Abstract
The Cucumber necrosis virus (CNV) particle is a T=3 icosahedron consisting of 180 identical coat protein (CP) subunits. Plants infected with wild-type CNV accumulate a high number of T=3 particles, but other particle forms have not been observed. Particle polymorphism in several T=3 icosahedral viruses has been observed in vitro following the removal of an extended N-terminal region of the CP subunit. In the case of CNV, we have recently described the structure of T=1 particles that accumulate in planta during infection by a CNV mutant (R1+2) in which a large portion of the N-terminal RNA binding domain (R-domain) has been deleted. In this report we further describe properties of this mutant and other CP mutants that produce polymorphic particles. The T=1 particles produced by R1+2 mutants were found to encapsidate a 1.9-kb RNA species as well as smaller RNA species that are similar to previously described CNV defective interfering RNAs. Other R-domain mutants were found to encapsidate a range of specifically sized less-than-full-length CNV RNAs. Mutation of a conserved proline residue in the arm domain near its junction with the shell domain also influenced T=1 particle formation. The proportion of polymorphic particles increased when the mutation was incorporated into R-domain deletion mutants. Our results suggest that both the R-domain and the arm play important roles in the formation of T=3 particles. In addition, the encapsidation of specific CNV RNA species by individual mutants indicates that the R-domain plays a role in the nature of CNV RNA encapsidated in particles.
Many virus particles assemble into T=3 icosahedra that consist of 180 copies of identical coat protein (CP) subunits. The CPs of several of these viruses can be made to assemble into 60-subunit T=1 particles, a process that is facilitated by the removal of an extended N-terminal region of the CP (8, 9, 15, 23, 24, 35, 38). The N-terminal region of the subunit is flexible and thereby enables the CP to adopt the two major conformations required for T=3 particle formation. As T=1 particle formation does not require quasiequivalent interactions among its 60 subunits, removal of the extended N-terminal region shifts equilibrium away from T=3 particle assembly and toward the assembly of other structures such as T=1 icosahedra (8, 15, 22, 26, 35, 39). The N-terminal regions often contain many basic amino acids and are located primarily in the interior of the virion (33, 37) and therefore are generally accepted to anchor virion RNA within the virus particle.
Examples of T=3 viruses whose capsid proteins can be made to form T=1 particles include the plant viruses Brome mosaic virus (BMV) (6, 24), Turnip crinkle virus (TCV) (38), Tomato bushy stunt virus (TBSV) (15), Southern bean mosaic virus (36), and the related Sesbania mosaic virus (SeMV) (35). There are also several naturally occurring T=1 particles such as the satellite particles associated with helper viruses. However, in these cases, the T=1 particles are assembled from distinct CPs encoded by the satellite RNA genome (2, 17).
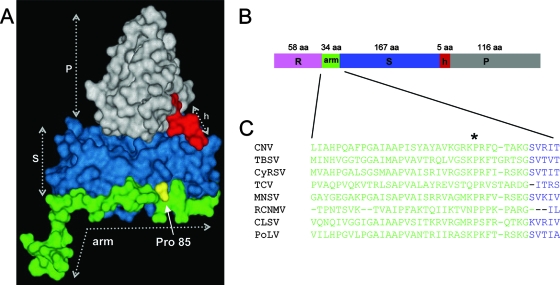
Cucumber necrosis virus (CNV) is a member of the Tombusvirus genus in the family Tombusviridae. Recent cryoelectron microscopy studies have shown that the CNV particle is a T=3 icosahedron whose structure is highly similar to that of TBSV, the type member of the Tombusvirus genus (21). Each of the 180 CP subunits that comprise the CNV particle consists of three major domains: an N-terminal 58-amino-acid (aa) R-domain, a shell domain (S-domain), and a protruding domain (P-domain) (Fig. 1). The R-domain, which is highly basic, is present within the interior of the particle, where it binds RNA and in addition is likely to comprise part of the recently described highly structured inner shell of the CNV virion (21). The S-domain forms the outer shell, and the P-domain projects from the surface of the particle. A flexible 34-aa arm, which also lies within the particle interior, connects the R- and S-domains. The CNV CP subunit exists in three conformationally distinct states termed A, B, and C, each repeated 60 times in the virion. The CP arm is ordered in the C subunit but is relatively disordered in the A and B subunits. The R-domain is disordered in the A, B, and C subunits. It has been noted that a conserved proline (Pro) residue (Pro85 in CNV) (Fig. 1) exists near the junction of the arm and S-domains of several related small spherical plant RNA viruses (4). It has recently been suggested that this residue may be involved in the ability of the arm to adopt the two arm conformations required for quasiequivalent interactions among the subunits in T=3 particles (19). The amino-terminal portion of three C-subunit arms interact at the particle threefold axis to form a β-annulus which contributes to, but is not necessary for, the integrity of the capsid and overall particle stability (14, 16, 21). Recent studies have demonstrated that specific regions of the R-domain and arm are responsible for targeting the CP to specific subcellular locations, possibly as sites for virus assembly/disassembly (30).
FIG. 1.
(A)Tertiary structure of the CNV CP C subunit with ordered arm, S-, and P-domains, with the hinge (h) indicated. The structure is based on homology modeling using the X-ray crystal structure of the TBSV CP subunit (27). The location of the conserved Pro85 residue is also shown. The disordered R-domain is not shown. (B) The CP linear structure is shown, using the same color scheme as in panel A. The number of amino acids in each domain is given. (C) Alignment of the arm region and the junction at the S-domain of several members of the Tombusviridae family, demonstrating conservation of the Pro residue (Pro85 in CNV) (asterisk) in this region. CNV, TBSV, and Cymbidium ringspot virus (CyRSV) are tombusviruses, TCV and Melon necrotic spot virus (MNSV) are carmoviruses, Red clover necrotic mosaic virus (RCNMV) is a dianthovirus, and CLSV (Cucumber leaf spot virus) and PoLV (Pothos latent virus) are aureusviruses.
T=1 particles have not been observed in wild-type (WT) CNV particle preparations (unpublished observations). In this report, we show that deletion of specific R-domain sequences as well as mutation of a conserved Pro residue in the CNV arm results in the accumulation of T=1 particles as well as particles intermediate in size between T=3 and T=1 particles in infected plants (intermediately sized [IS] particles). In addition, each R-domain mutant encapsidates specific, less-than-full-length CNV RNA species. These observations along with the previously described function for the R and arm regions in subcellular targeting (30) suggest a highly orchestrated role for the CNV arm and R-domain sequences in particle assembly, morphology, and the nature of the encapsidated RNA.
MATERIALS AND METHODS
Construction of R-domain mutants.
CNV mutants R1+2, R3(−), RΔ2-58, and RΔ32-58 were constructed by deletion mutagenesis of a CNV cDNA clone encompassing the CP open reading frame by use of methods similar to those previously described (18). Following mutagenesis, an EcoRI/NcoI fragment that encompasses the mutated R-domain region was cloned into the corresponding site of an infectious CNV cDNA clone. The Pro85Gly mutant was similarly constructed as previously described (18). The double mutants R1+2/Pro85Gly and R(3-)Pro85/Gly were constructed by substitution of Gly for Pro at arm residue 85 in mutants R1+2 and R3(−) also by use of site-directed mutagenesis (18). All clones were sequenced to confirm the intended mutations.
Transcript inoculation and purification of mutant particles.
Approximately 1.5 μg of T7 polymerase runoff transcripts of mutant cDNA clones was used to inoculate four leaves of Nicotiana benthamiana as previously described (20). Particles were purified from plants at 5 to 7 days postinoculation by differential centrifugation as follows. Infected leaves were ground in 2 volumes of 100 mM sodium acetate (pH 5.0) containing 5 mM β-mercaptoethanol and allowed to stand on ice for 30 to 60 min. The slurry was then filtered with Miracloth (Calbiochem) and centrifuged at 8,000 × g in a GSA rotor. The supernatant was adjusted to 8% polyethylene glycol (molecular weight, 8,000; Sigma) and stirred at 4°C for 1 to 2 h. Virus was pelleted by centrifugation at 8,000 × g in a GSA rotor, resuspended in 10 mM sodium acetate, pH 5.0, and centrifuged at 20,000 × g in an Eppendorf microcentrifuge to pellet any insoluble material. The supernatant was then subjected to high-speed centrifugation (40,000 rpm [145,000 × g] for 2.5 h in a Ti 50.2 rotor) at 4°C and then resuspended in 10 mM sodium acetate, pH 5.0. Particle concentration was assessed spectrophotometrically and then confirmed by agarose gel electrophoresis through a 1% agarose gel buffered in Tris-borate, pH 8.2 (TB). Known concentrations of WT CNV particles (as determined by spectrophotometry) were used as mass standards. Gels were stained with ethidium bromide (EtBr) in the presence of TB buffer containing 10 mM EDTA (18) to visualize virion RNA and in Coomassie blue to visualize particle protein. The total mass of WT or mutant virus particles obtained per gram of infected leaves was used to calculate the percent virus yield.
Sucrose gradient fractionation: R1+2 particles.
Sucrose gradient fractionation was used to separate T=1 from T=3 particles obtained from mutant R1+2. Gradients were prepared by layering 10 ml each of 40%, 30%, 20%, and 10% sucrose (wt/vol) in 10 mM sodium acetate buffer, pH 5.0. Gradients were allowed to stand overnight at 4 C. Total R1+2 particle preparations were layered on the top of the gradient and then centrifuged in an SW28 rotor at 27,500 rpm (100,000 × g) for 3 h. The opalescent material forming the two bands was collected with a syringe, diluted three times in 10 mM sodium acetate, pH 5.0 and subjected to one more round of high-speed centrifugation (140,000 × g for 2.5 h). Pellets were resuspended in 10 mM sodium acetate, pH 5.0, and stored at 4C.
Virion RNA extraction and electrophoresis.
Virion RNA was extracted from virus particles by use of phenol saturated with 1× virion RNA extraction buffer (50 mM Tris-HCl, pH 7.8, 25 mM EDTA, and 0.25% sodium dodecyl sulfate). Virus (10 to 50 μg) in 100 μl of virion extraction buffer was vigorously vortexed in the presence of 200 μl of buffered phenol and followed by extraction of the aqueous phase with chloroform-isoamyl alcohol (24:1). RNA was analyzed by electrophoresis through a 1% agarose gel buffered in 40 mM Tris-borate, pH 8.2, containing 1 mM EDTA and visualized by staining with EtBr or SybrSafe (Invitrogen).
RT-PCR and sequence analysis.
Reverse transcription-PCR (RT-PCR) was conducted on RNA extracted from sucrose gradient-purified R1+2 T=1 particles by use of primers corresponding to the 5′ and 3′ terminal regions of CNV genomic RNA (gRNA). The 668-nucleotide (nt) defective RNA was directly sequenced following gel purification of the major RT-PCR species. The remaining defective RNAs were cloned into pGEM-T prior to sequence analysis. Sequences were obtained using an Applied Biosystems 310 genetic analyzer.
TEM.
Mutant particle samples were applied to a collodion-covered 300-mesh copper grid and allowed to adsorb for 2 to 3 min. Samples were washed with sterile, deionized water and then stained with 2% uranyl acetate. Particles were viewed at magnifications of ×27,000 to ×80,000 in a JEOL 100CX transmission electron microscope (TEM) operated at 80 kV.
Polyacrylamide gel electrophoresis analysis and Western blotting of mutant CP.
Virus particles were denatured and electrophoresed through NuPage Bis-Tris gels (Invitrogen) essentially according to the manufacturer's recommended conditions. Proteins were blotted onto polyvinylidene difluoride (PVDF) membranes (Bio-Rad) and probed with a mixture of three polyclonal antibodies known to be specific to the CNV S- and P domains (41) as well as the CNV arm and R-domains. A peroxidase-labeled goat anti-rabbit antibody (Sigma) was then used with the ECL Plus kit and Hyperfilm MP (GE Healthcare) to visualize bound antibodies.
Extraction of virus particles from agarose gels.
Virus particles were electrophoresed through a 2% agarose-TB gel at 100 V for 2 h. Gels were then stained with SybrSafe (Invitrogen) stain in the presence of 10 mM EDTA in order to visualize the viral RNA. Gel slices containing T=3 or T=1 particles were cut from the gel and then ground in a 1.5-ml tube by use of a minipestle in the presence of 1/4× denaturing LDS buffer (Invitrogen). Samples were allowed to sit at room temperature for 1 h and then heated to 42°C for 20 min. Samples were then centrifuged at 20,000 × g for 10 min. The supernatant was concentrated down to 1/4 the volume (i.e., to a 1× concentration of LDS buffer) by use of a Speed-Vac (Savant). Samples were then heated to 70°C for 15 min and loaded onto a 4 to 12% NuPage Bis-Tris gel for analysis as described above.
Northern blot analysis of viral RNA.
Virion RNA was extracted as described above and electrophoresed on a denaturing agarose gel. Following electrophoresis, RNA was blotted to Zetaprobe membranes (Bio-Rad) via capillary transfer overnight in 10 mM sodium hydroxide. Blots were then incubated overnight with a mixture of two randomly primed 32P-labeled probes derived from the CNV 5′ terminal 447 nt or the 3′ terminal 428 nt. The washed blot was exposed and visualized using the Cyclone Plus storage phosphor system (Perkin Elmer).
RESULTS AND DISCUSSION
Production of T=1 particles occurs during infection by a CNV CP R-domain deletion mutant.
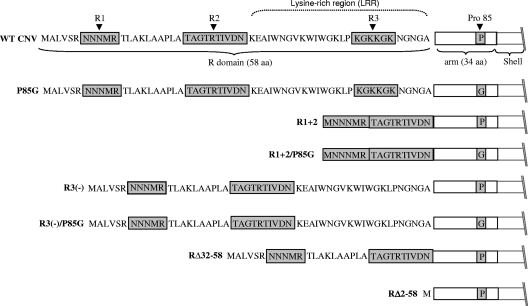
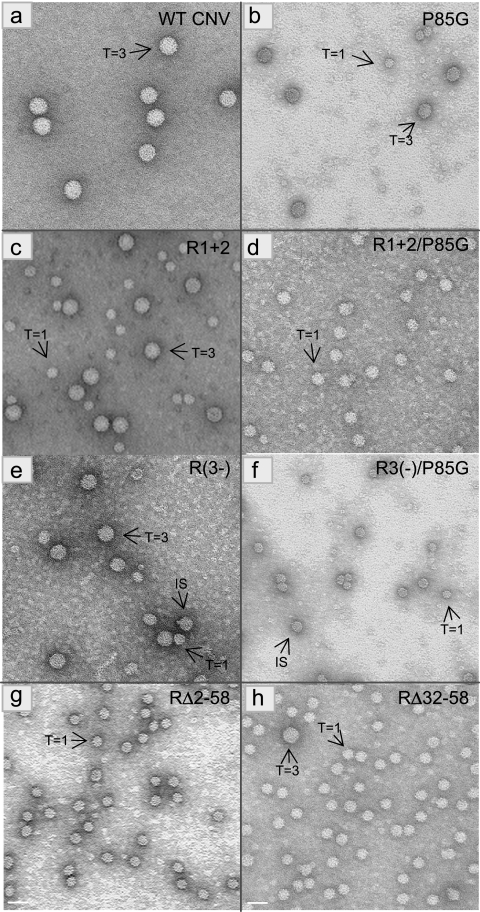
The amino acid sequence of the N-terminal region of an engineered CNV CP deletion mutant, R1+2, is shown in Fig. 2. R1+2 contains 16 aa (aa 7 to 11 and 22 to 31) of the 58-aa WT CNV R-domain. Leaves of plants inoculated with transcripts containing the R1+2 CP mutant developed CNV symptoms; however, the yield of particles was only approximately 2.8% that of WT CNV (Table 1). Particles extracted from plants infected with this mutant were examined by TEM and found to consist of two distinctly sized spherical particles (Fig. 3A, panel 2). The larger particles are similar in size and gross morphology to WT CNV T=3 particles (34 nm) and represent approximately 40% of the total number of particles. (Table 1 and Fig. 3A, panels 1 and 2). The other particles are smaller spheres with an estimated size of 23 nm and represent the remaining 60% of the particles (Table 1 and Fig. 3A, panel 2). Recent cryoelectron microscopy studies have shown that the smaller particles have T=1 icosahedral symmetry. Their structure is described in detail by Katpally et al. (21).
FIG. 2.
Summary of R-domain and arm mutant structures. The top of the diagram shows the 58-aa sequence of the WT CNV CP R-domain along with the downstream 34-aa arm region (open box) and the beginning of the S-domain. Shaded regions in the R-domain correspond to previously identified sequences found to be important for particle accumulation (unpublished data). The C-terminal LRR is indicated above the R-domain sequence. R and arm region mutants are shown beneath. Sequences retained in each mutant are shown. The presence of a proline (P) or glycine (G) at aa position 85 in the arm region of the various mutants is indicated in the shaded box.
TABLE 1.
Properties of particles and RNA found in CNV CP mutants
| Mutanta | %WT CNV yieldb | T=3:IS:T=1 ratioc | Mass of CP (kDa)d
|
Size (kb) of RNA species found in particles of CNV mutantse
|
||
|---|---|---|---|---|---|---|
| Predicted | Observed | Predicted | Observed | |||
| WT CNV | 100 | 1:0:0 | 41 | 41 | 4.7 (2.09) | 4.7 |
| P85G | 0.4 | 1:0:1.5 | 41 | 41, 34, 31 | 4.7 | 4.7, 0.7 |
| R1+2 | 2.8 | 1:0:1.5 | 36 | 36, 34 | 4.58 (1.97) | 1.9, 1.4, 0.9 |
| R1+2/P85G | 5.8 | 1:0:10 | 36 | 36, 34 | 4.58 | 0.6-0.75 |
| R3(−) | 14.9 | 7:7:1 | 40 | 40, 38, 34, 31 | 4.68 (2.08) | 2.1, 1.2-1.5, 0.6-0.75 |
| R3(−)/p85G | 0.7 | 0:1:1.4 | 40 | 40, 34, 31 | 4.68 | 0.6 |
| RΔ32-58 | 6.5 | >95% T=1 | 38 | 38, 37, 34 | 4.62 (2.02) | 2.0, 0.6 |
| RΔ2-58 | 0.6 | >95% T=1 | 34 | 34 | 4.53 | 0.6-0.75 |
See Fig. 2 for description of mutants.
Percent WT CNV yield indicates the yield of mutant particles as a percentage of that of WT CNV. Equivalent masses of infected leaves were used, and the values shown are the average yields in mass obtained from two independent experiments.
The approximate T=3:IS:T=1 particle ratio was assessed by TEM of uranyl acetate-stained particles. At least 250 particles were examined using several different areas of one or more TEM grids. In the case of P85G, particle ratios were obtained only from preparations that contained T=1 particles (see text).
Predicted masses are based on the known mass of the CNV CP (40) minus the mass of the amino acids deleted in each construct. Observed masses were estimated following denaturing polyacrylamide gel electrophoresis as described for Fig. 6 and 8. Numbers in italics correspond to the approximate sizes of deleted CP species found in the various mutants. Underlined numbers correspond to CP species that are enriched in IS or T=1 particles of the indicated mutant (Fig. 8). The 38-kDa species was observed only for IS particles of R3(−).
The known sizes of WT CNV gRNA (40) and the predicted sizes of the gRNA of each of the mutants are shown in kilobases. For mutants observed to encapsidate an RNA species similar in size to CNV sgRNA1 (2.09 kb) (34), the predicted size of the sgRNA1 for that mutant is shown in parenthesis. Observed sizes correspond to the approximate sizes of the predominant RNA species extracted from particle preparations. Species that may correspond to encapsidated sgRNA1 are shown in italics. Sizes were estimated following denaturing agarose gel electrophoresis as described in Fig. 7. The 0.7 kb RNA of P85G was not always present in particle preparations (see text).
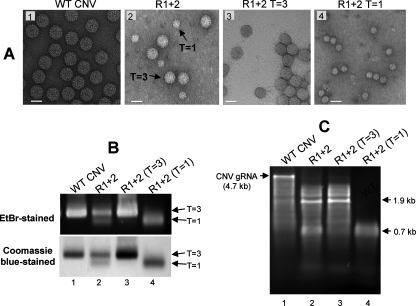
FIG. 3.
Analysis of R1+2 particles and particle RNA. (A) TEM of particles extracted from leaf tissue infected with the indicated mutant. Particles were stained with 2% uranyl acetate and photographed at a magnification of ×80,000. The bars correspond to approximately 34 nm. (B) Agarose gel of total particles from plants infected with WT CNV (lane 1) and R1+2 infections (lane 2). Lanes 3 and 4 contain the faster- and slower-migrating species, respectively, of sucrose gradient-purified R 1+2 particles. The gels were stained with EtBr or Coomassie blue as indicated. The positions of the two major virion species are shown with arrows. (C) Agarose gel of RNA extracted from total R1+2 particles (lane 2) or sucrose gradient-fractioned particles as in panel B. The numbers on the right correspond to the approximate sizes of the major RNA species.
Further analysis of R1+2 particles by agarose gel electrophoresis showed the presence of two electrophoretically distinct species (Fig. 3B, lane 2). The slower-migrating species likely corresponds to the T=3 particles observed by TEM, as it comigrates with WT CNV particles (Fig. 3B, lane 1), whereas the faster-migrating species corresponds to the smaller previously described T=1 particles. The observation that these species stain with both EtBr and Coomassie blue indicates that they contain both RNA and protein components.
Sucrose gradient fractionation was used to further characterize the two species of particles (Fig. 3A and C). As expected, TEM showed that the faster-sedimenting particles are similar to CNV T=3 virions (Fig. 3A, panel 3) and that the slower-sedimenting ones are similar to T=1 particles (Fig. 3A, panel 4).
RNA extracted from unfractionated particles was found to contain two major RNA species of approximately 1.9 kb and 0.7 kb as well as several additional minor RNA species ranging in size from approximately 1.2 kb to 3.4 kb (Fig. 3C, lane 2). Full-length gRNA was also observed but only at barely detectable levels (Fig. 3C, lane 2). RNA extracted from sucrose gradient-fractionated particles showed that the 1.9-kb RNA as well as the less abundant higher- and lower-molecular-weight RNAs are associated with T=3 particles, whereas the 0.7-kb RNA is primarily associated with T=1 particles (Fig. 3C, lanes 3 and 4, respectively). Together, these observations indicate that the R-domain sequences that are deleted in mutant R1+2 influence the accumulation of T=3 particles in CNV-infected plants. In addition, the deleted R-domain sequences affect the size of RNA species that are encapsidated.
R1+2 T=1 particles encapsidate DI-like RNA species.
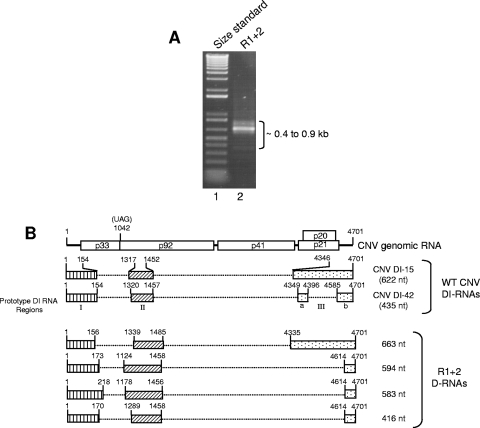
Several tombusviruses have been shown to generate defective interfering RNAs (DI RNAs) during infection (10, 28, 32). These RNAs range in size from approximately 0.4 to 0.9 kb and typically retain portions of the 5′ and 3′ termini of viral gRNA. The small RNA species present in R1+2 particles prompted us to determine if these species represent CNV DI RNAs. Accordingly, RT-PCR analysis was conducted using primers specific to the 5′ and 3′ termini of CNV gRNA. As is shown in Fig. 4A, a prominent RT-PCR product of approximately 0.7 kb is evident along with several less prominent 0.4- to 0.9-kb species. These species were cloned and their sequences aligned with the sequences of CNV gRNA as well as those of previously described CNV DI RNAs (11). Figure 4B shows a diagrammatic representation of the relationship of one cloned 663-nt species (D-663) to CNV gRNA and two previously described prototypical CNV DI RNAs (DI-15 and DI-42) (11). D-663 shows features similar to that of DI RNAs, i.e., the presence of three major nucleotide sequence domains (I, II, and III) that are similar in size and genomic position to CNV prototype DI RNAs (Fig. 4B). The sequences and structures of three additional cloned RT-PCR products consisting of 594 nt, 583 nt, and 416 nt were determined (Fig. 4B). These species also share features in common with tombusvirus DI RNAs. However, they are also distinctive in that they lack region IIIa and contain a shorter region IIIb. In addition, region II of the 594-nt DI-like RNA and both regions I and II of the 583-nt RNAs are long in comparison to previously described CNV DI RNAs (11). Confirmation that the 0.7-kb RNAs indeed consist of CNV-related RNAs was confirmed by probing a Northern blot of R1+2 virion RNA with a 32P-labeled cDNA probe corresponding to the 5′ and 3′ termini of CNV RNA (see Fig. 7A and B, lane 7).
FIG. 4.
Characterization of CNV DI-like RNAs in T=1 particles of mutant R1+2. (A) EtBr-stained agarose gel showing RT-PCR products of RNA purified from T=1 particles of R1+2 (lane 2). RT-PCR primers corresponded to the 5′ and 3′ termini of CNV gRNA. Lane 1 contains a molecular size standard (1-kb Plus DNA ladder; Invitrogen). The approximate sizes of the RT-PCR products are indicated. (B) Structure of R1+2 DI-like RNAs. The structure of CNV genome RNA is shown at the top along with the structures of two previously described prototype CNV DI RNAs (11). The three major regions of the CNV genome that are retained in the prototype DI RNAs (I, II, and III) are depicted by the boxed regions in CNV DI-15 and DI-42. Region III of DI-42 is divided into subregions IIIa and IIIb as previously described (11). The bottom shows structures of DI-like RNAs deduced from nucleotide sequence analysis. The shading patterns used correspond to those of the prototype DI-like RNAs. Numbers at the 5′ and 3′ termini of the boxes indicate the nucleotide positions at the boundaries of the box region relative to CNV gRNA. The size in nucleotides of each DI or DI-like RNA is shown on the right. D-RNA, defective RNA.
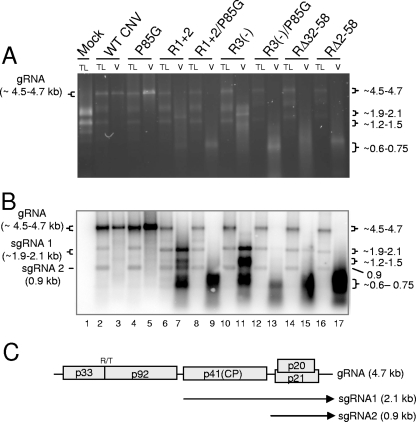
FIG. 7.
Agarose gel (A) and Northern blot analysis (B) of total leaf RNA extracts and virion RNAs of the indicated mutants. Leaves infected with each of the mutants were ground in liquid nitrogen. Total leaf RNA was extracted from one portion of the frozen mixture, and virions were extracted from another portion. Equal amounts of total leaf RNA (TL) or RNA extracted from virions (V) were loaded onto a denaturing agarose gel and stained with EtBr. A duplicate gel was blotted and hybridized with a mixture of two probes, one corresponding to the 5′ terminal 447 nt and the other to the 3′ terminal 428 nt of CNV gRNA. (C) Genome structure of WT CNV RNA showing the sizes and origins of sgRNA 1 (2.1 kb) and sgRNA 2 (0.9 kb). The values on the left in panel A correspond to the known size of gRNA in WT CNV virions and total leaf RNA extracts (4.7 kb) as well as the range of sizes predicted for the gRNA of the various mutants (summarized in Table 1). The values on the right in panel A correspond to the approximate sizes of the RNA species detected in particles of the various mutants (Table 1). The values on the left in panel B correspond to the known and predicted sizes of gRNA and sgRNA1 and -2 that are detected in blots of WT and mutant-infected total leaf RNA extracts. The range of sizes shown for gRNA and sgRNA2 reflects the sizes of the various mutant constructs (Table 1). The size of the sgRNA2 is not affected in the mutants. The values on the right in panel B correspond to the approximate sizes of various species detected in blotted total leaf and virion RNA extracts (Table 1). Sizes were estimated using the Invitrogen 0.24- to 9.5-kb RNA ladder as well as the known sizes of CNV gRNA, sgRNA1, and sgRNA2.
As described above, RNA extracted from R1+2 particles (Fig. 3C, lane 2) contains a predominant 1.9-kb species as well as lower levels of higher- and lower-molecular-weight RNAs. Northern blot analysis of these virion RNAs indicates that the 1.9-kb RNA is derived from the CNV genome, as it hybridizes to a 32P-labeled cDNA probe specific to CNV RNA (see Fig. 7A and B, lane 7). Its length is similar to the predicted size (1.85 kb) of a CP subgenomic RNA (sgRNA) that would be produced from the genome of the R1+2 mutant. RT-PCR analysis and sequencing have confirmed that this species does indeed correspond to the R1+2 CP sgRNA (unpublished data).
Unlike most other characterized DI RNAs, the DI-like RNAs in R1+2 particles were not found to be present at high levels in infected leaves (see Fig. 7A and B, lane 6). It is possible that these particular DI-like RNAs are not readily replicated by the CNV replicase. Alternatively, encapsidation of these DI-like RNAs within T=1 particles (which is not observed in WT CNV infections) may interfere with their replication. With respect to the DI-like RNAs lacking region IIIa, the low level of accumulation is consistent with the previous observation that TBSV DI RNA mutants in which region IIIa is deleted replicate at only approximately 10% the efficiency of prototypical TBSV DI RNAs (31).
As described in more detail below with regard to other R-domain mutants, it is possible that CP sequences that are absent in the R1+2 mutant normally play a role in the recognition of CNV sequences specifically found in full-length gRNA. In vitro assays assessing a role in such interactions with DI RNA and various gRNA sequences would be valuable in assessing this possibility.
Other R-domain mutants generate polymorphic particles during infection.
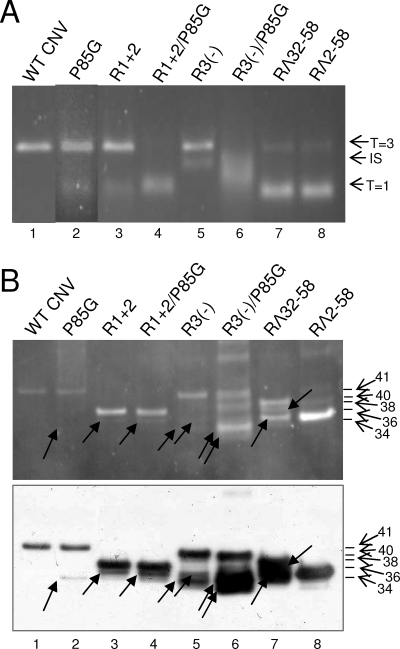
Further deletion mutants were analyzed for particle polymorphism as described above. Mutant RΔ32-58, which lacks the C-terminal 26 aa of the R-domain (Fig. 2), was found by TEM to produce primarily T=1-sized particles (>95%) but also produced a low level of T=3 particles (Table 1 and Fig. 5). Agarose gel electrophoresis showed that the major particle species (Fig. 6, lanes 7) migrates similarly to that of the T=1 particles of R1+2, reinforcing the conclusion that mutant RΔ32-58 produces primarily T=1 particles. Analysis of RΔ2-58 particles which lack nearly the complete R-domain (Fig. 2) showed that more than 95% of the particles observed by TEM appear to be T=1 particles (Table 1 and Fig. 5, panel g). In addition, RΔ2-58 particles comigrate with R1+2 T=1 particles following agarose gel electrophoresis (Fig. 6A, compare lane 8 with lane 3, respectively). TEM analysis of another mutant, R3(−), which lacks the highly basic 6 aa in region III of the R-domain (Fig. 2), produced a mixture of three particle sizes. The largest particles are similar in size to WT CNV T=3 and the smallest are similar in size to T=1 particles. IS particles discernibly smaller than T=3 particles were also evident (Fig. 5, panel e). Based on size, it is possible that these particles are similar to the 120-subunit “pseudo-T=2” particles observed in other studies (22, 25). The ratio of the T=3, IS, and T=1 particle types was determined to be approximately 7:7:1 (Table 1). Examination of R3(−) particles by agarose gel electrophoresis showed a prominent species that comigrates with CNV T=3 particles (Fig. 6, lanes 5). Another species that migrates at a position intermediate to that of either T=3 or T=1 particles was also observed (Fig. 6, lanes 5). This species likely corresponds to the IS particles observed by TEM analysis.
FIG. 5.
TEM of particles extracted from leaves infected with the indicated mutants. Particles were stained with 2% uranyl acetate and photographed at a magnification of ×80,000. The T=3 or T=1 morphology of particles is shown with arrows. The bar corresponds to approximately 34 nm. Table 1 contains a summary of the various particle types observed for each mutant.
FIG. 6.
Analysis of particles obtained from plants infected with single and double P85G and R-domain mutants. (A) Approximately 1.5-μg portions of purified virions were electrophoresed through a 2% TB-agarose gel. The gel was stained with SybrSafe (Invitrogen). The arrows on the right indicate possible particle morphologies as assessed by TEM (Fig. 5). (B) Virions of the indicated mutants were denatured with 1× LDS buffer, electrophoresed through 4 to 12% NuPage-MES (morpholineethanesulfonic) gels, and blotted to PVDF membranes. Membranes were then treated with SYPRO-RUBY blot stain and visualized by epifluorescence (top). The blot (bottom) was probed using a mixture of three CNV antibodies known to be specific to the arm and the R- and S-domains (18) (unpublished data). The mass of WT CNV CP and the predicted molecular masses of the CPs of the mutants are indicated by the arrows and summarized in Table 1. Arrows within the blot point to the major truncated proteins detected in each mutant (Table 1). Standards (not shown) were SeeBlue+2 and Mark 12 (Invitrogen).
Although it seems likely that the small particles found in RΔ2-58 and RΔ32-58 possess T=1 symmetry, as we have shown for R1+2 small particles, more-detailed analyses are required to confirm their precise morphology. Nevertheless, our finding that CNV R-domain mutants generate polymorphic particles supports the notion that R-domain sequences play a crucial role in determining T=3 capsid morphology.
The capsid proteins of several T=3 viruses have been shown to be capable of assembling into T=1 particles and other such polymorphic structures in vitro (6, 15, 23, 24, 35, 36, 38). Many of these studies were conducted using CP in which the N-terminal region was proteolytically cleaved. In the case of SeMV, T=1 particles were generated in bacterial cells via the expression of truncated CP sequences (35), and in the case of TBSV, T=1 particles were produced during the expression of CP R-domain mutants in insect cells (15). In BMV and TCV, T=1 particles are formed in vitro following trypsin treatment of T=3 particles (7, 13) and can also be formed spontaneously following cleavage by exogenous proteases present in virus particle preparations (24). Our studies with CNV differ from these in that CNV polymorphic particle production was induced in planta during virus infection. Such studies should therefore assist in future work aimed at elucidating the in vivo assembly process. In addition, as suggested in a previous study, specific CNV CP sequences along with specific host factors have been implicated in the CNV assembly/disassembly pathway (41). It is anticipated that the studies described here should facilitate further analyses of these aspects of the assembly process. In vivo assembly studies have recently also been reported by Calhoun et al. (6). These studies showed that particular BMV CP deletion mutants that can produce polymorphic particles in plants cannot do so under in vitro conditions, suggesting that host factors play a role in BMV assembly.
The R3(−) IS particles may be similar to the SeMV “pseudo-T=2” particles (particles formed from 12 pentamers of A/B dimers) proposed by Lokesh et al. (25). The SeMV CP mutants that produced the “pseudo-T=2” particles lack a highly basic (arginine-rich) motif in the R-domain. A similar highly basic sequence (KGKKGK) is absent in R3(−) CP. In BMV, particles consisting of 12 pentamers of A/B dimers were formed in yeast upon the coexpression of CP and an engineered CP mRNA. In these studies, polymorphic particle formation was linked to the size of the encapsidated CP mRNA (22). Similar results were obtained in BMV mutant-infected (6) and agroinfiltrated (1) plants. R3(−) particles encapsidate several specific less-than-full-length CNV RNA species (Fig. 7, lane 11). Thus, the polymorphic particles in R3(−) may similarly be a result of RNA size-induced polymorphism; however, further work is required to determine the size of RNA in IS particles and to ascertain the basis for the formation of these particles.
CNV R-domain mutants selectively encapsidate a variety of CNV-related RNA species during infection.
RNAs extracted from purified particles of each of the R-domain mutants described above were analyzed by agarose gel electrophoresis (Fig. 7A) and Northern blotting using a CNV-specific probe (Fig. 7B). It can be seen that each mutant encapsidates distinctly sized CNV-related RNAs that differ significantly in size and relative proportion from what is seen for WT CNV virion RNA (Table 1 and Fig. 7A and B, compare lanes 7, 11, 15, and 17 with lane 3). Specifically, RNA extracted from WT CNV particles consists primarily of the 4.7-kb gRNA (Fig. 7A and B, lane 3) and very low levels (undetectable in Fig. 7) of the 2.1-kb CP sgRNA (sgRNA1) and the 0.9-kb sgRNA (sgRNA2) (Fig. 7C). In contrast, each of the R-domain mutants contain little or no detectable gRNA and disproportionately higher levels of a variety of lower-molecular-weight RNAs ranging in size from ∼0.6 to 2.1 kb (Table 1 and Fig. 7A and B, lanes 7, 11, 15, and 17). The 1.9- to 2.1-kb species present in virions of R3(−) and RΔ32-58 may correspond to the CP sgRNA (sgRNA1) (Fig. 7C) (as shown for the R1+2 1.9-kb species [see above]), as their sizes are similar to the sizes of the sgRNA1s predicted for each mutant, i.e., 2.05 kb and 2.0 kb, respectively Table 1. Similarly, based on size, the 0.9-kb species in R1+2 and RΔ32-58 virions may correspond to encapsidated sgRNA2; however, this requires confirmation. As with RNA extracted from the T=1 particles of R1+2, the small RNA species (0.6 to 0.75 kb) present in particles of the remaining mutants may represent DI-like RNAs; however, further analyses are required to verify this assumption. The origins of the 1.2- to 1.5-kb RNAs present in particles of R1+2 and R3(−) are not known. Similarly sized RNAs have not previously been observed to occur at high levels in CNV infections.
Northern blot analysis of total RNA extracted from leaves infected with WT CNV and each of the R-domain mutants was conducted to address the basis for the distinctive profile of RNA species present in virions of each mutant. As can be seen in Fig. 7B, WT CNV-infected leaves contain, as expected, three main RNA species with sizes of 4.7 kb, 2.1 kb, and 0.9 kb (Fig. 7B, lane 2), which correspond to gRNA, sgRNA1, and sgRNA2, respectively (Fig. 7C). Total leaf RNA extracts of each R-domain mutant contain similarly sized RNAs (Fig. 7B, lanes 6, 10, 14, and 16); moreover, the relative proportions of the three RNAs are similar to what is observed for WT CNV-infected leaves.
As shown in Table 1, particles of the various R-domain mutants accumulate to levels much lower than that of WT CNV, with relative yields ranging from 0.6% for RΔ2-58 to 14.9% for R3(−). This is despite the observation that the levels of gRNA (Fig. 7) (and CP [Fig. 8]) present in total leaf extracts are approximately equivalent to those of WT CNV. Thus, the distinctive profile of the RNA species present in particles of each of the mutants is likely due to the CP of the R-domain mutants lacking regions critical for efficient capsid assembly and/or the accommodation of specific CNV RNAs.
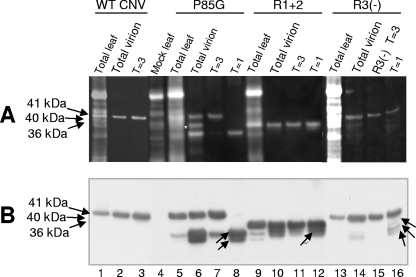
FIG. 8.
Denaturing polyacrylamide gel electrophoresis of protein extracted from infected leaves (TL), total virions (V), and gel-purified particles of P85G, R1+2 and R3(−). WT CNV-, P85G-, R1+2-, and R3(−)-infected leaves were ground in liquid nitrogen. A portion of the ground material was used to extract total protein and another portion was used for total virion extraction. Virions were electrophoresed through a 2% agarose-TB gel, and particles corresponding to T=3, IS, and T=1 particles were purified from gel slices. (A) Samples were denatured with 1× LDS buffer, electrophoresed through 4 to 12% NuPage-MES gels, and blotted to PVDF membranes. Membranes were then treated with SYPRO-RUBY blot stain and visualized by epifluorescence. (B) Western analysis of the blot shown in panel A using a mixture of CNV antibodies specific to the R-, arm, and S-domains. The predicted sizes of CNV CP (41 kDa) as well as those of full-length P85G (41 kDa), R1+2 (36 kDa), and R3(−) (40 kDa) are shown on the left. Arrows within the blot point to truncated proteins found within T=1 virions (Table 1). The white asterisk indicates the position of an unidentified nonviral protein that copurified with P85G particles. Size standards (not shown) were SeeBlue+2 and Mark 12 (Invitrogen).
As with the N-terminal regions of other T=3 viruses, the CNV R-domain is highly positively charged: 11 of the 58 R-domain amino acids are arginine (three residues) or lysine (eight residues) (Fig. 2). The C-terminal 26 aa of the R-domain contains seven lysine residues, making it a particularly lysine-rich region (LRR) (Fig. 2). The R-domains of R1+2, RΔ32-58, and R2Δ58 are each missing the complete LRR and retain only two (R1+2), four (RΔ32-58), and no (RΔ2-58) basic amino acids (Fig. 2). R3(−) lacks the highly basic KGKKGK sequence present within the LRR. Our observation that particles of the various R-domain mutants do not encapsidate full-length RNA may reflect, in part, the inability of particles to stably accommodate full-length RNA due to the inability of the capsid to neutralize the negatively charged phosphates of the RNA. It is also possible that the deleted regions in the R-domain mutants normally play a role in the selection of full-length CNV RNA during assembly. Other factors, such as the volume of the interior of capsid, are also likely to play a role in the size of encapsidated RNA. This is most obvious in the case of T=1 particles, which are unlikely to have an internal volume sufficient to accommodate RNA that is greater than approximately 1 kb in length. Physical constraints that impose an upper limit on the amount and/or length of encapsidated RNA species have been demonstrated for TCV (29).
The absence of full-length RNA in particles of R3(−) suggests that the KGKKGK sequence in particular could play an important role in the encapsidation of full-length CNV RNA, perhaps via its ability to specifically recognize a region in CNV gRNA that is not represented in the RNAs encapsidated by R3(−). Positively charged regions in the amino-terminal regions of the CPs of several plant and animal viruses are well known to be involved in packaging of viral RNA sequences (30).
Despite the absence of the KGKKGK sequence in the various mutants, Northern blot analysis shows that CNV RNA species represent the majority of encapsidated RNAs (Fig. 7A and B). In addition, host RNAs do not appear to be encapsidated, since blots probed with radiolabeled cDNA to total uninfected leaf RNA do not give detectable hybridization signals (unpublished observations). As the various mutants lack large portions of the R-domain and RΔ2-58 lacks the complete R-domain, CP sequences outside of the R-domain are likely to play roles in the specificity of encapsidation, presumably by recognition of sequences present specifically in the RNAs encapsidated by these mutants. Other icosahedral viruses, such as TCV (29), Southern bean mosaic virus (12), Red clover necrotic mosaic virus (3), Turnip yellow mosaic virus (5), and BMV (30), have been shown to contain specific packaging signals.
The possibility that the KGKKGK sequence may specifically select full-length CNV gRNA is particularly intriguing when one considers that the KGKKGK sequence is present in triplicate near the base of the CNV β-annulus, a structure present exclusively in the T=3 particle and suggested to contribute to T=3 particle assembly (13, 16, 21). The hypothesized interaction may ensure that predominantly full-length RNA is encapsidated and, at the same time, promote the preferential assembly of T=3 particles.
A Pro-Gly substitution in the CNV CP arm domain affects particle polymorphism.
In a previous study, we showed that a CNV CP arm mutant (P85G) in which a Gly residue is substituted for a Pro residue at aa 85 (Fig. 1 and 2) results in a dramatic decrease in particle accumulation (approximately 0.4% that of WT CNV) (Table 1) (18). Pro85 is a highly conserved residue in the tombusvirus CP arm and, due to its location near the arm/S junction (Fig. 1), has been postulated to play a role in determining the two main orientations of CP subunit arm (A/B subunit and C subunit) in particles (4, 18). TEM of several P85G particle preparations have indicated that both T=3 and T=1 icosahedra are produced during infection (Fig. 5) with a ratio of approximately 1:1.5 (Table 1). Agarose gel electrophoresis of P85G particles shows the presence of two bands (Fig. 6, lanes 2), one that comigrates with WT CNV T=3 particles (Fig. 6, lanes 1) and another other that comigrates with the T=1 particles of R1+2 (Fig. 6, lanes 3). Analysis of RNA extracted from virions shows that full-length P85G gRNA is encapsidated (Fig. 7B, lane 5). Some preparations of P85G have also been found to contain low levels of virus-related RNA in the range of 0.6 to 0.75 kb (data not shown). The presence of T=1 particles in P85G mutants suggests that Pro85 is important for the maintenance of T=3 morphology.
Further dissection of CNV CP sequences involved in determining particle morphology.
To further investigate the roles of the R and arm regions as factors influencing particle polymorphism, “double” CP mutants were created by incorporating the R1+2 or R3(−) mutations into P85G mutants [mutants R1+2/P85G and R3(−)/P85G, respectively] (Fig. 2). As described above, the ratio of T=3 to T=1 particles in the “single” R1+2 particle preparations is ∼1:1.5, as is that of P85G (Table 1). However, TEM of R1+2/P85G particles (Fig. 5, panel d) indicates that the level of T=1 particles is significantly higher, with a T=3-to-T=1 ratio of approximately 1:10 (Table 1). A similar shift in the ratio of polymorphic particles was observed for R3(−)/P85G double mutants; that is, whereas R3(−) particle preparations consist primarily of an approximately equal mixture of T=3 and IS icosahedra, the double mutant consists of no detectable T=3 particles and an ∼1:1.4 ratio of IS to T=1 particles (Table 1 and Fig. 5, panel f). A shift in the relative migration of particles of the single and double mutants is observed following agarose gel electrophoresis (Fig. 6A), reinforcing observations made using TEM analyses. In particular, whereas slower-migrating species are predominantly observed in P85G and R1+2 particle preparations (Fig. 6A, lanes 2 and 3, respectively), preparations of the corresponding double mutant consist primarily of the faster-migrating particles (Fig. 6A, lane 4). Similarly, whereas R3(−) particle preparations consist primarily of slower-migrating T=3 and IS particles (Fig. 6A, lane 5), R3(−)/P85G preparations consist of a broad band of faster-migrating species in the range of that observed for IS and T=1 particles (Fig. 6A, lane 6). EtBr-stained agarose gels and Northern blot analyses of RNA extracted from particles of R1+2/P85G and R3(−)/P85G show a strong shift toward encapsidation of smaller RNA species in the range from 0.6 to 0.75 kb (Fig. 7A and B, compare lanes 5, 7, and 9 and lanes 5, 11, and 13, respectively). Together, these observations reinforce the conclusion that both the R-domain and P85G play important roles in the assembly of T=3 particles.
The observation that the double mutants have a greater effect in reducing accumulation of T=3 particles than do the respective single mutants suggest that the mechanisms responsible for P85G-induced and R-domain mutant-induced polymorphism are different. We suggest that Pro85 may assist in T=3 particle formation by facilitating the structural transitions in the arm that are known to be required for quasiequivalent subunit interactions in T=3 particle assembly. In contrast, the R-domain may facilitate T=3 particle formation through its proposed role in formation of the highly structured inner core of the CNV particle, as has been suggested by Katpally et al. (21). In addition, as described above, the R-domain KGKKGK sequence may play a role in coordinating the assembly of RNA at the base of the β-annulus, which is present exclusively in the T=3 particle. The role of the R-domain sequence in the assembly of T=3 particles has been noted for other viruses, such as SeMV and TBSV (15, 25). In both cases, a short highly basic region of the R-domain immediately N terminal to the CP region that forms the β-annulus at the T=3 particle threefold axis promoted T=3 particle assembly.
Particles of P85G and R-domain mutants contain truncated CP species that are preferentially associated with T=1 and IS particles.
An analysis of the CP subunits of arm and R-domain mutants was conducted using denaturing polyacrylamide gel electrophoresis and Western blotting. It can be seen in Fig. 6B that particle preparations of P85G, R1+2, R1+2/P85G, R3(−), R3(−)/P85G, and RΔ32-58 each contain a CP subunit of the expected length. However, truncated CP species that range in size between approximately 31 and 34 kDa are also observed (Table 1). RΔ2-58 particles were found to consist primarily of the full-length 34-kDa protein (Fig. 6B, lane 8), and truncated CP was not detected in WT CNV particles (Fig. 6B, lane 1).
Experiments were conducted to determine if the truncated CP species shown in Fig. 6B are differentially represented in T=3, IS, or T=1 particles of P85G, R1+2, and R3(−). Particles were electrophoresed through an agarose gel, and bands corresponding to T=3 and T=1 particles of P85G and R1+2 were excised. For R3(−), T=3 and IS particles were separately excised. Particle protein was extracted from the gel slices, denatured, and then electrophoresed through a denaturing polyacrylamide gel. Figure 8A and B (lane 6) show that a 41-kDa and an approximately 32-kDa species which both react to CNV CP-specific antibodies are present in P85G particle preparations. As well, an additional CNV CP-specific species of ∼31 kDa that may have escaped detection in the experiment described for Fig. 6 is also present. We note that in this particular experiment, a protein of approximately 36 kDa is also observed (Fig. 8A, lane 6). However, this protein did not react to the mixture of CNV antibodies used in this experiment (Fig. 8B, lane 6), nor was it observed following extraction of CP from T=3 or T=1 particles from agarose gels (Fig. 8, compare lanes 6 and 8). This protein therefore likely corresponds to a host protein that copurified with this specific P85G preparation. Figure 8 (lane 7) shows that T=3 particles extracted from agarose gel slices of P85G particles are highly enriched for the 41-kDa species but also contain small amounts of the ∼32-kDa species. In contrast, only the ∼32-kDa and ∼31-kDa species are extracted from T=1 particles (Fig. 8, lane 8). Similarly, the T=3 particles of R1+2 consist primarily of the full-length 36-kDa protein (Fig. 8, lane 11), whereas the T=1 particles, although containing the full-length protein, are slightly enriched for the truncated 34-kDa protein (Fig. 8, lane 12). In the case of R3(−), T=3 particles consist primarily of the full-length 40-kDa species (Fig. 8, lane 15), whereas the excised gel piece containing IS particles is enriched for faster-migrating species of approximately 36 kDa, 34 kDa, and 32 kDa (Fig. 8, lane 16; see Table 1 for a summary).
It is known that the R and arm regions of the CNV CP are sensitive to proteolytic cleavage, as is true for several other T=3 viruses (18). These observations suggest that T=1 particles may form most efficiently from mutant CP subunits that have undergone cleavage in vivo.
We note that the level of truncated CP in P85G particle preparations is variable, ranging from just detectable (Fig. 6B, lane 2) to highly abundant (Fig. 8, lane 6). A similar variation has been observed in the level of T=1 particles that are produced (data not shown). In such cases, it was found that the introduced Gly codon in a portion of the viral RNA population had undergone mutation to a Ala or Val codon.
The observations that CNV T=1 particles can be formed in plants from R-domain mutants and that they are composed exclusively of pentamers (21) suggest that pentamers can be intermediates in the assembly of CNV T=3 particles during infection. In the case of TCV, trimers of C/C dimers have been suggested to initiate T=3 particle assembly (38). More recently, however, pentamers of A/B dimers have been suggested to be more likely assembly intermediates for T=3 virus formation (33, 39). Our finding that CNV R-domain mutants produce a variety of particle types that encapsidate virus-related RNA species during infection may provide future opportunities to investigate the nature of the assembly initiation structure in vivo and the possibility that dual but preferential assembly pathways exist.
Finally, as alluded to above, we have recently found that specific regions of the CNV arm and R-domain act as transit peptides during virus infection, resulting in the targeting of the CP to specific subcellular organelles and likely requiring specific interaction with host components known to assist in subcellular targeting (reference 41 and unpublished observations). Thus, it is possible that the arm and R-domain serve to anchor CP sequences to these organelles and thereby specifically facilitate the assembly and/or disassembly of T=3 particles. Future studies should assist in substantiating this hypothesis as well as the specific roles of the R and arm regions in T=3 particle assembly in plants.
Acknowledgments
We thank Steve Orban for providing technical assistance during the course of this project.
Footnotes
Published ahead of print on 21 November 2007.
REFERENCES
- 1.Annamalai, P., and A. L. Rao. 2005. Replication-independent expression of genome components and capsid protein of brome mosaic virus in planta: a functional role for viral replicase in RNA packaging. Virology 33896-111. [DOI] [PubMed] [Google Scholar]
- 2.Ban, N., and A. McPherson. 1995. The structure of satellite panicum mosaic virus at 1.9 A resolution. Nat. Struct. Biol. 2882-890. [DOI] [PubMed] [Google Scholar]
- 3.Basnayake, V. R., T. L. Sit, and S. A. Lommel. 2006. The genomic RNA packaging scheme of Red clover necrotic mosaic virus. Virology 345532-539. [DOI] [PubMed] [Google Scholar]
- 4.Bergdoll, M., M. H. Remy, C. Cagnon, J. M. Masson, and P. Dumas. 1997. Proline-dependent oligomerization with arm exchange. Structure 5391-401. [DOI] [PubMed] [Google Scholar]
- 5.Bink, H. H., J. Schirawski, A. L. Haenni, and C. W. Pleij. 2003. The 5′-proximal hairpin of turnip yellow mosaic virus RNA: its role in translation and encapsidation. J. Virol. 777452-7458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Calhoun, S. L., J. A. Speir, and A. L. Rao. 2007. In vivo particle polymorphism results from deletion of a N-terminal peptide molecular switch in brome mosaic virus capsid protein. Virology 364407-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cuillel, M., B. Jacrot, and M. Zulauf. 1981. A T=1 capsid formed by the protein of brome mosaic virus in the presence of trypsin. Virology 11063-72. [DOI] [PubMed] [Google Scholar]
- 8.Dong, X. F., P. Natarajan, M. Tihova, J. E. Johnson, and A. Schneemann. 1998. Particle polymorphism caused by deletion of a peptide molecular switch in a quasiequivalent icosahedral virus. J. Virol. 726024-6033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Erickson, J. W., A. M. Silva, M. R. Murthy, I. Fita, and M. G. Rossmann. 1985. The structure of a T = 1 icosahedral empty particle from southern bean mosaic virus. Science 229625-629. [DOI] [PubMed] [Google Scholar]
- 10.Finnen, R. L., and D. M. Rochon. 1995. Characterization and biological activity of DI RNA dimers formed during cucumber necrosis virus coinfections. Virology 207282-286. [DOI] [PubMed] [Google Scholar]
- 11.Finnen, R. L., and D. M. Rochon. 1993. Sequence and structure of defective interfering RNAs associated with cucumber necrosis virus infections. J. Gen. Virol. 741715-1720. [DOI] [PubMed] [Google Scholar]
- 12.Hacker, D. L. 1995. Identification of a coat protein binding site on southern bean mosaic virus RNA. Virology 207562-565. [DOI] [PubMed] [Google Scholar]
- 13.Harrison, S. C. 1983. Virus structure: high-resolution perspectives. Adv. Virus Res. 28175-240. [DOI] [PubMed] [Google Scholar]
- 14.Harrison, S. C., and A. Jack. 1975. Structure of tomato bushy stunt virus. Three-dimensional X-ray diffraction analysis at 16 A resolution. J. Mol. Biol. 97173-191. [DOI] [PubMed] [Google Scholar]
- 15.Hsu, C., P. Singh, W. Ochoa, D. J. Manayani, M. Manchester, A. Schneemann, and V. S. Reddy. 2006. Characterization of polymorphism displayed by the coat protein mutants of tomato bushy stunt virus. Virology 349222-229. [DOI] [PubMed] [Google Scholar]
- 16.Hui, E., and D. Rochon. 2006. Evaluation of the roles of specific regions of the Cucumber necrosis virus coat protein arm in particle accumulation and fungus transmission. J. Virol. 805968-5975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jones, T. A., and L. Liljas. 1984. Structure of satellite tobacco necrosis virus after crystallographic refinement at 2.5 A resolution. J. Mol. Biol. 177735-767. [DOI] [PubMed] [Google Scholar]
- 18.Kakani, K., R. Reade, and D. Rochon. 2004. Evidence that vector transmission of a plant virus requires conformational change in virus particles. J. Mol. Biol. 338507-517. [DOI] [PubMed] [Google Scholar]
- 19.Kakani, K., M. Robbins, and D. Rochon. 2003. Evidence that binding of cucumber necrosis virus to vector zoospores involves recognition of oligosaccharides. J. Virol. 773922-3928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kakani, K., J. Y. Sgro, and D. Rochon. 2001. Identification of specific cucumber necrosis virus coat protein amino acids affecting fungus transmission and zoospore attachment. J. Virol. 755576-5583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Katpally, U., K. Kakani, R. Reade, K. Dryden, D. Rochon, and T. J. Smith. 2007. Structures of T=1 and T=3 particles of cucumber necrosis virus: evidence of internal scaffolding. J. Mol. Biol. 365502-512. [DOI] [PubMed] [Google Scholar]
- 22.Krol, M. A., N. H. Olson, J. Tate, J. E. Johnson, T. S. Baker, and P. Ahlquist. 1999. RNA-controlled polymorphism in the in vivo assembly of 180-subunit and 120-subunit virions from a single capsid protein. Proc. Natl. Acad. Sci. USA 9613650-13655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kumar, A., V. S. Reddy, V. Yusibov, P. R. Chipman, Y. Hata, I. Fita, K. Fukuyama, M. G. Rossmann, L. S. Loesch-Fries, T. S. Baker, and J. E. Johnson. 1997. The structure of alfalfa mosaic virus capsid protein assembled as a T=1 icosahedral particle at 4.0- Å resolution. J. Virol. 717911-7916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Larson, S. B., R. W. Lucas, and A. McPherson. 2005. Crystallographic structure of the T=1 particle of brome mosaic virus. J. Mol. Biol. 346815-831. [DOI] [PubMed] [Google Scholar]
- 25.Lokesh, G. L., T. D. Gowri, P. S. Satheshkumar, M. R. Murthy, and H. S. Savithri. 2002. A molecular switch in the capsid protein controls the particle polymorphism in an icosahedral virus. Virology 292211-223. [DOI] [PubMed] [Google Scholar]
- 26.Lucas, R. W., Y. G. Kuznetsov, S. B. Larson, and A. McPherson. 2001. Crystallization of Brome mosaic virus and T = 1 Brome mosaic virus particles following a structural transition. Virology 286290-303. [DOI] [PubMed] [Google Scholar]
- 27.Olson, A. J., G. Bricogne, and S. C. Harrison. 1983. Structure of tomato busy stunt virus. IV. The virus particle at 2.9 A resolution. J. Mol. Biol. 17161-93. [DOI] [PubMed] [Google Scholar]
- 28.Panavas, T., J. Pogany, and P. D. Nagy. 2002. Analysis of minimal promoter sequences for plus-strand synthesis by the Cucumber necrosis virus RNA-dependent RNA polymerase. Virology 296263-274. [DOI] [PubMed] [Google Scholar]
- 29.Qu, F., and T. J. Morris. 1997. Encapsidation of turnip crinkle virus is defined by a specific packaging signal and RNA size. J. Virol. 711428-1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rao, A. L. 2006. Genome packaging by spherical plant RNA viruses. Annu. Rev. Phytopathol. 4461-87. [DOI] [PubMed] [Google Scholar]
- 31.Ray, D., and K. A. White. 1999. Enhancer-like properties of an RNA element that modulates tombusvirus RNA accumulation. Virology 256162-171. [DOI] [PubMed] [Google Scholar]
- 32.Ray, D., and K. A. White. 2003. An internally located RNA hairpin enhances replication of Tomato bushy stunt virus RNAs. J. Virol. 77245-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reddy, V. S., and J. E. Johnson. 2005. Structure-derived insights into virus assembly. Adv. Virus Res. 6445-68. [DOI] [PubMed] [Google Scholar]
- 34.Rochon, D. M., and J. C. Johnston. 1991. Infectious transcripts from cloned cucumber necrosis virus cDNA: evidence for a bifunctional subgenomic mRNA. Virology 181656-665. [DOI] [PubMed] [Google Scholar]
- 35.Sangita, V., G. L. Lokesh, P. S. Satheshkumar, C. S. Vijay, V. Saravanan, H. S. Savithri, and M. R. Murthy. 2004. T=1 capsid structures of Sesbania mosaic virus coat protein mutants: determinants of T=3 and T=1 capsid assembly. J. Mol. Biol. 342987-999. [DOI] [PubMed] [Google Scholar]
- 36.Savithri, H. S., and J. W. Erickson. 1983. The self-assembly of the cowpea strain of southern bean mosaic virus: formation of T=1 and T=3 nucleoprotein particles. Virology 126328-335. [DOI] [PubMed] [Google Scholar]
- 37.Schneemann, A. 2006. The structural and functional role of RNA in icosahedral virus assembly. Annu. Rev. Microbiol. 6051-67. [DOI] [PubMed] [Google Scholar]
- 38.Sorger, P. K., P. G. Stockley, and S. C. Harrison. 1986. Structure and assembly of turnip crinkle virus. II. Mechanism of reassembly in vitro. J. Mol. Biol. 191639-658. [DOI] [PubMed] [Google Scholar]
- 39.Tang, J., J. M. Johnson, K. A. Dryden, M. J. Young, A. Zlotnick, and J. E. Johnson. 2006. The role of subunit hinges and molecular “switches” in the control of viral capsid polymorphism. J. Struct. Biol. 15459-67. [DOI] [PubMed] [Google Scholar]
- 40.Tremaine, J. H. 1972. Purification and properties of cucumber necrosis virus and a smaller top component. Virology 48582-590. [DOI] [PubMed] [Google Scholar]
- 41.Xiang, Y., K. Kakani, R. Reade, E. Hui, and D. Rochon. 2006. A 38-amino-acid sequence encompassing the arm domain of the Cucumber necrosis virus coat protein functions as a chloroplast transit peptide in infected plants. J. Virol. 807952-7964. [DOI] [PMC free article] [PubMed] [Google Scholar]