Abstract
After fusion of the human immunodeficiency virus type 1 (HIV-1) envelope with the host cell membrane, the HIV-1 core enters the cell cytoplasm. Core components are then restructured to form the reverse transcription complex (RTC); the biochemical details of this process are currently unclear. To investigate early RTC formation, we characterized the endogenous reverse transcription activity of virions, which was less efficient than reverse transcription during cell infection and suggested a requirement for a cell factor. The addition of detergent to virions released reverse transcriptase and capsid, and reverse transcription products became susceptible to the action of exogenous nucleases, indicating virion disruption. Disruption was coincident with the loss of the endogenous reverse transcription activity of virions, particularly late reverse transcription products. Consistent with this observation, the use of a modified “spin thru” method, which uses brief detergent exposure, also disrupted virions. The addition of lysates made from mammalian cell lines (Jurkat, HEK293T, and NIH 3T3 cells) to virions delipidated by detergent stimulated late reverse transcription efficiency. A complex with reverse transcription activity that was slower sedimenting than virions on a velocity gradient was greatly stimulated to generate full-length reverse transcription products and was associated with only relatively small amounts of capsid. These experiments suggest that cell factors are required for efficient reverse transcription of HIV-1.
Early in human immunodeficiency virus type 1 (HIV-1) replication, the core enters the cytoplasm after cell fusion of the viral envelope. The core then undergoes restructuring, leading to the formation of the reverse transcription complex (RTC) during a process commonly referred to as uncoating (31). The viral proteins, including matrix, Vpr, integrase, reverse transcriptase (RT), genomic RNA (reviewed in reference 44), and possibly host cell proteins (30) such as Gemin2 (22), are thought to constitute part of the RTC.
The role of capsid in RTC formation is unclear. Functional RTCs isolated from cells infected with HIV-1 particles lack capsid (14), and only trace amounts were reported to be associated with preintegration complexes (PIC) (28). Deoxyribonucleotides added to HIV-1 virions are taken up and incorporated into reverse transcription products, a process referred to as endogenous reverse transcription (ERT) (sometimes referred to as natural endogenous reverse transcription) (24, 47, 49, 50). ERT in HIV-1, which can be initiated without added detergent, differs from ERT in other retroviruses, which require nonionic detergent or compounds that can partially or completely solubilize the virion envelope (6, 19, 32). Detergent-free ERT is reported to partially disrupt the structure of the core (48). The loss of capsid prior to RTC formation and the disruption of cores during ERT suggest that the virion-derived core was restructured postentry. In support of this idea, an “uncoating” activity that released capsid from HIV-1 cores and was required for RT activation was attributed to a cellular factor (5). Hence, capsid is thought to dissociate soon after entry and, therefore, is generally not considered part of the HIV-1 RTC.
Other evidence suggests a more complex model than simply a requirement for the release of capsid for RTC formation. First, the release of capsid is not a prerequisite for RT activation, as reverse transcription occurs in intact virions during ERT. Second, RTCs were found, by using immunofluorescent microscopy, to be associated with capsid (27). Experiments with capsid mutants suggest that the HIV-1 core must remain intact for some period of time after virus entry (16, 17). Hence, it is possible that capsid may be associated with RTC, at least in the initial period postentry, but the association is weak and easily disrupted by biochemical procedures. A requirement for a regulated process of uncoating has been invoked to reconcile the lack of evidence for a physical association of capsid with the RTC, contrary to the genetic and microscopic data (10, 36). In other words, core structure is either wholly or partially maintained to complete some replication step, after which disassembly is completed.
HIV particles with irregular core morphologies (38) and altered stabilities (15) are unable to undergo reverse transcription in cells. For some of these mutants, the amount of RT in the core was less than in wild-type particles and there was greater retention of capsid (39). The above findings suggest two possible interpretations: either mutant core is defective for uncoating, which results in a block to reverse transcription or, alternatively, the maintenance of a specific core structure is required for reverse transcription. In support of the latter interpretation, virions treated with the nonhydrolyzable ATP analogue ATP-γ-S had irregular core morphology and were deficient in ERT and in reverse transcription in infected cells (21). The maintenance of a specific core structure in the cytoplasm may support the RTC complex and protect the RNA genome and product cDNA from host cell degradation.
Cell proteins may assist the uncoating or reverse transcription process (5, 29). Consistent with this idea, other host cell proteins are known to regulate early replication. For example, a number of host restriction factors which can block retroviral infection have now been characterized (reviewed in references 9 and 34). Target cell cyclophilin A is important for efficient HIV-1 replication by possibly blocking an unidentified restriction factor (25, 33, 37). A number of host proteins are known to be associated with the RTC/PIC. The barrier-to-autointegration factor, high-mobility-group protein, and LEDGEF/p75 assist integration (reviewed in reference 43). PML and Ini-1 are recruited by the PIC (42), and the latter stimulates transcription (4); and the survival motor neuron-interacting protein 1, Gemin2, modulates reverse transcription (22). In addition, HIV-1 infection of somatic cell mutants was blocked at an early stage of replication (18), and additional functions will likely be attributed to other host cell proteins. Recent work with two different cell systems suggests the existence of a host cell factor requirement (13, 35). In vitro reverse transcription systems also suggest a cell factor requirement during reverse transcription (24). Hence, cell factors may be required to complete early replication.
In this paper, we describe an in vitro reconstitution system to study RTC activity and to determine the effects of exogenous cell factors. As a starting point, HIV-1 virion preparations highly active for ERT activity were treated with detergents and the effect on reverse transcription was observed. Exposure to the nonionic detergent Triton X-100 reduced ERT activity in parallel with envelope removal and disruption of the core structure. When mammalian cell lysates were added to virions in which the envelope had been removed by mild detergent treatment, endogenous synthesis of late products was increased, suggesting that a cell factor assists the core's immature RTC. A complex was formed that sedimented more slowly than whole virions on a velocity gradient. These observations suggest that one or more cell factors may assist core uncoating.
MATERIALS AND METHODS
Cell lines and virus culture.
The MAGI CXCR4-expressing cell line (National Institutes of Health [NIH] AIDS research and reference reagent program) was grown in Dulbecco's modified Eagle's medium supplemented with 10% heat-inactivated newborn bovine serum, penicillin-streptomycin, glutamine, 0.2 mg/ml geneticin, 0.1 mg/ml hygromycin B, and 1 μg/ml puromycin. All other cells were grown in RPMI 1640 supplemented with 10% newborn bovine serum and penicillin-streptomycin. All cell lines were incubated at 37°C in 5% CO2 (standard conditions). A stock of HIVNL4.3 (2) was generated by transfection of the corresponding proviral DNA using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) into HEK293T cells according to the manufacturer's recommendations. Cell culture supernatants were removed at 48 h posttransfection and centrifuged (200 × g, 10 min), and the supernatant was filtered (0.45 μm) and stored in 1-ml aliquots at −80°C. To generate a MAGI cell line-derived virus stock, cells were plated to give approximately 50% confluence and then incubated overnight. The cells were infected with HIVNL4.3 (250 ng p24) and incubated under standard conditions for 2 h. The culture medium was then removed, and the cells were washed four times with sterile phosphate-buffered saline (PBS). Fresh medium was then added (Dulbecco's modified Eagle's medium with 10% fetal calf serum, 0.2 mg/ml G418, 0.1 mg/ml hygromycin, 1 μg/ml puromycin, and 1% penicillin-streptomycin), and the cells were incubated under standard conditions for 6 days. The supernatant was then harvested, centrifuged (1,000 rpm, 10 min), and filtered (0.45 μm). Virus was concentrated by ultracentrifugation (100,000 × g, 2 h, 4°C) on a 20% sucrose cushion, and the pellet was resuspended overnight in 1/10 of the culture volume of sterile PBS and stored in 50-μl aliquots at −80°C until needed. The plasmids used as described above were made available through the NIH AIDS research and reference reagent program.
ERT assays.
Reverse transcription products were generated by the addition of sucrose cushion-purified virus particles (equivalent to 10 ng p24) to a mixture containing 10 mM Tris, pH 7.4, 10 mM MgCl2, and 200 μM of each deoxynucleoside triphosphate in RPMI 1640 medium (final volume of 50 μl) for up to 18 to 20 h at 37°C; DNase I (500 U/ml), Triton X-100, and other optional additives were included as indicated in the text at the concentrations shown. When detergent was used, it was the last component added. A no-nucleotide control reaction mixture was always included. If relevant, lysate or fractions were added to 1/10 of the final volume unless indicated (5 to 20 μg of total protein). Products were extracted, once with an equal amount of phenol:chloroform:isoamyl alcohol (25:24:1) and once with chloroform. The extracts were ethanol precipitated, washed with 70% ethanol, dried, and resuspended in 100 μl of 0.1 mM EDTA. Purified reaction products (5 μl) were added to the reaction mixture containing 0.4 μM of each primer, Sybr green I, 30 U/ml platinum Taq polymerase, 20 mM Tris-HCl, pH 8.4, 50 mM KCl, 3 mM MgCl2, 200 μM of each deoxynucleoside triphosphate, 20 U/liter uracil-N-glycosylase (Invitrogen, Carlsbad, CA) in a final volume of 15 μl. A no-DNA control (5 μl of 0.1 mM EDTA, pH 8.0) was also included. The standard primer sets used for amplification were as follows: for strong-stop DNA, forward primer (5′-dGGTCTCTCTGGTTAGACCA-3′) and reverse primer (5′-dAAGCAGTGGGTTCCCTAGTTAG-3′); for first-strand transfer DNA, forward primer (5′-dAGCAGCTGCTTTTTGCCTGTACT) and reverse primer (5′-dACACAACAGACGGGCACACAC); for full-length minus-strand DNA, forward primer (5′-dCAAGTAGTGTGTGCCCGTCTGTT) and reverse primer (5′-dCCTGCGTCGAGAGAGCTCCTCTGG); and for second-strand transfer DNA, forward primer (5′-dAGCAGCTGCTTTTTGCCTGTACT) and reverse primer (5′-dCCTGCGTCGAGAGAGCTCCTCTGG). The mixtures were subjected to 1 cycle of 2 min at 50°C and 2 min at 95°C and 40 cycles of 15 s at 95°C and 30 s at 65°C on a Rotor-Gene 3000 thermocycler (Corbett) set to collect Sybr fluorescent signal after the 65°C step. The copy number (proviral equivalents) was determined by reference to a standard curve prepared by dilution of plasmid pNL4.3. A no-nucleotide control was always included, and either the results were negligible or the data were discarded.
Cell infection.
MAGI cells (2 × 105) were added to the wells of 6-well plates, with triplicate wells for each sample. An uninfected control was also included. The cells were incubated at 37°C overnight. A control sample of HIVNL4.3 was heat inactivated at 80°C for 15 min. HIVNL4.3 particles, either infectious or heat-inactivated control virus (virus equivalent to 200 ng p24), were added to the cells in duplicate, and they were incubated for 18 h. The cells were washed with PBS three times and then trypsinized and resuspended in 1 ml PBS. The cells were then centrifuged (14,000 rpm, 1 min), washed once with PBS, and recentrifuged. The cells were resuspended in TE (250 μl; 10 mM Tris, pH 7.4, 10 mM EDTA). These mixtures were diluted 1:10 in 0.1 mM EDTA for the real-time PCR. Cytoplasmic DNA was prepared by the method of Hirt (23). Briefly, lysis buffer (250 μl; 10 mM Tris, pH 7.4, 10 mM EDTA, 1.2% sodium dodecyl sulfate, 100 μg/ml proteinase K) was added and the lysate incubated at 37°C for 2 h. A concentration of 5 M NaCl (125 μl) was added, and the mixture was incubated overnight on ice. The lysate was centrifuged (14,000 rpm, 10 min). The supernatant was extracted, once with phenol:chloroform:isoamyl alcohol (25:24:1; 250 μl) and once with chloroform (250 μl). The extracts were ethanol precipitated, washed with 70% ethanol, dried, and resuspended in 100 μl of 0.1 mM EDTA. Reverse transcription products were analyzed by quantitative PCR as described above.
Detergent treatment and p24 and RT colorimetric assays.
Triton X-100 detergent was added at room temperature to virions at the concentrations indicated in the text. The samples were then immediately diluted to 10 ml in PBS and centrifuged at 100,000 × g for 2 h (Beckman Sw41Ti rotor; 28,000 rpm, 4°C). Prior to centrifugation, the supernatant and pellet fractions were assayed for p24 antigen by using a RETROtek HIV-1 p24 antigen enzyme-linked immunosorbent assay (ELISA) (Zeptometrix) according to the manufacturer's instructions. For RT analysis, after the addition of detergent to virions, an RT colorimetric assay was used to measure RT activity (Roche) released from virions using a poly(A):oligo(dT)15 template. The manufacturer's instructions were followed, with the exception that a detergent-free reaction buffer, which was identical to the supplied buffer in all other respects, was used.
“Spin thru” method.
The “spin thru” method was performed as described previously with a minor modification (1): the equilibrium gradient described above was used, but an additional layer (200 μl) containing either 15% OptiPrep and 0.03% (vol/vol) Triton X-100 or no detergent (no-detergent control) was placed above the gradient. A second buffer layer (200 μl) containing 10% OptiPrep was laid above the Triton-containing layer, and the sample was laid on top.
Preparation of human cell lysates and S100 and P100 fractions.
Cells grown to near confluence were washed once with sterile PBS. Adherent cells were induced to shed by the addition of 1 mM EDTA in sterile PBS, followed by incubation for 20 min. Cold PBS (five times the packed cell pellet volumes) was added to the dishes, and the cells were gently resuspended. The cells were centrifuged (1,000 rpm, 10 min), the supernatant was discarded, and the pellet was suspended in lysis buffer (five times the packed cell pellet volume of 10 mM Tris, pH 7.4, 1.5 mM MgCl2, 10 mM KCl, 1× complete protease inhibitor cocktail [Roche], and 0.5 mM β-mercaptoethanol). Protease inhibitor cocktail was omitted in extracts prepared for protease digestion experiments. The cells were centrifuged as described above, the supernatant was discarded, and the pellet was resuspended in two times the packed cell pellet volume of lysis buffer. The cells were lysed with 10 rapid strokes of a Dounce homogenizer, 1 ml at a time. The lysate was cleared by centrifugation in a refrigerated microfuge (4°C, 12,000 rpm, 10 min). One-half of the crude preparation was removed and stored at −80°C. The remaining part of the preparation was centrifuged at 100,000 × g for 1 h in an Sw60Ti rotor. The supernatant (S100) was removed for storage at −80°C. The pellet (P100) was resuspended overnight in the original volume of lysis buffer and stored at −80°C. To prepare a dialyzed fraction, the crude lysate was dialyzed for 4 h against 1,000 times the volume of lysis buffer using a membrane with a molecular mass cutoff of 3,500 Da, with the buffer being changed at 2 h. The protein concentration was measured by using a commercially available Bradford assay (Bio-Rad, CA). The final protein concentration varied with the cell line and from preparation to preparation but was typically 1 mg/ml for a crude Jurkat lysate and 0.5 mg/ml for the S100 and P100 fractions, 4 mg/ml for the 293T crude lysate, 2 mg/ml for the NIH 3T3 crude lysate, and 3 mg/ml for the Vero crude lysate.
Protease and nuclease treatment of cell lysates.
Lysates were treated with either proteinase K (S100 fraction; 20 μg/ml in 1 mM CaCl2, 10 mM Tris, pH 7.4, and 1 mM dithiothreitol) or RNase A (crude lysate; 50 μg/ml) for 1 h at 37°C. Protease treatment was stopped by the addition of inhibitor (2 mM EGTA and 4 mM Pefabloc) followed by incubation at 37°C for 2 h, and RNase treatment was stopped by the addition of RNasin (2 U/μl). The control reaction mixtures contained lysate only, lysate with either protease/RNase or inhibitor, or lysate with protease/RNase and inhibitor added simultaneously without incubation at 37°C (no-incubation control). Treated samples and controls (20-μl volume) were added to standard ERT reaction mixtures as described above, and the reaction products were extracted and analyzed by quantitative real-time PCR.
Equilibrium density and velocity gradient ultracentrifugation.
For equilibrium density gradients, OptiPrep (Axis-Shield) was diluted from 60% to 20% in 5% steps in buffer (5 mM Tris, pH 7.4, 20 mM NaCl, 1 mM MgCl2, and 0.5 mM β-mercaptoethanol). OptiPrep was added in layers of 400 μl, starting with the 60% layer at the bottom and progressively increasing up to 20%. The layers were allowed to diffuse at room temperature for 4 h to form a continuous density gradient. The sample (400 μl) was placed on top of the gradient and centrifuged (Beckman Sw60Ti; 34,000 rpm, 4°C, 20 h). Fractions (400 μl) were collected from the top of the tube. The fractions were assayed for endogenous RT activity, RT activity, and p24 antigen, and the density of each fraction was determined by weighing part of the sample (100 μl). Velocity gradient ultracentrifugation was performed as described above but using 28% to 14% OptiPrep in 2% steps with a 60% cushion at the bottom, and the gradient was centrifuged for 2 h at 34,000 rpm in an Sw60Ti rotor.
RESULTS
Intravirion reverse transcription is inefficient in comparison to reverse transcription in cell infection.
Our laboratory and others have previously studied the property of HIV-1 virions of undergoing detergent-free ERT (24, 47, 49, 50). For this work, to improve sensitivity, we used a concentrated virion preparation produced by ultracentrifugation over a sucrose cushion. These virus stocks, which were highly active for reverse transcription, were then characterized.
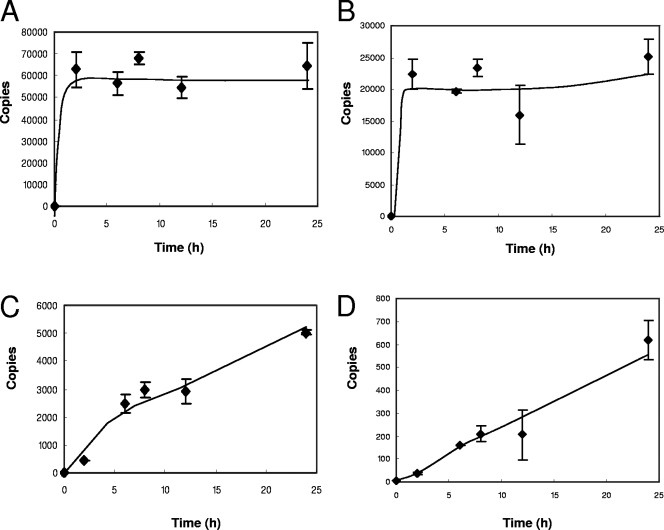
A time course of the detergent-free ERT reaction was performed up to 24 h. The reaction was initiated by the addition of nucleotides to the virions, and the reaction products were analyzed by quantitative real-time PCR. The generation of negative-strand strong-stop and first-strand transfer DNA products was completed after approximately 2 h (Fig. 1A and B), whereas downstream reverse transcription products, such as full-length minus-strand and second-strand transfer DNAs, gave linear reaction kinetics for up to 24 h (Fig. 1C and D). The efficiency of the first-strand transfer step relative to the amount of strong-stop DNA produced reached a maximum of 55%, which was higher than we reported previously (24), possibly due to improved methods using concentrated virus preparations. The efficiencies at 24 h of the generation of DNA products further downstream were successively lower, at 10% and 1.4% for full-length minus-strand and second-strand transfer products, respectively. The production of positive-strand RT products by virions has been reported previously in blood and semen of HIV-infected patients (49), for long-term cultures of simian immunodeficiency virus (12), and after ERT (11). Hence, our data confirm the results of previous studies showing that the virion and its core have all the biochemical activities necessary for the completion of early HIV-1 positive-strand synthesis to the second-strand transfer step.
FIG. 1.
The kinetics of in vitro natural ERT. Reactions were initiated by the addition of deoxyribonucleotides and terminated at the 0-, 2-, 6-, 8-, 12-, and 24-h time points. Reaction products were purified and detected by quantitative real-time PCR for (A) negative-strand strong-stop, (B) first-strand transfer, (C) full-length minus-strand, and (D) second-strand transfer DNAs. Error bars indicate standard deviations (n = 3).
Using the capsid levels measured in our virus preparations and the number of Gag proteins per virion given in recent reports (8), we were able to estimate the number of virions per reaction. The number of copies of strong-stop products was then used to calculate the number of active virions, which was approximately 10% of particles under these reaction conditions. Interestingly, this result is similar to those in recent reports of the proportion of infectious particles in HIV-1 preparations (3, 41) and suggests that particles that are competent for ERT constitute the infectious population of particles.
We then compared the ERT reaction results with the results for reverse transcription in infected cells. Cells were infected and cytoplasmic DNAs purified at 18 h postinfection. For all products, reverse transcription during cell infection was more efficient (Table 1). This was apparent from the high levels of late reverse transcription products, i.e., full-length and second-strand transfer DNAs, relative to the levels of negative-strand strong-stop DNA. This suggests some contribution of the cell environment to the process of reverse transcription; whether this was related to a cell factor(s) or uncoating of the virion core is undetermined. In other words, the virion core is an immature complex which most likely requires a cytoplasmic environment to restructure to form a bona fide RTC.
TABLE 1.
HIV-1 reverse transcription reaction efficiency
| Viral sample | Percentage ± standard deviation of strong-stop DNA of:
|
||
|---|---|---|---|
| Jump DNA (U3-R-U5) | Full-length DNA (R-UTR) | Second-strand transfer DNA (U3-R-U5-UTR) | |
| Intracellular | |||
| Uninfected | 0 | 0 | 0 |
| Heat inactivated | 0 | 0 | 0 |
| Infecteda | 115 ± 15 (n = 3) | 45 ± 6 (n = 3) | 40 ± 6 (n = 3) |
| Intravirionb | 55 ± 11 (n = 22) | 10 ± 3 (n = 8) | 1.4 ± 0.7 (n = 21) |
Measured 18 h postinfection.
Measured 18 h postinitiation.
The effect of detergent on endogenous RT activity.
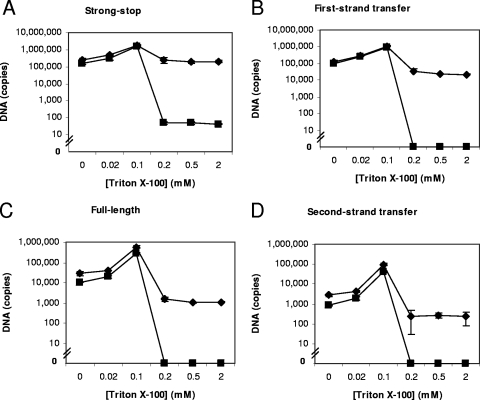
To develop an in vitro system for reverse transcription, a variety of detergents were tested for their effect on ERT activity (data not shown). ERT reactions can contain detergent (7, 19, 20, 40) or be detergent-free (47). Triton X-100 was found to be the most suitable as it was the least detrimental to ERT activity. Figure 2 shows the results of reactions performed using a range of detergent concentrations with exogenous DNase I as an indicator of virion integrity; reactions without DNase I were also performed. Regardless of whether DNase I was present, at the optimal 0.1 mM (0.006%) concentration, reactions with Triton X-100 produced seven times more copies of strong-stop DNA than the no-detergent control (Fig. 2). At or below a 0.1 mM concentration of Triton X-100, the detergent presumably permeabilized the virion to nucleotides, increasing ERT activity. Hence, virion ERT activity is stable at or below this concentration of Triton X-100.
FIG. 2.
The effect of the addition of Triton X-100 on the virion. The ERT reaction was conducted with 0, 0.02, 0.1, 0.2, 0.5, or 2 mM Triton X-100 with (▪) or without (⧫) exogenous DNase I for 18 to 20 h at 37°C. Reaction products were extracted and detected by quantitative real-time PCR. Negative-strand strong-stop, first-strand transfer, full-length minus-strand, and second-strand transfer products were detected, and copy numbers are shown. Error bars indicate standard deviations (n = 3). The data shown are representative of the results of experiments conducted in duplicate.
In the absence of DNase I, the deleterious effects of increasing the detergent beyond the optimal concentration offset the beneficial effects of increased virion permeability, leading to a reduction in signal. The addition of RNase inhibitors to detergent-treated virions did not improve the reaction efficiency (data not shown), suggesting that template degradation was not the cause of the loss of signal; however, the possibility remains that nuclease activity was responsible for the loss of late reverse transcription products.
Loss of ERT activity is coincident with disruption of the virion.
Experiments were also carried out in the presence of DNase I to examine the accessibility of reverse transcription products to degradation after the disruption of virions with detergent (Fig. 2). A difference in signal was observed between reactions with and without DNase I when the Triton X-100 was added at 0.2 mM and greater concentrations.
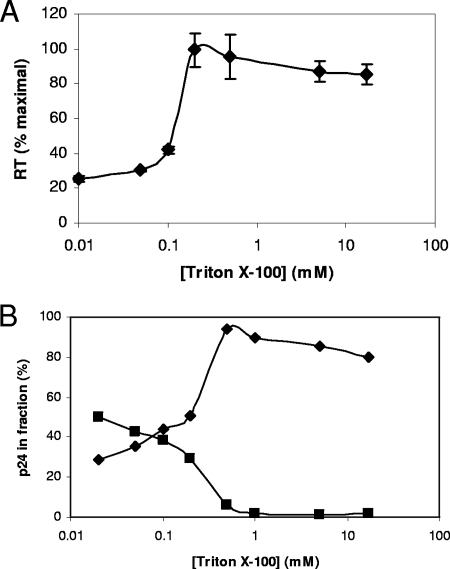
To further explore the disruption of virions with detergent, Triton X-100 was added to virus over a range of concentrations and disruption was measured by the ability of solubilized RT to utilize poly(A):oligo(dT)15 homopolymeric template (Fig. 3A). RT is contained within the viral core; hence, these data demonstrate that the core was disrupted at between 0.1 and 0.2 mM Triton X-100 and, also, that RT was not functionally inactivated by Triton X-100 at these concentrations. In a complementary experiment, virions were also exposed to detergent and particulate material was removed by ultracentrifugation. The amount of capsid remaining in the supernatant, as measured by p24 ELISA, revealed that the detergent did indeed solubilize capsid (Fig. 3B), with a sharp transition that occurred between 0.2 and 0.5 mM Triton X-100. A concomitant decrease in p24 in the pellet fraction was also observed. Considering all the above data, it appears that virions, including the core, are disrupted by an approximately 0.2 mM concentration of Triton X-100.
FIG. 3.
Physical disruption of the HIV-1 virion by detergent. (A) Detergent was added to virions at a range of concentrations, and the mixture was assayed for RT using homopolymeric templates. Error bars indicate standard deviations (n = 3). (B) Virions exposed to detergent were sedimented by ultracentrifugation, and the amount of p24 was measured in the supernatant (⧫) and pellet (▪) fractions. The data shown are representative of the results of experiments conducted in duplicate.
The solubilization of the core with increasing detergent concentrations was coincident with loss of ERT activity (Fig. 2). At concentrations which would completely disrupt the virion (0.5 mM Triton X-100), ERT synthesis of reverse transcription products dropped 1.2 times (strong-stop cDNA), 6 times (first-strand transfer product), and 28 times (full-length minus-strand product) compared to the levels in those samples with no detergent (no DNase I reactions) (Fig. 2). These data suggest that the physically disruptive effects of detergent do not primarily affect initiation but, rather, later steps in reverse transcription. They indicate that low concentrations of detergent increase endogenous activity, while at higher concentrations, cores disassemble with loss of endogenous activity.
HIV-1 virions are disrupted by a “spin thru” method.
To further study the effects of removing the envelope, we compared our findings above with the results of the use of a method which is reported to be less denaturing to virions, commonly referred to as the “spin thru” technique (1, 26). From our observations regarding Triton X-100 described above, we were interested in determining the effect on virions delipidated by using this method. Virions were passed briefly through a detergent-containing layer to minimize the deleterious effects of the detergent. The detergent layer (0.03% Triton X-100) was prepared to have a density that was less than that of intact virus (15% OptiPrep). This concentration of detergent has been used by others performing this method (1). This detergent layer was placed over a continuous OptiPrep density gradient of 20% to 60% lacking detergent. A buffer layer of 10% OptiPrep with no detergent was also placed between the sample and the detergent layer to minimize detergent exposure. After 20 h of ultracentrifugation, fractions were removed and assayed for RT activity on a homopolymer RNA template and for p24 by ELISA. ERT reactions were also performed in the presence of DNase I to monitor virion disruption by determining the nuclease susceptibilities of the reverse transcription products.
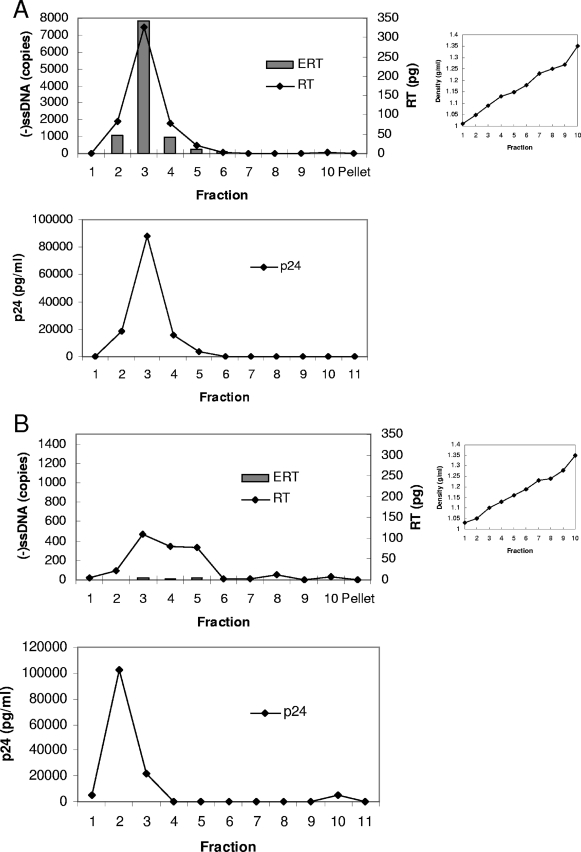
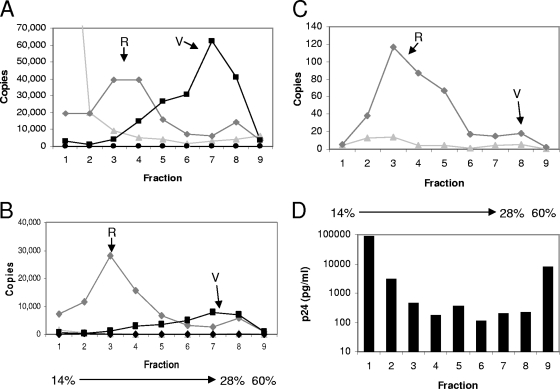
Control samples with virions that were subjected to the “spin thru” method but without a detergent layer had peak capsid, RT, and ERT activities in the same fraction (Fig. 4A, fraction 3), which had a buoyant density of 1.09 g/ml. This density was slightly less than that reported for HIV-1 in sucrose but may be due to the different separation medium used. The fractions from the sample with the detergent layer were assayed for ERT activity. Strong-stop products were reduced, most likely due to increased susceptibility of the products to DNase I digestion, but there was only minor reduction of RT enzyme activity on a homopolymeric template (Fig. 4B). Hence, exposure of the virion to 0.03% (0.5 mM) Triton X-100, even for a presumably brief period, denatured the immature intravirion RT complex without directly disrupting the biochemical activity of the RT enzyme itself. The RT sedimented in a broad peak that was partly denser than capsid protein, possibly indicating association with a denser nucleoprotein complex. Capsid protein was present in a sharp peak that was less dense after detergent exposure. This dissociation of peak capsid protein and RT activities and the susceptibility of strong-stop products to DNase I digestion after detergent exposure indicate disruption of the basic structure of the virion. Interestingly, there was a minor peak in both capsid and RT in the more-dense detergent-treated fractions (fractions 8 to 10, densities 1.25 to 1.3 g/ml) where cores were expected (1), indicating that some free core may have been produced; however, we were unable to detect ERT activity in this fraction. This experiment provides further evidence that relatively low concentrations of Triton X-100 disrupt the virion structure and ERT activity after only brief exposure.
FIG. 4.
Effect of “spin thru” method on HIV-1 ERT activity and structure. Fractions were prepared by the modified “spin thru” method, and their nascent ERT activity and capsid (p24) and RT activities were measured. Data for preparations without (A) or with (B) a 0.03% (vol/vol) Triton X-100 layer are shown. (−)ssDNA, negative-strand strong-stop DNA. The data shown are representative of the results of two experiments.
Mammalian cell lysates increase the generation of late reverse transcription product generation at concentrations of Triton X-100 which are disruptive to the virion.
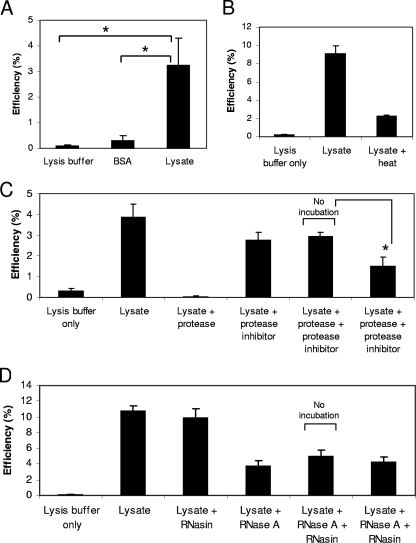
To reconstitute RTC formation in vitro, a cell lysate prepared from the Jurkat human T cell line was added to detergent-treated virions. To delipidate virions, a concentration of 0.2 mM Triton X-100 was chosen to minimize the deleterious effects of detergent, as this was the lowest concentration which permeabilized the viral envelope (Fig. 3B). The addition of lysate resulted in stimulation of the ability of the ERT reaction to generate late reverse transcription products in comparison with the generation of these products in lysis buffer and bovine serum albumin (BSA) controls at equivalent concentrations (Fig. 5A) in at least 10 independent experiments. Interestingly, the amount of strong-stop DNA generated was similar irrespective of the addition of lysate or detergent (data not shown), indicating that only late reverse transcription DNA synthesis, i.e., negative-strand full-length and second-strand transfer DNAs, was affected by the addition of lysate.
FIG. 5.
The effect on the ERT reaction of adding cell lysate. ERT reaction mixtures containing virions with 0.2 mM Triton X-100 were mixed with either untreated or treated Jurkat cell lysate or control (lysis buffer or BSA) and incubated for 18 to 20 h at 37°C, and nascent products were extracted and detected by quantitative real-time PCR. The reaction efficiency was calculated as the fraction of second-strand transfer products generated relative to strong-stop DNA products, expressed as a percentage. The experiments performed compared Jurkat lysate with lysis buffer control and (A) BSA (100-μg/ml final concentration) control, (B) heat inactivation (90°C for 30 min), (C) proteinase K (20 μg/ml, 1 h, 37°C), and (D) RNase A (50 μg/ml, 1 h, 37°C). Error bars indicate standard deviations (n = 3). An asterisk indicates a P value between the results of two treatments of <0.05 by Student's t test. The data shown are representative of the results of experiments conducted in duplicate.
The factor(s) in the Jurkat cell lysate that increased the generation of late reverse transcription products was heat labile, suggesting that it was a protein (Fig. 5B). To further explore the possibility that the factor(s) was proteinaceous, lysate was treated with protease and digestion stopped by using a protease inhibitor. The effect of the treated lysate was determined by ERT reaction. To demonstrate that residual protease was not carrying over into the ERT reaction, a no-incubation control which contained both protease and inhibitor added simultaneously without incubation at 37°C for 1 h was included. ERT reactions with protease-treated lysates were significantly less efficient (P < 0.05) than those with the no-incubation control, and these results suggested that the factor(s) was a protein (Fig. 5C). We were unable to completely stop the stimulatory effects of lysate by proteinase K digestion. This may reveal a limitation of the assay system, as the addition of higher concentrations of proteinase K resulted in reduced efficiency in the no-incubation control also, suggesting an inability of the inhibitor to block proteinase K activity at higher concentrations (data not shown). Hence, we were constrained to use the proteinase K concentration shown, which had only a partial effect. To investigate the involvement of a host cellular RNA (Fig. 5D), lysate was similarly treated with RNase A. Comparison with an equivalent no-incubation control provided no evidence for the involvement of a cellular RNA under these conditions.
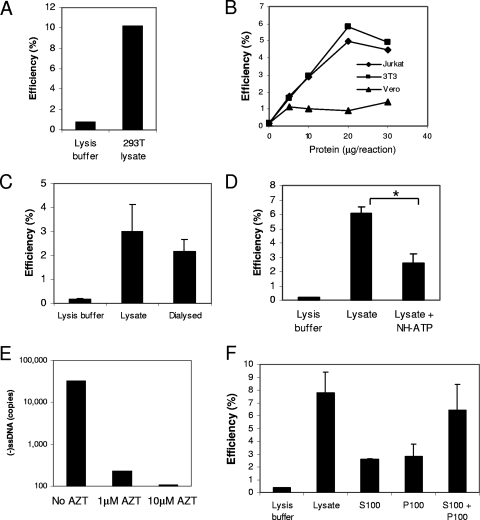
The activity was not specific to T cells, as a lysate prepared from HEK293T cells was also able to stimulate production of late cDNAs (Fig. 6A). The factor may be common to mammalian cells as it was present in mouse (NIH 3T3) as well as human (Jurkat and HEK293T) cells but, interestingly, not African green monkey (Vero) cells (Fig. 6B); the latter may be complicated by TRIM5α activity in these cells (46). A dialysate of the crude lysate retained most of the stimulatory activity, indicating that the factor(s) responsible has a molecular mass greater than 3.5 kDa (Fig. 6C). The nonhydrolyzable ATP analogue ATP-γ-S partially inhibited the activity (Fig. 6D), suggesting that an ATP-dependent step may be involved. The activity is also sensitive to 3′-azido-3′-deoxythymidine triphosphate (AZT; Calbiochem) (Fig. 6E), indicating genuine reverse transcription. The supernatant and pellet fractions from the ultracentrifugation (S100 and P100 fractions, respectively) had reduced activities compared to that of the crude lysate; however, when the fractions were combined, the activity was completely reconstituted, suggesting that the activity may be separable into at least two components (Fig. 6F). Alternatively, the centrifugation conditions were unable to completely sediment the activity into the pellet fraction.
FIG. 6.
Further characterization of activity in cell lysates. Reactions of ERT reaction mixtures containing virions with 0.2 mM Triton X-100 and mixed with either cell lysate, fraction, or lysis buffer control were performed and analyzed as described in the legend to Fig. 5. The reaction mixtures contained (A) a lysate prepared from HEK293T cells; (B) lysates prepared from Jurkat, NIH 3T3, or Vero cells; (C) a dialyzed fraction in comparison with a crude lysate; (D) 1 mM of the nonhydrolyzable ATP analogue ATP-γ-S (NH-ATP); (E) AZT (1 and 10 μM) and additional deoxynucleotides (200 μM dATP, CTP, and GTP and 10 μM dTTP); and (F) Jurkat S100, P100, or combined S100 and P100 fractions. (−)ssDNA, negative-strand strong-stop DNA. Error bars indicate standard deviations (n = 3). An asterisk indicates a P value between the results of two treatments of <0.05 by Student's t test. The data shown are representative of the results of experiments conducted in duplicate, with the exception of the experiment whose results are shown in panel D, which was performed once.
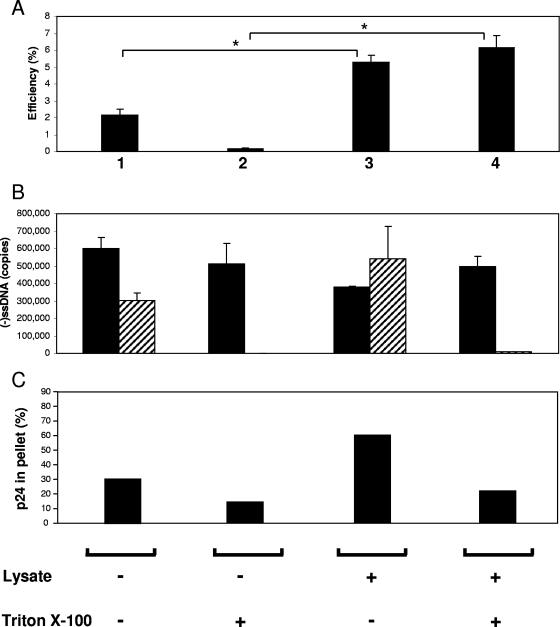
We then compared reactions containing cell lysate with and without detergent treatment. When cell lysate was added, reactions without detergent showed a modest, twofold stimulation of late cDNA products (Fig. 7A, lane 1 compared to lane 3). This was shown subsequently to be due to stabilization of virions by the lysate during the overnight incubation, as determined by an increase in the amount of sedimentable p24 (Fig. 7C). In contrast, when 0.2 mM Triton X-100 was present, there was a 36-fold difference in second-strand transfer product generation when cell lysate was added (Fig. 7A, lane 2 compared to lane 4). To determine the state of virions exposed to detergent in the presence of cell lysate, identical reactions with DNase I added were conducted in parallel. In the absence of detergent, DNase I led to minimal changes in strong-stop DNA, consistent with the presence of intact virions. In reactions with added detergent, the strong-stop DNA product was nearly completely digested (Fig. 7B), indicating that the virion was indeed disrupted. The disruption of virions by detergent when lysate is not present normally results in a reduction in the ability to generate late reverse transcription products (Fig. 2 and 4). However, the addition of cell lysate induced detergent resistance in the immature RTC present in the core of the disrupted virion, enabling the synthesis of late reverse transcription products.
FIG. 7.
Nuclease digestion of detergent-treated virions during ERT. (A) ERT reaction mixtures were prepared either with or without Jurkat lysate and either with or without 0.2 mM Triton X-100 and incubated for 18 to 20 h at 37°C. The efficiency of production of second-strand transfer products was determined by quantitative PCR as described in the Fig. 5 legend. (B) Identical reactions conducted in parallel were digested with DNase I (500 U/ml), and negative-strand strong-stop DNA [(−)ssDNA] products generated in these reactions were measured (hatched columns). These results are presented alongside the results for strong-stop products from the reaction mixtures not containing DNase I (filled columns). Error bars indicate standard deviations (n = 3). An asterisk indicates a P value between the results of two treatments of <0.05 by Student's t test. The data shown are representative of the results of experiments conducted in duplicate. (C) Reactions parallel to those described for panel A were performed and subjected to ultracentrifugation (100,000 × g, 2 h), and the amount of p24 in the pellet was determined as a percentage of total p24 in the sample. +, present; −, absent.
It was conceivable that lysate or detergent addition inhibited DNase I activity. To exclude this possibility, 106 copies of a pNL43 proviral HIV-1 plasmid DNA were added to identical mock-ERT reaction mixtures. DNase I activity was adequate to remove nearly all the plasmid, indicating that cell lysate did not inhibit DNase I activity (data not shown) and that virions had been delipidated by 0.2 mM Triton X-100. The possibility that the effect of the cell lysate was due to the addition of an endogenous DNA-dependent DNA polymerase activity can be eliminated due to the absolute requirement for an RNA-dependent DNA polymerase activity to generate the target for our second-strand transfer primer set. A contribution by endogenous RNA-dependent DNA polymerase activity was eliminated by determination of the RT activity of Jurkat lysate. The RT activity of the virions added to the ERT reaction mixture was more than 2,500 times greater than that in the cell lysate as determined by the colorimetric RT assay (data not shown). Therefore, all of the above results, demonstrating stabilization and enhancement of RT activity in vitro, suggest that one or more cell factors may have a similar role when core is restructured to form the RTC in vivo.
Addition of a human cell lysate induces the formation in vitro of a complex with efficient reverse transcription activity.
Virions were mixed with cell lysate prepared from 293T cells, treated with detergent, and incubated overnight with nucleotides under standard ERT reaction conditions. Cell lysates prepared from 293T cells were used in these experiments as they consistently produced highly concentrated active fractions. To minimize the detrimental effects of detergent, virions were again delipidated with a low concentration of Triton X-100 (0.2 mM). The reaction products were then separated on a 14%-to-28% OptiPrep velocity gradient, and reverse transcription products were detected in the fractions. When lysate was mixed with detergent-treated virions, a complex associated with strong-stop reverse transcription products was detected (peak R in Fig. 8A). The generation of full-length minus-strand transcription products was greatly increased by the addition of lysate (Fig. 8B). The full-length DNA synthesis efficiency relative to the level of strong-stop DNA product in fraction 3 was 71%, which compares with 45% efficiency observed for full-length products during cell infection after 18 h (Table 1). This was in contrast to the results for detergent-treated virions not incubated with lysate, where the strong-stop DNA synthesis remained in the top fraction (fraction 1) and had no full-length DNA. In addition, a complex with the same sedimentation properties was capable of nascent reverse transcription product generation after fractionation (Fig. 8C), indicating that the complex is capable of synthesizing nascent full-length DNA (the relatively low signal in Fig. 8C is due to the small amounts of material used [5 μl of a 400-μl fraction]). These data demonstrate that a highly efficient RTC-like complex can be reconstituted in vitro. This complex was associated with only 0.5% of the total capsid in the virus preparation that was used (Fig. 8D), which is consistent with in vivo observations of low amounts of capsid in the RTC.
FIG. 8.
Reconstitution of the RTC in vitro. (A, B) Modified ERT reaction mixtures containing detergent-treated (0.2 mM Triton X-100) virions either with (⧫) or without (▴) HEK293T lysate or untreated virions with lysate (▪) were incubated overnight with nucleotides. A no-nucleotide control (•) was also included. Samples were then subjected to velocity gradient ultracentrifugation (14%-to-28% OptiPrep with a 60% cushion at 100,000 × g for 2 h). Fractions (0.4 ml) were removed from the top of the gradient, and nucleic acids were extracted and detected using primers specific for (A) strong-stop or (B) full-length reverse transcription products by real-time PCR. Virion (V) and in vitro-reconstituted (R) RTC data are indicated. (C) As described above, but complexes were formed for 2 h and then fractionated as described above. Fractions (5 μl of each) were assayed for nascent ERT activity, and full-length products were detected by real-time PCR. Data for samples containing detergent-treated virions with (⧫) or without (▴) lysate are shown. (D) p24 levels in velocity gradient fractions of the sample containing detergent-treated virions and HEK293T cell lysate. The data shown are representative of the results of experiments conducted in duplicate.
DISCUSSION
These data confirm and extend previous work on intravirion reverse transcription in which we noted reduced strand transfer efficiency in ERT compared with the efficiency in cell infection (24). Improvements to methodology meant that we were able to measure later events in the HIV reverse transcription pathway (Fig. 1). This technical advance confirmed that the enzymatic activities sufficient for reverse transcription are present in the virion-derived cores. However, these experiments revealed that intravirion reverse transcription was inefficient in comparison with reverse transcription in cell infection, as exemplified by the poor efficiency of the production of intravirion late reverse transcription products (Table 1). This observation suggests a requirement for a contribution from the host cell environment for optimal HIV-1 reverse transcription, as was previously shown for avian sarcoma and leukosis virus (ASLV) (29). These data (Fig. 2 and 3) also suggested that core structure may need to be maintained for efficient reverse transcription; this is consistent with observations of an optimal stability for core which predicts that core remains intact for some time postentry for efficient reverse transcription (17).
The addition of Triton X-100 at concentrations (0.5 mM) which completely disrupted virions reduced the generation of late cDNA products to a greater extent than early products. Whether this was due to the loss of a virion component(s), such as RT, or the exposure of core template RNA and product DNA to nucleases is unknown at present. One possibility suggested by these data is that capsid may remain associated with the RTC for some part of the reverse transcription pathway. The core may provide a scaffold for the RTC and increase the effective concentration of components important for the steps of reverse transcription, facilitating strand transfers and the efficiency of the overall reaction. As reverse transcription can be induced to occur in the virion, the core itself does not sterically constrain polymerase elongation. We recently reported the isolation without detergent of HIV-1 cores that were capable of reverse transcription in the absence of an uncoating activity (45). However, the elongation of viral DNA by RT could result in shedding of capsid, as suggested by the effect of detergent-free ERT on virion morphology (48).
Consistent with the observations of the effects of detergent, the exposure of virions to Triton X-100 (0.5 mM, or 0.03%) through the modified “spin thru” method also resulted in their disruption. The reduction of early (strong-stop DNA) products was most likely due to digestion by exogenous DNase I and not to direct inhibition of the enzymatic activity of the RT enzyme, which remained active in colorimetric assays using homopolymeric templates. In addition, this concentration of detergent was much lower than that used in lysis buffers used in commercial RT assays (0.5% Triton X-100). The efficiency of isolation of cores from HIV-1 particles has reportedly been improved by “spin thru” methods, and RT activity on exogenous templates has been detected in such fractions (39). For example, cores prepared by the “spin thru” method were recently used to demonstrate an uncoating activity in cell lysates (5). As we did not show core isolation, we cannot comment on the use of the method for this purpose. However, it was apparent that virions were sufficiently disrupted by brief exposure to 0.03% Triton X-100 as to make ERT products susceptible to exogenous nuclease digestion.
Cell lysates prepared from Jurkat, HEK293T, and NIH 3T3 cells greatly increased ERT efficiency, i.e., the fraction of second-strand transfer products relative to strong-stop DNA, expressed as a percentage (Fig. 6A and B). The factor(s) responsible is most likely proteinaceous in nature and was partially inhibited by nonhydrolyzable ATP. Whether a protein kinase or other ATP binding protein is required is unknown. This activity was segregated into soluble and insoluble components, each with partial activity, that when combined reconstituted the full activity (Fig. 6F). The level of improved reverse transcription efficiency was consistently as high as or higher than the reverse transcription efficiency observed in cell infection with respect to full-length HIV-1 DNA synthesis. For example, reverse transcription in cells was about 45% for full-length DNA synthesis (Table 1), in comparison with the peak fraction of the reconstituted material separated on the velocity gradient, which had an efficiency of 71% for this product. In addition, ERT reactions with added lysate were consistently more efficient compared to the reactions in controls. Importantly, we ruled out the possibility that a low-level cellular RNA-dependent DNA polymerase activity was capable of the observed robust DNA synthesis. Overall, these data indicate that cell factors were required for efficient reverse transcription.
We are the first to report an in vitro reconstitution assay for HIV-1 incorporating cellular factors. We hypothesize that one or more cellular activities are required to form the reverse transcription elongation complex. The activity described here differed from that reported to stimulate ASLV reverse transcription (29). First, a cytoplasmic factor was reported to stimulate ASLV RT initiation, which was not seen in this study. Second, ASLV RT elongation was stimulated by a nuclear extract, whereas HIV-1 RT elongation was stimulated by a cytoplasmic factor(s). The cell factor(s) may do this in one of two ways. The recent identification of a host cell uncoating activity in a lysate prepared from activated T cells has provided weight to the suggestion that shedding of capsid is a prerequisite for reverse transcription (5). The first possibility, then, is that the cell lysate may have an activity that uncoats the core and/or regulates the formation of the RTC. Our observation of low amounts of capsid associated with the reconstituted complex is consistent with this model. However, as loss of core structure leads to loss of the ability to generate late reverse transcription products (Fig. 2 and 3), another hypothesis is that the cell stimulatory factor stabilizes the whole or part of the core. Alternatively, the cell factor may provide some essential function for the RTC by becoming part of the complex. Gemin2 may become part of the RTC and stimulates both early and late reverse transcription products (22). The activity we have identified differs from that of Gemin2 in that it specifically stimulates late reverse transcription. Understanding the precise mechanism of this novel activity will require further experimentation.
It has been suggested that uncoating and reverse transcription proceed by the ordered disassembly of the core (10, 36), and cell factors may assist this process. Our observation that a mammalian cell factor(s) enables the reconstitution of an efficient RTC from delipidated virions is consistent with this model of uncoating. Our in vitro reconstitution system will enable us to address the many unanswered questions regarding uncoating and early RTC formation. We are in the process of identifying what factor(s) is responsible for this effect. The identification of such factor(s) would reveal a novel therapeutic target against HIV-1.
Acknowledgments
We thank Andreas Suhrbier for useful suggestions about the manuscript.
This work was supported by the Australian National Health and Medical Research Council, project grant 298926.
Footnotes
Published ahead of print on 28 November 2007.
REFERENCES
- 1.Accola, M. A., Å. Öhagen, and H. G. Göttlinger. 2000. Isolation of human immunodeficiency virus type 1 cores: retention of Vpr in the absence of p6gag. J. Virol. 746198-6202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adachi, A., H. E. Gendelman, S. Koenig, T. Folks, R. Willey, A. Rabson, and M. A. Martin. 1986. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J. Virol. 59284-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andreadis, S., T. Lavery, H. E. Davis, J. M. Le Doux, M. L. Yarmush, and J. R. Morgan. 2000. Toward a more accurate quantitation of the activity of recombinant retroviruses: alternatives to titer and multiplicity of infection. J. Virol. 743431-3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ariumi, Y., F. Serhan, P. Turelli, A. Telenti, and D. Trono. 2006. The integrase interactor 1 (INI1) proteins facilitate Tat-mediated human immunodeficiency virus type 1 transcription. Retrovirology 347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Auewarakul, P., P. Wacharapornin, S. Srichatrapimuk, S. Chutipongtanate, and P. Puthavathana. 2005. Uncoating of HIV-1 requires cellular activation. Virology 33793-101. [DOI] [PubMed] [Google Scholar]
- 6.Bishop, D. H., R. Ruprecht, R. W. Simpson, and S. Spiegelman. 1971. Deoxyribonucleic acid polymerase of Rous sarcoma virus: reaction conditions and analysis of the reaction product nucleic acids. J. Virol. 8730-741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boone, L. R., and A. M. Skalka. 1981. Viral DNA synthesized in vitro by avian retrovirus particles permeabilized with melittin. I. Kinetics of synthesis and size of minus- and plus-strand transcripts. J. Virol. 37109-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Briggs, J. A., M. N. Simon, I. Gross, H. G. Krausslich, S. D. Fuller, V. M. Vogt, and M. C. Johnson. 2004. The stoichiometry of Gag protein in HIV-1. Nat. Struct. Mol. Biol. 11672-675. [DOI] [PubMed] [Google Scholar]
- 9.Cullen, B. R. 2006. Role and mechanism of action of the APOBEC3 family of antiretroviral resistance factors. J. Virol. 801067-1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dismuke, D. J., and C. Aiken. 2006. Evidence for a functional link between uncoating of the human immunodeficiency virus type 1 core and nuclear import of the viral preintegration complex. J. Virol. 803712-3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dornadula, G., S. Yang, R. J. Pomerantz, and H. Zhang. 2000. Partial rescue of the Vif-negative phenotype of mutant human immunodeficiency virus type 1 strains from nonpermissive cells by intravirion reverse transcription. J. Virol. 742594-2602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dornadula, G., H. Zhang, O. Bagasra, and R. J. Pomerantz. 1997. Natural endogenous reverse transcription of simian immunodeficiency virus. Virology 227260-267. [DOI] [PubMed] [Google Scholar]
- 13.Dueck, M., and J. Guatelli. 2007. Evidence against a direct antiviral activity of the proteasome during the early steps of HIV-1 replication. Virology 3611-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fassati, A., and S. P. Goff. 2001. Characterization of intracellular reverse transcription complexes of human immunodeficiency virus type 1. J. Virol. 753626-3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fitzon, T., B. Leschonsky, K. Bieler, C. Paulus, J. Schroder, H. Wolf, and R. Wagner. 2000. Proline residues in the HIV-1 NH2-terminal capsid domain: structure determinants for proper core assembly and subsequent steps of early replication. Virology 268294-307. [DOI] [PubMed] [Google Scholar]
- 16.Forshey, B. M., and C. Aiken. 2003. Disassembly of human immunodeficiency virus type 1 cores in vitro reveals association of Nef with the subviral ribonucleoprotein complex. J. Virol. 774409-4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Forshey, B. M., U. von Schwedler, W. I. Sundquist, and C. Aiken. 2002. Formation of a human immunodeficiency virus type 1 core of optimal stability is crucial for viral replication. J. Virol. 765667-5677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gao, G., and S. P. Goff. 1999. Somatic cell mutants resistant to retrovirus replication: intracellular blocks during the early stages of infection. Mol. Biol. Cell 101705-1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garapin, A. C., J. P. McDonnell, W. Levinson, N. Quintrell, L. Fanshier, and J. M. Bishop. 1970. Deoxyribonucleic acid polymerase associated with Rous sarcoma virus and avian myeloblastosis virus: properties of the enzyme and its product. J. Virol. 6589-598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gilboa, E., S. Goff, A. Shields, F. Yoshimura, S. Mitra, and D. Baltimore. 1979. In vitro synthesis of a 9 kbp terminally redundant DNA carrying the infectivity of Moloney murine leukemia virus. Cell 16863-874. [DOI] [PubMed] [Google Scholar]
- 21.Gurer, C., A. Höglund, S. Höglund, and J. Luban. 2005. ATPγS disrupts human immunodeficiency virus type 1 virion core integrity. J. Virol. 795557-5567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hamamoto, S., H. Nishitsuji, T. Amagasa, M. Kannagi, and T. Masuda. 2006. Identification of a novel human immunodeficiency virus type 1 integrase interactor, Gemin2, that facilitates efficient viral cDNA synthesis in vivo. J. Virol. 805670-5677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hirt, B. 1967. Selective extraction of polyoma DNA from infected mouse cell cultures. J. Mol. Biol. 26365-369. [DOI] [PubMed] [Google Scholar]
- 24.Hooker, C. W., and D. Harrich. 2003. The first strand transfer reaction of HIV-1 reverse transcription is more efficient in infected cells than in cell-free natural endogenous reverse transcription reactions. J. Clin. Virol. 26229-238. [DOI] [PubMed] [Google Scholar]
- 25.Keckesova, Z., L. M. J. Ylinen, and G. J. Towers. 2006. Cyclophilin A renders human immunodeficiency virus type 1 sensitive to Old World monkey but not human TRIM5α antiviral activity. J. Virol. 804683-4690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kotov, A., J. Zhou, P. Flicker, and C. Aiken. 1999. Association of Nef with the human immunodeficiency virus type 1 core. J. Virol. 738824-8830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McDonald, D., M. A. Vodicka, G. Lucero, T. M. Svitkina, G. G. Borisy, M. Emerman, and T. J. Hope. 2002. Visualization of the intracellular behavior of HIV in living cells. J. Cell Biol. 159441-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miller, M. D., C. M. Farnet, and F. D. Bushman. 1997. Human immunodeficiency virus type 1 preintegration complexes: studies of organization and composition. J. Virol. 715382-5390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Narayan, S., and J. A. Young. 2004. Reconstitution of retroviral fusion and uncoating in a cell-free system. Proc. Natl. Acad. Sci. USA 1017721-7726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nermut, M. V., and A. Fassati. 2003. Structural analyses of purified human immunodeficiency virus type 1 intracellular reverse transcription complexes. J. Virol. 778196-8206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nisole, S., and A. Saib. 2004. Early steps of retrovirus replicative cycle. Retrovirology 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Novak, U., R. Friedrich, and K. Moelling. 1979. Elongation of DNA complementary to the 5′ end of the avian sarcoma virus genome by the virion-associated RNA-dependent DNA polymerase. J. Virol. 30438-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sokolskaja, E., L. Berthoux, and J. Luban. 2006. Cyclophilin A and TRIM5α independently regulate human immunodeficiency virus type 1 infectivity in human cells. J. Virol. 802855-2862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sokolskaja, E., and J. Luban. 2006. Cyclophilin, TRIM5, and innate immunity to HIV-1. Curr. Opin. Microbiol. 9404-408. [DOI] [PubMed] [Google Scholar]
- 35.Srichatrapimuk, S., and P. Auewarakul. 2007. Resistance of monocyte to HIV-1 infection is not due to uncoating defect. Virus Res. 126277-281. [DOI] [PubMed] [Google Scholar]
- 36.Stremlau, M., M. Perron, M. Lee, Y. Li, B. Song, H. Javanbakht, F. Diaz-Griffero, D. J. Anderson, W. I. Sundquist, and J. Sodroski. 2006. Specific recognition and accelerated uncoating of retroviral capsids by the TRIM5α restriction factor. Proc. Natl. Acad. Sci. USA 1035514-5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stremlau, M., B. Song, H. Javanbakht, M. Perron, and J. Sodroski. 2006. Cyclophilin A: an auxiliary but not necessary cofactor for TRIM5alpha restriction of HIV-1. Virology 351112-120. [DOI] [PubMed] [Google Scholar]
- 38.Tang, S., T. Murakami, B. E. Agresta, S. Campbell, E. O. Freed, and J. G. Levin. 2001. Human immunodeficiency virus type 1 N-terminal capsid mutants that exhibit aberrant core morphology and are blocked in initiation of reverse transcription in infected cells. J. Virol. 759357-9366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tang, S., T. Murakami, N. Cheng, A. C. Steven, E. O. Freed, and J. G. Levin. 2003. Human immunodeficiency virus type 1 N-terminal capsid mutants containing cores with abnormally high levels of capsid protein and virtually no reverse transcriptase. J. Virol. 7712592-12602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Temin, H. M., and S. Mizutani. 1970. RNA-dependent DNA polymerase in virions of Rous sarcoma virus. Nature 2261211-1213. [DOI] [PubMed] [Google Scholar]
- 41.Thomas, J. A., D. E. Ott, and R. J. Gorelick. 2007. Efficiency of human immunodeficiency virus type 1 postentry infection processes: evidence against disproportionate numbers of defective virions. J. Virol. 814367-4370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Turelli, P., V. Doucas, E. Craig, B. Mangeat, N. Klages, R. Evans, G. Kalpana, and D. Trono. 2001. Cytoplasmic recruitment of INI1 and PML on incoming HIV preintegration complexes: interference with early steps of viral replication. Mol. Cell 71245-1254. [DOI] [PubMed] [Google Scholar]
- 43.Van Maele, B., K. Busschots, L. Vandekerckhove, F. Christ, and Z. Debyser. 2006. Cellular co-factors of HIV-1 integration. Trends Biochem. Sci. 3198-105. [DOI] [PubMed] [Google Scholar]
- 44.Warrilow, D., and D. Harrich. 2007. HIV-1 replication from after cell entry to the nuclear periphery. Curr. HIV Res. 5293-299. [DOI] [PubMed] [Google Scholar]
- 45.Warrilow, D., D. Stenzel, and D. Harrich. 2007. Isolated HIV-1 core is active for reverse transcription. Retrovirology 477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yap, M. W., S. Nisole, C. Lynch, and J. P. Stoye. 2004. Trim5α protein restricts both HIV-1 and murine leukemia virus. Proc. Natl. Acad. Sci. USA 10110786-10791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang, H., G. Dornadula, P. Alur, M. A. Laughlin, and R. J. Pomerantz. 1996. Amphipathic domains in the C terminus of the transmembrane protein (gp41) permeabilize HIV-1 virions: a molecular mechanism underlying natural endogenous reverse transcription. Proc. Natl. Acad. Sci. USA 9312519-12524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang, H., G. Dornadula, J. Orenstein, and R. J. Pomerantz. 2000. Morphologic changes in human immunodeficiency virus type 1 virions secondary to intravirion reverse transcription: evidence indicating that reverse transcription may not take place within the intact viral core. J. Hum. Virol. 3165-172. [PubMed] [Google Scholar]
- 49.Zhang, H., G. Dornadula, and R. J. Pomerantz. 1996. Endogenous reverse transcription of human immunodeficiency virus type 1 in physiological microenviroments: an important stage for viral infection of nondividing cells. J. Virol. 702809-2824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang, H., G. Dornadula, and R. J. Pomerantz. 1998. Natural endogenous reverse transcription of HIV-1. J. Reprod. Immunol. 41255-260. [DOI] [PubMed] [Google Scholar]