Abstract
Amiloride derivatives are known blockers of the cellular Na+/H+ exchanger and the epithelial Na+ channel. More recent studies demonstrate that they also inhibit ion channels formed by a number of viral proteins. We previously reported that 5-(N-ethyl-N-isopropyl)amiloride (EIPA) modestly inhibits intracellular replication and, to a larger extent, release of human rhinovirus 2 (HRV2) (E. V. Gazina, D. N. Harrison, M. Jefferies, H. Tan, D. Williams, D. A. Anderson and S. Petrou, Antiviral Res. 67:98-106, 2005). Here, we demonstrate that amiloride and EIPA strongly inhibit coxsackievirus B3 (CVB3) RNA replication and do not inhibit CVB3 release, in contrast to our previous findings on HRV2. Passaging of plasmid-derived CVB3 in the presence of amiloride generated mutant viruses with amino acid substitutions in position 299 or 372 of the CVB3 polymerase. Introduction of either of these mutations into the CVB3 plasmid produced resistance to amiloride and EIPA, suggesting that they act as inhibitors of CVB3 polymerase, a novel mechanism of antiviral activity for these compounds.
The Picornaviridae are small, nonenveloped, positive-sense RNA viruses. They are currently divided into nine genera, three of which are significant causes of human disease, the Enteroviruses, Rhinoviruses, and Hepatoviruses (26). The picornaviral genome is approximately 7,500 nucleotides long and directs synthesis of a ∼240-kDa polyprotein containing one structural (P1) and two nonstructural (P2 and P3) domains. The polyprotein is proteolytically cleaved by viral proteases 2A, 3C, and 3CD into three to four capsid proteins and a number of nonstructural proteins. Most of the nonstructural proteins, including the 3C and 3CD proteases, are used in viral RNA replication (reviewed in reference 2). The 2B, 2BC, and 2C proteins from the P2 domain are involved in intracellular membrane rearrangement and formation of replication complexes. The P3 proteins most directly involved in RNA synthesis are 3B (VPg), used as a protein primer for both positive- and negative-strand RNA synthesis, and 3Dpol, an RNA-dependent RNA polymerase. Other P3 proteins (3AB, 3C, and 3CD) are also involved in RNA synthesis.
Amiloride and its derivates are known blockers of the cellular Na+/H+ exchanger and the epithelial Na+ channel (reviewed in reference 14). In addition, amiloride derivatives, principally 5-(N,N-hexamethylene)amiloride (HMA), have been shown to inhibit ion channels formed by the proteins of human immunodeficiency virus (HIV), hepatitis C virus, coronavirus, and dengue viruses (7, 13, 18, 19, 27). We have previously reported that amiloride, 5-(N-ethyl-N-isopropyl)amiloride (EIPA) and benzamil inhibit propagation of human rhinovirus 2 (HRV2; a Rhinovirus) in HeLa cells and that the antiviral activity is unlikely to be due to the inhibition of the cellular Na+/H+ exchanger or the epithelial Na+ channel (9).
Here, we demonstrate that these compounds inhibit replication of coxsackievirus B3 (CVB3; an Enterovirus) in HeLa cells to a greater extent than seen previously for HRV2 and via a significantly different mechanism. We show that amiloride and EIPA specifically inhibit CVB3 RNA replication, and mutations conferring resistance to both compounds are located close to the active center of the RNA-dependent RNA polymerase.
MATERIALS AND METHODS
Chemicals, media, cells, and viruses.
Amiloride, EIPA, HMA, benzamil, actinomycin D, and guanidine hydrocholoride (GHCl) were purchased from Sigma-Aldrich. Amiloride was dissolved in dimethyl sulfoxide; EIPA and HMA were dissolved in ethanol; and benzamil, actinomycin D, and GHCl were dissolved in water. Final concentrations of the solvents in culture medium did not exceed 0.08% (dimethyl sulfoxide) and 0.25% (ethanol), levels at which they had no antiviral effects. [5,6-3H]uridine and [35S]methionine-cysteine mix were purchased from Amersham.
HeLa T cells (picornavirus-susceptible HeLa line) were obtained from the Victorian Infectious Diseases Reference Laboratory (Melbourne, Australia) and maintained in minimal essential medium (MEM; Gibco), supplemented with 5% heat-inactivated fetal bovine serum (FBS; Thermo Electron).
CVB3 (Nancy strain) and HRV2 were obtained from the American Type Culture Collection and propagated in HeLa T cells.
Virus infections.
HeLa T cells in 24-well plates (Falcon) were incubated with CVB3 at the indicated multiplicity of infection (MOI) for 1 h at 37°C in MEM containing 1% FBS, except in the protein labeling experiments, where Dulbecco's modified Eagle medium (DMEM; Gibco) was used. After virus adsorption the cells were washed once, supplied with fresh medium, and further incubated at 37°C for the time periods indicated in the figure legends (calculated starting from the time of virus addition to the cells).
Metabolic activity assay.
Metabolic activity of cells was measured using the indicator Alamar blue (Serotec) that fluoresces as a result of its chemical reduction by living cells. Cells in 24-well plates were washed with MEM and incubated with 10% Alamar blue in MEM for 20 min at 34°C. Fluorescence was then measured using a Wallac 1420 Victor plate reader (Perkin-Elmer Life Sciences) with 530-nm excitation and 590-nm emission filters.
Analysis of RNA synthesis.
HeLa T cells in 24-well plates were infected with CVB3 or HRV2 at an MOI of 10 PFU/cell. After 1 h of virus adsorption at 37°C (CVB3) or 34°C (HRV2), the cells were washed once and supplied with fresh medium containing the indicated compounds (see Fig. 3) and 20 μCi/ml of [5,6-3H]uridine. The cells were further incubated at 37°C for 8 h (CVB3) or at 34°C for 9 h (HRV2), and then cytoplasmic RNA was extracted using an RNeasy Kit (Qiagen) according to the instructions of the manufacturer. The RNA was electrophoresed through a formaldehyde-1% agarose gel and transferred to a Hybond-N membrane (Amersham) by capillary action for subsequent autoradiography.
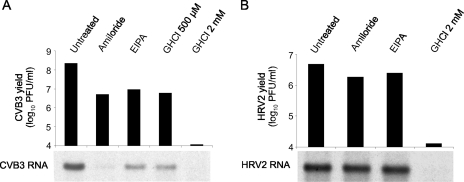
FIG. 3.
Effects of amiloride, EIPA, and guanidine on viral RNA yield in a single replication cycle. HeLa cells were infected with CVB3 (A) or HRV2 (B) at an MOI of 10. After virus adsorption, the cells were labeled with [5,6-3H]uridine and treated with 400 μM amiloride, 25 μM EIPA, or GHCl (500 μM and 2 mM) or left untreated until 9 h (CVB3) or 10 h (HRV2) postinfection. Cytoplasmic RNA was then extracted and analyzed by formaldehyde-agarose gel electrophoresis and autoradiography. Virus titers in cell lysates were determined in parallel. The data are from one representative experiment of three. Contrast and brightness of the images were increased slightly by using Adobe Photoshop CS.
For pulse-labeling of CVB3 RNA, HeLa T cells were infected at an MOI of 10 PFU/cell or mock infected and then incubated in MEM-1% FBS. At the indicated time points (see Fig. 4) the incubation medium was replaced with MEM-1% FBS, actinomycin D (1 μg/ml) and, when indicated, the compounds, and the cells were incubated at 37°C for 30 min. Medium was then replaced with the same medium supplemented with 80 μCi/ml of [5,6-3H]uridine, and the incubation continued for another 30 min. The cytoplasmic RNA was then extracted and processed as described above, except for the time course of CVB3 RNA synthesis, where RNA was extracted using Trizol (Invitrogen) following the manufacturer's instructions and then quantitated using a Wallac 1450 Microbeta liquid scintillation counter (Perkin-Elmer Life Sciences).
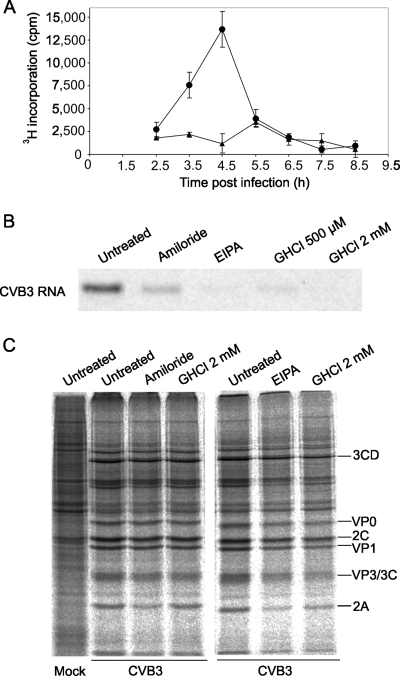
FIG. 4.
CVB3 RNA and protein syntheses in the presence of amiloride and EIPA. (A) Time course of CVB3 RNA synthesis. HeLa cells were infected with CVB3 at an MOI of 10 or mock infected. At the indicated times the cells were labeled with [5,6-3H]uridine in the presence of actinomycin D for 30 min. RNA was then extracted and 3H incorporation was quantitated using a scintillation counter. •, infected cells; ▴, mock-infected cells. The values are averages ± standard errors of the mean. (B and C) Effects of compounds on viral RNA and protein syntheses. The cells were infected as described above. The indicated compounds were added to the cultures at 4 h postinfection, and 30 min later the cells were labeled with [5,6-3H]uridine (B) or [35S]methionine-cysteine (C) for 30 min in the same medium. Cytoplasmic RNA was then extracted and analyzed by formaldehyde-agarose gel electrophoresis and autoradiography (B). Alternatively, the cells were lysed, and the proteins were analyzed by SDS-polyacrylamide gel electrophoresis, followed by visualization on a phosphorimager. Positions of select CVB3 proteins are marked on the right in panel C. Contrast and brightness of images in panels B and C were increased slightly by using Adobe Photoshop CS.
Analysis of protein synthesis.
HeLa T cells in 24-well plates were infected with CVB3 in DMEM-1% FBS at an MOI of 10 or mock infected. At 4 h postinfection the medium was replaced with methionine-cysteine-free DMEM (Gibco) containing 1% FBS and compounds, where indicated, and incubated at 37°C for 30 min. Medium was then replaced with the same medium supplemented with 80 μCi/ml of [35S]methionine-cysteine and further incubated for 30 min. The wells were then washed, and the cells were lysed in 0.5% Nonidet P-40, 100 mM Tris-HCl, and 100 mM NaCl (pH 7.5). Aliquots of clarified lysates were electrophoresed in sodium dodecyl sulfate (SDS)—10% polyacrylamide gels, and the gels were dried and visualized using a phosphorimager (Fujix Bas 2000; Fujitsu).
Generation of amiloride-resistant mutants.
A p53CB3/T7 plasmid, containing the full genome of the CVB3 Nancy strain, was a kind gift of Frank van Kuppeveld (24, 25). The plasmid was linearized using SalI and transcribed using a RiboMax Large Scale RNA Production System T7 (Promega) following the manufacturer's instructions. HeLa T cells in six-well plates (Falcon) were transfected with 5 μg/well of transcribed RNA using DMRIE-C reagent (1,2-dimyristyloxypropyl-3-dimethyl-hydroxy ethyl ammonium bromide and cholesterol; Gibco), according to the manufacturer's instructions, and incubated at 37°C for ∼48 h until all cells were lysed. The virus in the culture supernatant was amplified in one passage and used for generation of amiloride-resistant mutants. Selection conditions were as follows: each virus passage was performed by infecting HeLa T cells at an MOI of 0.01 for 1 h, followed by washing and incubation in MEM-1% FBS containing 400 μM amiloride or no compound for 48 h. Virus yields were then quantified by plaque assay. After 13 passages resistance was achieved, and six isolates of amiloride-treated virus and two isolates of passaged untreated virus were plaque purified twice.
Analysis of mutations.
To identify the resistance-conferring mutations, HeLa cells in 175-cm2 flasks were infected with each of the eight virus isolates at an MOI of 1, and infection was allowed to continue for 24 h. Cells were then lysed by freeze-thawing, and viruses were purified from clarified cell lysates by centrifugation through a 30% sucrose cushion at 140,000 × g for 6 h at 4°C in a P28S rotor (Beckman). Virus pellets were then resuspended in 10 mM Tris-HCl, 10 mM KCl, and 1.5 mM MgCl2 (pH 7.4); viral capsids were digested with proteinase K (1.5 mg/ml) in the presence of 1% SDS and RNase inhibitor (Applied Biosystems). Viral RNA was then isolated via phenol-chloroform extraction and reverse transcribed using the TaqMan reverse transcription kit with random hexamer oligonucleotides (Applied Biosystems) following the manufacturer's instructions. The resulting cDNA was amplified by PCR using Platinum Taq DNA Polymerase High Fidelity (Invitrogen) to produce eight products which spanned the entire P2 and P3 regions of the viral genome (Table 1). Each PCR product was then gel purified using a QIAquick gel extraction kit (Qiagen) and sequenced in both directions employing the same primers as for the PCR.
TABLE 1.
Primers used to amplify and sequence the P2 and P3 coding regions
| PCR fragment | Orientation | Sequence | 5′ Position (nucleotide) |
|---|---|---|---|
| Product 1 | Forward | 5′-AAACTCAGGTGCCAAGCGGTATGC-3′ | 2694 |
| Reverse | 5′-CCTCTTAGTGAGCACGACCACAGCA-3′ | 3414 | |
| Product 2 | Forward | 5′-GAACGTGAACTTCCAACCCAGCG-3′ | 3231 |
| Reverse | 5′-ATTACGGAATCCCTATGGCTGAACG-3′ | 4016 | |
| Product 3 | Forward | 5′-GGTGAGGAACCACGATGACCTGATC-3′ | 3915 |
| Reverse | 5′-CGTCTCCTTGTTCTGCCAAATGGT-3′ | 4602 | |
| Product 4 | Forward | 5′-TTAATTGGAAGGTCGCTTGC-3′ | 4465 |
| Reverse | 5′-CACATTTGTGTCAGTGGCTG-3′ | 5238 | |
| Product 5 | Forward | 5′-TGCTCAAATCGGTAGACAGTGAGGC-3′ | 5105 |
| Reverse | 5′-CCATCAGGGCTTCTCAGCAGCA-3′ | 5862 | |
| Product 6 | Forward | 5′-CTTAGCCAAGGAGGAAGTGG-3′ | 5631 |
| Reverse | 5′-AGAAGGTAGCGAAAGGAAAG-3′ | 6410 | |
| Product 7 | Forward | 5′-CGGTTACCCATATGTTGCAC-3′ | 6261 |
| Reverse | 5′-CTGAAGCTGGTAAGGGTTAC-3′ | 6947 | |
| Product 8 | Forward | 5′-AAAGTGTACAAAGGGATTGACTTGG-3′ | 6847 |
| Reverse | 5′-ATTCGGTGCGGAAAAAAAAAAA-3′ | 7392 |
Cloning of CVB3 mutants.
EcoRI-XbaI and XbaI-SalI fragments of the p53CB3/T7 plasmid containing the 2A and 3D coding regions, respectively, were individually subcloned into a pLitmus 28i plasmid (Stratagene). Site-directed mutagenesis was then performed on the resulting plasmids using a QuickChange Site-Directed Mutagenesis Kit (Stratagene) to introduce the D48G mutation in the 2A protein (to yield 2A-D48G) and the S299T or A372V mutation in the 3Dpol protein (to yield 3D-S299T or 3D-A372V, respectively), using the following mutagenic primer pairs: 5′-GCACATGGATGTGGTATTATAGCCAG-3′ and 5′-CTGGCTATAATACCACATCCATGTGC-3′; 5′-GTACCAGTATTTTCAACACAATGATTAAC-3′ and 5′-GTTAATCATTGTGTTGAAAATACTGGTAC-3′; or 5′-CCTGGACCAACGTCACTTTCCTAAAGAGG-3′ and 5′-CCTCTTTAGGAAAGTGACGTTGGTCCAGG-3′, respectively (mutated nucleotides are underlined). The mutated EcoRI-XbaI fragment and two mutated XbaI-SalI fragments were fully sequenced and individually back-cloned into the p53CB3/T7 plasmid, replacing the original fragments, to create the 2A-D48G, 3D-S299T, and 3D-A372V CVB3 mutants, respectively. A double mutant 2A-D48G/3D-A372V was created by replacing both the EcoRI-XbaI and XbaI-SalI fragments of the p53CB3/T7 plasmid with the corresponding mutated fragments. The mutations were confirmed by sequencing, and mutant viruses were then produced via transfection as described above.
RESULTS
Anti-CVB3 activity and cytotoxicity.
Antiviral activity of amiloride and its derivatives was examined on multiple cycles of CVB3 replication, similar to the previous study on HRV2 (9). HeLa cells were infected with CVB3 (Nancy strain) at an MOI of 0.01 PFU/cell and incubated in the presence of HMA, EIPA, benzamil, or amiloride over a range of concentrations or left untreated for 48 h. The cells were then lysed into the culture supernatants by freeze-thawing, and virus yield in each sample was quantitated via plaque assay. In parallel, the compounds were tested for cytotoxicity by incubating mock-infected cells for 48 h with the same concentrations of the compounds and then measuring cell metabolism using the fluorescent indicator Alamar blue.
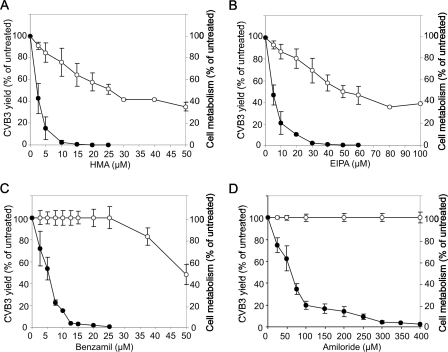
EIPA, benzamil, and amiloride had stronger antiviral activity against CVB3 than previously seen against HRV2, with 50% inhibitory concentrations of 2, 10, and 60 μM, respectively (Fig. 1). In comparison, 50% inhibition of HRV2 reproduction was achieved at 7, 40, and 120 μM concentrations of EIPA, benzamil, and amiloride, respectively (9). HMA has not been tested against HRV2; its activity against CVB3 was similar to that of EIPA (Fig. 1). HMA and EIPA inhibited cell metabolism by 50% at a 25 μM concentration, benzamil achieved 50% inhibition at 100 μM, and amiloride was noncytotoxic up to 1 mM (Fig. 1 and data not shown).
FIG. 1.
Antiviral activity and cytotoxicity of HMA, EIPA, benzamil, and amiloride. HeLa cells infected with CVB3 (MOI of 0.01) or mock infected were treated with various concentrations of HMA (A), EIPA (B), benzamil (C), and amiloride (D) for 48 h or left untreated. Infected cells were then lysed into culture medium by freeze-thawing, and virus yields were quantified by plaque assay. Metabolism of mock-infected cells was measured using the fluorescent indicator Alamar blue. •, CVB3 yield; ○, cell metabolism. The data points are averages ± standard errors of the mean from three independent experiments.
Time-of-addition studies.
To determine which stage(s) of the CVB3 replication cycle is affected by the compounds, we examined the dependence of the antiviral effects of EIPA and amiloride on the time of their addition during a single replication cycle.
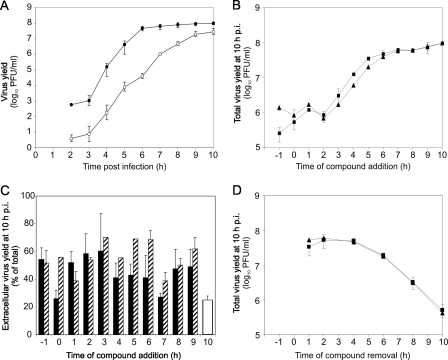
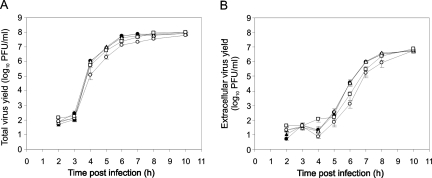
The time courses of CVB3 production and release were measured by infecting HeLa cells at an MOI of 1 and collecting cell lysates and culture supernatants at 1-h intervals, starting from 2 h postinfection (calculated from the moment of virus addition to the cells). The CVB3 titers in cell lysates and culture supernatants were then measured by plaque assay, and virus production (total virus yield) was quantitated as a sum of intra- and extracellular titers. Progeny virus production started at 3 h postinfection, with virus release starting at approximately the same time (Fig. 2A). Both virus production and release continued until 10 h postinfection, at which time ∼25% of the progeny virus was in the culture medium (Fig. 2A).
FIG. 2.
(A) Time courses of CVB3 production and release. HeLa cells were infected with CVB3 at an MOI of 1. At the indicated times culture supernatants were collected, cells were lysed in equal volumes of fresh medium, and intra- and extracellular virus titers were measured by plaque assay. Total virus yield was quantitated as a sum of intra- and extracellular titers. •, total virus yield; ○, extracellular virus yield. (B and C) Time-of-addition assay. The cells were infected as above, and 25 μM EIPA or 400 μM amiloride was added to the culture medium at the indicated times. At 10 h postinfection virus yields were measured as above. White bar, untreated virus. (D) Time-of-removal assay. The cells were infected in the presence of 25 μM EIPA or 400 μM amiloride. At various times postinfection culture supernatants were replaced with compound-free medium, and the infection was allowed to continue until 10 h postinfection when total virus yields were measured. Effects of EIPA (▴) and amiloride (▪) on CVB3 production are shown in panels B and D. The effects of EIPA (striped bars) and amiloride (solid bars) on CVB3 release are shown in panel C. The data points are averages ± standard errors of the mean from three independent experiments with duplicate samples.
To examine the dependence of the antiviral effects of EIPA and amiloride on the time of addition, HeLa cells were infected with CVB3 at an MOI of 1, and 25 μM EIPA or 400 μM amiloride was added to the culture medium 1 h prior to infection, at the time of CVB3 addition, or at 1-h intervals thereafter. When the compounds were added before infection, the virus inoculum was pretreated with each compound for 1 h. At 10 h postinfection cell lysates and culture supernatants were collected, and virus titers were measured. Concentrations of 25 μM for EIPA and 400 μM for amiloride were not cytotoxic under these conditions, with metabolism of uninfected cells after 11 h of treatment being 99% ± 5% and 100% ± 5% of untreated cells, respectively.
The results showed that both EIPA and amiloride strongly inhibited intracellular virus production: addition of the compounds between 1 and 2 h postinfection (after virus entry and prior to the start of progeny virus production) caused an 81-fold (EIPA) and 93-fold (amiloride) average reduction in total virus yield (Fig. 2B). Addition of the compounds at later times, during the course of progeny virus production, resulted in a time-dependent decrease of the antiviral effect (Fig. 2B). EIPA and amiloride did not inhibit CVB3 release from the infected cells: the proportion of progeny virus released into the culture medium was on average twofold higher in the presence of the compounds than in the untreated sample, independent of the time of compound addition (Fig. 2C).
To assess whether the antiviral effects of amiloride and EIPA were reversible, HeLa cells were infected in the presence of the compounds, which were then removed from the culture medium at various times postinfection. Virus replication was then allowed to continue until 10 h postinfection. When the compounds were removed at 1, 2, or 4 h postinfection, CVB3 production at 10 h postinfection was equivalent to that of the untreated cultures (Fig. 2D). Exposure to the compounds beyond 4 h caused progressive reduction in CVB3 yields (Fig. 2D).
The above data demonstrated that the predominant antiviral effect of EIPA and amiloride on CVB3 is the reversible inhibition of the intracellular virus replication (i.e., RNA replication, protein synthesis/processing, or virus assembly). Benzamil also predominantly affected this stage of the CVB3 replication cycle (data not shown), suggesting that the three compounds are likely to have the same mechanism of action.
Amiloride and EIPA reduce CVB3 RNA production in a single replication cycle.
To determine which stage of intracellular virus production (RNA replication, protein synthesis/processing, or virus assembly) is inhibited by amiloride and EIPA, we first assessed the effects of the compounds on CVB3 RNA production in a single replication cycle.
HeLa cells were infected with CVB3 at an MOI of 10 and treated with 400 μM amiloride or 25 μM EIPA starting from 1 h postinfection, or cells were left untreated. Treatment with GHCl (500 μM and 2 mM), a known inhibitor of picornaviral RNA replication (3), was used as a control. The cells were labeled with [5,6-3H]uridine starting from 1 h postinfection. At 9 h postinfection cytoplasmic RNA was extracted from the cells and analyzed by formaldehyde-agarose gel electrophoresis and autoradiography. Virus titers in cell lysates were determined in parallel.
GHCl at 2 mM caused a ∼10,000-fold reduction in CVB3 yield, as expected. The effect of 500 μM GHCl was more modest: a ∼25-fold reduction in virus yield, which was equal to the effect of EIPA at this MOI and slightly lower than the effect of amiloride (∼45-fold reduction) (Fig. 3A). The effects of the three compounds on accumulation of 3H-labeled CVB3 RNA corresponded to their effects on virus yield: 500 μM GHCl and EIPA reduced the amount of viral RNA to a similar extent, amiloride had a stronger effect, and 2 mM GHCl reduced the RNA yield to an undetectable level (Fig. 3A).
Some ion transport blockers are known to inhibit cellular nucleoside transport, in which case the reduction in incorporation of [3H]uridine into RNA does not reflect the reduction in RNA synthesis (17). To exclude the possibility that EIPA or amiloride may inhibit uridine transport, we performed [5,6-3H]uridine labeling of HRV2-infected cells, since we have previously shown that 25 μM EIPA causes only approximately a twofold reduction in HRV2 production (9). The experimental conditions were similar to those with CVB3, except the incubation was conducted at 34°C until 10 h postinfection.
Treatment with either 400 μM amiloride or 25 μM EIPA had no effect on the yield of 3H-labeled HRV2 RNA, consistent with only ∼2.5-fold and ∼2-fold reductions in virus yield, respectively (Fig. 3B). This result demonstrated that the compounds did not affect [3H]uridine transport, confirming that amiloride and EIPA reduce CVB3 RNA production.
Published data suggest that GHCl does not directly affect assembly of enteroviruses (23, 28). The above results showed a comparable reduction of CVB3 titers and viral RNA yields between amiloride, EIPA, and 500 μM GHCl, which implied that amiloride and EIPA do not inhibit virus assembly. This narrowed the potentially affected stages of CVB3 replication to RNA replication, protein synthesis, or protein processing.
Amiloride and EIPA inhibit CVB3 RNA replication.
Picornavirus RNA replication and protein synthesis are coupled processes, and inhibition of one of them indirectly inhibits the other. A standard method to determine which of the two processes is inhibited directly is to add the inhibitor and pulse-labeled viral RNA and proteins at the time in the replication cycle when sufficient amounts of RNA and proteins have been produced to allow continuation of one process independent of the other.
The time course of CVB3 RNA synthesis was determined at an MOI of 10 PFU/cell. CVB3 RNA synthesis peaked at 4.5 h postinfection, after which it rapidly declined, returning to background levels around 1 h later (Fig. 4A). Based on this, the effects of amiloride and EIPA on CVB3 RNA and protein synthesis were examined at 4.5 h postinfection. The compounds were added to infected cells at 4 h postinfection, and 30 min later the cells were pulse-labeled with [5,6-3H]uridine or [35S]methionine-cysteine in the presence of the compounds for a further 30 min. Treatment with GHCl, which inhibits enteroviral RNA replication but not protein synthesis (3, 4), was used as a control.
Amiloride and EIPA inhibited CVB3 RNA replication, with the levels of inhibition comparable to that of 500 μM GHCl (Fig. 4B). Amiloride was less effective than EIPA under these conditions, most likely due to its lower lipophilicity and therefore slower entry into the cell (14). No significant inhibition of overall viral protein synthesis or proteolytic cleavage by amiloride or EIPA was observed (Fig. 4C).
Identification of mutations conferring resistance to amiloride.
The above results suggested that amiloride and its derivatives may inhibit a viral protein involved in CVB3 RNA replication. To test this hypothesis, HeLa cells were transfected with CVB3 (Nancy) RNA produced from the p53CB3/T7 plasmid (24, 25), and the resulting virus was passaged in the presence of 400 μM amiloride or without treatment. Amiloride rather than EIPA was chosen for passaging due to its low toxicity. After 13 passages, virus yields in amiloride-treated cultures became similar to those in untreated cultures. At that stage, six isolates of amiloride-passaged virus as well as two isolates of untreated virus were plaque purified. To confirm amiloride resistance of amiloride-selected isolates, HeLa cells were infected with the six viruses at an MOI of 0.01 and incubated with 400 μM amiloride for 48 h or left untreated. Virus yields in the samples were then measured by plaque assay. The results showed that all isolates of amiloride-selected viruses had similar levels of resistance to amiloride, with virus yields in the presence of the compound being on average 45% of the yields in its absence (data not shown).
To identify mutations conferring resistance, the P2-P3 genomic regions encoding the nonstructural proteins of the six amiloride-resistant isolates and two untreated isolates were reverse transcribed and sequenced. The results are presented in Table 2. All amiloride-resistant isolates had amino acid substitutions within the 3Dpol: three isolates (A2, A5, and A6) had a serine-to-threonine substitution at amino acid position 299 (3D-S299T mutation), while the other isolates (A1, A3, and A4) had an alanine-to-valine substitution at amino acid position 372 (3D-A372V mutation) (Table 2). In addition, each of the isolates that contained the 3D-A372V mutation also contained the aspartic acid-to-glycine substitution at amino acid position 48 within the 2A protein (2A-D48G mutation). The 2A-D48G mutation was also present in one of three isolates containing the 3D-S299T mutation (A5). None of these mutations was present in the two untreated isolates (U1 and U2). A number of other mutations were identified; however, these were either silent mutations or amino acid substitutions that were present in only one amiloride-resistant isolate and therefore considered random (Table 2).
TABLE 2.
Amino acid substitutions in the P2 and P3 domains of CVB3 variants isolated after passaging in the presence of amiloride or without treatment
| CVB3 protein | Amino acid substitution | CVB3 isolate(s)a |
|---|---|---|
| 2A | Y8H | U1 |
| 2A | D48G | A1, A3, A4, A5 |
| 2A | I110T | U2, A6 |
| 2B | N26D | A6 |
| 3A | I54L | A1 |
| 3D | S299T | A2, A5, A6 |
| 3D | A372V | A1, A3, A4 |
U1 and U2, isolates of passaged untreated CVB3; A1 to A6, isolates of CVB3 passaged in the presence of 400 μM amiloride.
To determine whether any of the three common mutations were responsible for the amiloride-resistant phenotype, the p53CB3/T7 plasmid was mutated to generate three viruses containing single mutations, 3D-S299T, 3D-A372V, and 2A-D48G, and a double mutant virus containing the 3D-A372V and 2A-D48G mutations. To assess the replicative fitness of the resulting mutant viruses, we measured the kinetics of their production and release compared to the wild-type virus at an MOI of 1. The kinetics and magnitude of production and release of 3D-S299T and 3D-A372V viruses were identical to the wild-type virus (Fig. 5A and B). The replication rate of the 2A-D48G virus was somewhat slower; however, by 10 h postinfection total and extracellular virus yields reached those of wild-type virus. The double mutant had an intermediate phenotype between 2A-D48G and wild-type viruses (Fig. 5A and B). These results showed that the mutations had no major effects on CVB3 viability.
FIG. 5.
Effects of mutations upon the kinetics of CVB3 replication. HeLa cells were infected with wild-type (•), 3D-S299T (▴), 3D-A372V (▵), 2A-D48G (○), or 2A-D48G/3D-A372V (□) viruses at an MOI of 1. At the indicated times culture supernatants were collected, cells were lysed in equal volumes of fresh medium, and intra- and extracellular virus titers were measured by plaque assay. Total virus yield was quantitated as a sum of intra- and extracellular titers. (A) Total virus yield. (B) Extracellular virus yield. The data are averages ± standard errors of the mean from three independent experiments.
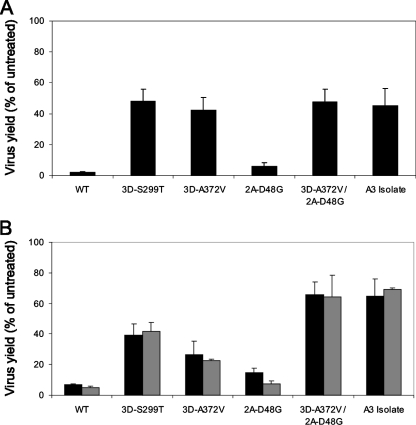
Susceptibility of the mutant viruses to 400 μM amiloride was assessed over multiple replication cycles at an MOI of 0.01. Wild-type plasmid-derived virus and amiloride-resistant isolate A3, containing the 2A-D48G and 3D-A372V mutations, were used as controls. The results showed that each of the mutations in 3Dpol conferred resistance to amiloride. While the wild-type virus yield in the presence of the compounds was 2% of the untreated isolates, the yields of the 3D-S299T and 3D-A372V viruses were much higher (48% and 42% of the untreated isolates, respectively), similar to the yield of the A3 isolate (45% of the untreated isolates) (Fig. 6A). This is a 20- to 24-fold increase in amiloride resistance compared to the wild-type virus. In contrast, the D48G mutation in the 2A protein had a small effect upon resistance, with the yield of the 2A-D48G virus in the presence of amiloride (6% of the untreated isolates) close to that of wild-type virus, and the yield of the 2A-D48G/3D-A372V virus (47% of the untreated isolates) was close to that of the 3D-A372V virus (Fig. 6A).
FIG. 6.
Effect of mutations upon CVB3 susceptibility to amiloride and EIPA. HeLa cells were infected with the indicated viruses at an MOI of 0.01 (A) or 1 (B) and treated with 400 μM amiloride (black bars) or 25 μM EIPA (gray bars) or left untreated. Total virus yields were measured at 48 h (A) or 10 h (B) postinfection. (A) Effect of amiloride on virus yields over multiple replication cycles. (B) Effect of amiloride or EIPA on virus yields in a single replication cycle. WT, wild-type plasmid-derived CVB3; A3 isolate, amiloride-resistant isolate of CVB3 generated by passaging in the presence of 400 μM amiloride. The data are averages ± standard errors of the mean from three independent experiments.
To examine whether amiloride-resistant mutations conferred cross-resistance to EIPA, we assessed susceptibility of the mutant viruses to 400 μM amiloride and 25 μM EIPA in a single replication cycle. These conditions were chosen to avoid cytotoxicity of EIPA over long incubation times. The cells were infected at an MOI of 1 and incubated with the compounds starting from 1 h postinfection. Total virus yields were measured at 10 h postinfection. Virus yields were similar in the presence of amiloride or EIPA for each virus (7% and 5% of the untreated isolates for wild-type, 39% and 42% for 3D-S299T, 27% and 22% for 3D-A372V, 15% and 8% for 2A-D48G, 65% and 64% for 2A-D48G/3D-A372V, and 65% and 69% for A3, respectively) (Fig. 6B). These results showed that amiloride-resistant mutations indeed conferred cross-resistance to EIPA.
Interestingly, the effects of mutations on resistance to amiloride in a single replication cycle were somewhat different from those seen in multiple replication cycles. While in multiple replication cycles the 3D-S299T, 3D-A372V, 2A-D48G/3D-A372V, and A3 viruses had similar levels of resistance (Fig. 6A), in a single replication cycle only the double mutant 2A-D48G/3D-A372V was equal to the A3 isolate containing the same mutations, as expected, while the 3D-S299T and 3D-A372V mutants were less resistant (Fig. 6B). The 2A-D48G virus was again the least resistant among the mutants. The double mutant 2A-D48G/3D-A372V replicated in the presence of amiloride or EIPA to a higher titer than the combined titers of the corresponding single mutants (Fig. 6B).
DISCUSSION
We have previously shown that amiloride and its derivatives, EIPA and benzamil, inhibit reproduction of HRV2 in HeLa cells and that the antiviral activity of these compounds is unlikely to be due to inhibition of their normal cellular targets (9). Here, we demonstrate that these compounds have a stronger antiviral effect against CVB3 than against HRV2 but with the same order of antiviral potency: EIPA > benzamil > amiloride. Despite this apparent similarity, the mechanisms of antiviral activity are significantly different between the two picornaviruses.
The antiviral activity of amiloride and EIPA against CVB3 was due to the inhibition of RNA replication, while no effect of the compounds upon HRV2 RNA replication was observed. Additionally, our previous data showed an inhibitory effect of EIPA on HRV2 release (9), whereas the release of CVB3 was not inhibited by EIPA or amiloride.
To examine the mechanism of inhibition of CVB3 RNA replication, we generated amiloride-resistant CVB3 variants and sequenced the nonstructural regions of the genomes. Amiloride-resistant CVB3 isolates had either an S299T mutation in 3Dpol or a combination of two mutations: A372V in 3Dpol and D48G in the 2A protein (one isolate with the S299T mutation also had the D48G mutation). Both 3Dpol mutations, when individually introduced into the CVB3 genome, produced resistance to amiloride equal to that of the amiloride-passaged isolate (A3) in multiple replication cycles, indicating that no other mutations, including any unidentified mutations outside of the genomic region sequenced, were necessary to produce the resistant phenotype. The mutations conferred resistance to not only amiloride but also EIPA, confirming that amiloride and EIPA have the same mechanism of action.
Serine 299 resides within the structural motif B of the catalytic palm domain of 3Dpol (1, 12, 16, 21). It is adjacent to N298 (N297 in poliovirus), which is located in the ribose-binding pocket of the polymerase active site and plays a crucial role in the selection of ribonucleoside triphosphates over deoxynucleoside triphosphates (10, 11, 15). A372 resides within the structural motif E of 3Dpol, which has been implicated in helping to position the 3′ end of the primer strand during RNA elongation (1). The location of both S299T and A372V mutations within structural motifs involved in the catalytic activity indicates that amiloride and EIPA may bind within or close to the active site of the 3Dpol.
S299 is highly conserved within the Enterovirus genus, with only 5% of 211 analyzed isolates having a threonine at the structurally homologous position. In contrast, alanine is less prevalent (14%) than valine (86%) at the position corresponding to A372 of CVB3 Nancy 3Dpol. HRV2 3Dpol has both threonine and valine in positions corresponding to S299 and A372 of CVB3 3Dpol, which may explain the lack of inhibition of HRV2 RNA replication by amiloride and EIPA.
The 2A-D48G mutation had only minimal effects upon amiloride resistance in multiple replication cycles, which is surprising because this mutation was present in four out of six amiloride-resistant isolates, and D48 is a highly conserved amino acid within the enteroviruses (present in 96% of 211 isolates; glycine has not been reported at this position). This mutation appeared, however, to have a more pronounced effect in a single replication cycle. The combination of 3D-A372V and 2A-D48G mutations produced a virus that replicated in the presence of amiloride or EIPA to a higher titer than the combined titers of the single mutants. This implies a potential synergistic effect of both mutations in a single replication cycle. Elucidation of the effect of the D48G mutation on the functions of the 2A protein is needed to explain the observed differences in the effects of the mutations between single and multiple replication cycles.
Apoptosis is a common release pathway for CVB3 and rhinoviruses (5, 6, 20). However, differences between CVB3 and HRV2 in the sensitivity of virus release to Ca2+ influx suggest that their release mechanisms are not identical (9, 22). The contrasting effects of EIPA on CVB3 and HRV2 release observed in our studies may reflect differences in their release mechanisms. Further studies are required to elucidate the details of the mechanism of picornavirus release, and amiloride derivatives may be useful tools in these studies.
The amiloride derivatives have been shown to inhibit ion channels formed by transmembrane proteins of HIV, hepatitis C virus, coronavirus, and dengue viruses (7, 13, 18, 19, 27). HMA has been shown to inhibit HIV type 1 replication in cultured macrophages and coronavirus replication in L929 cells when used at concentrations similar to those effective against CVB3 in this study; the effect is attributed to inhibition of the ion channels formed by the Vpu or E proteins, respectively (8, 27). Our data represent the first examples of antiviral activity of amiloride derivates not due to inhibition of a viral ion channel. The location of mutations conferring CVB3 resistance to amiloride and EIPA, close to the active center of CVB3 polymerase, indicates that these compounds may act as nonnucleoside polymerase inhibitors, a novel mechanism of activity for these compounds.
Acknowledgments
We thank Frank van Kuppeveld for providing the p53CB3/T7 plasmid and Brett Cromer for helpful discussions.
This work was supported in part by Select Vaccines, Ltd., Melbourne, Victoria, Australia.
Footnotes
Published ahead of print on 21 November 2007.
REFERENCES
- 1.Appleby, T. C., H. Luecke, J. H. Shim, J. Z. Wu, I. W. Cheney, W. Zhong, L. Vogeley, Z. Hong, and N. Yao. 2005. Crystal structure of complete rhinovirus RNA polymerase suggests front loading of protein primer. J. Virol. 79277-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bedard, K. M., and B. L. Semler. 2004. Regulation of picornavirus gene expression. Microbes Infect. 6702-713. [DOI] [PubMed] [Google Scholar]
- 3.Caliguiri, L. A., and I. Tamm. 1968. Action of guanidine on the replication of poliovirus RNA. Virology 35408-417. [DOI] [PubMed] [Google Scholar]
- 4.Caliguiri, L. A., and I. Tamm. 1968. Distribution and translation of poliovirus RNA in guanidine-treated cells. Virology 36223-231. [DOI] [PubMed] [Google Scholar]
- 5.Carthy, C. M., B. Yanagawa, H. Luo, D. J. Granville, D. Yang, P. Cheung, C. Cheung, M. Esfandiarei, C. M. Rudin, C. B. Thompson, D. W. Hunt, and B. M. McManus. 2003. Bcl-2 and Bcl-xL overexpression inhibits cytochrome c release, activation of multiple caspases, and virus release following coxsackievirus B3 infection. Virology 313147-157. [DOI] [PubMed] [Google Scholar]
- 6.Deszcz, L., J. Seipelt, E. Vassilieva, A. Roetzer, and E. Kuechler. 2004. Antiviral activity of caspase inhibitors: effect on picornaviral 2A proteinase. FEBS Lett. 56051-55. [DOI] [PubMed] [Google Scholar]
- 7.Ewart, G. D., K. Mills, G. B. Cox, and P. W. Gage. 2002. Amiloride derivatives block ion channel activity and enhancement of virus-like particle budding caused by HIV-1 protein Vpu. Eur. Biophys. J. 3126-35. [DOI] [PubMed] [Google Scholar]
- 8.Ewart, G. D., N. Nasr, H. Naif, G. B. Cox, A. L. Cunningham, and P. W. Gage. 2004. Potential new anti-human immunodeficiency virus type 1 compounds depress virus replication in cultured human macrophages. Antimicrob. Agents Chemother. 482325-2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gazina, E. V., D. N. Harrison, M. Jefferies, H. Tan, D. Williams, D. A. Anderson, and S. Petrou. 2005. Ion transport blockers inhibit human rhinovirus 2 release. Antiviral Res. 6798-106. [DOI] [PubMed] [Google Scholar]
- 10.Gohara, D. W., J. J. Arnold, and C. E. Cameron. 2004. Poliovirus RNA-dependent RNA polymerase (3Dpol): kinetic, thermodynamic, and structural analysis of ribonucleotide selection. Biochemistry 435149-5158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gohara, D. W., S. Crotty, J. J. Arnold, J. D. Yoder, R. Andino, and C. E. Cameron. 2000. Poliovirus RNA-dependent RNA polymerase (3Dpol): structural, biochemical, and biological analysis of conserved structural motifs A and B. J. Biol. Chem. 27525523-25532. [DOI] [PubMed] [Google Scholar]
- 12.Hansen, J. L., A. M. Long, and S. C. Schultz. 1997. Structure of the RNA-dependent RNA polymerase of poliovirus. Structure 51109-1122. [DOI] [PubMed] [Google Scholar]
- 13.Kim, C. G., V. Lemaitre, A. Watts, and W. B. Fischer. 2006. Drug-protein interaction with Vpu from HIV-1: proposing binding sites for amiloride and one of its derivatives. Anal. Bioanal. Chem. 3862213-2217. [DOI] [PubMed] [Google Scholar]
- 14.Kleyman, T. R., and E. J. Cragoe, Jr. 1988. Amiloride and its analogs as tools in the study of ion transport. J. Membr. Biol. 1051-21. [DOI] [PubMed] [Google Scholar]
- 15.Korneeva, V. S., and C. E. Cameron. 2007. Structure-function relationships of the viral RNA-dependent RNA polymerase: fidelity, replication speed, and initiation mechanism determined by a residue in the ribose-binding pocket. J. Biol. Chem. 28216135-16145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Love, R. A., K. A. Maegley, X. Yu, R. A. Ferre, L. K. Lingardo, W. Diehl, H. E. Parge, P. S. Dragovich, and S. A. Fuhrman. 2004. The crystal structure of the RNA-dependent RNA polymerase from human rhinovirus: a dual function target for common cold antiviral therapy. Structure 121533-1544. [DOI] [PubMed] [Google Scholar]
- 17.Plagemann, P. G., and C. Woffendin. 1987. Effects of Ca2+-channel antagonists on nucleoside and nucleobase transport in human erythrocytes and cultured mammalian cells. Biochim. Biophys. Acta 928243-250. [DOI] [PubMed] [Google Scholar]
- 18.Premkumar, A., C. R. Horan, and P. W. Gage. 2005. Dengue virus M protein C-terminal peptide (DVM-C) forms ion channels. J. Membr. Biol. 20433-38. [DOI] [PubMed] [Google Scholar]
- 19.Premkumar, A., L. Wilson, G. D. Ewart, and P. W. Gage. 2004. Cation-selective ion channels formed by p7 of hepatitis C virus are blocked by hexamethylene amiloride. FEBS Lett. 55799-103. [DOI] [PubMed] [Google Scholar]
- 20.Si, X., H. Luo, A. Morgan, J. Zhang, J. Wong, J. Yuan, M. Esfandiarei, G. Gao, C. Cheung, and B. M. McManus. 2005. Stress-activated protein kinases are involved in coxsackievirus B3 viral progeny release. J. Virol. 7913875-13881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thompson, A. A., and O. B. Peersen. 2004. Structural basis for proteolysis-dependent activation of the poliovirus RNA-dependent RNA polymerase. EMBO J. 233462-3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van Kuppeveld, F. J., J. G. Hoenderop, R. L. Smeets, P. H. Willems, H. B. Dijkman, J. M. Galama, and W. J. Melchers. 1997. Coxsackievirus protein 2B modifies endoplasmic reticulum membrane and plasma membrane permeability and facilitates virus release. EMBO J. 163519-3532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vance, L. M., N. Moscufo, M. Chow, and B. A. Heinz. 1997. Poliovirus 2C region functions during encapsidation of viral RNA. J. Virol. 718759-8765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van Ooij, M. J., D. A. Vogt, A. Paul, C. Castro, J. Kuijpers, F. J. van Kuppeveld, C. E. Cameron, E. Wimmer, R. Andino, and W. J. Melchers. 2006. Structural and functional characterization of the coxsackievirus B3 CRE(2C): role of CRE(2C) in negative- and positive-strand RNA synthesis. J. Gen. Virol. 87103-113. [DOI] [PubMed] [Google Scholar]
- 25.Wessels, E., D. Duijsings, R. A. Notebaart, W. J. Melchers, and F. J. van Kuppeveld. 2005. A proline-rich region in the coxsackievirus 3A protein is required for the protein to inhibit endoplasmic reticulum-to-Golgi transport. J. Virol. 795163-5173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Whitton, J. L., C. T. Cornell, and R. Feuer. 2005. Host and virus determinants of picornavirus pathogenesis and tropism. Nat. Rev. Microbiol. 3765-776. [DOI] [PubMed] [Google Scholar]
- 27.Wilson, L., P. Gage, and G. Ewart. 2006. Hexamethylene amiloride blocks E protein ion channels and inhibits coronavirus replication. Virology 353294-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wimmer, E., C. U. Hellen, and X. Cao. 1993. Genetics of poliovirus. Annu. Rev. Genet. 27353-436. [DOI] [PubMed] [Google Scholar]