [Google Scholar]
- 27.Ono, E., S. Yoshino, K. Amagai, S. Taharaguchi, C. Kimura, J. Morimoto, M. Inobe, T. Uenishi, and T. Uede. 2004. Enhanced resistance to herpes simplex virus type 1 infection in transgenic mice expressing a soluble form of herpesvirus entry mediator. Virology 320267-275. [DOI] [PubMed] [Google Scholar]
- 28.Sali, A., and T. L. Blundell. 1993. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 234779-815. [DOI] [PubMed] [Google Scholar]
- 29.Sarrias, M. R., J. C. Whitbeck, I. Rooney, C. F. Ware, R. J. Eisenberg, G. H. Cohen, and J. D. Lambris. 2000. The three HveA receptor ligands, gD, LT-alpha and LIGHT bind to distinct sites on HveA. Mol. Immunol. 37665-673. [DOI] [PubMed] [Google Scholar]
- 30.Shimojima, M., T. Miyazawa, Y. Ikeda, E. L. McMonagle, H. Haining, H. Akashi, Y. Takeuchi, M. J. Hosie, and B. J. Willett. 2004. Use of CD134 as a primary receptor by the feline immunodeficiency virus. Science 3031192-1195. [DOI] [PubMed] [Google Scholar]
- 31.Smith, C. A., T. Farrah, and R. G. Goodwin. 1994. The TNF receptor superfamily of cellular and viral proteins: activation, costimulation, and death. Cell 76959-962. [DOI] [PubMed] [Google Scholar]
- 32.Strausberg, R. L., E. A. Feingold, L. H. Grouse, J. G. Derge, R. D. Klausner, F. S. Collins, L. Wagner, C. M. Shenmen, G. D. Schuler, S. F. Altschul, B. Zeeberg, K. H. Buetow, C. F. Schaefer, N. K. Bhat, R. F. Hopkins, H. Jordan, T. Moore, S. I. Max, J. Wang, F. Hsieh, L. Diatchenko, K. Marusina, A. A. Farmer, G. M. Rubin, L. Hong, M. Stapleton, M. B. Soares, M. F. Bonaldo, T. L. Casavant, T. E. Scheetz, M. J. Brownstein, T. B. Usdin, S. Toshiyuki, P. Carninci, C. Prange, S. S. Raha, N. A. Loquellano, G. J. Peters, R. D. Abramson, S. J. Mullahy, S. A. Bosak, P. J. McEwan, K. J. McKernan, J. A. Malek, P. H. Gunaratne, S. Richards, K. C. Worley, S. Hale, A. M. Garcia, L. J. Gay, S. W. Hulyk, D. K. Villalon, D. M. Muzny, E. J. Sodergren, X. Lu, R. A. Gibbs, J. Fahey, E. Helton, M. Ketteman, A. Madan, S. Rodrigues, A. Sanchez, M. Whiting, A. Madan, A. C. Young, Y. Shevchenko, G. G. Bouffard, R. W. Blakesley, J. W. Touchman, E. D. Green, M. C. Dickson, A. C. Rodriguez, J. Grimwood, J. Schmutz, R. M. Myers, Y. S. Butterfield, M. I. Krzywinski, U. Skalska, D. E. Smailus, A. Schnerch, J. E. Schein, S. J. Jones, and M. A. Marra. 2002. Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences. Proc. Natl. Acad. Sci. USA 9916899-16903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Unutmaz, D., V. N. KewalRamani, and D. R. Littman. 1998. G protein-coupled receptors in HIV and SIV entry: new perspectives on lentivirus-host interactions and on the utility of animal models. Semin. Immunol. 10225-236. [DOI] [PubMed] [Google Scholar]
- 34.Whitbeck, J. C., S. A. Connolly, S. H. Willis, W. Hou, C. Krummenacher, M. Ponce de Leon, H. Lou, I. Baribaud, R. J. Eisenberg, and G. H. Cohen. 2001. Localization of the gD-binding region of the human herpes simplex virus receptor, HveA. J. Virol. 75171-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang, B., S. Jin, J. Jin, F. Li, and R. C. Montelaro. 2005. A tumor necrosis factor receptor family protein serves as a cellular receptor for the macrophage-tropic equine lentivirus. Proc. Natl. Acad. Sci. USA 1029918-9923. [DOI] [PMC free article] [PubMed] [Google Scholar]