Abstract
The NS5A protein of hepatitis C virus (HCV) plays an important but undefined role in viral RNA replication. NS5A has been proposed to be a three-domain protein, and the crystal structure of the well-conserved amino-terminal domain I has been determined. The remaining two domains of NS5A, designated domains II and III, and their corresponding interdomain regions are poorly understood. We have conducted a detailed mutagenesis analysis of NS5A domains II and III using the genotype 1b HCV replicon system. The majority of the mutants containing 15 small (8- to 15-amino-acid) deletions analyzed were capable of efficient RNA replication. Only five deletion mutations yielded lethal phenotypes, and these were colinear, spanning a 56-amino-acid region within domain II. This region was further analyzed by combining triple and single alanine scanning mutagenesis to identify individual residues required for RNA replication. Based upon this analysis, 23 amino acids were identified that were found to be essential. In addition, two residues were identified that yielded a small colony phenotype while possessing only a moderate defect in RNA replication. These results indicate that the entire domain III region and large portions of domain II of the NS5A protein are not required for the function of NS5A in HCV RNA replication.
Hepatitis C virus (HCV) is the sole member of the Hepacivirus genus of the Flaviviridae family of enveloped, positive-strand RNA viruses (23). HCV is capable of establishing persistent infections in humans, and long-term infection with the virus is often associated with chronic liver disease, hepatocellular carcinoma, and a host of extrahepatic disease states. HCV infection is remarkably widespread, with estimates suggesting nearly 3% of the world's population has been infected with this virus, although the surveillance data are far from complete (1).
The HCV genome consists of an RNA molecule of approximately 9.6 kb in size that contains a single, large open reading frame flanked by structured 5′ and 3′ nontranslated regions (NTRs). Viral proteins are translated as part of a large polyprotein precursor via an internal ribosome entry site within the 5′ NTR. The 10 HCV proteins are organized in the polyprotein in the order NH2-C-E1-E2-p7-NS2-NS3-NS4A-NS4B-NS5A-NS5B-COOH (15). Following a complex series of polyprotein cleavage events catalyzed by host and viral proteases, the mature, nonstructural (NS) proteins assemble into a membrane-associated HCV RNA replicase complex. The putative replicase has been imaged in cell lines harboring HCV replicons and has been shown to contain all of the NS proteins, as well as actively replicating viral RNA (12). The NS3, NS4A, NS4B, NS5A, and NS5B proteins comprise the minimal viral protein components required for RNA replication in subgenomic HCV replicon systems, with the remaining HCV proteins being dispensable for replication at least in this context (3, 26). The functions of the majority of these essential replicase proteins have been at least partially defined. The NS3 protein contains the viral serine protease activity responsible for much of the polyprotein processing as well as an RNA helicase activity that is likely involved in genome replication (9, 13, 14, 16, 38). The NS4A protein serves as a cofactor for the activities of NS3 and is important in tethering NS3 to cellular membranes (9, 33). The NS4B protein plays a direct role in the remodeling of host cell membranes, presumably to generate the site for viral replicase assembly (7). The NS5B protein serves as the viral RNA-dependent RNA polymerase (6). No definitive function for NS5A in the RNA replicase complex has yet been identified; however, considerable progress has been made in recent years with respect to the characterization of NS5A, and this protein is now known to be an absolutely required component of the viral replicase (3, 26).
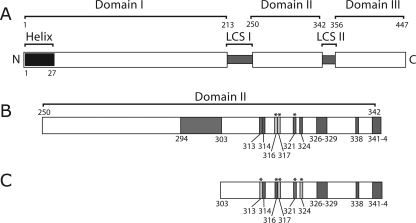
NS5A is a large (56- to 58-kDa) hydrophilic phosphoprotein organized in three domains (I, II, and III), each of which is separated by repetitive low-complexity sequences (see Fig. 1A) (40). The amino-terminal region of NS5A, designated domain I, contains the first 213 amino acids of the protein. This region contains an amino-terminal membrane-anchoring sequence (5, 34, 36). Four essential cysteine residues within domain I collectively bind to a single structural zinc ion, and mutation of these residues results in the complete inhibition of RNA replication (40). The structure of the majority of domain I has recently been determined by X-ray crystallography, revealing a novel protein fold (41). Based on similar zinc binding properties, domain I of the bovine viral diarrhea virus NS5A protein is believed to be similar in structure to HCV NS5A; however, this remains to be verified experimentally (42). The crystal structure of domain I is that of a dimer, and the interface between protein molecules that generates the dimer is characterized by a large, basic groove that has been proposed to serve as a site of RNA binding. NS5A possesses an RNA binding activity, although the relevant binding partner has yet to be determined, and the relationship between the proposed RNA binding groove and the RNA binding activity observed remains unclear (17).
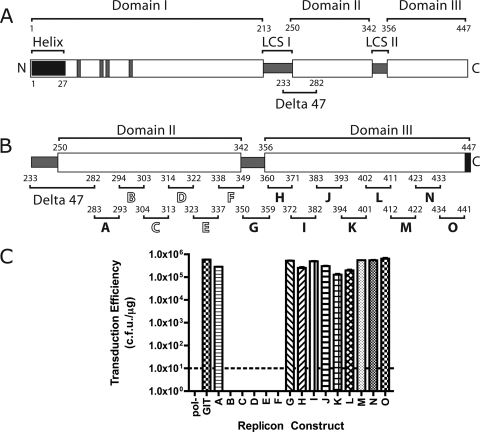
FIG. 1.
(A) Schematic representation of the hepatitis C virus NS5A protein. NS5A has been proposed to consist of three domains (labeled domains I, II, and III) with domains separated by low-complexity sequences (labeled LCS I and II). The position of the amino-terminal amphipathic helix membrane anchor is shown (labeled helix). The four cysteine residues corresponding to the NS5A zinc coordination site are indicated by gray bars in the domain I region of the protein. The location of the previously described Δ47 (Delta 47) deletion is shown. The amino (N) and carboxyl (C) termini of NS5A are shown. All numbers refer to the amino acid number of the Con1 isolate of NS5A 1b, with the amino terminus of the mature protein designated as amino acid one. (B) Schematic of deletions within domains II and III of NS5A. The domain II and III regions of NS5A are shown in greater detail from panel A. Deletions are labeled A through O. Numbers above each deletion letter indicate the amino acids deleted, and the black bars approximate the locations of the deletions on the domain organization model of NS5A. Deletions indicated with solid black letters are deletions that are dispensable for HCV RNA replication. Deletions labeled with outline block white letters are not viable in the HCV replicon system. The six carboxyl-terminal amino acids of NS5A are indicated by a black block, indicating that these residues were not tested for their role in RNA replication in this work. The designations for domains II and III and Δ47 are as described above for panel A. (C) RNA replication fitness of replicons bearing deletions within NS5A domains II and III. Graph presenting transduction efficiency of replicon RNAs in units of CFU/μg of RNA. The pol− designation refers to a Con1/SG-neo replicon RNA bearing a nonfunctional RNA-dependent RNA polymerase sequence that serves as a negative control for replication experiments. The transduction efficiency of the parental Con1/SG-neo GIT replicon (labeled GIT) is shown. The letters A through O on the abscissa refer to the analysis of the individual deletion mutants shown in panel B. The dashed line indicates the limit of detection of the assay.
NS5A domains II and III are far less conserved among HCV genotypes than domain I is and are far less well understood. A large deletion of 47 amino acids, referred to as Δ47 hereafter, was isolated as a replicon adaptive mutation, suggesting that at least portions of domain II are dispensable for RNA replication (3). Liang et al. attempted to determine a nuclear magnetic resonance structure for domain II but found this region contained low secondary structure content and probable conformational variability (21). Domain III of NS5A apparently possesses a great deal of inherent flexibility, as it tolerates large heterologous insertions, such as green fluorescent protein, and still retains its activity during RNA replication (2, 24, 31, 32). Domain III contains a large number of potential phosphoacceptor sites, most likely involved in NS5A hypophosphorylation, and mutations of these sites do not disrupt RNA replication (2). Domains II and III, the intervening low-complexity sequence regions, and to a lesser extent, domain I, have been identified as sites of interaction between NS5A and several cellular proteins, but the relevance of this to HCV replication and pathogenesis remains to be determined (for a review, see reference 43).
Despite recent progress with respect to NS5A research, the function of this enigmatic protein in the viral replication complex remains elusive. While some insight has been gained regarding the features of domain I of NS5A, the properties of domains II and III are poorly understood. The roles of these variable regions of NS5A in RNA replication or in some other activity in the viral life cycle remain to be elucidated. Results from sequence alignment analysis of NS5A protein coding regions from a diverse set of HCV genotypes indicate that small blocks of well-conserved amino acids can be found within these regions. These results imply the presence of selective pressure for their maintenance and further suggest that these regions play an important role in some aspect of the HCV life cycle.
The gap in our knowledge of NS5A domain II and III function led us to investigate their importance for HCV RNA replication. Subgenomic replicons provide an excellent system for assessing the importance of these regions in HCV RNA replication (3, 27). In these systems, HCV RNAs containing the viral NTRs and the coding sequence for the minimal replicase protein components are linked to a reporter or selectable gene expression cassette, such as a neomycin resistance cassette. This permits a rapid and sensitive determination of RNA replication efficiency based on the expression of the reporter gene or selection of colonies harboring replicons resistant to the drug G418 (referred to as G418 transduction). We have generated a series of 15 small deletions starting at the carboxyl terminus of the previously reported Δ47 adaptive mutation (amino acids 233 to 282) and spanning all of domains II and III, except for the C-terminal six amino acids (which were not altered to avoid potential interference with polyprotein processing). Of these deletions, all but five colinear deletions in domain II were viable, indicating that all of domain III and at least part of domain II are dispensable for HCV RNA replication. The region of NS5A corresponding to the five lethal deletion mutations was targeted by alanine scanning mutagenesis to generate a nearly complete map of NS5A residues required for RNA replication. Our initial mapping efforts were conducted in the context of a replicon bearing three adaptive mutations, two in NS3 and one in NS4B, allowing evaluation of mutations in the context of a wild-type NS5A coding sequence. The potential for genetic incompatibility among different classes of adaptive mutations and our directed NS5A mutations obliged us to analyze all mutations producing a lethal or near-lethal phenotype in the context of a second replicon genetic backbone, that of the S2204I replicon system (3). We also analyzed the RNA replication capacity of all lethal or near lethal mutants by quantitative real-time reverse transcriptase PCR in transient-replication assays to ensure that the observed defects in the G418 transduction assay represented defects in HCV RNA replication. Finally, we examined NS5A protein stability and phosphorylation state of these defective mutants using transient translation and Western blotting. These data provide the first single amino acid resolution map of residues in NS5A domains II and III required for RNA replication and provides a template for future studies in determining the function(s) of NS5A in the HCV life cycle.
MATERIALS AND METHODS
Cloning and in vitro mutagenesis.
All mutagenesis was performed using the QuikChange II (Stratagene) in vitro mutagenesis system following the manufacturer's standard protocols. Mutations were generated in the shuttle vector pSL1180GIT, which contained the BsrG1-MfeI fragment of the Con1/SG-neo GIT replicon encompassing the NS5A coding sequence and surrounding region. The Con1/SG-neo GIT replicon is identical to the subgenomic replicons described previously (3), with the exception of the presence of two adaptive mutations in NS3 (E1202G and T1280I) and one in NS4A (K1846T), thereby allowing analysis of NS5A mutants on a wild-type NS5A backbone (25). Once generated in the pSL1180GIT shuttle vector, mutations of interest were subcloned by moving the BsrG1-MfeI fragment of pSL1180GIT containing the desired mutation into the parental Con1/SG-neo GIT replicon plasmid. All mutations generated by QuikChange mutagenesis were confirmed by DNA sequence analysis of the final replicon plasmids. For analysis of replication in the context of the Con1/SG-neo S2204I replicon, mutations were subcloned from Con1/Sg-neo GIT or pSL1180GIT into a replicon plasmid harboring the S2204I change. Mutations were moved such that the final plasmid contained the S2204I mutation and the desired mutation, and not any adaptive mutations from the GIT vector. In some instances, S2204I series mutants were generated by QuikChange mutagenesis in the vector pSL1180S2204I, a construct analogous to pSL1180GIT but with the S2204I mutation and not the E1202G, T1280I, and K1846T mutations found in GIT clones. In the numbering of NS5A amino acid residues in this article, the first serine residue of NS5A is residue number one. Mutations of amino acids in NS5A are designated by the single-letter amino acid code of the parental sequence, the residue number in NS5A, and the altered amino acid present in the mutant construct; for example, a change of tryptophan 329 of NS5A to an alanine residue would be designated as W329A. Conversion of this numbering scheme to the amino acid numbering scheme of the complete Con1 isolate 1b genotype HCV polyprotein requires the addition of 1,972 to the numbers used in this article.
Cell culture.
Huh-7.5 (human hepatoma) cells were maintained in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 10% fetal bovine serum, nonessential amino acids, 100 U/ml penicillin, and 100 μg/ml streptomycin at 37°C in 5% CO2. Cells were passaged every 3 days at a split ratio of 1:3 after treatment with 0.05% trypsin and 0.02% EDTA. All experiments were performed using cells between passage numbers 28 to 60 expanded from an original seed stock of the Huh-7.5 cell clone described previously (4).
In vitro RNA transcription.
In vitro RNA transcription of HCV replicon RNAs was performed as described previously (3). Cesium chloride density gradient-purified Con1/SG-neo GIT DNA was linearized with ScaI (New England Biolabs), followed by phenol-chloroform extraction and ethanol precipitation. RNA transcripts were synthesized at 37°C for 1.5 h in a 50-μl reaction mixture containing 40 mM Tris-HCl (pH 7.9), 10 mM NaCl, 12 mM MgCl2, 2 mM spermidine, 10 mM dithiothreitol, 3 mM each ribonucleoside triphosphate, 100 U of RNasin (Roche), 100 U T7 RNA polymerase (Epicenter Technologies), and 3 μg of linearized DNA template. Following transcription, 10 U of DNase I (Ambion) was added, and reaction mixtures were incubated at 37°C for 20 min to remove the DNA template. RNA was then purified using the Qiagen RNeasy kit following the manufacturer's instructions (Qiagen). The optional “on column” DNase treatment described for the Qiagen RNeasy kit was used to further remove residual DNA template. RNA yield was determined by quantification with a UV spectrophotometer. The quality of RNA was further confirmed by agarose gel electrophoresis.
Electroporation of HCV replicons and G418 selection.
Experiments using HCV replicons were performed essentially as described previously, with minor modifications (3). Briefly, subconfluent Huh-7.5 cells at were trypsinized, washed twice in ice-cold phosphate-buffered saline, and resuspended at a concentration of 2.5 × 107 cells/ml in ice-cold RNase-free phosphate-buffered saline. A 400-μl aliquot of cells was then mixed with 1 μg of HCV replicon RNA and electroporated using a BTX square wavelength electroporator in a 0.2-mm-gap cuvette (five 99-microsecond pulses, 900 V, 1.1-second time interval). Following a 10-min recovery period at room temperature, cells were diluted in 9.6 ml of complete medium and plated into 100-mm dishes. Cells were plated at 1 × 106, 1 × 105, and 1 × 104 cells at a fixed overall cell density of 1 × 106 cells/ml, with the remaining cell density consisting of cells electroporated with a replication-deficient Con1/SG-neo GIT pol− RNA. Selection of replicon-containing colonies was performed 2 days postelectroporation by the addition of complete medium supplemented with 1 mg/ml G418. Selection was carried out for a period of 2 weeks postelectroporation with fresh medium containing G418 added to the cells every 4 days. Following selection, cells were fixed with 7% formaldehyde and stained with 1% crystal violet in 50% ethanol. G418-resistant colonies were then counted and used to calculate transduction efficiency in units of CFU per microgram of input RNA. All experimental results presented are the means of three independent RNA preparations delivered by three independent electroporations. Error bars represent the standard errors of the means.
Transient-replication assay by real-time reverse transcriptase PCR.
For transient-replication assays, cells were electroporated with 1 μg of HCV RNA as described above, and following recovery of electroporated cells in Dulbecco's modified Eagle's medium, the cells were plated at a density of 5 × 105 cells per well on a six-well dish. At various time points postelectroporation, cells were washed twice in phosphate-buffered saline, and RNAs were then extracted and purified using the Qiagen RNeasy kit following the manufacturer's instructions. Extracted, purified RNAs were quantified by UV absorbance, and 100 nanograms of total RNA was used for real-time reverse transcriptase PCR. All real-time PCR experiments were performed using a Roche LightCycler 480 using HCV-specific primers and probe as described previously (18). Values are normalized using the human glyceraldehyde-3-phosphate dehydrogenase endogenous control primer set (catalog no. 4326317E; Applied Biosystems).
Transient expression of HCV proteins and Western blotting.
Transient expression of the HCV polyprotein was accomplished using transient transfection of replicon plasmid DNA, followed by infection with a recombinant vaccinia virus expressing T7 RNA polymerase, vTF7-3 (10). Briefly, 5 × 105 cells were infected with vTF7-3 at a multiplicity of infection of 10 for 30 min at room temperature and then transfected with 1 μg of replicon plasmid DNA using Fugene 6 transfection reagent. Twenty-four hours postinfection, cells were washed and lysed in cold NETN buffer (50 mM Tris-HCl [pH 8.0], 120 mM NaCl, 1 mM EDTA, 0.5% NP-40) supplemented with Roche complete protease inhibitor cocktail. Lysates were then centrifuged at 16,000 × g for 20 min at 4°C. Following centrifugation, the soluble fractions were mixed with sodium dodecyl sulfate-polyacrylamide gel electrophoresis sample buffer, boiled for 2 min, and separated on sodium dodecyl sulfate-8% polyacrylamide gels prior to electroblotting onto nitrocellulose membranes. For immunological detection of NS5A, monoclonal antibody 9E10 was used as described previously (22).
RESULTS
Analysis of replication phenotypes of HCV RNAs bearing deletions in NS5A domains II and III.
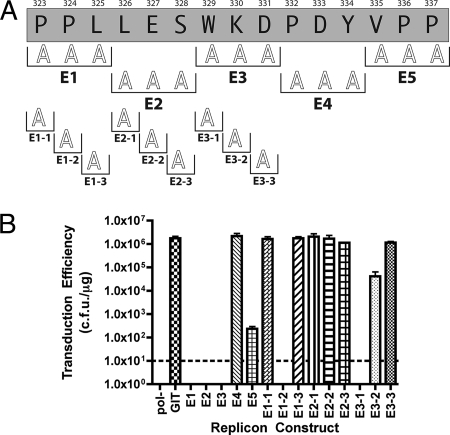
As an initial approach to identify which regions of domains II and III are required for HCV RNA replication, we constructed a series of 15 small (8- to 15-amino-acid) deletions within these regions in the context of the HCV 1b subgenomic replicon system. These deletions were colinear such that they spanned the entire domain II and III regions starting at the carboxyl terminus of the previously described Δ47 adaptive mutation (3) and proceeding within six amino acids of the carboxyl terminus of the NS5A protein. These last six amino acids of NS5A were not deleted or otherwise altered to ensure proper proteolytic processing of NS5A from the HCV polyprotein. The deletions were designated A through O and are shown schematically on the domain II and III region of NS5A in Fig. 1B. The replication fitness of each individual deletion construct was determined using the Con1/SG-neo GIT replicon system. Results of this analysis, presented in terms of G418 transduction efficiency in units of CFU per microgram of RNA transfected, are shown in Fig. 1C. Replication fitness in this experiment, and in all subsequent experiments, is shown compared to a parental Con1/SG-neo GIT replicon (labeled GIT in all figures) bearing no deletion and the Con1/SG-neo GIT pol− replicon (designated pol−), a negative, non-replication-competent control bearing a lethal lesion in the NS5B RNA-dependent RNA polymerase. The results shown in Fig. 1C are the means of three independent preparations of each RNA electroporated into Huh-7.5 cells, with the standard error of the mean used for generation of error bars. Figure 1C clearly illustrates the robust replication of the parental Con1/SGneo GIT replicon (5.80 × 105 CFU/μg) and the lack of replication of the pol− control RNA (0 CFU/μg). Replicons bearing deletion A (amino acids 283 to 293) displayed a replication phenotype similar to that of the wild-type control (2.80 × 105 CFU/μg), indicating that this region of NS5A was not required for HCV RNA replication. Replicons bearing deletions B (amino acids 294 to 303), C (amino acids 304 to 313), D (amino acids 314 to 322), E (amino acids 323 to 337), and F (amino acids 338 to 349) were defective, yielding no G418-resistant colonies. These data suggest that deletions B to F may contain one or more residues essential for HCV RNA replication. The remaining deletions, deletions G through O, produced no obvious defect, with transduction efficiencies close to that observed for the parent. These data indicate that the region of NS5A corresponding to deletions G through O, some 91 amino acids, does not contain any specific residues required for HCV RNA replication.
Alanine scanning of lethal deletions.
Based upon this low-resolution deletion mapping study, the importance of individual amino acids for RNA replication within the lethal deletions was probed by alanine scanning mutagenesis. This was accomplished by constructing triple alanine scanning mutations for each deleterious deletion. Mutants were designated with the parental deletion letter code and a number as a unique identifier (for example, B1 is the first triple alanine mutation of the B deletion region). Triple alanine mutants that displayed an impaired G418 replication phenotype were then scanned by single alanine mutagenesis in an attempt to identify the specific residue(s) responsible for the observed phenotype.
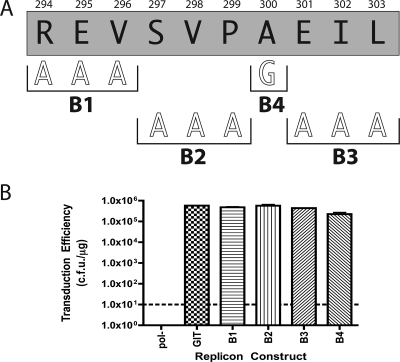
Triple alanine mutational analysis was initially performed on deletion B (Fig. 2). The mutant designated B1 (R294A, E295A, and V296A) yielded a transduction efficiency of 4.8 × 105 CFU/μg, indicating that these three amino acids did not contribute significantly to the RNA replication activity of NS5A. Similarly, mutants B2 (S297A, V298A, and P299A) and B3 (E301, I302, and L303) did not display any discernible defect, with transduction efficiencies of 5.7 × 105 and 2.24 × 105 CFU/μg, respectively. An alanine residue present within the wild-type NS5A sequence of the B deletion region of NS5A was substituted with glycine to determine its importance. Mutant B4, which possessed a glycine residue in place of alanine at amino acid residue 300, displayed a transduction efficiency of 4.36 × 105 CFU/μg. Clearly, none of the triple alanine substitutions in the B deletion region nor the single alanine-to-glycine mutation resulted in any discernible defect in this assay. On the basis of these results, we concluded that the B deletion region was important for RNA replication but contained no specific amino acid side chain requirements for this process.
FIG. 2.
(A) Sequence of the B deletion from Fig. 1B showing additional alanine scanning mutagenesis performed. Black letters within the gray shaded box correspond to the single-letter code amino acid sequence of deletion B. Numbers above the shaded box refer to the amino acid number of the NS5A protein. White outline block letters indicate the amino acid substitutions introduced at the positions indicated by the black horizontal brackets. Bold black letters under the horizontal brackets refer to the designation for the clone containing the amino acid substitution above the brackets. For example, the boldface letter B1 refers to a triple alanine mutation of R294, E295, and V296. (B) Graph of transduction efficiency for each replicon construct shown in panel A. Graph labeling and controls are as described in the legend to Fig. 1C. The letters B1 through B4 correspond to the designations in panel A.
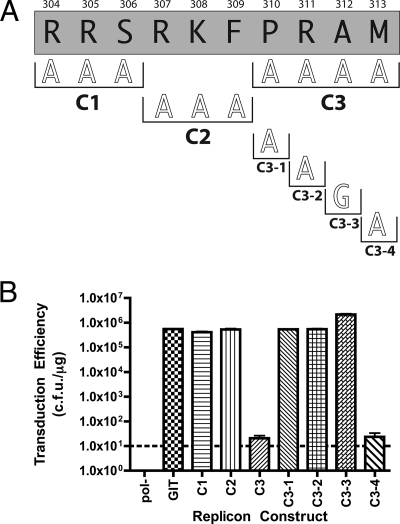
A similar approach was employed to examine the region of NS5A corresponding to deletion C (Fig. 3). Replicons bearing the C1 mutation (R304A, R305A, and S306A) produced a wild-type phenotype with a transduction efficiency of 4.13 × 105 CFU/μg. Similarly, replicons with the C2 mutation (R307A, K308A, and F309A) displayed replication levels similar to that of the parental control (5.33 × 105 CFU/μg). Interestingly, mutant C3 (P310A, R311A, A312A, and M313A) yielded a transduction efficiency of only 20 CFU/μg, close to the detection limit of the replicon assay (10 CFU/μg). Therefore, we generated individual amino acid substitutions within the C3 region. Mutants C3-1 (P310A), C3-2 (R311A), and C3-3 (A312G) yielded transduction efficiencies of 5.40 × 105 CFU/μg, 5.46 × 105 CFU/μg, and 2.15 × 106 CFU/μg, respectively, indicating that none of these residues is essential for HCV RNA replication. Interestingly, mutant C3-3 had a reproducible fourfold increase in RNA replication relative to the parent, suggesting that this mutation may confer a weak adaptive change similar to those originally described by Blight et al. (3). The final mutation generated in the C3 region was C3-4 (M313A), and replicons bearing this change exhibited a transduction efficiency of only 23 CFU/μg. This result is similar to the phenotype observed for the parental C3 mutant, suggesting that residue M313 was likely responsible for the C3 defect.
FIG. 3.
(A) Sequence of the C deletion from Fig. 1B showing additional alanine scanning mutagenesis performed. Numbering and clone designation conventions are as described in the legend to Fig. 2A. C1, C2, and C3 refer to triple alanine mutation of the designated residues. C3-1 through C3-4 designate individual alanine or glycine mutations of residues above the black brackets. (B) Graph of transduction efficiency for each replicon construct shown in panel A. Graph labeling and controls are as described in the legend to Fig. 1C. The letters C1 through C3 and C3-1 through C3-4 correspond to the designations in panel A.
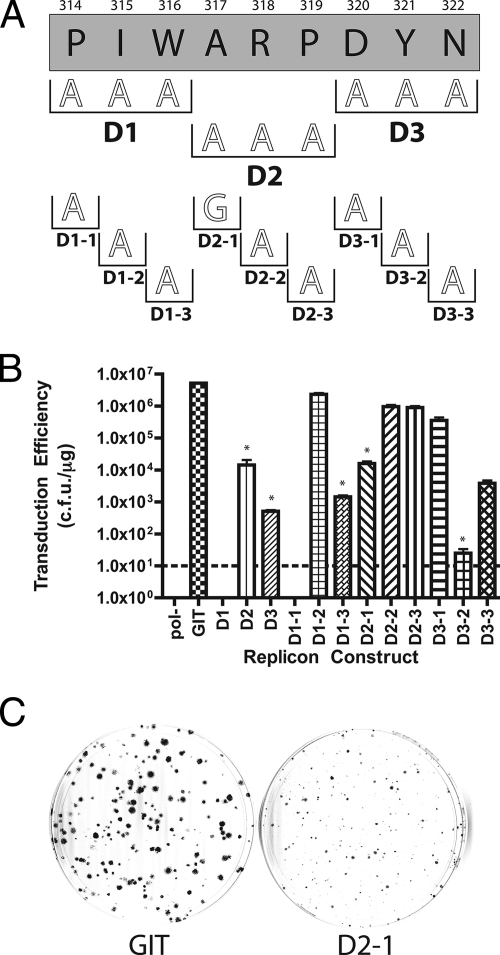
A series of three triple alanine mutations, D1 (P314A, I315A, and W316A), D2 (R318A and P319A), and D3 (D320A, Y321A, and N322A) were constructed to identify important residues within the lethal D series deletion mutant (Fig. 4). The D1 mutant was lethal, with no colonies observed in the replicon assay. The transduction efficiencies of the D2 (1.43 × 104 CFU/μg) and D3 (5.13 × 102 CFU/μg) mutants were considerably lower than that of the parental control (5.19 × 106 CFU/μg), but the mutants were still competent for RNA replication. Interestingly, the D2 and D3 mutant replicons produced a small colony phenotype. Colonies for these mutants were approximately 1/5 to 1/10 the size of the colonies generated by the parental replicon (Fig. 4C). Using single amino acid mutagenesis, we first investigated the lethal phenotype seen for replicons with the D1 mutation. Mutations D1-1 (P314A), D1-2 (I315A), and D1-3 (W316A) were generated, and the corresponding replicons were tested for replication competence. The D1-1 mutant was lethal, yielding no colonies. The D1-2 mutant had a replication phenotype similar to that of the parental replicon (2.32 × 106 CFU/μg). The D1-3 replicon was moderately impaired but retained the ability to confer G418 resistance (1.43 × 103 CFU/μg). These data suggest that the proline residue at position 314 is important for HCV RNA replication.
FIG. 4.
(A) Sequence of the D deletion from Fig. 1B showing additional alanine scanning mutagenesis performed. Numbering and clone designations are as described in the legends to Fig. 2A and 3A. (B) Graph of transduction efficiency for each replicon construct shown in panel A. Graph labeling and controls are as described in the legend to Fig. 1C. Asterisks above column bars indicate replicons that showed a small colony phenotype in the replication assay. (C) Representative colony phenotypes for normal and small colony mutants. Crystal violet-stained colonies following G418 selection. GIT represents 104 cells plated in a 100-mm dish for the parental strain GIT replicon. D2-1 represents 106 cells plated in a 100-mm dish of a small colony phenotype mutant to give approximately the same number of colonies as the GIT control.
Mutations D2-1 (A317G), D2-2 (R318A), and D2-3 (P319A) were generated to investigate the small colony phenotype observed for the D2 mutant. As expected, replicons bearing D2-1 (1.60 × 104 CFU/μg), D2-2 (9.63 × 105 CFU/μg), and D2-3 (8.93 × 105 CFU/μg) were all competent for G418 transduction. However, only replicons with the D2-1 mutation yielded the small colony phenotype. Individual mutations D3-1 (D320A), D3-2 (Y321A), and D3-3 (N322A) were generated and analyzed. Replicons with D3-1 (3.53 × 105 CFU/μg) and D3-3 (3.83 × 103 CFU/μg) were moderately impaired but did produce G418-resistant colonies of normal size. D3-2, with a transduction efficiency of only 25 CFU/μg, was severely impaired and produced small colonies that resembled those of the D3 parent. The D series deletion and subsequent alanine mutants therefore identified one residue required for RNA replication (P314) and two residues (A317 and Y321) leading to a small colony phenotype.
The next series of mutations were generated to analyze the importance of residues within the lethal E deletion (Fig. 5). A series of five triple alanine mutations were generated spanning the residues in the original E deletion, including E1 (P323A, P324A, and L325A), E2 (L326A, E327A, and S328A), E3 (W329A, K330A, and D331A), E4 (P332A, D333A, and Y334A), and E5 (V335A, P336A, and P337A). Replicons containing mutations E1, E2, and E3 were not viable and yielded no G418-resistant colonies. Replicons harboring the E4 and E5 mutations were both replication competent, although to various degrees, with E4 exhibiting a wild-type phenotype (2.18 × 106 CFU/μg) and E5 exhibiting a severely impaired but replication-competent phenotype (2.35 ×102 CFU/μg). Single alanine mutations were generated for each residue in the E1, E2, and E3 triple alanine mutants to identify specific residues responsible for their lethal phenotypes. The E1 series mutants, E1-1 (P323A) and E1-3 (L325A), were replication competent with transduction efficiencies of 1.65 × 106 CFU/μg and 1.76 × 106 CFU/μg, respectively. The E1-2 mutant was lethal for RNA replication in the GIT backbone. The E2 series individual alanine mutants E2-1 (L326A) (2.07 × 106 CFU/μg), E2-2 (E327A) (1.68 × 106 CFU/μg), and E2-3 (S328A) (1.15 × 106 CFU/μg) yielded a wild-type G418 transduction phenotype. Similar to the B deletion, the E2 region of NS5A is important for RNA replication but contained no specific amino acid side chain requirements for this process.
FIG. 5.
(A) Sequence of the E deletion from Fig. 1B showing additional alanine scanning mutagenesis performed. Numbering and clone designations are as described in the legends to Fig. 2A and 3A. (B) Graph of transduction efficiency for each replicon construct shown in panel A. Graph labeling and controls are as described in the legend to Fig. 1C.
The final mutations generated for the E series mapping were E3-1 (W329A), E3-2 (K330A), and E3-3 (D331A). Replicons bearing the E3-1 mutation were nonviable, indicating that W329 is an essential residue for replication. Replicons bearing E3-2 (4.13 × 104 CFU/μg) and E3-3 (1.14 × 106 CFU/μg) were replication competent but to differing degrees, with E3-2 moderately impaired and E3-3 fully competent for replication. Collectively, our data for the E region of NS5A implicate amino acids P324, S328, and W329 and the residues within the E2 triple alanine mutant (L326, E327, and S328) as residues important for HCV RNA replication.
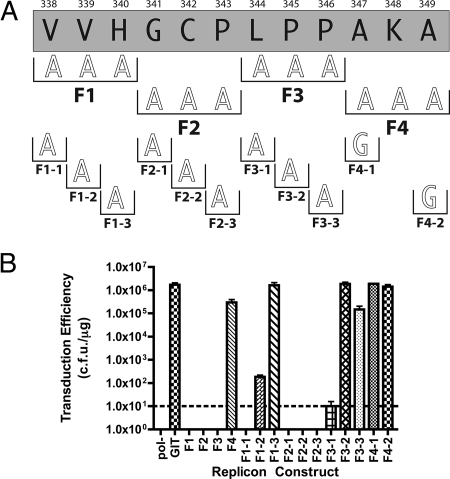
The final lethal deletion mutation analyzed was region F (Fig. 6). Again, we first generated triple alanine mutations spanning the original deletion. These mutations were designated F1 (V338A, V339A, and H340A), F2 (G341A, C342A, and P343A), F3 (L344A, P345A, and P346A), and F4 (K348A). Replicons containing F1, F2, and F3 were not viable, indicating that important residues were contained in all three regions. The replicon containing F4 yielded a transduction efficiency of 2.99 × 105 CFU/μg, a very moderate level of impairment, indicating that K348 is not essential for replication. The parental Con1/SG-neo GIT replicon yielded a transduction efficiency of 1.70 × 106 CFU/μg in this experiment. Individual residues required for replication within the lethal F1, F2, and F3 deletions were then scanned by single alanine mutagenesis. The F1-1 mutation (V338A) generated replicons with a lethal phenotype. The F1-2 (V339A) mutant was severely impaired, yielding a transduction efficiency of 1.83 × 102 CFU/μg. F1-3 (H340A) generated a wild-type phenotype (1.62 × 106 CFU/μg), indicating that H304 is not essential. In contrast, individual alanine substitutions spanning the F2 triple alanine mutation, F2-1 (G341A), F2-2 (C342A), and F2-3 (P343A), were all lethal, demonstrating the importance of each of these residues. The F3-1 mutant (L344A) displayed only a moderate defect (1.48 × 105 CFU/μg). Finally, the two alanine residues present in the F4 region of the wild-type NS5A sequence were substituted with glycine residues. These mutations, designated F4-1 (A347G) and F4-2 (A349G), were generated, and the corresponding replicons were assayed for replication fitness. F4-1 (1.86 × 106 CFU/μg) and F4-2 (1.41 × 106 CFU/μg) produced results very similar to the parental replicon, indicating that these residues have little impact on HCV RNA replication.
FIG. 6.
(A) Sequence of the F deletion from Fig. 1B showing additional alanine scanning mutagenesis performed. Numbering and clone designations are as described in the legends to Fig. 2A and 3A. (B) Graph of transduction efficiency for each replicon construct shown in panel A. Graph labeling and controls are as described in the legend to Fig. 1C.
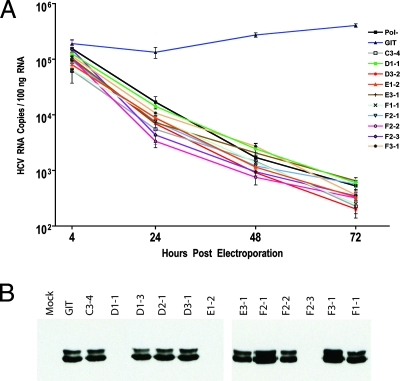
With a total of 10 individual amino acid mutants lethal or near lethal (less than 50 CFU/μg) for RNA replication in the G418 transduction assay, we proceeded to test the replication fitness of these mutants in a transient RNA replication assay using quantitative real-time reverse transcriptase PCR. This assay was chosen to allow a direct measure of HCV RNA replication, so as to eliminate some of the limitations of the G418 transduction assay. The G418 transduction assay measures the ability of a replicon RNA to initiate RNA replication, persist as a stable nontoxic replicating element, and generate RNA levels and neomycin phosphotransferase II translation product that are sufficient to allow the transduced cell to grow into a scorable colony. Although the number of colonies observed in these assays is often referred to as a measure of RNA replication fitness, it is usually not clear what step in the process of generating a G418-resistant colony is affected by an engineered mutation. It is possible that mutants generating low levels of G418 resistance actually represent revertants or pseudorevertants, evolved during the extend course of G418 selection, that restore replicative fitness. The transient-replication assay avoids many of these potential pitfalls and allows a view of the kinetics of RNA replication, which often can be used to detect reversions. Our analysis of the lethal or near lethal mutants in this transient-replication assay, compared to a parental GIT replicon and a defective pol− replicon, is presented in Fig. 7A. The data clearly demonstrate the lack of replication of our panel of mutants over a 72-h period compared to a replication-competent GIT replicon and a pol− replicon control.
FIG. 7.
Secondary analysis of selected mutants. (A) Transient RNA replication analysis of selected mutants by real-time reverse transcriptase PCR assay of HCV RNA. Values shown are normalized to glyceraldehyde-3-phosphate dehydrogenase mRNA and expressed as the number of HCV RNA copies per 100 ng of total RNA. Only the replication-competent GIT RNA was capable of producing detectable RNA replication in this experiment. (B) Transient protein expression of various NS5A mutants analyzed by Western blotting with an anti-NS5A monoclonal antibody. The two phosphoforms of NS5A are clearly visible.
We next investigated whether the defects seen in our replication assays could be linked to differences in NS5A stability or phosphorylation state for our individual alanine mutants. We used a recombinant vaccinia virus expressing T7 RNA polymerase coupled with transient transfection of our mutant replicon plasmids, which are driven by a T7 promoter. After allowing expression of the viral polyprotein to proceed for 24 h, cells were lysed in the presence of detergent, and lysates were centrifuged at high speed to separate soluble material. The soluble fraction was then used for Western blotting and probed with an NS5A-specific monoclonal antibody believed to recognize an epitope in domain III (T. L. Tellinghuisen and C. M. Rice, unpublished data). The results of this analysis are shown in Fig. 7B. Some obvious defects in protein translation, stability, or solubility were observed for D1-1 (P314A), E1-2 (P324A), and F2-3 (P343A), as no NS5A was detected in these samples. NS3 and NS5B could readily be detected in these samples, confirming our DNA sequence analysis data showing that these mutations did not disrupt the HCV open reading frame (data not shown). It is interesting to note that all of these residues represent mutation of proline residues, although the significance of this observation is not clear. The remaining mutants had no visible defect in NS5A detection. No differences in the phosphorylation state of NS5A were apparent in this analysis, with all of the mutant NS5A proteins detected possessing levels of the NS5A phosphoforms similar to that of the parental GIT replicon.
The well documented issue of genetic incompatibility among different classes of adaptive mutations (8, 25), combined with mutations we have generated in NS5A, a “hot spot” for adaptive mutations (3), made it of interest to analyze our lethal mutations in a different replicon genetic background. The low efficiency of replication of the wild-type Con1 strain of HCV in cell culture eliminates the possibility of testing our panel of mutations in this nonadapted replicon system (3, 27). We therefore elected to test our mutations in the context of the S2204I-bearing replicon (3). This replicon bears a single adaptive mutation in the NS5A protein coding sequence at a site not modified in our mapping experiments. The results of this analysis are summarized in Table 1. We also included the two moderately impaired replicons that produced a small colony phenotype in GIT in this analysis. A number of interesting differences exist when mutations are analyzed in both the GIT and S2204I backgrounds (Fig. 8B and C). Several mutations show increased replication in the S2204I background, including C3-4 (M313A) which increased from 20 CFU/μg to 1.20 × 104 CFU/μg, E1-2 (P324A) which changed from a lethal phenotype in GIT to a moderately impaired phenotype in S2204I (6.75 × 105 CFU/μg), and F2-1 (G341A) which was also lethal in GIT and displayed a very low level of replication in S2204I (10 CFU/μg). It is interesting to note that C3-4 (M313A) and E1-2 (P324A), although showing an increase in replication fitness in S2204I relative to GIT, changed the colony size phenotype from normal in GIT to small in S2204I. This is in contrast to replicons that had a small colony phenotype in GIT with a moderate level of impaired replication, D1-3 (W316A) and D2-1 (A317G) and retained this small colony phenotype in S2204I. The situation was not as simple as S2204I simply increasing replication of our mutants, as D1-3 (W316A) showed a decreased replication fitness in S2204I (20 CFU/μg) relative to that seen in GIT (1.43 × 103 CFU/μg). Aside from these changes, the bulk of the mutants analyzed displayed the same or very similar phenotypes in both genetic backgrounds (Fig. 8C).
TABLE 1.
Results of analysis of mutations in the context of the GIT and S2204I-bearing replicon
| Constructa | GIT replicationb | GIT colony size | S2204I replicationb | S2204I colony size | Western blot | Real-time PCR (in GIT) |
|---|---|---|---|---|---|---|
| GIT | 1.6 × 106-2.18 × 106 | Normal | NAc | NA | Normal | Replication |
| S2204I | NA | NA | 3.64 × 106 | Normal | NDd | ND |
| C3-4 (M313A) | 23 | Normal | 1.20 × 104 | Small | Normal | No replication |
| D1-1 (P314A) | 0 | 0 | Defect | No replication | ||
| D1-3 (W316A) | 1.43 × 103 | Small | 20 | Small | Normal | No replication |
| D2-1 (A317G) | 1.60 × 104 | Small | 1.00 × 104 | Small | Normal | ND |
| D3-2 (Y321A) | 25 | Small | 20 | Small | Normal | No replication |
| E1-2 (P324A) | 0 | 6.75 × 105 | Small | Defect | No replication | |
| E3-1 (W329A) | 0 | 0 | Normal | No replication | ||
| F1-1 (V338A) | 0 | 0 | Normal | No replication | ||
| F2-1 (G341A) | 0 | 10 | Normal | Normal | No replication | |
| F2-2 (C342A) | 0 | 0 | Normal | No replication | ||
| F2-3 (P343A) | 0 | 0 | Defect | No replication | ||
| F3-1 (L344A) | 10 | Normal | 10 | Normal | Normal | No replication |
Clone designations used in manuscript (single-letter amino acid code of the wild-type residue, amino acid position in the NS5A protein, and the amino acid residue present in the mutant).
Replication as determined by G418 transduction assay (in CFU per microgram of input RNA).
NA, not applicable.
ND, not determined.
FIG. 8.
(A) Schematic representation of the hepatitis C virus NS5A protein. Labeling is essentially as described in the legend to Fig. 1A. (B) Enlargement of the domain II region of NS5A indicating the locations of residues determined to be important in RNA replication. The large dark gray box from 294 to 303 indicates the position of the lethal deletion B1. Additional small dark gray boxes indicate the locations of other residues that produced lethal or near lethal phenotypes in the replicon system assays. Light gray boxes designate residues that produced a small colony phenotype in the replicon system assay yet still produced significant levels of colony formation. Asterisks indicate positions that yielded small colony phenotypes independent of G418 transduction efficiency. Numbers labeling these boxes refer to the amino acid number of the highlighted residue in the NS5A protein. (C) Enlargement of the region in domain II containing residues analyzed in the context of the S2204I replicon. Residues shaded in dark gray or light gray designate replication phenotypes as described above for panel B. Asterisks indicate replicons producing a small colony phenotype. Regions not highlighted in gray were not analyzed in the S204I replicon.
DISCUSSION
The data presented here provide a comprehensive map of amino acids within NS5A domains II and III that are dispensable or required for HCV RNA replication in the context of a genotype 1b GIT replicon. By constructing a series of deletion and alanine scanning mutations, a total of 23 amino acids in domain II have now been identified that are required for efficient G418 transduction in the GIT subgenomic replicon system (Fig. 8). These residues include a 10-amino-acid deletion (deletion B) and a triple alanine mutant (mutant E2) in which no specific required residues could be identified by individual residue alanine scanning. Perhaps these mutations represent a disruption of the spacing of important replication elements flanking these regions or the disruption of an important secondary structural element that is required for the global fold of NS5A. Since all of the residues within these regions can be individually substituted with alanine, no essential side chain interactions are required for the function of this region. Also included in our list of 23 essential residues are 10 positions that produce a lethal or near lethal phenotype when changed to alanine. An additional two amino acids within this domain generate a small colony phenotype while only moderately impairing replication, suggesting that these residues, although not absolutely required for replication, are important. The data clearly indicate that, in the context of small individual deletions, domain III is not required for RNA replication. Previous analysis of the codon substitution rates among HCV isolates suggest no conserved RNA elements exist in these regions of the HCV genome, suggesting our mutations are functioning at the protein level (37). Collectively, these data, in conjunction with our previous studies pertaining to domain I of NS5A, suggest that the essential RNA replicase activities of NS5A lie within domains I and II.
Our results agree with several previous studies indicating the flexibility of NS5A domain III. In 2004, Moradpour et al. first described a transposon-based screening effort that identified at least two sites in domain III that tolerated insertion of green fluorescent protein (32). A deletion/mutation study of phosphoacceptor sites present in domain III indicated that although residues in domain III were responsible for modulating NS5A hyperphosphorylation, these mutants were viable in the replicon system (2). On the basis of these results, in 2005, Appel et al. identified another independent site within domain III that was tolerant of the insertion of fluorescent fusion proteins (2). More recently, a second transposon and deletion analysis study identified additional regions in domains II and III that could tolerate insertion of green fluorescent protein without disruption of replicon viability (24). In this later study, the two regions tolerant of insertions were quite large (68 to 74 amino acids), suggesting that substantial portions of domains II and III of NS5A could be altered by insertional mutagenesis and not deleteriously affect replication (24). Deletions generated in this previous study indicated that portions of domains II and III were dispensable for replication but did not provide a complete map of which residues are essential. The viable deletion and insertion mutants identified by Liu et al. (24) outline the locations of important residues that were identified in our study. These previous studies, in conjunction with the work presented in this paper, highlight the extreme plasticity of domain III and to a lesser extent domain II.
The residues we have shown to be essential for RNA replication in the context of the GIT replicon are, not surprisingly, largely conserved among the 35 HCV genotype reference sequences in the HCV Los Alamos sequence database, with some notable exceptions (19). Positions mutated in clones D1-1 (P314), D3-2 (Y321), E1-2 (P324), E2-3 (S328), E3-1 (W329), F2-1 (G341), and F2-2 (C342) are conserved among all of the HCV genotype reference sequences. The C3-4 (M313) position is present as a methionine in some, but not all, genotype 1b isolates. The majority of reference sequences have a leucine at this position, with one isolate having a phenylalanine. The similar sizes of methionine and leucine side chains suggest that the size of the residue at this position might be more important than the presence of a specific side chain, perhaps involving a hydrophobic contact. The F1-1 position (V338) is a valine in the reference sequences of genotype 1a, 1b, 1c, and some 2a and 2c sequences. In other reference strains, this position is a leucine, threonine, or glutamine. It is interesting to note that all the HCV reference sequences have a valine residue at position 338 or 339, perhaps indicating that some differences exist in the spacing of residues in this region within HCV isolates. F2-3 (P343) is conserved among genotype 1a, 1b, 1c, and 5a sequences but is present as an alanine in all other isolates. It is therefore interesting that an alanine at this position is not tolerated in either the GIT or S2204I background but is the most common residue at this position among all of the HCV isolates. Position F3-1 (L344) is a leucine in the majority of the reference sequences, but valine and isoleucine are also present at this position, suggesting a small hydrophobic residue is required at this location. Similarly, residues that generate a small colony phenotype with only a moderate impairment of RNA replication are conserved among all of the current HCV genotype reference sequences, as is that of the small colony phenotype residue D3-2, which severely impairs RNA replication.
We investigated the small colony phenotype phenomenon, thinking that these colonies harbored replicons bearing revertants or pseudorevertants that partially restored replication. We therefore cloned colonies from these experimental samples and subjected the viral RNA in these clones to sequence analysis. In all cases, we were able to identify the correct input mutation, indicating that no same site reversion had occurred. Further sequencing of the viral polyprotein failed to identify any additional mutations in these clones, indicating that these mutants are capable of replication without additional mutations, albeit at a level lower than that of the wild type.
Perhaps the most surprising result in our analysis is the striking differences of the importance of some amino acid residues in different cell culture-adapted replicons. Although the majority of our mutations yielded similar phenotypes in both the GIT and S2204I replicons, some notable differences existed (Fig. 8B and C). These include fairly dramatic increases in replication fitness in S2204I, such as in the case of C3-4 (M313A), E1-2 (P324A), and to a lesser extent F2-1 (G341A). In the case of the most dramatic increases, C3-4 and E1-2, the S2204I replicons exhibited a small colony phenotype, indicating that these mutants were less than ideal for replication in the context of the S2204I clone. What controls whether a replicon generates a lethal or severely impaired phenotype with normal colony size in one replicon background and a more efficient replication yet small colony phenotype in another background is not known. This is particularly confusing when the observation that some mutants had small colony phenotypes in both backgrounds is considered. The mechanism that governs the generation of these colony phenotypes remains to be determined. It is interesting to note that the E1-2 (P324A) protein was not detectable in Western blots of transiently expressed protein in GIT and was incompetent for replication in this context yet still retained replication competence in S2204I, suggesting that the genetic backbone used might influence the stability of at least some NS5A mutations. Perhaps less dramatic variations of this concept control replication fitness and colony morphology in our other mutants. Further complicating the issue is the fact that D1-3 (W316A) was considerably more fit in the context of the GIT replicon than in S2204I. Clearly, the adaptive mutation context in which a mutation is analyzed can have a significant impact on the experimental results, at least in the case of some mutations. Unfortunately, our understanding of the mechanism by which adaptive mutations increase RNA replication is incomplete, making interpretation of these results difficult. The majority of our mutations yielded the same or similar phenotypes in both replicon genetic backgrounds, validating our mapping experimental results. It is also important to note that although there were some dramatic differences in replication fitness and colony morphology for some mutants when comparing genetic backgrounds, in no case did any mutant from our panel display a wild-type phenotype in either background.
A large number of cellular protein interactions with NS5A map to regions that we have shown to be nonessential for RNA replication in this study. The vast majority of these interactions have not been shown to have any effect on HCV RNA replication (for a review, see reference 29). Many of these proteins interact with the tandem polyproline motifs found spanning deletions F and G and include Grb2, Lyn, Hck, Fyn, and Lck (28, 39). In the case of residues in deletion F, only one proline (P343) residue and one leucine (L344) residue appear to be essential for replicon function in our experiments, with several remaining residues implicated in these interactions viable in our experiments. The mutant with deletion G, spanning the majority of the polyproline motif and deleting residues known to be important for the aforementioned interactions, is also viable in our experiments. This is perhaps not surprising, as mutation of these residues by MacDonald et al. (30) disrupted interaction with these cellular factors but failed to hinder HCV RNA replication. Cellular proteins have also been mapped to other larger interaction surfaces in domain III, including p53 and human TATA box-binding protein-associated factor II, proteins proposed to be involved in the avoidance of apoptosis in HCV-infected cells (20). We made no attempt in our experiments to monitor the inhibition of p53 apoptosis and therefore cannot comment on the importance of these interactions in light of our mutants, save for saying that no mutants with deletions in domain III showed any decreased cell viability in our assays. Our deletions in domain III overlap at least a portion of the interaction sites mapped for the HCV core protein (11), but our subgenomic replicons do not express HCV core, making interpretation of the importance of the NS5A-core interaction impossible in our experiments. Recently, Pfeiffer and Kirkegaard (35) identified a mutation in domain III that conferred limited resistance to ribavirin. Additionally, deletions in this region were important in establishing at least a low level of HCV replication in nonhepatic cell lines of human and mouse origin, although the mechanism behind this observation is not known (47). Collectively, these NS5A interactions may participate in other aspects of the HCV life cycle that cannot be addressed with the replicon system but yet may be critical for replication, persistence, and disease processes in vivo.
Our data provide a map of amino acid residues in NS5A domains II and III essential for RNA replication in the context of a genotype 1b GIT replicon. Using a combination of deletion and alanine scanning mutagenesis, a total of 23 amino acids in domain II have now been identified that are required for efficient G418 transduction in the GIT subgenomic replicon system, with domain III not essential in this process. We have identified a number of interesting mutations that confer very different phenotypes in the context of different adaptive mutations. This essential residue map provides a working template for planning additional studies to probe the role of domain II and III residues in RNA replication and other aspects of the HCV life cycle.
Acknowledgments
We thank Matthew Evans, Brett Lindenbach, Shihyun You, Christopher Jones, and Joseph Marcotrigiano for scientific discussions related to this article and Kathleen Hefferon for editing the manuscript.
T.L.T. was supported, in part, by a fellowship from the National Institutes of Health Ruth L. Kirschstein National Research Service Award (5F32 AI51820-03) and a Career Development Award (1 K22 AI067645-01) granted through the National Institute of Allergy and Infectious Disease. Additional financial support for this work came from grant 5 R01 CA57973-12 from the National Institutes of Health and the Greenberg Medical Research Institute (C.M.R.).
Footnotes
Published ahead of print on 21 November 2007.
REFERENCES
- 1.Anonymous. 1997. Hepatitis C: global prevalence. Wkly. Epidemiol. Rec. 72341-344. [PubMed] [Google Scholar]
- 2.Appel, N., T. Pietschmann, and R. Bartenschlager. 2005. Mutational analysis of hepatitis C virus nonstructural protein 5A: potential role of differential phosphorylation in RNA replication and identification of a genetically flexible domain. J. Virol. 793187-3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blight, K. J., A. A. Kolykhalov, and C. M. Rice. 2000. Efficient initiation of HCV RNA replication in cell culture. Science 2901972-1974. [DOI] [PubMed] [Google Scholar]
- 4.Blight, K. J., J. A. McKeating, and C. M. Rice. 2002. Highly permissive cell lines for hepatitis C virus genomic and subgenomic RNA replication. J. Virol. 7613001-13014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brass, V., E. Bieck, R. Montserret, B. Wolk, J. A. Hellings, H. E. Blum, F. Penin, and D. Moradpour. 2002. An amino-terminal amphipathic alpha-helix mediates membrane association of the hepatitis C virus nonstructural protein 5A. J. Biol. Chem. 2778130-8139. [DOI] [PubMed] [Google Scholar]
- 6.Choo, Q.-L., G. Kuo, A. J. Weiner, L. R. Overby, D. W. Bradley, and M. Houghton. 1989. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science 244359-362. [DOI] [PubMed] [Google Scholar]
- 7.Egger, D., B. Wolk, R. Gosert, L. Bianchi, H. E. Blum, D. Moradpour, and K. Bienz. 2002. Expression of hepatitis C virus proteins induces distinct membrane alterations including a candidate viral replication complex. J. Virol. 765974-5984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Evans, M. J., C. M. Rice, and S. P. Goff. 2004. Genetic interactions between hepatitis C virus replicons. J. Virol. 7812085-12089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Failla, C., L. Tomei, and R. DeFrancesco. 1994. Both NS3 and NS4A are required for proteolytic processing of hepatitis C virus nonstructural proteins. J. Virol. 683753-3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fuerst, T. R., E. G. Niles, F. W. Studier, and B. Moss. 1986. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc. Natl. Acad. Sci. USA 838122-8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goh, P. Y., Y. J. Tan, S. P. Lim, S. G. Lim, Y. H. Tan, and W. J. Hong. 2001. The hepatitis C virus core protein interacts with NS5A and activates its caspase-mediated proteolytic cleavage. Virology 290224-236. [DOI] [PubMed] [Google Scholar]
- 12.Gosert, R., D. Egger, V. Lohmann, R. Bartenschlager, H. E. Blum, K. Bienz, and D. Moradpour. 2003. Identification of the hepatitis C virus RNA replication complex in Huh-7 cells harboring subgenomic replicons. J. Virol. 775487-5492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grakoui, A., D. W. McCourt, C. Wychowski, S. M. Feinstone, and C. M. Rice. 1993. A second hepatitis C virus-encoded proteinase. Proc. Natl. Acad. Sci. USA 9010583-10587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grakoui, A., D. W. McCourt, C. Wychowski, S. M. Feinstone, and C. M. Rice. 1993. Characterization of the hepatitis C virus-encoded serine proteinase: determination of proteinase-dependent polyprotein cleavage sites. J. Virol. 672832-2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grakoui, A., C. Wychowski, C. Lin, S. M. Feinstone, and C. M. Rice. 1993. Expression and identification of hepatitis C virus polyprotein cleavage products. J. Virol. 671385-1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hijikata, M., H. Mizushima, T. Akagi, S. Mori, N. Kakiuchi, N. Kato, T. Tanaka, K. Kimura, and K. Shimotohno. 1993. Two distinct proteinase activities required for the processing of a putative nonstructural precursor protein of hepatitis C virus. J. Virol. 674665-4675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang, L., J. Hwang, S. D. Sharma, M. R. Hargittai, Y. Chen, J. J. Arnold, K. D. Raney, and C. E. Cameron. 2005. Hepatitis C virus nonstructural protein 5A (NS5A) is an RNA-binding protein. J. Biol. Chem. 28036417-36428. [DOI] [PubMed] [Google Scholar]
- 18.Jones, C. T., C. L. Murray, D. K. Eastman, J. Tassello, and C. M. Rice. 2007. Hepatitis C virus p7 and NS2 proteins are essential for production of infectious virus. J. Virol. 818374-8383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuiken, C., K. Yusim, L. Boykin, and R. Richardson. 2005. The Los Alamos hepatitis C sequence database. Bioinformatics 21379-384. [DOI] [PubMed] [Google Scholar]
- 20.Lan, K. H., M. L. Sheu, S. J. Hwang, S. H. Yen, S. Y. Chen, J. C. Wu, Y. J. Wang, N. Kato, M. Omata, F. Y. Chang, and S. D. Lee. 2002. HCV NS5A interacts with p53 and inhibits p53-mediated apoptosis. Oncogene 214801-4811. [DOI] [PubMed] [Google Scholar]
- 21.Liang, Y., C. B. Kang, and H. S. Yoon. 2006. Molecular and structural characterization of the domain 2 of hepatitis C virus non-structural protein 5A. Mol. Cells 2213-20. [PubMed] [Google Scholar]
- 22.Lindenbach, B. D., M. J. Evans, A. J. Syder, B. Wolk, T. L. Tellinghuisen, C. C. Liu, T. Maruyama, R. O. Hynes, D. R. Burton, J. A. McKeating, and C. M. Rice. 2005. Complete replication of hepatitis C virus in cell culture. Science 309623-626. [DOI] [PubMed] [Google Scholar]
- 23.Lindenbach, B. D., and C. M. Rice. 2001. Flaviviridae: the viruses and their replication, p. 991-1041. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed., vol. 1. Lippincott-Raven Publishers, Philadelphia, PA. [Google Scholar]
- 24.Liu, S., I. H. Ansari, S. C. Das, and A. K. Pattnaik. 2006. Insertion and deletion analyses identify regions of non-structural protein 5A of hepatitis C virus that are dispensable for viral genome replication. J. Gen. Virol. 87323-327. [DOI] [PubMed] [Google Scholar]
- 25.Lohmann, V., S. Hoffmann, U. Herian, F. Penin, and R. Bartenschlager. 2003. Viral and cellular determinants of hepatitis C virus RNA replication in cell culture. J. Virol. 773007-3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lohmann, V., F. Korner, A. Dobierzewska, and R. Bartenschlager. 2001. Mutations in hepatitis C virus RNAs conferring cell culture adaptation. J. Virol. 751437-1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lohmann, V., F. Korner, J. O. Koch, U. Herian, L. Theilmann, and R. Bartenschlager. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285110-113. [DOI] [PubMed] [Google Scholar]
- 28.Macdonald, A., K. Crowder, A. Street, C. McCormick, and M. Harris. 2004. The hepatitis C virus NS5A protein binds to members of the Src family of tyrosine kinases and regulates kinase activity. J. Gen. Virol. 85721-729. [DOI] [PubMed] [Google Scholar]
- 29.Macdonald, A., and M. Harris. 2004. Hepatitis C virus NS5A: tales of a promiscuous protein. J. Gen. Virol. 852485-2502. [DOI] [PubMed] [Google Scholar]
- 30.Macdonald, A., S. Mazaleyrat, C. McCormick, A. Street, N. J. Burgoyne, R. M. Jackson, V. Cazeaux, H. Shelton, K. Saksela, and M. Harris. 2005. Further studies on hepatitis C virus NS5A-SH3 domain interactions: identification of residues critical for binding and implications for viral RNA replication and modulation of cell signalling. J. Gen. Virol. 861035-1044. [DOI] [PubMed] [Google Scholar]
- 31.McCormick, C. J., S. Maucourant, S. Griffin, D. J. Rowlands, and M. Harris. 2006. Tagging of NS5A expressed from a functional hepatitis C virus replicon. J. Gen. Virol. 87635-640. [DOI] [PubMed] [Google Scholar]
- 32.Moradpour, D., M. J. Evans, R. Gosert, Z. Yuan, H. E. Blum, S. P. Goff, B. D. Lindenbach, and C. M. Rice. 2004. Insertion of green fluorescent protein into nonstructural protein 5A allows direct visualization of functional hepatitis C virus replication complexes. J. Virol. 787400-7409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pang, P. S., E. Jankowsky, P. J. Planet, and A. M. Pyle. 2002. The hepatitis C viral NS3 protein is a processive DNA helicase with cofactor enhanced RNA unwinding. EMBO J. 211168-1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Penin, F., V. Brass, N. Appel, S. Ramboarina, R. Montserret, D. Ficheux, H. E. Blum, R. Bartenschlager, and D. Moradpour. 2004. Structure and function of the membrane anchor domain of hepatitis C virus nonstructural protein 5A. J. Biol. Chem. 27940835-40843. [DOI] [PubMed] [Google Scholar]
- 35.Pfeiffer, J. K., and K. Kirkegaard. 2005. Ribavirin resistance in hepatitis C virus replicon-containing cell lines conferred by changes in the cell line or mutations in the replicon RNA. J. Virol. 792346-2355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sapay, N., R. Montserret, C. Chipot, V. Brass, D. Moradpour, G. Deleage, and F. Penin. 2006. NMR structure and molecular dynamics of the in-plane membrane anchor of nonstructural protein 5A from bovine viral diarrhea virus. Biochemistry 452221-2233. [DOI] [PubMed] [Google Scholar]
- 37.Smith, D. B., and P. Simmonds. 1997. Characteristics of nucleotide substitution in the hepatitis C virus genome: constraints on sequence change in coding regions at both ends of the genome. J. Mol. Evol. 45238-246. [DOI] [PubMed] [Google Scholar]
- 38.Tai, C.-L., W.-K. Chi, D.-S. Chen, and L.-H. Hwang. 1996. The helicase activity associated with hepatitis C virus nonstructural protein 3 (NS3). J. Virol. 708477-8484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tan, S.-L., H. Nakao, Y. He, S. Vijaysri, P. Neddermann, B. L. Jacobs, B. J. Mayer, and M. G. Katze. 1999. NS5A, a non-structural protein of hepatitis C virus, binds growth factor receptor-bound protein 2 adaptor protein in a src homology 3 domain/ligand-dependent manner and perturbs mitogenic signaling. Proc. Natl. Acad. Sci. USA 965533-5538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tellinghuisen, T. L., J. Marcotrigiano, A. E. Gorbalenya, and C. M. Rice. 2004. The NS5A protein of hepatitis C virus is a zinc metalloprotein. J. Biol. Chem. 27948576-48587. [DOI] [PubMed] [Google Scholar]
- 41.Tellinghuisen, T. L., J. Marcotrigiano, and C. M. Rice. 2005. Structure of the zinc-binding domain of an essential component of the hepatitis C virus replicase. Nature 435374-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tellinghuisen, T. L., M. S. Paulson, and C. M. Rice. 2006. The NS5A protein of bovine viral diarrhea virus contains an essential zinc-binding site similar to that of the hepatitis C virus NS5A protein. J. Virol. 807450-7458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tellinghuisen, T. L., and C. M. Rice. 2002. Interaction between hepatitis C virus proteins and host cell factors. Curr. Opin. Microbiol. 5419-427. [DOI] [PubMed] [Google Scholar]
- 44.Wakita, T., T. Pietschmann, T. Kato, T. Date, M. Miyamoto, Z. Zhao, K. Murthy, A. Habermann, H. G. Krausslich, M. Mizokami, R. Bartenschlager, and T. J. Liang. 2005. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat. Med. 11791-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yi, M., R. A. Villanueva, D. L. Thomas, T. Wakita, and S. M. Lemon. 2006. Production of infectious genotype 1a hepatitis C virus (Hutchinson strain) in cultured human hepatoma cells. Proc. Natl. Acad. Sci. USA 1032310-2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhong, J., P. Gastaminza, G. Cheng, S. Kapadia, T. Kato, D. R. Burton, S. F. Wieland, S. L. Uprichard, T. Wakita, and F. V. Chisari. 2005. Robust hepatitis C virus infection in vitro. Proc. Natl. Acad. Sci. USA 1029294-9299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhu, Q., J. T. Guo, and C. Seeger. 2003. Replication of hepatitis C virus subgenomes in nonhepatic epithelial and mouse hepatoma cells. J. Virol. 779204-9210. [DOI] [PMC free article] [PubMed] [Google Scholar]