Abstract
The Moloney murine leukemia virus (MMLV) belongs to the Retroviridae family of enveloped viruses, which is known to acquire minute amounts of host cellular proteins both on the surface and inside the virion. Despite the extensive use of retroviral vectors in experimental and clinical applications, the repertoire of host proteins incorporated into MMLV vector particles remains unexplored. We report here the identification of host proteins from highly purified retroviral vector preparations obtained by rate-zonal ultracentrifugation. Viral proteins were fractionated by one-dimensional sodium dodecyl sulfate-polyacrylamide gel electrophoresis, in-gel tryptic digested, and subjected to liquid chromatography/tandem mass spectrometry analysis. Immunogold electron microscopy studies confirmed the presence of several host membrane proteins exposed at the vector surface. These studies led to the identification of 27 host proteins on MMLV vector particles derived from 293 HEK cells, including 5 proteins previously described as part of wild-type MMLV. Nineteen host proteins identified corresponded to intracellular proteins. A total of eight host membrane proteins were identified, including cell adhesion proteins integrin β1 (fibronectin receptor subunit beta) and HMFG-E8, tetraspanins CD81 and CD9, and late endosomal markers CD63 and Lamp-2. Identification of membrane proteins on the retroviral surface is particularly attractive, since they can serve as anchoring sites for the insertion of tags for targeting or purification purposes. The implications of our findings for retrovirus-mediated gene therapy are discussed.
The Moloney murine leukemia virus (MMLV) is a simple prototypical virus of the Retroviridae family of enveloped RNA viruses. The hallmark of this family lies in their ability to reverse transcribe their genome from RNA to double-stranded DNA and integrate into a host chromosome. After integration, the retroviral DNA (or provirus) is stably maintained in the cell and is transmitted to the progeny during cell division, like any regular cellular gene. This ability of retroviruses to stably integrate into the host cell genome is precisely what motivated the development of retroviral gene transfer vectors. In spite of the widespread use of retroviral vectors in both experimental and clinical studies, many fundamental aspects of the MMLV biology remain unclear. In particular, the exact composition of retroviral particles, in terms of host proteins that are incorporated into the virions, remains to be determined.
As is the case for most RNA viruses, the MMLV heavily relies on host cell functions for its replication since it only carries minimal genetic information. Its genome contains three coding domains that give rise to the Gag, Gag-Pol, and Env polyprotein precursors, which are proteolyzed into nine individual proteins during virus maturation. Gag is the most abundant polyprotein in the virion, representing about 3/4 of the total virion protein content. Gag polyprotein (Pr65Gag) is cleaved during maturation into three structural proteins: matrix (p15MA), capsid (p30CA), and nucleocapsid (p10NC). Gag also codes for the p12 protein, which possesses no clear function and therefore is as yet unnamed. Encapsulated within the MMLV particle are the three pol-encoded viral enzymes essential for viral replication, namely, the reverse transcriptase (RT), the integrase (IN), and the protease (PR). Retroviral particles possess a lipid membrane derived from the producer cell in which is embedded the viral Env protein composed of the surface (SU) and transmembrane (TM) subunits in wild-type MMLV. In the case of retroviral vectors, the wild-type Env protein is frequently replaced by the envelope protein of other viruses, such as the vesicular stomatitis virus (VSV) glycoprotein (VSV-G). Additionally, retroviruses are known to incorporate minute amounts of host cellular proteins on the surface and inside the virion (40). Host proteins can be incorporated into viral particles either randomly (simply because they are present at the site of virus budding) or specifically (because they interact with any of the virus constituents). Some of these molecules have been shown to fully retain their function on the virions and contribute to their pathogenicity (31, 38, 41).
Extensive work has been performed to identify the host proteins incorporated into the retrovirus human immunodeficiency virus type 1 (HIV-1), the etiological agent of AIDS, by use of a variety of techniques. The most recent studies exploit the capability of liquid chromatography/tandem mass spectrometry (LC-MS/MS) to identify multiple proteins in these highly complex mixtures (14, 57). By use of this approach, 253 host proteins were identified in HIV-1 macrophage-derived virions, including 33 proteins previously described for HIV-1 preparations produced by other cell types (14). Studies have shown that MMLV particles acquire actin and the actin-binding protein moesin (35, 44, 46). Ubiquitin, another abundant cellular protein, was found in the virions not only as free ubiquitin but also as monoubiquitinated Gag proteins (42, 43). More recently, endophilin-2, a component of the host cell's endocytic machinery involved in clathrin-mediated endocytosis, was found to interact with the MA domain of the MMLV Gag polyprotein and to be packaged into MMLV-like particles along with clathrin and α-adaptin (AP-2) (70). To the best of our knowledge, no host surface protein has been identified for MMLV particles to date. However, the colocalization of the Gag polyprotein of MMLV and late endosomal markers (i.e., CD63, Lamp-1, CD82) in 293 HEK cells has been shown (60). These results also suggested that 293-derived MMLV used the late endosome/exosome sorting pathway to exit the cell, as do HIV-1 macrophage-derived virions (36, 49).
The identification of host cell-derived proteins incorporated into retrovirus particles has proved to be difficult due to the presence of contaminating cell membrane vesicles in purified preparations. These membrane vesicles have a density similar to that of virions and were found to copurify with them when standard equilibrium (isopycnic) density centrifugation techniques were used (9, 22). In a previous study, we reported the development of a rate-zonal ultracentrifugation strategy that rendered highly purified retroviral vector preparations free of any significant cell membrane vesicle contamination and thus suitable for the study of both interior and exterior virus particle proteins (58). The high level of purity achieved by this method was demonstrated using a variety of techniques. In this study, we report the identification of host proteins on the retroviral particles purified by rate-zonal ultracentrifugation using MS analysis and immunogold electron microscopy.
MATERIALS AND METHODS
Retrovirus vector production.
The retrovirus used in this study is a MMLV-derived retrovirus vector produced by the 293-GPG packaging cell line (39). This cell line, a generous gift from J. Galipeau (Lady Davis Institute for Medical Research, Montreal, QC, Canada), generates a VSV-G-pseudotyped retrovirus vector encoding a fusion protein between the herpes simplex virus thymidine kinase (TK) protein and the green fluorescent protein (48). Suspension-adapted cell cultures were maintained in calcium-free Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (HyClone, Logan, UT) and tetracycline (1 μg/ml; Fisher Scientific, Nepean, ON, Canada). Vector stocks were produced by omitting tetracycline from the culture medium, thus inducing VSV-G expression as described before (59). A pool of crude virus supernatants from the different days of harvest was clarified by microfiltration and concentrated 20-fold by ultra/diafiltration (59).
Retrovirus vector purification.
Concentrated virus supernatant stocks were purified by rate-zonal ultracentrifugation followed by size exclusion chromatography as previously described (58). Briefly, retrovirus particles were isolated by carefully layering 3 ml of 20-fold-concentrated virus stock on top of a 10 to 30% continuous iodixanol gradient (34 ml). Ultracentrifugation was carried out in a Beckman ultracentrifuge using a SW28 rotor and spinning at 100,000 × g for 4 h at 4°C. Three fractions were collected by puncturing the bottom of the tube: bottom fraction (12.5 ml), virus-containing fraction (10 ml), and top fraction (14.5 ml). Prior to chromatography, concentration of the virus-containing iodixanol fraction was carried out by ultra/diafiltration in a 10-ml stirred cell unit using a YM 100-kDa membrane (Millipore, Etobicoke, ON, Canada). Retentates were diafiltered against 10 ml of cold phosphate-buffered saline (PBS), and the final retentate volume was adjusted to achieve a 10-fold concentration. Virions were further purified by size exclusion chromatography using an XK 16/40 glass column packed with Sepharose CL-4B (Amersham Biosciences, Piscataway, NJ) as described previously (58). The virus eluted in the column voided volume (voided volume = 23 ml). Virus-containing fractions were pooled and concentrated 10-fold using a 10-ml stirred cell unit as described above.
Subtilisin treatment.
Subtilisin treatment of a 10-fold concentrated iodixanol gradient virus fraction was carried out as previously described (45) using a 5× stock solution containing 25 mg/ml of subtilisin (Fluka Biochemika, Buchs, Switzerland) in 100 mM Tris-HCl, 5 mM CaCl2 (pH 8) for a final concentration of 5 mg of protease/ml. The reaction was stopped by the addition of 5 μg/ml phenylmethylsulfonyl fluoride (Fluka BioChemika). Digested retrovirus preparations were then subjected to size exclusion chromatography as described in “Retrovirus vector purification” above.
Fractionation of purified virus preparations by 1D gel electrophoresis.
Purified virus samples were mixed 3:1 with 4× NuPage sample buffer (Invitrogen Life Technologies, Burlington, ON, Canada) containing 50 mM dithiothreitol and heated at 70°C for 10 min. Proteins were fractionated by electrophoresis on Novex 4 to 12% Tris-glycine precast minigels (Invitrogen) run under reducing conditions using NuPage Tris-glycine running buffer (Invitrogen) for 90 min at 125 V. Protein bands were visualized by silver staining (61). Band intensity images were obtained using a Kodak digital science image station 440cf equipped with the Kodak digital science one-dimensional (1D) image analysis software (Eastman Kodak, Rochester, NY).
In-gel digestion and LC-MS/MS.
Protein bands were excised from the gel and subjected to in-gel tryptic digestion on a robotic MassPrep Workstation (Micromass; Waters Corp., Milford, MA). Analysis of tryptic digests was performed by LC-MS/MS. Peptide separation was achieved by loading the samples onto a Zorbax 300SB-C18 trapping column (Agilent Technologies, Mississauga, ON, Canada) followed by a Biobasic C18 analytical column (New Objective, Woburn, MA) using an Agilent 1100 series nano-high-performance liquid chromatography system (Agilent Technologies). The eluted peptides were electrosprayed into a QTRAP 4000 hybrid triple quadrupole linear ion trap mass spectrometer (ABI-Sciex, Concord, ON), and the three most intense precursor ions from each survey scan (MS scan) were selected and subjected to fragmentation by collision-induced dissociation and detection (MS/MS scan). Peak lists of the MS/MS data were generated with a Mascot v16b11 script from ABI-Sciex and submitted to Mascot v1.9 (Matrix Science, London, United Kingdom) for protein identification by searching against NCBI nonredundant databases of human or virus protein sequences. Mascot searches were performed using carbamidomethyl-cysteine as a fixed modification and oxidation of methionines as a variable modification with a single trypsin miscleavage allowed. The allowed mass search tolerances were 1.5 Da for precursor m/z and 0.8 Da for MS/MS fragment m/z. An in-house-developed program (CellMapBase) was used to cluster and group peptides and proteins based on peptides identified at the 95% confidence level by Mascot so as to generate a nonredundant minimal list of identified proteins (30).
Negative and immunogold staining of MMLV vectors.
For negative-stain electron microscopy, retroviral particles were mounted onto electron microscope grids by direct sedimentation of purified iodixanol gradient samples by use of a Beckman Airfuge ultracentrifuge and the A-100 fixed angle microrotor as previously described (2). The grids were negatively stained with uranyl acetate and examined in a transmission electron microscope at the Armand-Frappier Institute (Laval, QC, Canada) (1). The presence of various host surface antigens on the retroviral vector membrane was investigated by indirect immunogold labeling using the following primary antibodies: mouse anti-HMFG1 monoclonal antibody (MAb) (EDM45) from NeoMarkers, mouse anti-CD9 MAb (TS9) and mouse anti-CD29 MAb (BD15) from Diaclone, rabbit polyclonal anti-CD63 (H193), mouse anti-LAMP-2 MAb (H4B4) and mouse anti-CD81 MAb (1.3.3.22) from Santa Cruz Biotechnology Inc. (Santa Cruz, CA), and mouse anti-CD9P-1 MAb, which was a generous gift from Eric Rubinstein (Hôpital Paul Brousse, Villejuif, France). For immunogold electron microscopy studies, retroviral particles mounted onto electron microscope grids as described previously, dried, and rinsed with PBS. Nonspecific reactive sites on the grids were blocked with 1% ovalbumin (5 min, room temperature [RT]). The grids were incubated with a primary antibody for 1 h at RT and washed three times with PBS (5 to 10 min). Subsequently, the grids were exposed to 1% ovalbumin (5 min, RT) and incubated with appropriate secondary antibodies for 1 h at RT. Secondary antibodies used were 10-nm gold-conjugated goat anti-mouse or goat anti-rabbit from BB International (Cardiff, United Kingdom). Following incubation with a secondary antibody, the grids were rinsed with PBS and water and dried at RT. Finally, the grids were negatively stained with uranyl acetate and examined in a transmission electron microscope.
RESULTS
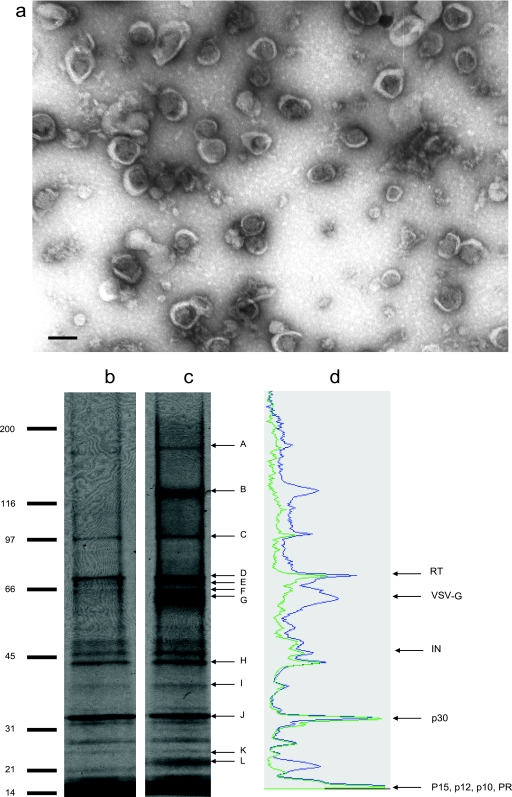
The retrovirus used in this study is a VSV-G-pseudotyped MMLV-derived retrovirus vector encoding a fusion protein between the herpes simplex virus TK protein and the green fluorescent protein as the vector transgene. This vector was produced using the 293-GPG packaging cell line. To explore the composition of the retroviral vector particles, harvested supernatants containing active vector particles were subjected to a series of purification steps. A complete description of this method and a thorough characterization of the purified samples obtained using this method were previously reported (58). Electron microscopy analysis of the purified virus preparation used in the present study showed numerous roughly spherical mature type C retrovirus particles in a size range of 80 to 120 nm in diameter, free of protein aggregates (Fig. 1a). The particles appeared uniform in shape, except for the presence of some broken particles. Interestingly, the round condensed viral core inside the virions is clearly visualized in the transmission electron photograph, providing further evidence of the lack of contamination with “empty” membrane vesicles. Therefore, these purified preparations were found to be suitable for the study of the whole MMLV vector proteome by LC-MS/MS.
FIG. 1.
Fractionation of purified retroviral vector preparations by 1D gel electrophoresis. (a) Negative-stain electron microscopy photograph of retrovirus particles purified by rate-zonal ultracentrifugation at a magnification of ×127,500. Virions were negatively stained using uranyl acetate staining. Bar, 100 nm. (b and c) Purified virus preparations with (b) and without (c) subtilisin treatment were fractionated on a 4 to 12% Tris-glycine polyacrylamide gel (Invitrogen) run under reducing conditions and visualized by silver staining. Protein bands from the gel shown in panel c were excised and subjected to in-gel tryptic digestion prior to MS/MS analysis. Bands containing statistically significant peptide identifications (bands A to L) are indicated for gel c. (d) Band intensity profiles of gel lanes b (green) and c (blue) superimposed. Arrows indicate migration positions of MMLV virus-encoded proteins. This image was obtained using Kodak digital science 1D image analysis software. Band intensity is plotted as arbitrary units.
Fractionation of purified retrovirus preparations by 1D gel electrophoresis and subtilisin digestion analysis.
Subtilisin is a serine protease commonly used by HIV-1 researchers to study the location of cellular proteins in retrovirus particles (40, 45, 46). This nonspecific protease is able to digest all proteins located on the surface of the virions as well as proteins found in contaminating cell membrane vesicles, whereas proteins located inside the virions are protected from digestion by the viral lipid envelope. In addition, the loss in protein content after subtilisin treatment is considered a good indication of the amount of cell membrane vesicle contaminating a virus preparation.
Consistent with previous observations, the 1D-SDS-PAGE silver-stained profile of retrovirus vector particles showed multiple bands with a wide range of molecular masses (Fig. 1). Examination of complete (Fig. 1c) versus subtilisin-digested (Fig. 1b) viral particles was performed. Very little protein content was removed from the virions by subtilisin proteolytic digestion, further substantiating the lack of contamination with membrane vesicles. Importantly, the complete removal of VSV-G surface protein (band G) indicates that the enzymatic treatment was efficient. Figure 1d shows the intensity profile of the protein bands resolved on the SDS-PAGE gel lanes shown in Fig. 1b (green) and 1c (blue). In addition to the virus-encoded protein bands, which alone account for over 90% of the viral protein content as determined by Coomassie blue staining in a previously reported study (58), several additional nonidentified protein bands were visualized by silver staining in the purified virus preparations. SDS-PAGE analysis of subtilisin-treated and untreated virus preparations indicated that most of these bands were resistant to enzymatic digestion. Therefore, most nonviral bands observed corresponded to proteins located inside the virions. On the other hand, aside from the VSV-G protein band (band G), at least two other bands were clearly susceptible to proteolytic digestion by subtilisin. The bands migrating at ∼23 kDa (band L) and 130 kDa (band B) would represent membrane cellular proteins incorporated into the virions. Other protein bands show some degree of susceptibility to subtilisin treatment, particularly those in the high-molecular-mass range (Fig. 1d). Protein bands from Fig. 1c were excised and subjected to in-gel tryptic digestion prior to MS/MS analysis. Bands containing statistically significant peptide identifications are indicated in Fig. 1c with letters A to L.
Proteomic analyses of highly purified MMLV vector particles.
Protein bands from non-subtilisin-treated purified samples (Fig. 1c) were excised from the gel and subjected to in-gel tryptic digestion followed by LC-MS/MS analysis in order to identify both interior and exterior host proteins incorporated into MMLV particles. All clearly visible bands were cut out of the gel, with the exception of the nonresolved band migrating at the dye front. The tandem MS data were submitted to the Mascot search engine for protein identification by comparison against NCBI nonredundant databases of 3,466,531 human protein sequences or 238,744 virus protein sequences. A total of 12 of the 17 bands analyzed by MS showed statistically significant peptide identifications (those peptides identified at a confidence level of greater than 95%). Multiple identifications per band were achieved, resulting in a total of 6 viral proteins and 25 host proteins identified. Protein identifications were considered only for peptides exhibiting appropriate statistics and tryptic sequences, with a single trypsin miscleavage allowed. The majority of the proteins were identified with more than one statistically significant peptide. However, 10 host proteins were identified with single statistically significant peptides. These peptide sequences were validated by rigorous visual inspection of their MS/MS fragmentation spectra. All showed good correlation between expected y or b fragment ions and spectrum peaks of greatest intensity and signal-to-noise ratio. In addition, these 10 host proteins were previously identified for different retroviral and/or exosome particles.
Virus-encoded proteins in retroviral vector particles.
Virus-encoded proteins found by tandem MS are shown in Table 1. The presence of p30CA in band J, VSV-G in band G, and RT in band D at their expected molecular masses was established. Peptides of these proteins were also found in other bands at slightly higher or lower molecular mass (Table 1). The band migrating at 64 kDa (band G) also contained uncleaved Gag polyprotein precursor (Pr65Gag) at the expected molecular mass as determined by the identification of two peptides, one corresponding to the p30CA domain and the other one sharing p30CA and p10NC sequences. Only a fragment of the viral IN (∼46 kDa) was detected at ∼38 kDa (band I). The remaining MMLV structural proteins (p15MA, p10NC, and p12) and the viral PR were not detected by MS. This result was expected since, according to their molecular masses, reactivity to anti-MMLV (58), and resistance to subtilisin digestion (Fig. 1), they are present in the nonresolved band comigrating with the dye front that was excluded from this study. Curiously, a band migrating at ∼43 kDa (band H) was found to contain TK protein (corresponding to the vector transgene), suggesting that this protein may also be encapsulated into virions.
TABLE 1.
Virus-encoded proteinsa
| Band (mol. mass [kDa]) | Protein name | Mol. mass (kDa)b | Accession no.c | Virus origin | No. of peptidesd | Peptide sequence(s) |
|---|---|---|---|---|---|---|
| D (72) | RT | 74,655 | NP-955591 | MMLV | 9 (K, 2) | AELIALTQALK, ALLQTLGNLGYR, ATSTPVSIK, EFLGTAGFCR, ETVMGQPTPK, LDPVAAGWPPCLR, LNVYTDSR, MVAAIAVLTK, QAPLIIPLK |
| G (64) | Pr65Gag (p30) | 60,727 | NP-057934 | MMLV | 1 | QLLLAGLQNAGR |
| Pr65Gag (p30-p10) | 60,727 | NP-057934 | MMLV | 1 | LLATVVSGQK | |
| VSV-G | 55,655 (G, 67) | NP-955548 | VSV | 8 (F, 1; I, 1) | AIQADGWMCHASK, ILDYSLCQ ETWSK, MVGMISGTTTER, QIYTDIEMNR, SFTPSVEQCK, SNYFAYETGGK, VDIAAPILSR, KIYTDIEMNR | |
| H (43) | TK | 40,790 | NP-044624 | HSV-1e | 1 | TTTTQLLVALGSR |
| I (38) | IN | 46,338 | NP-955592 | MMLV | 1 | LLEEIFPR |
| J (32) | p30CA | 30,609 | NP-955585 | MMLV | 2 (H, 1; I, 2; K, 2) | GITQGPNESPSAFLER, QLLLAGLQNAGR |
Peptide mass lists were submitted for search against the NCBI nr database (virus taxonomy) using the Mascot search engine (v1.9).
Calculated molecular mass based on the amino acid sequence. The mass of the posttranslationally modified form is given in parentheses (G, glycosylated).
NCBI reference sequence accession number.
The number of peptides identified for the same protein in other bands is indicated in parentheses.
HSV-1, herpes simplex virus type 1.
Host-associated proteins in retroviral vector particles.
A total of 25 cellular proteins were identified in MMLV vector particles by LC-MS/MS analyses (Table 2), including clathrin, moesin, actin, endophilin-2, and ubiquitin, which have been described previously as parts of MMLV virions (35, 42-44, 46, 70). It is noteworthy that twenty host proteins identified in this work had not yet been described as MMLV constituents, 16 of the 25 cellular proteins identified were previously found in proteomic analysis of HIV-1 macrophage-derived virions, and 16 of the 25 proteins identified were previously described to be associated with exosome particles (Table 2). Importantly, all host proteins were found at their expected molecular masses, with the exception of beta-tubulin, for which only a fragment of the full-length protein (∼48 kDa) was detected at ∼24 kDa (band K). The presence of ubiquitin in different bands is consistent with the potential presence of ubiquitylated Gag products, as previously reported (42, 43). In addition to their full-length form, fragments of heat shock protein 70 (Hsc70), polyadenylate-binding protein (PABP), and HIC2 protein were detected in faster-migrating bands, probably as a result of proteolytic degradation.
TABLE 2.
Host-associated proteinsa
| Band (mol. mass [kDa]) | Protein nameb | Mass (kDa)c | Accession no.d | Reference(s) for protein appearance ase:
|
Locationf | Functionh | No. of peptidesg | Peptide sequence(s) | |
|---|---|---|---|---|---|---|---|---|---|
| Exoe | Retrof | ||||||||
| A (188) | Clathrin heavy chain 1 | 191,615 | Q00610 | 55, 71 | 14 | C | Endocytosis | 2 | IVLDNSVFSEHR, KFNALFAQGNYSEAAK |
| B (130) | Integrin β1 | 88,465 (G, 130) | P05556 | 15, 56 | 14 | M | Cell adhesion | 2 | SAVTTVVNPK, TVMPYISTTPAK |
| CD9P-1 (EWI-F) | 98,556 (G, 135) | Q5VVU9 | M | Functions ascribed to CD81 and CD9 | 6 | AQDGDFIFSK, EGEPFELR, LDTVGSDAYR, MPDSTLPGSR, SVLALTHEGR, TANDAVELHIK | |||
| C (97) | AIP-1/Alix | 96,023 | Q8WUM4 | 33, 67 | 14, 63 | C | LE trafficking | 4 | CSDIVFAR, LLDEEEATDNDLR, STPVNVPISQK, TMQGSEVVNVLK |
| Glycogen phosphorylase | 97,092 | P11217 | C | Carbohydrate metabolism | 3 | VAAAFPGDVDR, VIFLENYR, VLYPNDNFFEGK | |||
| LAMP-2 (CD107b) | 44,961 (G, 100-110) | P13473 | 55, 67 | C | LE marker | 1 | IPLNDLFR | ||
| D (72) | CD81P-3 (EWI-2) | 65,034 (G, 70) | Q969P0 | 33 | M | Functions ascribed to CD81 and CD9 | 1 | STLQEVVGIR | |
| Moesin | 67,820 | P26038 | 27, 33, 71 | 14, 46 | C | Membrane organazation | 1 | QEAEEAKEALLQASR | |
| E (70) | Hsc70 | 70,898 | P11142 | 33, 68, 71 | 14, 24, 57 | C | Chaperone | 1 (J, 1) | DAGTIAGLNVLR, TTPSYVAFTDTER |
| PABP | 70,671 | P11940 | C | RNA binding protein | 3 (H, 3) | ALDTMNFDVIK, FGPALSVK, FSPAGPILSIR, NFGEDMDDER, SGVGNIFIK | |||
| G (64) | HIC2 | 66,156 | Q96JB3 | N | Transcriptional repressor | 1 (I, 1; J, 1) | QLLLQLNQQR | ||
| H (43) | Actin | 41,737 | P60709 | 33, 68, 71 | 14, 35, 46 | Cytoskeleton | Cell motility | 4 | AGFAGDDAPR, EITALAPSTMK, GYSFTTTAER, SYELPDGQVITIGNER |
| CD63 (LAMP-3) | 25,637 (G, 40-60) | Q5TZP3 | 18, 71 | 14, 22, 36, 38 | C, M | LE marker | 1 | VMSEFNNNFR | |
| Endophilin-2 | 41,490 | Q99961 | C | Vesicle recycling | 1 | QAVQILDELAEK | |||
| MFG-E8 | 43,123 (G, 40-45) | Q08431 | 21, 68 | M | Cell adhesion | 1 | EVTGIITQGAR | ||
| ILF2 | 43,062 | Q12905 | N | Transcription regulator | 2 | ILITTVPPNLR, QPLALNVAYR | |||
| I (38) | GAPDH | 36,053 | P04406 | 33, 71 | 14, 44 | C, N | Glycolysis | 1 | GALQNIIPASTGAAK |
| Poly(rC)-binding protein | 37,498 | Q15365 | 14 | N | Nuclear export | 3 | IANPVEGSTDR, INISEGNCPER, LVVPASQCGSLIGK | ||
| Ubiquitin | 8,565 | P62988 | 52 | 14, 42, 43, 57 | C, N | UPS/protein trafficking | 2 (H, 1; K, 1; L, 1) | IQDKEGIPPDQQR, TITLEVEPSDTIENVK | |
| Vacuolar ATP synthase | 40,329 | P61421 | 21 | 14 | C | Vesicular acidification | 3 | AYLESFYK, LLFEGAGSNPGDK, LYPEGLAQLAR | |
| K (24) | Peroxiredoxin 1 | 22,110 | Q06830 | 14 | C | Redox regulation | 2 | LVQAFQFTDK, QITVNDLPVGR | |
| Tubulin beta | 47,767 | Q5JP53 | 27, 67, 71 | 14 | Microtubules | Microtubules | 1 | IMNTFSVVPSPK | |
| Histone H4 | 11,367 (A, 11-35) | P62805 | 67 | 14, 57 | N | Compact DNA | 1 | ISGLIYEETR | |
| Rab 7 | 23,490 | P51149 | 55, 67 | 14 | C | LE trafficking | 1 | VIILGDSGVGK | |
| Rac3 | 21,379 | P60763 | 14 | C | Signaling | 2 | AVLCPPPVK, YLECSALTQR | ||
Peptide mass lists were submitted for search against the NCBI nr database (Homo sapiens taxonomy) using Mascot search engine (v1.9).
Protein synonyms are provided in parentheses. Proteins in boldface were previously found in MMLV.
Calculated mass based on the amino acid sequence. The posttranslationally modified form mass is provided in parentheses (G, glycosylated; A, acetylated).
UniProt reference sequence accession number.
Exo, proteins were previously described in these references as being in exosomes. Retro, host proteins were previously described in these references as being in other retroviral particles (mainly HIV-1 virions).
Membrane refers to plasma membrane. Cytoplasm includes proteins associated with intracellular membranes. C, cytoplasm; N, nucleus; M, plasma membrane.
The numbers of peptides identified for the same protein in other bands are indicated in parentheses.
LE, late endosome; UPS, ubiquitin-proteosome system.
Consistent with the subtilisin digestion analysis described earlier, most of the host proteins identified by LC-MS/MS (19 out of 25) corresponded to intracellular proteins presumably located inside the virions (Table 2). These include proteins involved in various cytoplasmic systems such as cytoskeleton (actin, moesin), microtubules (beta-tubulin), signaling (Rac3), metabolism (peroxiredoxin-1, GAPDH [glyceraldehyde-3-phosphate dehydrogenase], glycogen phosphorylase), chaperones (HSc70), ubiquitin-proteosome system/protein trafficking (ubiquitin), and vesicle trafficking system-related proteins (Rab 7, vacuolar ATP synthase, endophilin-2, clathrin, AIP-1/Alix). Additionally, the purified virion preparations contained several nuclear proteins including histone 4, poly(rC)-binding protein, ILF2, HIC-2, and PABP. Six out of 25 proteins identified in MMLV preparations were host membrane proteins. Their corresponding bands were visualized mainly in the high-molecular-mass range of the SDS-PAGE gel. These bands show some degree of susceptibility to subtilisin treatment (Fig. 1d). The most clearly subtilisin-digested band (band B) contained two host membrane proteins: integrin β1 (CD29) and CD9-partner 1 (CD9P-1). Additionally, LC-MS/MS analysis revealed the presence of membrane proteins in bands C (LAMP-2), D (CD81-partner 3), and H (CD63 and HMFG-E8).
Confirmation of host proteins on retroviral vectors' surfaces by immunogold labeling.
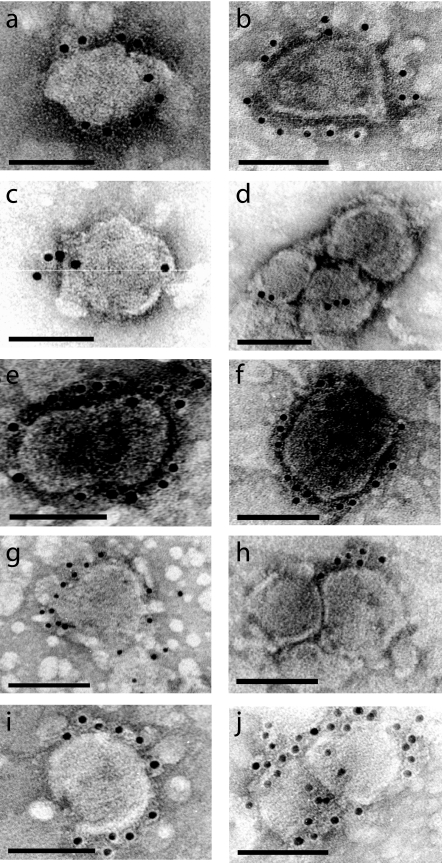
Due to potential contamination with cell membrane, host proteins on the virus surface have been the most challenging to conclusively identify (40, 41). In contrast, the identification of host proteins that reside inside the virions has been straightforward following subtilisin treatment of purified retroviral preparations. Immunogold labeling experiments were carried out to confirm the presence of the host surface proteins found by MS on the MMLV vector surface. Specific antibodies against human CD29 (integrin β1), CD9P-1, LAMP-2, CD63, and HMFG-E8 were employed. In all five immunogold labeling experiments, gold particles at the membrane of typical dense viral particle structures were observed (Fig. 2). In most cases, a dense retroviral core with an irregular boundary was readily distinguishable. For most antibodies tested, the immunogold reaction was strong and localized at the viral surface. The average number of gold particles observed per virion was 13 for CD29 (integrin β1) (n = 5), 20 for LAMP-2 (n = 4), 8 for CD63 (n = 3), and 13 for HMFG-E8 (n = 11) experiments. Immunogold labeling using anti-CD9P-1 MAb showed a weaker reaction with only three gold particles per virion (n = 4) but well-localized labeling at the viral surface. Omitting the use of a primary antibody completely abolished the detection of colloidal gold on the virus surface, indicating that no unspecific binding of secondary gold-labeled antibody occurred (data not shown). Gold particles were remarkably radially distributed on the outside surface of the virions. Staining was evenly distributed in well-dispersed particles but weaker or nonexistent in between viral particles, forming aggregates most probably due to antibody inaccessibility to these areas.
FIG. 2.
Confirmation of host surface proteins on retroviral vectors' surfaces by immunogold labeling. Purified retrovirus vector preparations were subjected to immunogold labeling studies to confirm the presence of several host membrane proteins identified by MS on the virus surface. Primary antibodies employed were mouse anti-CD29 (integrin β1) MAb (a and b), mouse anti-CD9P-1 MAb (c and d), mouse anti-LAMP-2 MAb (e and f), rabbit polyclonal anti-CD63 (g and h), and mouse anti-HMFG-E8 MAb (i and j). Goat anti-mouse or goat anti-rabbit secondary antibodies conjugated with 10-nm colloidal gold were used for the detection. Virions were negatively stained using uranyl acetate. Bar, 100 nm.
Identification of tetraspanins CD9 and CD81 in retroviral vectors by immunogold labeling.
SDS-PAGE analysis of subtilisin-treated and nontreated virions (Fig. 1) showed clear digestion of two non-virally encoded bands, one at ∼130 kDa (band B) and a second one at ∼23 kDa (band L), suggesting the presence of membrane proteins in these two bands at least. As expected from subtilisin analysis, host membrane proteins (integrin β1, CD9P-1) were identified by MS in band B, and the presence of these proteins on the virus surface was confirmed by immunogold labeling (Fig. 2). However, there was no identification of membrane proteins by MS in the ∼23-kDa band (band L). This intriguing result encouraged additional studies to assess the presence of membrane proteins in this band.
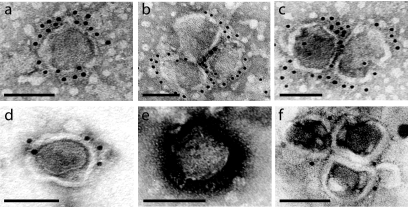
Among the proteins identified by MS, CD9P-1 (EWI-F) and CD81P-3 (EWI-2) are known molecular partners of the tetraspanins CD9 and CD81 (10, 12, 13, 62). Interestingly, both tetraspanins have a molecular mass of ∼26 kDa, consistent with the approximate molecular mass of band L. Based on this evidence, we hypothesized that these two proteins were present on MMLV particles. Preliminary Western blot studies using anti-CD81 and anti-CD9 MAbs showed the presence of faint ∼24-kDa bands in the purified vector preparations (data not shown). The presence of tetraspanins CD9 and CD81 on the virus surface was confirmed by immunogold labeling electron microscopy studies. In all cases, an irregular viral core structure, typical of MMLV, was clearly visible inside the virions. Studies with the anti-CD81 MAb showed a strong specific labeling reaction at the surface of electron-dense MMLV structures (Fig. 3a, b, and c). On average, 16 gold particles were visualized at the surface of each virion (n = 15). The labeling was evenly distributed all around the virions. The reaction with anti-CD9 was less profuse but specific to the membrane of MMLV particles (Fig. 3d, e, and f). Each virion had on average four gold particles on the surface (n = 19). In this particular experiment, most particles observed were part of large viral aggregates.
FIG. 3.
Identification of tetraspanins CD9 and CD81 on MMLV particles by immunogold labeling. Purified retrovirus vector preparations were subjected to immunogold labeling studies to assess the presence of tetraspanins CD9 and CD81 on the virus membrane. Primary antibodies employed were mouse anti-CD81 MAb (a, b, and c) and mouse anti-CD9 MAb (d, e, and f). A goat anti-mouse antibody conjugated with 10-nm colloidal gold was used for the detection. Virions were negatively stained using uranyl acetate. Bar, 100 nm.
DISCUSSION
As gene therapy progresses into clinical trials, full characterization of the viral vectors becomes an important issue. Even though host proteins are incorporated in low quantities into the viral particles, they can be biologically active (41). Therefore, their presence in the vector particle may be relevant to fully understand the retroviral vector's biology. This area of research has received significant attention among HIV researchers. In the case of viral vectors for gene therapy, the identification of host proteins incorporated into the vector particles may be particularly attractive, since in addition to fostering a better understanding of the vector biology, this knowledge may allow the alteration of specific vector properties at our convenience. In this work, we have shown the capabilities of LC-MS/MS in combination with immunogold electron microscopy to characterize the proteomes of retroviral vector particles. Using this approach, we have identified a total 22 new host proteins not previously described in MMLV virions in addition to 5 host proteins already known to be incorporated into wild-type MMLV particles (Table 2). This represents an important step forward in the elucidation of retroviral vector composition.
Many of the proteins identified in this work were previously found, not only in proteomic analysis of family-related viruses such as HIV-1, but also in exosome particles. These findings provide further support to a growing body of evidence suggesting that MMLV is able to hijack the endocytic pathway to escape from multiple cell types (8, 50, 60). Since both virions and exosomes would be generated in the endosomal compartment and exit the cell by budding into endosome-derived membranes, it is not surprising that they would share part of their composition. We have found that 293-derived MMLV particles incorporate at least 18 proteins that were also found in exosomes of various origins (Table 2).
In addition to the numerous cytoplasmic proteins found in the virions (70%), we have successfully identified eight host proteins exposed on the viral vector surface as confirmed by electron microscopy studies. Identification of these membrane proteins is particularly attractive, since they could serve as interesting anchoring sites for the insertion of tags for purification, labeling, or targeting purposes. Engineering vectors by inserting tags or chemically modifying the virus-encoded Env protein structure frequently affects the viruses' ability to transduce cells (29, 53, 64). This has been attributed partly to the inability of the Env protein to provide fusogenic functions for viral entry once its structure has been modified. To circumvent this problem, host surface proteins rather than the viral Env protein could be tagged. To date, this approach has not been tested, likely due to our poor understanding of the MMLV membrane composition. It might be feasible to genetically modify one of the host membrane proteins found in this work to contain such tags without compromising virus infectivity.
We have identified three cellular adhesion proteins on MMLV vectors surface, namely, integrin β1 (CD29), CD9, and HMFG-E8. It is interesting that the same three adhesion proteins have previously been implicated as potential candidates for exosome targeting to cells (67, 68). Our findings may have important implications in the mechanism of vector attachment to target cells. For retroviruses, this initial step in the virus life cycle was for a long time believed to be mediated solely by the viral Env protein. However, the presence of cellular components on the MMLV surface responsible for promoting virus-cell attachment has been proposed (17, 53, 69). To date, these components remain unidentified. In contrast, several host-derived cell adhesion proteins have been identified on the well-characterized HIV-1 surface (CD44, LFA-1, -2, and -3, and ICAM-1 and -3) (3, 7, 38), and some of these proteins were shown to enhance virus attachment and infectivity (19, 20, 31, 47). As in the case of HIV-1, it seems possible that host-derived cell adhesion proteins on the MMLV lipid membrane play a role in virus attachment and/or transducibility to target cells. Overexpression of these cell adhesion proteins on the virus surface may result in an enhanced avidity of attachment and transduction efficiency of retroviral vectors. On the other hand, researchers attempting the production of tissue-specific retroviral vectors for in vivo applications may find it convenient to prevent the incorporation of these proteins into the virions. Vector particles with such broad adhesion properties may bind to the first population of cells encountered, being wasted in nonrelevant compartments of the body, which would render vector targeting strategies unsuccessful (53, 54). Additionally, it is worth mentioning that both fibronectin receptor subunit beta (integrin β1) and CD9 have a known affinity for fibronectin (11, 16, 32, 51). Fibronectin fragments have been shown to improve retrovirus gene transfer efficiency and thus fibronectin-coated plates are usually used in ex vivo human gene therapy protocols (25, 26, 34). Nevertheless, the exact molecular mechanism by which fibronectin enhances retrovirus-mediated gene transfer has not been elucidated. It is believed that fibronectin enhances retroviral transduction by acting as a molecular bridge between the virus and the cells (5, 26). Bajaj and collaborators have shown that retroviral vector transduction efficiency in the presence of fibronectin fragments correlates with the levels of integrin β1 expression on the target cells (6). However, the identity of the molecules responsible for fibronectin binding on the retroviral lipid membrane remains unclear. Based on our findings, we hypothesize that integrin β1 and CD9 cell adhesion molecules present on the virions contribute to fibronectin binding.
The retroviral vector lipid membrane was also found to be enriched in tetraspanin proteins CD9, CD81, and CD63. While the molecular functions of most tetraspanins remain unclear, members such as CD9 and CD81 have been implicated in a variety of cellular functions such as cell adhesion, cell fusion, and cell motility. It has been recently shown that HIV-1 assembles and buds at membrane regions that contain tetraspanins such as CD9, CD63 and CD81 (37). In addition, two reports suggested that CD9 and CD81 may be involved in the modulation of HIV-1-induced membrane fusion (23, 28).
Another important point to consider is the immunogenic potential of the host proteins exposed at the vector surface, since they could be responsible for viral vector inactivation in vivo. It has been demonstrated that host proteins on the retroviral surface can elicit an immune response by the observation that immunization with human major histocompatibility complex class I protein (a membrane protein acquired by HIV-1) can protect monkeys from infection by simian immunodeficiency virus produced by human cells much more efficiently than immunization with simian immunodeficiency virus virions produced by monkey cells (4). For in vivo gene therapy applications, it may be interesting to silence any gene that codes for immunogenic proteins in the producer cell line in an attempt to produce retroviral vectors that lack these antigens and thus have a lower immunogenic potential.
Efforts toward the production of improved retroviral vectors for gene therapy have been focused almost exclusively on the manipulation of virus-encoded genes. Little attention has been paid to the acquisition of cellular components by the retroviral particle, which is mainly determined by the producer cell line. A good example would be the adoption of human packaging cell lines in place of murine packaging cells expressing Gal (α1-3) terminal carbohydrates to avoid vector inactivation by the human complement (65, 66). The authors suggest that host cellular proteins incorporated into the retroviral vector particles may be interesting points of manipulation to improve vector properties. The strategies proposed in this work for retroviral vectors could be easily extended to the manipulation of lentiviral vectors/ which are likely to incorporate a subset of host membrane proteins similar to the ones described in this work when produced by 293 HEK cells. It is our hope that the work presented here opens new avenues to improve retroviral and lentiviral gene therapy vectors.
Acknowledgments
We thank Miguel Chillon for critical evaluation of the manuscript. The help of Robert Alain with electron microscopy studies and of Leonid Kriazheb with SDS-PAGE silver staining techniques is greatly appreciated.
Footnotes
Published ahead of print on 21 November 2007.
REFERENCES
- 1.Alain, R. 1997. Quantitation of virus particles by negative stain electron microscopy. Microsc. Today 9720. [Google Scholar]
- 2.Alain, R., F. Nadon, C. Seguin, P. Payment, and M. Trudel. 1987. Rapid virus subunit visualization by direct sedimentation of samples on electron microscope grids. J. Virol. Methods 16209-216. [DOI] [PubMed] [Google Scholar]
- 3.Arthur, L. O., J. W. Bess, Jr., R. C. Sowder II, R. E. Benveniste, D. L. Mann, J. C. Chermann, and L. E. Henderson. 1992. Cellular proteins bound to immunodeficiency viruses: implications for pathogenesis and vaccines. Science 2581935-1938. [DOI] [PubMed] [Google Scholar]
- 4.Arthur, L. O., J. W. Bess, Jr., R. G. Urban, J. L. Strominger, W. R. Morton, D. L. Mann, L. E. Henderson, and R. E. Benveniste. 1995. Macaques immunized with HLA-DR are protected from challenge with simian immunodeficiency virus. J. Virol. 693117-3124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Asada, K., T. Uemori, T. Ueno, K. Hashino, N. Koyama, A. Kawamura, and I. Kato. 1998. Enhancement of retroviral gene transduction on a dish coated with a cocktail of two different polypeptides: one exhibiting binding activity toward target cells, and the other toward retroviral vectors. J. Biochem. (Tokyo) 1231041-1047. [DOI] [PubMed] [Google Scholar]
- 6.Bajaj, B., S. Behshad, and S. T. Andreadis. 2002. Retroviral gene transfer to human epidermal keratinocytes correlates with integrin expression and is significantly enhanced on fibronectin. Hum. Gene Ther. 131821-1831. [DOI] [PubMed] [Google Scholar]
- 7.Bastiani, L., S. Laal, M. Kim, and S. Zolla-Pazner. 1997. Host cell-dependent alterations in envelope components of human immunodeficiency virus type 1 virions. J. Virol. 713444-3450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Basyuk, E., T. Galli, M. Mougel, J. M. Blanchard, M. Sitbon, and E. Bertrand. 2003. Retroviral genomic RNAs are transported to the plasma membrane by endosomal vesicles. Dev. Cell 5161-174. [DOI] [PubMed] [Google Scholar]
- 9.Bess, J. W., Jr., R. J. Gorelick, W. J. Bosche, L. E. Henderson, and L. O. Arthur. 1997. Microvesicles are a source of contaminating cellular proteins found in purified HIV-1 preparations. Virology 230134-144. [DOI] [PubMed] [Google Scholar]
- 10.Boucheix, C., and E. Rubinstein. 2001. Tetraspanins. Cell. Mol. Life Sci. 581189-1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Buck, C. A., and A. F. Horwitz. 1987. Integrin, a transmembrane glycoprotein complex mediating cell-substratum adhesion. J. Cell Sci. Suppl. 8231-250. [DOI] [PubMed] [Google Scholar]
- 12.Charrin, S., F. Le Naour, V. Labas, M. Billard, J. P. Le Caer, J. F. Emile, M. A. Petit, C. Boucheix, and E. Rubinstein. 2003. EWI-2 is a new component of the tetraspanin web in hepatocytes and lymphoid cells. Biochem. J. 373409-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Charrin, S., F. Le Naour, M. Oualid, M. Billard, G. Faure, S. M. Hanash, C. Boucheix, and E. Rubinstein. 2001. The major CD9 and CD81 molecular partner. Identification and characterization of the complexes. J. Biol. Chem. 27614329-14337. [DOI] [PubMed] [Google Scholar]
- 14.Chertova, E., O. Chertov, L. V. Coren, J. D. Roser, C. M. Trubey, J. W. Bess, Jr., R. C. Sowder II, E. Barsov, B. L. Hood, R. J. Fisher, K. Nagashima, T. P. Conrads, T. D. Veenstra, J. D. Lifson, and D. E. Ott. 2006. Proteomic and biochemical analysis of purified human immunodeficiency virus type 1 produced from infected monocyte-derived macrophages. J. Virol. 809039-9052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clayton, A., A. Turkes, S. Dewitt, R. Steadman, M. D. Mason, and M. B. Hallett. 2004. Adhesion and signaling by B cell-derived exosomes: the role of integrins. FASEB J. 18977-979. [DOI] [PubMed] [Google Scholar]
- 16.Cook, G. A., C. M. Longhurst, S. Grgurevich, S. Cholera, J. T. Crossno, Jr., and L. K. Jennings. 2002. Identification of CD9 extracellular domains important in regulation of CHO cell adhesion to fibronectin and fibronectin pericellular matrix assembly. Blood 1004502-4511. [DOI] [PubMed] [Google Scholar]
- 17.Davis, H. E., J. R. Morgan, and M. L. Yarmush. 2002. Polybrene increases retrovirus gene transfer efficiency by enhancing receptor-independent virus adsorption on target cell membranes. Biophys. Chem. 97159-172. [DOI] [PubMed] [Google Scholar]
- 18.Escola, J. M., M. J. Kleijmeer, W. Stoorvogel, J. M. Griffith, O. Yoshie, and H. J. Geuze. 1998. Selective enrichment of tetraspan proteins on the internal vesicles of multivesicular endosomes and on exosomes secreted by human B-lymphocytes. J. Biol. Chem. 27320121-20127. [DOI] [PubMed] [Google Scholar]
- 19.Fortin, J. F., R. Cantin, G. Lamontagne, and M. Tremblay. 1997. Host-derived ICAM-1 glycoproteins incorporated on human immunodeficiency virus type 1 are biologically active and enhance viral infectivity. J. Virol. 713588-3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fortin, J. F., R. Cantin, and M. J. Tremblay. 1998. T cells expressing activated LFA-1 are more susceptible to infection with human immunodeficiency virus type 1 particles bearing host-encoded ICAM-1. J. Virol. 722105-2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gatti, J. L., S. Metayer, M. Belghazi, F. Dacheux, and J. L. Dacheux. 2005. Identification, proteomic profiling, and origin of ram epididymal fluid exosome-like vesicles. Biol. Reprod. 721452-1465. [DOI] [PubMed] [Google Scholar]
- 22.Gluschankof, P., I. Mondor, H. R. Gelderblom, and Q. J. Sattentau. 1997. Cell membrane vesicles are a major contaminant of gradient-enriched human immunodeficiency virus type-1 preparations. Virology 230125-133. [DOI] [PubMed] [Google Scholar]
- 23.Gordon-Alonso, M., M. Yanez-Mo, O. Barreiro, S. Alvarez, M. A. Munoz-Fernandez, A. Valenzuela-Fernandez, and F. Sanchez-Madrid. 2006. Tetraspanins CD9 and CD81 modulate HIV-1-induced membrane fusion. J. Immunol. 1775129-5137. [DOI] [PubMed] [Google Scholar]
- 24.Gurer, C., A. Cimarelli, and J. Luban. 2002. Specific incorporation of heat shock protein 70 family members into primate lentiviral virions. J. Virol. 764666-4670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hacein-Bey, S., F. Gross, P. Nusbaum, C. Hue, Y. Hamel, A. Fischer, and M. Cavazzana-Calvo. 2001. Optimization of retroviral gene transfer protocol to maintain the lymphoid potential of progenitor cells. Hum. Gene Ther. 12291-301. [DOI] [PubMed] [Google Scholar]
- 26.Hanenberg, H., X. L. Xiao, D. Dilloo, K. Hashino, I. Kato, and D. A. Williams. 1996. Colocalization of retrovirus and target cells on specific fibronectin fragments increases genetic transduction of mammalian cells. Nat. Med. 2876-882. [DOI] [PubMed] [Google Scholar]
- 27.Hegmans, J. P., M. P. Bard, A. Hemmes, T. M. Luider, M. J. Kleijmeer, J. B. Prins, L. Zitvogel, S. A. Burgers, H. C. Hoogsteden, and B. N. Lambrecht. 2004. Proteomic analysis of exosomes secreted by human mesothelioma cells. Am. J. Pathol. 1641807-1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ho, S. H., F. Martin, A. Higginbottom, L. J. Partridge, V. Parthasarathy, G. W. Moseley, P. Lopez, C. Cheng-Mayer, and P. N. Monk. 2006. Recombinant extracellular domains of tetraspanin proteins are potent inhibitors of the infection of macrophages by human immunodeficiency virus type 1. J. Virol. 806487-6496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Katane, M., E. Takao, Y. Kubo, R. Fujita, and H. Amanuma. 2002. Factors affecting the direct targeting of murine leukemia virus vectors containing peptide ligands in the envelope protein. EMBO Rep. 3899-904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kearney, R., F. Blondeau, P. McPherson, A. Bell, F. Servant, M. Drapeau, S. de Grandpre, and J. J. Bergeron. 2005. Elimination of redundant protein identifications in high throughput proteomics. Conf. Proc. IEEE Eng. Med. Biol. Soc. 54803-4806. [DOI] [PubMed] [Google Scholar]
- 31.Liao, Z., J. W. Roos, and J. E. Hildreth. 2000. Increased infectivity of HIV type 1 particles bound to cell surface and solid-phase ICAM-1 and VCAM-1 through acquired adhesion molecules LFA-1 and VLA-4. AIDS Res. Hum. Retrovir. 16355-366. [DOI] [PubMed] [Google Scholar]
- 32.Longhurst, C. M., J. D. Jacobs, M. M. White, J. T. Crossno, Jr., D. A. Fitzgerald, J. Bao, T. J. Fitzgerald, R. Raghow, and L. K. Jennings. 2002. Chinese hamster ovary cell motility to fibronectin is modulated by the second extracellular loop of CD9. Identification of a putative fibronectin binding site. J. Biol. Chem. 27732445-32452. [DOI] [PubMed] [Google Scholar]
- 33.Mears, R., R. A. Craven, S. Hanrahan, N. Totty, C. Upton, S. L. Young, P. Patel, P. J. Selby, and R. E. Banks. 2004. Proteomic analysis of melanoma-derived exosomes by two-dimensional polyacrylamide gel electrophoresis and mass spectrometry. Proteomics 44019-4031. [DOI] [PubMed] [Google Scholar]
- 34.Moritz, T., P. Dutt, X. Xiao, D. Carstanjen, T. Vik, H. Hanenberg, and D. A. Williams. 1996. Fibronectin improves transduction of reconstituting hematopoietic stem cells by retroviral vectors: evidence of direct viral binding to chymotryptic carboxy-terminal fragments. Blood 88855-862. [PubMed] [Google Scholar]
- 35.Nermut, M. V., K. Wallengren, and J. Pager. 1999. Localization of actin in Moloney murine leukemia virus by immunoelectron microscopy. Virology 26023-34. [DOI] [PubMed] [Google Scholar]
- 36.Nguyen, D. G., A. Booth, S. J. Gould, and J. E. Hildreth. 2003. Evidence that HIV budding in primary macrophages occurs through the exosome release pathway. J. Biol. Chem. 27852347-52354. [DOI] [PubMed] [Google Scholar]
- 37.Nydegger, S., S. Khurana, D. N. Krementsov, M. Foti, and M. Thali. 2006. Mapping of tetraspanin-enriched microdomains that can function as gateways for HIV-1. J. Cell Biol. 173795-807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Orentas, R. J., and J. E. Hildreth. 1993. Association of host cell surface adhesion receptors and other membrane proteins with HIV and SIV. AIDS Res. Hum. Retrovir. 91157-1165. [DOI] [PubMed] [Google Scholar]
- 39.Ory, D. S., B. A. Neugeboren, and R. C. Mulligan. 1996. A stable human-derived packaging cell line for production of high titer retrovirus/vesicular stomatitis virus G pseudotypes. Proc. Natl. Acad. Sci. USA 9311400-11406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ott, D. E. 1997. Cellular proteins in HIV virions. Rev. Med. Virol. 7167-180. [DOI] [PubMed] [Google Scholar]
- 41.Ott, D. E. 2002. Potential roles of cellular proteins in HIV-1. Rev. Med. Virol. 12359-374. [DOI] [PubMed] [Google Scholar]
- 42.Ott, D. E., L. V. Coren, E. N. Chertova, T. D. Gagliardi, and U. Schubert. 2000. Ubiquitination of HIV-1 and MuLV Gag. Virology 278111-121. [DOI] [PubMed] [Google Scholar]
- 43.Ott, D. E., L. V. Coren, T. D. Copeland, B. P. Kane, D. G. Johnson, R. C. Sowder II, Y. Yoshinaka, S. Oroszlan, L. O. Arthur, and L. E. Henderson. 1998. Ubiquitin is covalently attached to the p6Gag proteins of human immunodeficiency virus type 1 and simian immunodeficiency virus and to the p12Gag protein of Moloney murine leukemia virus. J. Virol. 722962-2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ott, D. E., L. V. Coren, D. G. Johnson, B. P. Kane, R. C. Sowder II, Y. D. Kim, R. J. Fisher, X. Z. Zhou, K. P. Lu, and L. E. Henderson. 2000. Actin-binding cellular proteins inside human immunodeficiency virus type 1. Virology 26642-51. [DOI] [PubMed] [Google Scholar]
- 45.Ott, D. E., L. V. Coren, D. G. Johnson, R. C. Sowder II, L. O. Arthur, and L. E. Henderson. 1995. Analysis and localization of cyclophilin A found in the virions of human immunodeficiency virus type 1 MN strain. AIDS Res. Hum. Retrovir. 111003-1006. [DOI] [PubMed] [Google Scholar]
- 46.Ott, D. E., L. V. Coren, B. P. Kane, L. K. Busch, D. G. Johnson, R. C. Sowder II, E. N. Chertova, L. O. Arthur, and L. E. Henderson. 1996. Cytoskeletal proteins inside human immunodeficiency virus type 1 virions. J. Virol. 707734-7743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Paquette, J. S., J. F. Fortin, L. Blanchard, and M. J. Tremblay. 1998. Level of ICAM-1 surface expression on virus producer cells influences both the amount of virion-bound host ICAM-1 and human immunodeficiency virus type 1 infectivity. J. Virol. 729329-9336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Paquin, A., D. E. Jaalouk, and J. Galipeau. 2001. Retrovector encoding a green fluorescent protein-herpes simplex virus thymidine kinase fusion protein serves as a versatile suicide/reporter for cell and gene therapy applications. Hum. Gene Ther. 1213-23. [DOI] [PubMed] [Google Scholar]
- 49.Pelchen-Matthews, A., B. Kramer, and M. Marsh. 2003. Infectious HIV-1 assembles in late endosomes in primary macrophages. J. Cell Biol. 162443-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pelchen-Matthews, A., G. Raposo, and M. Marsh. 2004. Endosomes, exosomes and Trojan viruses. Trends Microbiol. 12310-316. [DOI] [PubMed] [Google Scholar]
- 51.Pierschbacher, M. D., and E. Ruoslahti. 1984. Variants of the cell recognition site of fibronectin that retain attachment-promoting activity. Proc. Natl. Acad. Sci. USA 815985-5988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pisitkun, T., R. F. Shen, and M. A. Knepper. 2004. Identification and proteomic profiling of exosomes in human urine. Proc. Natl. Acad. Sci. USA 10113368-13373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pizzato, M., E. D. Blair, M. Fling, J. Kopf, A. Tomassetti, R. A. Weiss, and Y. Takeuchi. 2001. Evidence for nonspecific adsorption of targeted retrovirus vector particles to cells. Gene Ther. 81088-1096. [DOI] [PubMed] [Google Scholar]
- 54.Pizzato, M., S. A. Marlow, E. D. Blair, and Y. Takeuchi. 1999. Initial binding of murine leukemia virus particles to cells does not require specific Env-receptor interaction. J. Virol. 738599-8611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Potolicchio, I., G. J. Carven, X. Xu, C. Stipp, R. J. Riese, L. J. Stern, and L. Santambrogio. 2005. Proteomic analysis of microglia-derived exosomes: metabolic role of the aminopeptidase CD13 in neuropeptide catabolism. J. Immunol. 1752237-2243. [DOI] [PubMed] [Google Scholar]
- 56.Rieu, S., C. Geminard, H. Rabesandratana, J. Sainte-Marie, and M. Vidal. 2000. Exosomes released during reticulocyte maturation bind to fibronectin via integrin alpha4beta1. Eur. J. Biochem. 267583-590. [DOI] [PubMed] [Google Scholar]
- 57.Saphire, A. C., P. A. Gallay, and S. J. Bark. 2006. Proteomic analysis of human immunodeficiency virus using liquid chromatography/tandem mass spectrometry effectively distinguishes specific incorporated host proteins. J. Proteome Res. 5530-538. [DOI] [PubMed] [Google Scholar]
- 58.Segura, M. M., A. Garnier, and A. Kamen. 2006. Purification and characterization of retrovirus vector particles by rate zonal ultracentrifugation. J. Virol. Methods 13382-91. [DOI] [PubMed] [Google Scholar]
- 59.Segura, M. M., A. Kamen, P. Trudel, and A. Garnier. 2005. A novel purification strategy for retrovirus gene therapy vectors using heparin affinity chromatography. Biotechnol. Bioeng. 90391-404. [DOI] [PubMed] [Google Scholar]
- 60.Sherer, N. M., M. J. Lehmann, L. F. Jimenez-Soto, A. Ingmundson, S. M. Horner, G. Cicchetti, P. G. Allen, M. Pypaert, J. M. Cunningham, and W. Mothes. 2003. Visualization of retroviral replication in living cells reveals budding into multivesicular bodies. Traffic 4785-801. [DOI] [PubMed] [Google Scholar]
- 61.Shevchenko, A., M. Wilm, O. Vorm, and M. Mann. 1996. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 68850-858. [DOI] [PubMed] [Google Scholar]
- 62.Stipp, C. S., T. V. Kolesnikova, and M. E. Hemler. 2001. EWI-2 is a major CD9 and CD81 partner and member of a novel Ig protein subfamily. J. Biol. Chem. 27640545-40554. [DOI] [PubMed] [Google Scholar]
- 63.Strack, B., A. Calistri, S. Craig, E. Popova, and H. G. Gottlinger. 2003. AIP1/ALIX is a binding partner for HIV-1 p6 and EIAV p9 functioning in virus budding. Cell 114689-699. [DOI] [PubMed] [Google Scholar]
- 64.Tai, C. K., C. R. Logg, J. M. Park, W. F. Anderson, M. F. Press, and N. Kasahara. 2003. Antibody-mediated targeting of replication-competent retroviral vectors. Hum. Gene Ther. 14789-802. [DOI] [PubMed] [Google Scholar]
- 65.Takeuchi, Y., F. L. Cosset, et al. 1994. Type C retrovirus inactivation by human complement is determined by both the viral genome and the producer cell. J. Virol. 688001-8007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Takeuchi, Y., C. D. Porter, K. M. Strahan, A. F. Preece, K. Gustafsson, F. L. Cosset, R. A. Weiss, and M. K. Collins. 1996. Sensitization of cells and retroviruses to human serum by (alpha 1-3) galactosyltransferase. Nature 37985-88. [DOI] [PubMed] [Google Scholar]
- 67.Thery, C., M. Boussac, P. Veron, P. Ricciardi-Castagnoli, G. Raposo, J. Garin, and S. Amigorena. 2001. Proteomic analysis of dendritic cell-derived exosomes: a secreted subcellular compartment distinct from apoptotic vesicles. J. Immunol. 1667309-7318. [DOI] [PubMed] [Google Scholar]
- 68.Thery, C., A. Regnault, J. Garin, J. Wolfers, L. Zitvogel, P. Ricciardi-Castagnoli, G. Raposo, and S. Amigorena. 1999. Molecular characterization of dendritic cell-derived exosomes. Selective accumulation of the heat shock protein hsc73. J. Cell Biol. 147599-610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Walker, S. J., M. Pizzato, Y. Takeuchi, and S. Devereux. 2002. Heparin binds to murine leukemia virus and inhibits Env-independent attachment and infection. J. Virol. 766909-6918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang, M. Q., W. Kim, G. Gao, T. A. Torrey, H. C. Morse III, P. De Camilli, and S. P. Goff. 2003. Endophilins interact with Moloney murine leukemia virus Gag and modulate virion production. J. Biol. 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wubbolts, R., R. S. Leckie, P. T. Veenhuizen, G. Schwarzmann, W. Mobius, J. Hoernschemeyer, J. W. Slot, H. J. Geuze, and W. Stoorvogel. 2003. Proteomic and biochemical analyses of human B cell-derived exosomes. Potential implications for their function and multivesicular body formation. J. Biol. Chem. 27810963-10972. [DOI] [PubMed] [Google Scholar]