Abstract
The Autographa californica multicapsid nucleopolyhedrovirus (AcMNPV) GP64 envelope glycoprotein is essential for virus entry and plays an important role in virion budding. An AcMNPV construct that contains a deletion of the gp64 gene is unable to propagate infection from cell to cell, and this defect results from both a severe reduction in the production of budded virions and the absence of GP64 on virions. In the current study, we examined GP64 proteins containing N- and C-terminal truncations of the ectodomain and identified a minimal construct capable of targeting the truncated GP64 to budded virions. The minimal budding and targeting construct of GP64 contained 38 amino acids from the mature N terminus of the GP64 ectodomain and 52 amino acids from the C terminus of GP64. Because the vesicular stomatitis virus (VSV) G protein was previously found to rescue infectivity of a gp64null AcMNPV, we also examined a small C-terminal construct of the VSV G protein. We found that a construct containing 91 amino acids from the C terminus of VSV G (termed G-stem) was capable of rescuing AcMNPV gp64null virion budding to wild-type (wt) or nearly wt levels. We also examined the display of chimeric proteins on the gp64null AcMNPV virion. By generating viruses that expressed chimeric influenza virus hemagglutinin (HA) proteins containing the GP64 targeting domain and coinfecting those viruses with a virus expressing the G-stem construct, we demonstrated enhanced display of the HA protein on gp64null AcMNPV budded virions. The combined use of gp64null virions, VSV G-stem-enhanced budding, and GP64 domains for targeting heterologous proteins to virions should be valuable for biotechnological applications ranging from targeted transduction of mammalian cells to vaccine production.
Baculoviruses are enveloped viruses that contain large circular double-stranded DNA genomes ranging from approximately 80 to 180 kbp (37). Baculoviruses such as Autographa californica multicapsid nucleopolyhedrovirus (AcMNPV) have been studied as agents for biological control of insect pests, as expression vectors for high-level production of heterologous proteins, and as transduction vectors and potential agents for human gene therapy (14, 21). AcMNPV produces two virion phenotypes during the infection cycle (1, 7). One virion phenotype, the occlusion-derived virus, is adapted for stability in the environment and serves to propagate infection from animal to animal through oral transmission and infection of the midgut epithelial cells. The occlusion-derived virus assembles in the nucleus and contains an envelope derived from the inner nuclear membrane (5). In contrast, the other virion phenotype, the budded virus (BV), is adapted for propagation of infection from cell to cell throughout the animal after infection is established in the midgut. BV assembles at the plasma membrane as nucleocapsids bud through that membrane and acquire the envelope. In the case of AcMNPV, the BV contains two virus-encoded envelope proteins, GP64 and Ac23 (2, 11, 17, 31, 43).
GP64 is the major envelope glycoprotein of AcMNPV BV. GP64 is a type I integral membrane protein that is highly abundant on the BV envelope. Along with the major capsid protein (VP39), GP64 comprises one of the most abundant virion proteins (27). Native GP64 is glycosylated, phosphorylated, and acylated and is found in the virion as a disulfide-linked homotrimer (28, 33, 39, 45). BV enter cells by receptor-mediated endocytosis, and functional studies indicate that GP64 is involved in two major steps in BV entry, i.e., virion attachment and membrane fusion (3, 10, 15). After binding and endocytosis, the fusion activity of GP64 is triggered by low pH in the endosome, leading to fusion of the BV envelope and the endosome membrane and release of the nucleocapsid into the cytoplasm. AcMNPV also encodes and expresses a homolog of baculovirus F proteins, envelope proteins that are found in all known lepidopteran baculoviruses. In the group II NPVs, such as Spodoptera exigua MNPV and Lymantria dispar MNPV, F proteins serve as membrane fusion proteins (12, 29-31, 41, 42), and pseudotyping studies have shown that they are functional homologs of AcMNPV GP64 (16). However, the F protein homolog (Ac23) found in AcMNPV (and other group I NPVs, such as Orgyia pseudotsugata MNPV [OpMNPV]) does not appear to be a functional fusion protein (17, 31), and deletion of the AcMNPV Ac23 gene has no substantial effect on virus production or infectivity in insect cell culture (17). In striking contrast, deletion of the AcMNPV gp64 gene is lethal, and no infectious virus is generated in the absence of GP64 (22, 27). In addition, a well-characterized anti-GP64 monoclonal antibody (MAb), AcV1, neutralizes viral infectivity (11, 40, 46).
A number of prior studies have examined protein display on AcMNPV BV, and almost all studies were performed in the presence of wild-type (wt) GP64 (4, 6, 8, 9, 18, 23, 24, 35, 36, 38, 44). While the presence of GP64 on virions will be advantageous for some applications, other applications, such as those requiring specific cell targeting or binding, may not be feasible in the presence of wt GP64 because of its ability to promiscuously mediate binding and entry. In addition, baculovirus virions may also be used for displaying proteins for vaccine production, and in those applications, it would be desirable to eliminate the abundant wt baculovirus envelope protein.
In prior studies of a gp64null virus, it was found that virion production is severely reduced in the absence of GP64, and virions that are produced are not infectious (27). Thus, GP64 is critical for both virion production and entry. Interestingly, these critical functions of GP64 can be replaced by pseudotyping gp64null AcMNPV with certain heterologous viral envelope proteins (16, 19). When the AcMNPV GP64 protein was replaced with F protein from S. exigua MNPV or L. dispar MNPV or with the vesicular stomatitis virus (VSV) G protein, infectious virions were produced, and these pseudotyped viruses could be propagated in Sf9 cells. However, not all viral envelope proteins can substitute for GP64 (16). Thus, although some viral proteins can substitute for the budding function of GP64, the lack of efficient budding by the gp64null virus presents a substantial constraint on the potential use of pseudotyped gp64null viruses in experimental systems and for biotechnological applications such as surface display, targeted transduction, and gene therapy.
To reconstitute efficient budding in the absence of GP64, we examined a truncated form of the VSV G protein. We previously found that VSV G-pseudotyped gp64null viruses replicated and that BV were generated at high titers in Sf9 cells (16, 19), suggesting that VSV G mediated efficient budding. In prior studies, it was shown that a truncated form of the VSV G protein was capable of reconstituting budding in a G-null VSV (34). This truncated VSV G protein was comprised of a small segment from the C-terminal portion of the ectodomain plus the transmembrane (TM) and cytoplasmic tail (CTD) domains of VSV G. In the current study, we generated a similar VSV G construct that contained an N-terminal c-Myc epitope plus 42 amino acids from the C-terminal portion of the ectodomain, 20 amino acids from the predicted TM domain, and 29 amino acids from the predicted CTD of the VSV G protein (called a G-stem construct). To determine if the G-stem construct could rescue the budding defect of gp64null AcMNPV, a gene encoding G-stem was cloned into gp64null AcMNPV. We found that expression of the VSV G-stem construct resulted in the efficient production of virions, thus rescuing the budding defect observed in the gp64null virus. We also examined methods for efficiently targeting proteins to BV produced in this manner. Using truncated forms of GP64, we mapped regions necessary for efficient targeting of GP64 to the virion. By fusing heterologous proteins either to portions of GP64 or to G-stem constructs, heterologous proteins were targeted to the gp64null virions. Thus, we found that the G-stem construct rescues virion budding in gp64null AcMNPV and can be used in combination with protein fusions to target proteins to gp64null AcMNPV virions.
MATERIALS AND METHODS
Expression of protein constructs in gp64null AcMNPV.
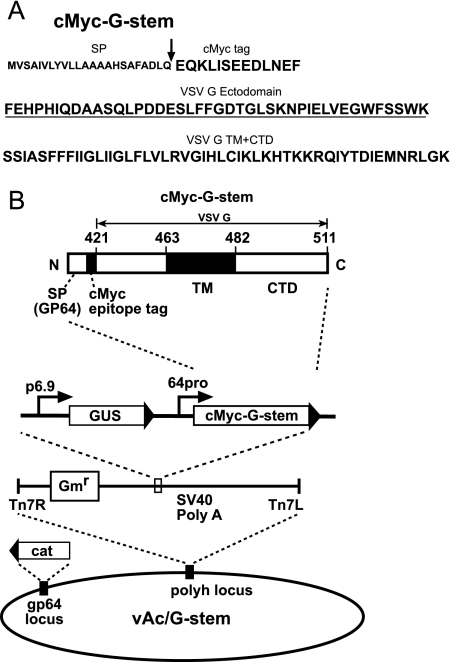
The gp64 gene of an AcMNPV bacmid (bMON14272; Invitrogen) was deleted from the AcMNPV genome as reported previously (16). The resulting bacmid was used to produce a gp64null virus, vAcgp64−, by propagating the virus in Sf9Op1D cells, which constitutively express the OpMNPV GP64 protein (20, 25). To express a VSV G-stem construct in the context of gp64null AcMNPV, we generated a donor plasmid construct designated pFBcMyc-G-stem. A truncated version of the VSV G protein that included 42 amino acids from the C terminus of the ectodomain plus the TM domain and the CTD was generated by PCR-mediated mutagenesis in the following manner. A forward primer with an EcoRI restriction site engineered into the 5′ end (5′-AAT GAATTC TTC GAA CAT CCT CAC ATT CAA-3′) was used in combination with a reverse primer that contained an XbaI site (5′-AA TCTAGA TTA CTT TCC AAG TCG GTT CAT CTC TAT-3′) to amplify the “stem” portion of the VSV G gene from a wt VSV G DNA template (pSM8141-VSV-G) (19). The PCR product was digested with EcoRI and XbaI, purified, and ligated into the EcoRI and XbaI sites of vector pdFB-gp64sig-cMyc, a pFastBac-derived plasmid containing the promoter, signal peptide, and signal cleavage site from the AcMNPV gp64 gene, followed by a c-Myc epitope tag and a cloning site (Fig. 1B) (46). The resulting construct was named pFBcMyc-G-stem. The truncated G-stem form of the VSV G gene carried by plasmid pFBcMyc-G-stem encodes an N-terminal c-Myc epitope tag linked by a Phe residue to the truncated VSV G-stem protein. Transposition of inserts from donor plasmids into the gp64null bacmid (14) and detection by gentamicin resistance and blue-white colony screening were performed as described in the Bac-to-Bac manual (Invitrogen) and were confirmed by PCR analysis and DNA sequencing. Cells stably expressing OpMNPV GP64 (cell line Sf9Op1D) (27, 32) were transfected with the bacmid DNA, and the resulting virus was harvested from cell supernatants and titrated on Sf9Op1D cells. The resulting virus was designated vAc/G-stem.
FIG. 1.
(A) Amino acid sequence of G-stem protein construct. The signal peptide (SP) derived from AcMNPV GP64 is shown in small bold letters, and the signal cleavage site is indicated by an arrow. The c-Myc tag, ectodomain, TM domain, and CTD sequences of the G-stem construct are shown in large bold letters, and ectodomain sequences derived from VSV G are underlined. The bottom line shows TM domain and CTD sequences. (B) Strategy for insertion of the VSV G-stem construct into the polyhedrin locus of a gp64null AcMNPV bacmid. The cassette inserted into the gp64null bacmid includes a p6.9 promoter-GUS reporter plus sequences encoding a truncated VSV G protein (VSV G-stem) under the control of the gp64 promoter (64pro). The VSV G-stem construct encodes the gp64 signal peptide, followed by a c-Myc tag and a truncated version of the VSV G protein that includes 42 amino acids of the C-terminal portion of the VSV G ectodomain plus the TM domain and the CTD of VSV-G. Numbers above the c-Myc-G-stem cassette (421 to 511) are amino acid sequence numbers for the VSV G protein. G-stem fusion protein gene cassettes were inserted by Tn7-based transposition into the polyhedrin locus of a gp64null AcMNPV bacmid derived from bMON14272.
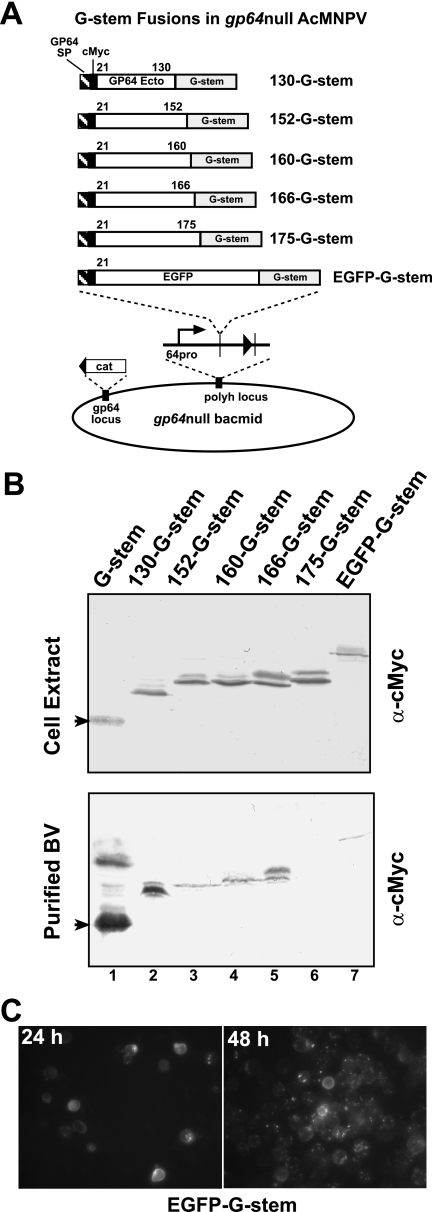
For analysis of the G-stem construct as a fusion partner for targeting heterologous proteins to AcMNPV BV, we constructed a series of plasmids in which C-terminal truncations of GP64 or the enhanced green fluorescent protein (EGFP) coding region was cloned in frame between the c-Myc epitope and VSV G-stem domains (see Fig. 4A) of plasmid pFBcMyc-G-stem. DNA fragments encoding C-terminally truncated portions of the GP64 ectodomain were PCR amplified from a wt AcMNPV DNA template. A single forward primer (EcoR57 Forward) with an EcoRI restriction site engineered into the 5′ end (5′-AA GAATTC GCG GAG CAC TGC AAC GCG-3′ [corresponding to a sequence 57 bp downstream of the gp64 start codon]) was used in combination with a downstream primer specific to each truncation. Each downstream primer also contained an EcoRI site engineered for in-frame insertion of the AcMNPV gp64 gene into vector pFBcMyc-G-stem. Each PCR product was digested with EcoRI, purified, and ligated into the EcoRI sites of vector pFBcMyc-G-stem to generate a truncated GP64 open reading frame (ORF) fused in frame at the N terminus of the VSV G-stem. To confirm the orientation of the insertions, each construct was analyzed by PCR and by sequencing. Recombinant viruses were produced from a gp64null AcMNPV bacmid as described above, and the viruses were designated vAc/130-G-stem, vAc/152-G-stem, vAc/160-G-stem, vAc/166-G-stem, vAc/175-G-stem, and vAc/EGFP-G-stem.
FIG. 4.
Display of GP64 peptides or EGFP on gp64null virions, using G-stem fusion proteins. (A) Strategy for insertion of VSV G-stem fusion protein gene constructs into the polyhedrin locus of the gp64null AcMNPV bacmid. Constructs encoding peptides derived from the GP64 protein ectodomain (labeled 130, 152, 160, 166, and 175) or EGFP were fused to the G-stem fusion protein and expressed under the control of the gp64 promoter. Each construct contains the GP64 signal peptide and cleavage site followed by the c-Myc epitope tag and either a portion of the GP64 ectodomain or the EGFP ORF. The cassettes were inserted into the bacmid as described in the legend to Fig. 1. (B) Western blot analysis of infected cell lysates and BV preparations. Cell extracts or purified BV preparations derived from Sf9 cells infected with the gp64null viruses expressing G-stem fusion constructs were examined for protein expression by using an anti-c-Myc antibody. (C) Immunofluorescence of Sf9 cells infected with the EGFP-G-stem virus. Micrographs show fluorescence from infected cells at 24 and 48 hpi.
Constructs encoding N-terminal truncations of the GP64 ectodomain were described previously (46) (see Fig. 3A). These constructs express proteins containing an N-terminal c-Myc epitope tag linked by a Phe residue to a portion of the GP64 ectodomain truncated at the N terminus of the mature GP64 protein.
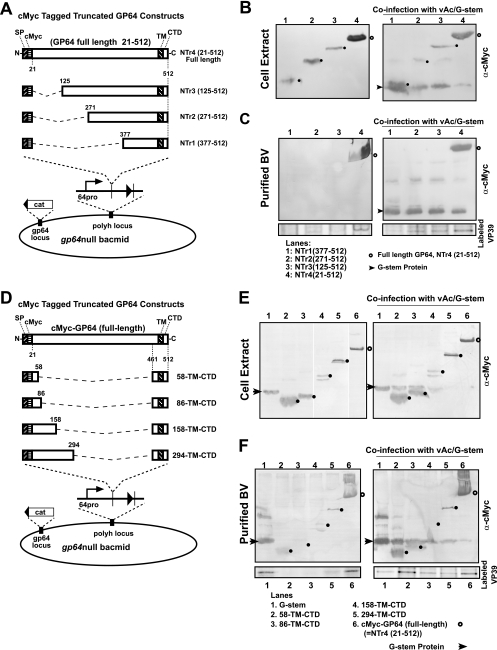
FIG. 3.
Analysis of expression, display, and budding by GP64 constructs containing truncations of the ectodomain. GP64 ectodomain truncations from either the N or C terminus and their insertion into gp64null AcMNPV are shown in panels A and D, and Western blot analyses of infected Sf9 cells or purified BV are shown in the panels on the right. (A) Truncations from the N terminus of the GP64 ectodomain are illustrated. Each construct contains the 20-amino-acid GP64 signal peptide followed by a c-Myc epitope tag. Various portions of the GP64 ectodomain downstream of amino acid 21 were deleted. (B) Western blot analysis of cell extracts from cells infected with viruses expressing fusion proteins containing a truncated GP64 protein (left) or coinfected with viruses expressing N-terminally truncated proteins and a virus expressing the G-stem protein (right). Small closed circles show the positions of the truncated proteins, and open circles show the position of the control construct, which contained the full-length GP64 protein ectodomain (right panel). (C) Western blot analysis of purified BV from cells infected with viruses expressing N-terminally truncated proteins (left) or coinfected with viruses expressing N-terminally truncated proteins and a virus expressing the VSV G-stem construct (right). The bottom panels show relative levels of VP39 detected from purified progeny virions labeled with [35S]methionine as described in Materials and Methods and detected by phosphorimager analysis of labeled proteins. (D) Strategy for construction of GP64 proteins with C-terminal ectodomain deletions. Each construct contained the GP64-stem domain and various portions of the GP64 ectodomain, as illustrated. The GP64-stem domain consists of amino acids 461 to 512 from the C terminus of the AcMNPV GP64 protein, consisting of 22 residues from the predicted GP64 ectodomain, 23 residues from the predicted GP64 TM domain, and the 7-residue CTD. Various portions of the GP64 ectodomain were fused to the GP64-stem such that in the mature protein, the stem construct was fused with a c-Myc epitope tag and 38 (residues 21 to 58), 66 (residues 21 to 86), 138 (residues 21 to 158), or 274 (residues 21 to 294) amino acids from the N terminus of the GP64 ectodomain. (E) Western blot analysis of cell extracts from cells infected with viruses expressing fusion proteins containing the GP64-stem and C-terminal deletions of the ectodomain (left). The right panel shows results from a similar experiment in which cells were coinfected with viruses expressing the same C-terminal GP64 ectodomain constructs and a virus expressing the VSV G-stem construct. (F) Western blot analysis of purified BV from cells infected with viruses expressing fusion proteins containing the GP64-stem and C-terminal deletions of the ectodomain (left) or coinfected with viruses expressing the same C-terminal GP64 ectodomain constructs and a virus expressing the VSV G-stem construct (right). The bottom panels show relative levels of VP39 detected from purified progeny virions labeled with [35S]methionine as described in Materials and Methods and detected by phosphorimager analysis of labeled proteins. Note that because of the low production of virions detected by Western blot analyses (see above), the construct containing 138 (residues 21 to 158) amino acids from the N terminus of the GP64 ectodomain was not included in the virion labeling experiment (panel F, lane 4, 158-TM-CTD).
Analysis of progeny virion production by [35S]methionine labeling.
Progeny virions from cells infected with vAcgp64−/Acgp64 (16), vAcgp64− (16), or vAc/G-stem were labeled with [35S]methionine in the following manner. Sf9 cells (1 × 107 cells) were plated in a T-25 flask (Corning Inc.), allowed to attach for 1 h, and then infected at a multiplicity of infection (MOI) of 10 for 1 h. At 29 h postinfection (hpi), the cells were starved by incubation in 3 ml methionine-free Grace's medium (Invitrogen) for 1 h, followed by the addition of 35S-EasyTag Express protein labeling mix (1,175.0 Ci/mmol; Perkin-Elmer) to a final concentration of 10 μCi/ml. At 37 hpi, unlabeled methionine was added to a final concentration of 10 mM, and cells were incubated at 27°C for an additional 48 h. Virions were harvested from supernatants and purified by being pelleted through a 25% sucrose cushion at 100,000 × g for 90 min at 4°C in a Beckman SW60 rotor. Virus pellets were resuspended in 300 μl phosphate-buffered saline (pH 6.2).
Construction of plasmids and baculoviruses encoding constructs containing a GP64-stem region.
A series of plasmids encoding the C-terminal GP64 region (the GP64-stem region) and various portions of the GP64 ectodomain was generated using the following strategy. First, DNA fragments containing various portions of the GP64 ORF were PCR amplified from a wt AcMNPV DNA template. The forward primer EcoR57 Forward was used in combination with a downstream primer specific for each truncation. Each downstream primer contained a KpnI site engineered for in-frame insertion of the AcMNPV gp64 gene into vector pFB-gp64sig-cmyc-TM-CTD, a pFastBac-derived plasmid containing the gp64 promoter and gene sequence encoding the signal peptide and cleavage site, a c-Myc tag, and a cloning site, followed by the coding region for 21 amino acids from the C terminus of the GP64 ectodomain plus the GP64 TM domain and CTD. Each PCR product was digested with EcoRI and KpnI and ligated into the EcoRI and KpnI sites of vector pFB-gp64sig-cmyc-TM-CTD to generate a series of constructs, with each containing the GP64-stem and various portions of the GP64 ectodomain (see Fig. 3D). Thus, each construct expresses a protein that contains an N-terminal c-Myc tag, a portion of the mature N-terminal region of the GP64 protein, and the so-called GP64-stem region. Each construct was confirmed by sequencing. Recombinant viruses were generated in a gp64null AcMNPV bacmid and produced in Sf9Op1D cells as described above. The viruses were designated vAc/58-TM-CTD, vAc/86-TM-CTD, vAc/158-TM-CTD, and vAc/294-TM-CTD (see Fig. 3D).
Coinfections.
For coinfection experiments, Sf9 cells (1 × 105 cells per well in six-well plates) were coinfected at a total MOI of 10 with equal amounts of the vAc/G-stem virus (MOI of 5) and one additional virus (also at an MOI of 5). Viruses used for coinfections included those illustrated in Fig. 3A and B and 5A. Progeny virions derived from coinfection experiments were purified and analyzed as described above.
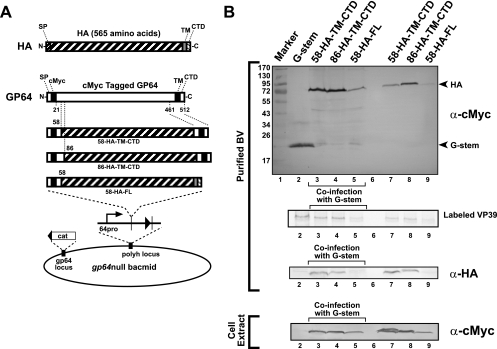
FIG. 5.
Targeting of HA fusions to AcMNPV BV and enhanced budding by G-stem. (A) Strategy for generating HA-GP64 fusions and insertion of chimeric HA constructs into the polyhedrin locus of a gp64null AcMNPV bacmid. Chimeric HA-GP64 protein constructs were generated by fusing the HA ectodomain (residues 18 to 528) with the GP64 signal peptide, various portions from the N terminus of the GP64 ectodomain, and the GP64-stem region or the native HA TM domain and CTD. Constructs were expressed under the control of the gp64 promoter. Chimeric HA-GP64 proteins each contained an N-terminal c-Myc epitope tag, either 38 or 66 residues of the GP64 ectodomain (at the N terminus), and a 91-amino-acid GP64-stem sequence at the C terminus (constructs 58-HA-TM-CTD and 86-HA-TM-CTD). Construct 58-HA-FL contained a c-Myc epitope tag, 38 amino acids from the N terminus of the GP64 ectodomain, and the HA ectodomain, TM domain, and CTD (residues 18 to 565). (B) Western blot analysis of BV preparations. Purified BV preparations derived from cells infected with viruses expressing HA fusion proteins (lanes 7, 8, and 9) or coinfected with viruses expressing HA fusion proteins and a virus expressing the VSV G-stem construct (lanes 3, 4, and 5) were examined for the presence of envelope protein constructs by use of an anti-c-Myc antibody (purified BV, top panel) or an anti-HA antibody (purified BV, bottom panel). VP39 from purified [35S]methionine-labeled progeny BV (purified BV, middle panel) was detected by phosphorimager analysis and used to more directly compare levels of progeny BV production. A control infection with only the virus expressing the c-Myc-tagged G-stem construct is shown in lane 2. Cell extracts from the above preparations were also examined for protein expression by using anti-c-Myc antibody (bottom).
Western blot analysis.
Cell lysates were prepared by washing cultured cells with phosphate-buffered saline and resuspending cells in NET buffer (20 mM Tris, pH 7.5, 150 mM NaCl, 0.5% deoxycholate, 1.0% Nonidet P-40, 1 mM EDTA) to which a protease inhibitor cocktail (Complete; Roche Applied Science) had been added according to the manufacturer's instructions. This NET buffer solution (500 μl) was added to 1 × 106 cells and incubated for 30 min at 4°C, and then nuclei were removed by pelleting at 4°C for 10 min at 18,000 × g. Virus purification was performed as described above. For Western blot analysis, 10 μl of the cell lysate or purified virus was mixed with 10 μl of 2× Laemmli buffer (125 mM Tris, 2% sodium dodecyl sulfate [SDS], 5% 2-mercaptoethanol, 10% glycerol, 0.001% bromophenol blue, pH 6.8) and heated to 100°C for 5 min prior to SDS-10% polyacrylamide gel electrophoresis (SDS-PAGE). Gels were blotted onto Immobilon-P membranes (Millipore) and blocked overnight at 4°C in TBST (25 mM Tris, pH 7.6, 150 mM NaCl, 0.1% Tween 20, 5% powdered milk). Blots were incubated for 1 h at room temperature with the following primary antibodies diluted in TBST: anti-c-Myc MAb (hybridoma supernatant) diluted 1:1,000, anti-VP39 MAb diluted 1:1,000, anti-hemagglutinin (anti-HA) (chicken polyclonal antiserum; a gift from Gary Whittaker) diluted 1:100, and anti-VSV G (MAb P5D4) diluted 1:10,000. After being washed three times in TBST, blots were incubated with a secondary antibody consisting of an alkaline phosphatase-conjugated goat anti-mouse or goat anti-chicken immunoglobulin G (IgG; Promega) at a dilution of 1:10,000. Western blots were processed as described earlier.
Construction of recombinant baculoviruses expressing chimeric HA proteins.
The portion of the influenza A/WSN/33 virus HA gene encoding the ectodomain (amino acids 18 to 528) was PCR amplified from plasmid pEWSN-HA (25). A forward primer (Kpn-HA Forward) with a KpnI restriction site engineered into the 5′ end (5′-AA GGTACC GAC ACA ATA TGT ATA GGC TAC CAT GCG AAC AAC TC-3′ [which includes a sequence immediately downstream of the HA signal peptide]) was used in combination with a downstream primer. The downstream primer (5′-AA GGTACC CTG ATA CAC CCC CAT TGA TTC CAA TTT C-3′) contained a KpnI site engineered for in-frame insertion of the HA gene into vectors pFB-gp64sig-cmyc-58-TM-CTD and pFB-gp64sig-cmyc-86-TM-CTD, which are pFastBac-derived plasmids containing the gp64 promoter and a sequence encoding the gp64 signal peptide and cleavage site, a c-Myc tag, 38 or 66 amino acids of the GP64 N-terminal ectodomain, and a KpnI cloning site, followed by 21 amino acids from the GP64 C-terminal ectodomain and the GP64 TM domain and GP64 CTD. The PCR product was digested with KpnI and ligated into the KpnI sites of vectors pFB-gp64sig-cmyc-58-TM-CTD and pFB-gp64sig-cmyc-86-TM-CTD to generate constructs containing the HA ectodomain and the GP64-stem. The resulting constructs were designated pFB-58-HA-TM-CTD and pFB-86-HA-TM-CTD, respectively. Thus, each construct expresses a protein that contains an N-terminal c-Myc tag, a portion of the mature N-terminal region of the GP64 protein, the HA ectodomain, and the GP64-stem region (see Fig. 5A). In addition, a construct containing the GP64 promoter and signal peptide combined with the HA ectodomain, TM domain, and CTD (HA amino acids 18 to 565) was also generated by first PCR amplifying the downstream portion of the HA gene from plasmid pEWSN-HA (25) and then inserting it into a pFastbac plasmid. The HA sequences were amplified using the same forward primer (Kpn-HA Forward) in combination with a downstream primer (5′-GAC AAGCTT catca GAT GCA TAT TCT GCA CTG CAA AGA CC-3′). The downstream primer contained a HindIII site engineered for insertion of the HA gene into vector pFB-gp64sig-cmyc-58-TM-CTD or pFB-gp64sig-cmyc-86-TM-CTD. The resulting plasmids, pFB-58-HA-FL and pFB-86-HA-FL, encode proteins that contain an N-terminal c-Myc tag, a portion of the mature N-terminal region of the GP64 protein, the HA ectodomain, and the HA TM domain and CTD region (see Fig. 5A).
Fluorescence microscopy.
Sf9 cells (1 × 105) seeded into six-well plates were infected at an MOI of 10 with the vAc/EGFP-G-stem virus expressing EGFP-G-stem and incubated for 24 or 48 h. Epifluorescence microscopy was performed with an inverted IX70 microscope (Olympus).
RESULTS
Rescue of gp64null AcMNPV budding by a VSV G-stem construct.
AcMNPV baculoviruses containing a knockout of the gp64 gene are unable to produce virions efficiently in the absence of the GP64 protein (27). To examine the requirements for virion budding, we initially generated a number of N-terminally truncated forms of the GP64 protein and examined them in budding assays, but we were unable to identify a truncated GP64 construct that rescued the budding defect. For rhabdovirus systems, it was previously shown that virion budding from G knockout viruses was dramatically reduced (20, 34). However, in the VSV system, a severely truncated form of the VSV G protein containing only a small portion of the C terminus of G rescues the VSV budding defect (34). Baculoviruses and rhabdoviruses are unrelated and represent vastly different viral infection systems. However, both bud from the plasma membrane, and we previously found that the VSV G protein was able to rescue infectivity of a gp64null baculovirus (16, 19). Therefore, we asked whether a similar small portion of the VSV G protein might restore efficient virion budding in gp64null AcMNPV.
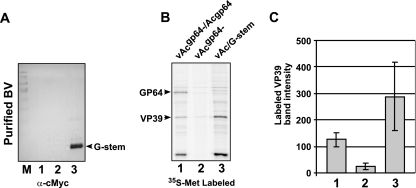
In the current study, we generated a truncated VSV G gene construct and inserted it into a gp64null AcMNPV genome under the control of the baculovirus AcMNPV gp64 promoter. The truncated VSV G construct contained an AcMNPV gp64 promoter and signal peptide, a c-Myc epitope tag (at the N terminus of the mature protein), 42 amino acids from the C terminus of the VSV G ectodomain (positions 421 to 463), and the predicted TM domain and CTD of VSV G (Fig. 1A). This N-terminally truncated VSV G construct which contains a c-Myc epitope tag was designated G-stem, and construction of the baculovirus bacmid expressing this construct is summarized in Fig. 1B. The resulting bacmid DNA was used to generate a virus, designated vAc/G-stem, by transfecting the bacmid DNA into a stable cell line (Sf9Op1D) that constitutively expresses the wt OpMNPV GP64 protein (32). To determine whether G-stem enhanced virion budding, we performed an AcMNPV budding assay (27) (Fig. 2). Sf9 cells were infected with control or G-stem-expressing gp64null viruses (all previously generated and titrated in Sf9Op1D cells), and progeny virions were metabolically labeled with [35S]methionine. Supernatants containing labeled progeny virions were collected, and virions were purified by being pelleted through 25% sucrose. In this assay, BV are isolated from the supernatant, and only progeny virions are labeled. Progeny virions were isolated in this manner from Sf9 cells infected with viruses vAcgp64− (a gp64null virus), vAcgp64−/Acgp64 (a gp64null virus that was repaired by reinserting a gp64 gene), and vAc/G-stem (a gp64null virus that expressed the G-stem construct). Purified virions were examined by SDS-PAGE, phosphorimager analysis, and Western blotting. Western blot analysis with an anti-c-Myc antibody showed that the G-stem construct was detected abundantly in virions generated from the vAc/G-stem virus (Fig. 2A, lane 3). Because VP39 (the major capsid protein) is highly abundant in virions and the quantity of VP39 per virion appears to be constant, the VP39 protein in virion preparations was used as an indicator of relative virion quantity. Comparisons of [35S]methionine-labeled VP39 bands from BV preparations derived from equivalent quantities of cell supernatants indicated that virion production from the gp64null virus expressing the G-stem construct (virus vAc/G-stem) was approximately 2.27 times higher than that of the virus expressing wt GP64 and about 11 times higher than that detected from the gp64null virus (Fig. 2B, lane 3 versus lanes 1 and 2, and Fig. 2C). The experiment shown in Fig. 2B was repeated three times with three independently labeled viral preparations, and these data were used to generate the graph in Fig. 2C. These quantitative data show clearly that expression of the VSV G-stem construct in the context of a gp64null baculovirus resulted in the rescue of the budding defect caused by the absence of the GP64 protein. Indeed, our preliminary measurements suggested that budding stimulated by the G-stem construct may exceed that from a virus expressing the wt GP64 protein. Thus, BV that contain no native GP64 protein can be generated efficiently using the G-stem construct.
FIG. 2.
Analysis of G-stem-mediated virion budding. (A) Western blot detection of G-stem protein in purified gp64null AcMNPV BV. BV were purified from Sf9 cells infected with either vAcgp64−/Acgp64 (a gp64null virus repaired by reinsertion of gp64; lane 1), vAcgp64− (a gp64null virus; lane 2), or vAc/G-stem (a gp64null virus in which the VSV G-stem construct was expressed; lane 3) and examined by Western blot analysis using an anti-c-Myc MAb. The arrowhead indicates the position of the c-Myc-tagged VSV G-stem protein. (B) Quantitative analysis of progeny BV production from Sf9 cells infected with vAcgp64−/Acgp64, vAcgp64−, or vAc/G-stem. Sf9 cells were infected with each virus, and progeny virions were labeled with [35S]methionine as described in Materials and Methods. Progeny virions were purified from infected cell supernatants by centrifugation through a sucrose cushion, followed by separation of virions in equilibrium sucrose density gradients. Virions derived from equivalent amounts of infected cell culture supernatants were electrophoresed in SDS-PAGE gels and examined by phosphorimager analysis of labeled proteins. The positions of GP64 and VP39 are indicated on the left. (C) Comparisons of integrated optical densities of the VP39 proteins from various preparations, as shown in panel B. Quantification data represent averages and standard deviations derived from three independently labeled BV preparations. Columns 1 to 3 represent VP39 levels from purified virion preparations for vAcgp64−/Acgp64, vAcgp64−, and vAc/G-stem, respectively.
Targeting and budding domains of GP64.
Although currently unknown, it is possible that domains necessary for efficient budding may be distinct and separate from those associated with targeting the protein to the virion. To examine this question with regard to GP64, we generated a series of gp64null baculoviruses that expressed GP64 proteins containing N- or C-terminal truncations of the ectodomain and examined the ability of each construct to either mediate budding or target to virions that were generated by G-stem-enhanced budding.
Analysis of N-terminal ectodomain truncations of GP64.
The strategy for the generation of N-terminal ectodomain truncation constructs and their insertion into the baculovirus genome is illustrated in Fig. 3A. Each GP64 construct contains the gp64 promoter region, the signal peptide, the cleavage site, a c-Myc epitope, various portions of the C terminus of the GP64 ectodomain, and the GP64 TM domain and CTD. Viruses expressing these N-terminally truncated GP64 constructs were propagated and titrated in Sf9Op1D cells (which express wt OpMNPV GP64) and then used to infect Sf9 cells at an MOI of 10. Although a number of constructs could not be expressed stably (not shown), the constructs with deletions of 104, 250, and 356 amino acids were all expressed stably and detected in infected cell lysates (Fig. 3B, left panel). Supernatants were harvested from infected cells at 72 hpi and examined for BV and the presence of the truncated GP64 constructs. Only the full-length GP64 construct was detected (Fig. 3C, left panel, lane 4). None of the N-terminally truncated GP64 constructs were detected in purified BV from the supernatants (Fig. 3C). These data suggested that either BV were not generated or the N-terminally truncated GP64 constructs were not targeted to BV. To separate these two possibilities, Sf9 cells were infected with each virus expressing an N-terminally truncated GP64 construct and coinfected with a virus expressing the VSV G-stem construct, a construct that rescues virion budding. In each case, virion production was rescued by the presence of the G-stem construct (Fig. 3C, right panel, arrowhead). However, only the full-length GP64 construct (and none of the N-terminally truncated GP64 proteins) was clearly detected in progeny virions (Fig. 3C, right panel, lane 4, open circle). These results were also confirmed by examining purified labeled progeny virions (Fig. 3C, bottom panels) as described previously. Thus, even the shortest N-terminal truncation examined (a construct that deleted amino acids 22 to 124) was not targeted to BV of AcMNPV.
Analysis of C-terminal ectodomain truncations of GP64.
To generate C-terminally truncated GP64 ectodomain constructs, various amounts of sequence upstream from amino acid 461 were removed from the GP64 ectodomain in a c-Myc-tagged GP64 construct (Fig. 3D). Each construct contained the GP64 signal peptide, the signal cleavage site, and a c-Myc epitope tag at the N terminus of the ectodomain and was anchored in the membrane by the 52-amino-acid GP64 C-terminal “stem” domain (GP64-stem; amino acids 461 to 512). GP64-stem included 22 residues from the predicted GP64 ectodomain, 23 residues from the predicted GP64 TM domain, and the 7-residue CTD. While some of the deletion constructs were not stably expressed (not shown), constructs containing either 38 (residues 21 to 58), 66 (residues 21 to 86), 138 (residues 21 to 158), or 274 (residues 21 to 294) amino acids from the N terminus of the GP64 ectodomain were expressed, and Western blots of infected cell extracts are shown in Fig. 3E (left panel). The gp64null viruses expressing these constructs were amplified and titrated in Sf9Op1D cells, and each virus was used to infect Sf9 cells either alone or by coinfection with the virus expressing the VSV G-stem protein as described earlier. Thus, each GP64 construct was expressed either alone or in the presence of VSV G-stem. Analysis of purified BV (Fig. 3F, left panel) showed that each construct was detected in AcMNPV virions, although some were detected at relatively low levels. Coinfections with the G-stem-expressing virus generally resulted in higher levels of detection of the GP64 constructs (Fig. 3F, left versus right panels). These data were also confirmed by examining purified labeled progeny virions (Fig. 3F, bottom panels) as described previously. Thus, each C-terminal GP64 ectodomain deletion stimulated virion budding, although none were found in virions at the same levels as the c-Myc-tagged wt GP64 construct (Fig. 3F, lanes 6). The observation that the smallest construct (vAc/58-TM-CTD) was found in purified virions indicates that the minimal sequence necessary for budding and targeting was mapped to a construct containing a combination of 38 amino acids from the N terminus of the mature GP64 protein and 52 amino acids from the C terminus (the so-called stem domain). In all cases, virion production appeared to be enhanced by the presence of the G-stem construct.
Construction and analysis of G-stem fusion proteins.
The results from the G-stem and GP64 studies described above suggested that it may be possible to display foreign proteins on the surfaces of gp64null AcMNPV BV by using a G-stem construct to mediate efficient budding and GP64 domains to target protein fusions to the BV. It is also possible that a heterologous protein fused with a combination of both GP64 and G-stem domains may be sufficient for both budding and targeting. To examine this possibility, we generated a series of constructs in which the N terminus of the GP64 protein was fused to the C-terminal 91-amino-acid G-stem construct (Fig. 4A). In addition, an epitope-tagged EGFP gene was also fused directly to the G-stem construct by inserting EGFP between the c-Myc epitope and the G-stem construct. Each construct was inserted into a gp64null baculovirus (Fig. 4A) as described previously and was examined for expression and targeting of the construct to BV (Fig. 4B).
For these studies, we generated constructs that displayed portions of the N terminus of GP64 (amino acids 21 to 130, 21 to 152, 21 to 160, 21 to 166, and 21 to 175), with each fused to the 91-amino-acid G-stem construct. Viruses expressing these constructs were propagated in Sf9Op1D cells, and expression of the G-stem fusions was detected by Western blot analysis of cell extracts or purified BV, using an anti-c-Myc antibody. Expression of all fusion constructs was detected in infected cell lysates (Fig. 4B, top panel, lanes 2 to 7). In BV preparations, all G-stem fusions were present, except for the construct containing amino acids 21 to 175 from GP64 (Fig. 4B, lower panel, lane 6). The G-stem construct alone (unfused) was found at much higher levels in BV preparations than were the fusion constructs (Fig. 4B, lower panel, compare lane 1 with lanes 2 to 7). Using the single c-Myc epitope present on each construct, these data show clear relative differences in detection of the different G-stem fusion constructs and confirm that most (all but one) of these fusion constructs were displayed on gp64null virions. Examination of cells infected with the virus expressing the EGFP-G-stem fusion by immunofluorescence microscopy (Fig. 4C) also further demonstrated expression of that construct in the infected cells. However, Western blot analysis of purified BV resulted in very low-level detection of the EGFP fusion, and fluorescence was not detected in purified virion preparations. Because most of the G-stem fusion protein constructs were found in the purified virion preparations, these studies indicate that for many proteins, G-stem fusions can be used to display foreign proteins on gp64null AcMNPV BV. Fusion of either portions of GP64 or EGFP with the G-stem construct permitted both the production of virions and display of the fusion construct on those virions in the absence of wt GP64.
Construction and analysis of HA-GP64 fusion proteins.
Examination of G-stem and GP64 domains necessary for budding and virion targeting suggested that these two concepts may be used effectively together. We next examined whether portions of the GP64 protein alone may be used to target foreign proteins to gp64null virions. To determine if the mapped targeting and budding domains of GP64 were sufficient for rescue of budding and targeting of a heterologous protein to the virion, we fused the ectodomain of influenza virus HA (A/WSN/33) between the C-terminal GP64-stem region and various portions of the N-terminal ectodomain (Fig. 5A). N-terminal fusions contained the GP64 signal peptide, a c-Myc epitope, and 38 or 66 amino acids from the N terminus of the GP64 ectodomain. The C terminus of each construct was comprised of either the 52-amino-acid GP64-stem (58-HA-TM-CTD and 86-HA-TM-CTD) (Fig. 5A) or the wt HA TM domain and CTD (Fig. 5A, 58-HA-FL). Each construct was inserted into a gp64null AcMNPV genome, and the resulting viruses were propagated in Sf9Op1D cells. Sf9 cells were then infected with each virus, either alone or in combination with a virus expressing the G-stem construct. In Western blots of purified BV preparations challenged with an anti-c-Myc antibody, the HA ectodomain fusions were detected abundantly (Fig. 5B, upper panel, HA). The identity of HA fusions was also confirmed by Western blot analysis with an anti-HA polyclonal antiserum (Fig. 5B, anti-HA). Coinfection of each of the HA fusion constructs with a virus expressing the G-stem construct resulted in higher levels of detection of the HA fusions, and in these constructs the G-stem was detected at lower levels. When they were expressed in the presence of the G-stem construct, two of the ectodomain fusions (58-HA-TM-CTD and 86-HA-TM-CTD) were detected at levels that appeared to be similar to the abundant expression of the G-stem construct alone. Although the HA construct that contained its own TM domain and CTD was expressed and detected on the purified BV, the levels were clearly lower than that of either construct containing the GP64 stem region (Fig. 5B, lane 5 versus lanes 3 and 4 and lane 9 versus lanes 7 and 8). Relative differences in BV production were observed more directly by examining the relative levels of the major capsid protein (VP39) in purified labeled BV preparations (Fig. 5B, labeled VP39, lane 5 versus lanes 3 and 4 and lane 9 versus lanes 7 and 8). Thus, the combined use of G-stem-stimulated budding and the GP64 targeting domain resulted in more efficient display of this heterologous protein on the AcMNPV BV. It is of special note that most prior studies of virion display have utilized protein expression from the very strong polyhedrin or p10 promoter (4, 6, 13, 18, 23, 26, 35, 44), whereas the constructs generated in the current study were generated with the native gp64 early/late promoter. Current studies are addressing the question of whether higher expression levels will lead to more abundance of the displayed protein on the cell surface after optimization of budding and targeting as described here.
DISCUSSION
In the current study, we examined envelope protein targeting to baculovirus virions and virion budding efficiency in AcMNPV-infected Sf9 cells. Our goals were to identify functional domains of the essential GP64 protein associated with targeting and budding and to develop tools for displaying heterologous proteins on the surfaces of gp64null virions. To examine the requirements for budding and protein targeting, we used the following two approaches: (i) truncated GP64 constructs were used to map GP64 regions necessary for targeting to virions and budding, and (ii) a VSV G-stem construct was used to independently rescue GP64 budding. In addition, we used the influenza virus HA protein as a model protein for analysis of display on gp64null AcMNPV virions.
We found that a 91-amino-acid C-terminal G-stem construct was capable of substituting for the budding function of the AcMNPV GP64 protein in the context of gp64null AcMNPV and that this construct rescued budding to near wt levels or higher. In addition, we found that targeting of GP64 to the virion required a portion (38 amino acids) of the N terminus of the GP64 ectodomain in combination with a small portion of the C terminus of GP64 (the 52-amino-acid GP64-stem domain). Thus, a mature protein consisting of only 90 amino acids from GP64 was targeted to the virion, and virion budding was detected when this construct was substituted for full-length GP64.
The observation that a VSV G-stem construct is capable of rescuing the budding defect of gp64null AcMNPV has important implications with regard to the mechanism of virion budding in these two virus groups and suggests that baculoviruses and rhabdoviruses may use similar mechanisms for virion budding. One possible mechanistic explanation is that an AcMNPV-encoded or -initiated budding complex is preassembled and subsequently triggered by the G-stem protein. Alternatively, the G-stem protein may somehow facilitate a budding process that is distinct from that normally utilized in AcMNPV budding, and that process is then utilized by AcMNPV.
In the absence of GP64, virion budding is severely reduced, which suggests that GP64 must first be targeted to the appropriate regions of the plasma membrane and then participate in the budding process. The requirement for GP64 in these two processes (targeting and budding) may not be mutually exclusive, since specific targeting to the site of budding may be required prior to the actual budding process. To address this problem and to separate these two issues experimentally, we used a VSV G-stem construct to rescue the budding defect caused by the gp64null virus and then mapped regions of the GP64 ectodomain necessary for GP64 targeting to the BV. Our data indicated that the N-terminal region of the GP64 ectodomain is necessary for GP64 targeting to the BV envelope. The C-terminal GP64 stem region also appears to serve a more specific role than simply anchoring the protein in the membrane. When results for various chimeric influenza virus HA fusion proteins were compared, we found that HA fusions that contained identical N-terminal regions (derived from GP64) but differed in the C-terminal stem regions (Fig. 5, 58-HA-TM-CTD versus 58-HA-FL) showed substantial differences in their localization to virions. Chimeras containing the GP64-stem were detected at higher levels than those containing the native HA stem region, suggesting a specific role for the GP64-stem in targeting or budding.
In future studies, it will be important to examine different combinations of the strategies outlined above and to determine precise quantitative differences in virion budding from gp64null viruses in the presence of G-stem alone or various G-stem or GP64-stem chimeras. In the current studies, we identified a solution to a substantial and important problem associated with the use of baculovirus gp64null viruses in research and biotechnology. Using VSV G-stem constructs to rescue budding in combination with chimeric constructs containing GP64 targeting domains, foreign proteins may be displayed effectively on gp64null virions.
Acknowledgments
We thank Gary Whittaker for providing plasmid pEWSN-HA, containing the HA gene from influenza virus A/WSN/33, and for polyclonal anti-HA antiserum. We also thank Joshua Huffer and Gerrit Heetderks for assistance with virus construction and for technical assistance.
This work was supported by a grant from the NIH (RO1 AI33657) and Boyce Thompson Institute project 1255.
Footnotes
Published ahead of print on 7 November 2007.
REFERENCES
- 1.Blissard, G. W. 1996. Baculovirus-insect cell interactions. Cytotechnology 2073-93. [DOI] [PubMed] [Google Scholar]
- 2.Blissard, G. W., and G. F. Rohrmann. 1989. Location, sequence, transcriptional mapping, and temporal expression of the gp64 envelope glycoprotein gene of the Orgyia pseudotsugata multicapsid nuclear polyhedrosis virus. Virology 170537-555. [DOI] [PubMed] [Google Scholar]
- 3.Blissard, G. W., and J. R. Wenz. 1992. Baculovirus GP64 envelope glycoprotein is sufficient to mediate pH-dependent membrane fusion. J. Virol. 666829-6835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boublik, Y., P. Di-Bonito, and I. M. Jones. 1995. Eukaryotic virus display: engineering the major surface glycoprotein of the Autographa californica nuclear polyhedrosis virus (AcNPV) for the presentation of foreign proteins on the virus surface. Nat. Biotechnol. 131079-1084. [DOI] [PubMed] [Google Scholar]
- 5.Braunagel, S. C., and M. D. Summers. 1994. Autographa californica nuclear polyhedrosis virus, PDV, and ECV viral envelopes and nucleocapsids: structural proteins, antigens, lipid and fatty acid profiles. Virology 202315-328. [DOI] [PubMed] [Google Scholar]
- 6.Ernst, W., R. Grabherr, D. Wegner, N. Borth, A. Grassauer, and H. Katinger. 1998. Baculovirus surface display: construction and screening of a eukaryotic epitope library. Nucleic Acids Res. 261718-1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Friesen, P. D., and L. K. Miller. 2001. Insect viruses, p. 599-628. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed., vol. 1. Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 8.Grabherr, R., W. Ernst, O. Doblhoff-Dier, M. Sara, and H. Katinger. 1997. Expression of foreign proteins on the surface of Autographa californica nuclear polyhedrosis virus. BioTechniques 22730-735. [DOI] [PubMed] [Google Scholar]
- 9.Grabherr, R., W. Ernst, C. Oker-Blom, and I. Jones. 2001. Developments in the use of baculoviruses for the surface display of complex eukaryotic proteins. Trends Biotechnol. 19231-236. [DOI] [PubMed] [Google Scholar]
- 10.Hefferon, K., A. Oomens, S. Monsma, C. Finnerty, and G. Blissard. 1999. Host cell receptor binding by baculovirus GP64 and kinetics of virion entry. Virology 258455-468. [DOI] [PubMed] [Google Scholar]
- 11.Hohmann, A. W., and P. Faulkner. 1983. Monoclonal antibodies to baculovirus structural proteins: determination of specificities by Western blot analysis. Virology 125432-444. [DOI] [PubMed] [Google Scholar]
- 12.IJkel, W. F. J., M. Westenberg, R. W. Goldbach, G. W. Blissard, J. M. Vlak, and D. Zuidema. 2000. A novel baculovirus envelope fusion protein with a proprotein convertase cleavage site. Virology 27530-41. [DOI] [PubMed] [Google Scholar]
- 13.Kitagawa, Y., H. Tani, C. K. Limn, T. M. Matsunaga, K. Moriishi, and Y. Matsuura. 2005. Ligand-directed gene targeting to mammalian cells by pseudotype baculoviruses. J. Virol. 793639-3652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kost, T. A., J. P. Condreay, and D. L. Jarvis. 2005. Baculovirus as versatile vectors for protein expression in insect and mammalian cells. Nat. Biotechnol. 23567-575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leikina, E., H. O. Onaran, and J. Zimmerberg. 1992. Acidic pH induces fusion of cells infected with baculovirus to form syncytia. FEBS Lett. 304221-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lung, O., M. Westenberg, J. M. Vlak, D. Zuidema, and G. W. Blissard. 2002. Pseudotyping Autographa californica multicapsid nucleopolyhedrovirus (AcMNPV): F proteins from group II NPVs are functionally analogous to AcMNPV GP64. J. Virol. 765729-5736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lung, O. Y., M. Cruz-Alvarez, and G. W. Blissard. 2003. Ac23, an envelope fusion protein homolog in the baculovirus Autographa californica multicapsid nucleopolyhedrovirus, is a viral pathogenicity factor. J. Virol. 77328-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Makela, A. R., H. Matilainen, D. J. White, E. Ruoslahti, and C. Oker-Blom. 2006. Enhanced baculovirus-mediated transduction of human cancer cells by tumor-homing peptides. J. Virol. 806603-6611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mangor, J. T., S. A. Monsma, M. C. Johnson, and G. W. Blissard. 2001. A gp64null baculovirus pseudotyped with vesicular stomatitis virus G protein. J. Virol. 752544-2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mebatsion, T., M. Konig, and K. K. Conzelmann. 1996. Budding of rabies virus particles in the absence of the spike glycoprotein. Cell 84941-951. [DOI] [PubMed] [Google Scholar]
- 21.Miller, L. K. (ed.). 1997. The baculoviruses. Plenum Press, New York, NY.
- 22.Monsma, S. A., A. G. P. Oomens, and G. W. Blissard. 1996. The GP64 envelope fusion protein is an essential baculovirus protein required for cell-to-cell transmission of infection. J. Virol. 704607-4616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mottershead, D., I. van der Linden, C. H. von Bonsdorff, K. Keinanen, and C. Oker-Blom. 1997. Baculoviral display of the green fluorescent protein and rubella virus envelope proteins. Biochem. Biophys. Res. Commun. 238717-722. [DOI] [PubMed] [Google Scholar]
- 24.Mottershead, D. G., K. Alfthan, K. Ojala, K. Takkinen, and C. Oker-Blom. 2000. Baculoviral display of functional scFv and synthetic IgG-binding domains. Biochem. Biophys. Res. Commun. 27584-90. [DOI] [PubMed] [Google Scholar]
- 25.Neumann, G., T. Watanabe, and Y. Kawaoka. 2000. Plasmid-driven formation of influenza virus-like particles. J. Virol. 74547-551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ojala, K., J. Koski, W. Ernst, R. Grabherr, I. Jones, and C. Oker-Blom. 2004. Improved display of synthetic IgG-binding domains on the baculovirus surface. Technol. Cancer Res. Treat. 377-84. [DOI] [PubMed] [Google Scholar]
- 27.Oomens, A. G. P., and G. W. Blissard. 1999. Requirement for GP64 to drive efficient budding of Autographa californica multicapsid nucleopolyhedrovirus. Virology 254297-314. [DOI] [PubMed] [Google Scholar]
- 28.Oomens, A. G. P., S. A. Monsma, and G. W. Blissard. 1995. The baculovirus GP64 envelope fusion protein: synthesis, oligomerization, and processing. Virology 209592-603. [DOI] [PubMed] [Google Scholar]
- 29.Pearson, M. N., C. Groten, and G. F. Rohrmann. 2000. Identification of the Lymantria dispar nucleopolyhedrovirus envelope fusion protein provides evidence for a phylogenetic division of the Baculoviridae. J. Virol. 746126-6131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pearson, M. N., and G. F. Rohrmann. 2002. Transfer, incorporation, and substitution of envelope fusion proteins among members of the Baculoviridae, Orthomyxoviridae, and Metaviridae (insect retrovirus) families. J. Virol. 765301-5304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pearson, M. N., R. L. Russell, and G. F. Rohrmann. 2001. Characterization of a baculovirus-encoded protein that is associated with infected-cell membranes and budded virions. Virology 29122-31. [DOI] [PubMed] [Google Scholar]
- 32.Plonsky, I., M. S. Cho, A. G. P. Oomens, G. W. Blissard, and J. Zimmerberg. 1999. An analysis of the role of the target membrane on the gp64-induced fusion pore. Virology 25365-76. [DOI] [PubMed] [Google Scholar]
- 33.Roberts, T. E., and P. Faulkner. 1989. Fatty acid acylation of the 67K envelope glycoprotein of a baculovirus Autographa californica nuclear polyhedrosis virus. Virology 172377-381. [DOI] [PubMed] [Google Scholar]
- 34.Robison, C. S., and M. A. Whitt. 2000. The membrane-proximal stem region of vesicular stomatitis virus G protein confers efficient virus assembly. J. Virol. 742239-2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tani, H., C. K. Limn, C. C. Yap, M. Onishi, M. Nozaki, Y. Nishimune, N. Okahashi, Y. Kitagawa, R. Watanabe, R. Mochizuki, K. Moriishi, and Y. Matsuura. 2003. In vitro and in vivo gene delivery by recombinant baculoviruses. J. Virol. 779799-9808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tani, H., M. Nishijima, H. Ushijima, T. Miyamura, and Y. Matsuura. 2001. Characterization of cell-surface determinants important for baculovirus infection. Virology 279343-353. [DOI] [PubMed] [Google Scholar]
- 37.Theilmann, D. A., G. W. Blissard, B. Bonning, J. Jehle, D. R. O'Reilly, G. F. Rohrmann, S. Thiem, and J. M. Vlak. 2005. Baculoviridae, p. 177-185. In H. V. Van Regenmortel, D. H. L. Bishop, M. H. Van Regenmortel, and C. M. Fauquet (ed.), Virus taxonomy: eighth report of the International Committee on Taxonomy of Viruses. Elsevier Academic Press, New York, NY.
- 38.Toivola, J., K. Ojala, P. O. Michel, M. Vuento, and C. Oker-Blom. 2002. Properties of baculovirus particles displaying GFP analyzed by fluorescence correlation spectroscopy. Biol. Chem. 3831941-1946. [DOI] [PubMed] [Google Scholar]
- 39.Volkman, L. E. 1986. The 64K envelope protein of budded Autographa californica nuclear polyhedrosis virus. Curr. Top. Microbiol. Immunol. 131103-118. [DOI] [PubMed] [Google Scholar]
- 40.Volkman, L. E., and P. A. Goldsmith. 1985. Mechanism of neutralization of budded Autographa californica nuclear polyhedrosis virus by a monoclonal antibody: inhibition of entry by adsorptive endocytosis. Virology 143185-195. [DOI] [PubMed] [Google Scholar]
- 41.Westenberg, M., F. Veenman, E. C. Roode, R. W. Goldbach, J. M. Vlak, and D. Zuidema. 2004. Functional analysis of the putative fusion domain of the baculovirus envelope fusion protein F. J. Virol. 786946-6954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Westenberg, M., H. Wang, W. F. IJkel, R. W. Goldbach, J. M. Vlak, and D. Zuidema. 2002. Furin is involved in baculovirus envelope fusion protein activation. J. Virol. 76178-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Whitford, M., S. Stewart, J. Kuzio, and P. Faulkner. 1989. Identification and sequence analysis of a gene encoding gp67, an abundant envelope glycoprotein of the baculovirus Autographa californica nuclear polyhedrosis virus. J. Virol. 631393-1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang, D. G., Y. C. Chung, Y. K. Lai, C. W. Lai, H. J. Liu, and Y. C. Hu. 2007. Avian influenza virus hemagglutinin display on baculovirus envelope: cytoplasmic domain affects virus properties and vaccine potential. Mol. Ther. 15989-996. [DOI] [PubMed] [Google Scholar]
- 45.Zhang, S. X., Y. Han, and G. W. Blissard. 2003. Palmitoylation of the Autographa californica multicapsid nucleopolyhedrovirus envelope glycoprotein GP64: mapping, functional studies, and lipid rafts. J. Virol. 776265-6273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhou, J., and G. W. Blissard. 2006. Mapping the conformational epitope of a neutralizing antibody (AcV1) directed against the AcMNPV GP64 protein. Virology 352427-437. [DOI] [PMC free article] [PubMed] [Google Scholar]