Abstract
Flock house virus (FHV), a bipartite RNA virus of insects and a member of the Nodaviridae family, shares viral replication features with the tripartite brome mosaic virus (BMV), an RNA virus that infects plants and is a member of the Bromoviridae family. In BMV and FHV, genome packaging is coupled to replication, a widely conserved mechanism among positive-strand RNA viruses of diverse origin. To unravel the events that modulate the mechanism of replication-coupled packaging, in this study, we have extended the transfer DNA (T-DNA)-based agroinfiltration system to express functional genome components of FHV in plant cells (Nicotiana benthamiana). Replication, intracellular membrane localization, and packaging characteristics in agroinfiltrated plant cells revealed that T-DNA plasmids of FHV were biologically active and faithfully mimicked complete replication and packaging behavior similar to that observed for insect cells. Synchronized coexpression of wild-type BMV and FHV genome components in plant cells resulted in the assembly of virions packaging the respective viral progeny RNA. To further elucidate the link between replication and packaging, coat protein (CP) open reading frames were precisely exchanged between BMV RNA 3 (B3) and FHV RNA 2 (F2), creating chimeric RNAs expressing heterologous CP genes (B3/FCP and F2/BCP). Coinfiltration of each chimera with its corresponding genome counterpart to provide viral replicase (B1+B2+B3/FCP and F1+F2/BCP) resulted in the expected progeny profiles, but virions exhibited a nonspecific packaging phenotype. Complementation with homologous replicase (with respect to CP) failed to enhance packaging specificity. Taken together, we propose that the transcription of CP mRNA from homologous replication and its translation must be synchronized to confer packaging specificity.
Flock house virus (FHV), a member of the family Nodaviridae, and brome mosaic virus (BMV), the type member of the Bromovirus genus, are multicomponent positive-strand RNA viruses of animals and plants, respectively (44, 52). FHV was first isolated from the New Zealand grass grub Costelytra zealandica (52). The 4.5-kb single-strand positive-sense RNA genome of FHV is divided between two capped and nonpolyadenylated RNAs copackaged into a single nonenveloped icosahedral virion with a T = 3 symmetry (50). Genomic RNA 1 (F1) is a 3,107-nucleotide (nt) sequence that encodes a 112-kDa RNA-dependent RNA polymerase (RdRp or protein A) that is necessary and sufficient for FHV RNA replication (8, 27, 42). In addition, F1 also encodes a 387-nt subgenomic RNA 3 (sgF3) sequence that corresponds to that of the 3′ terminus of F1 (16, 22). sgF3 encodes two proteins, b1 and b2. Protein b1 has no recognized function, and b2 is the designated suppressor of RNA silencing activity in Drosophila melanogaster S2 cells (33). Genomic RNA 2 (F2) is a 1,400-nt sequence that encodes the 43-kDa viral capsid protein (CP) precursor α required for the assembly of FHV provirions (49). Each provirion consists of 180 subunits of protein α arranged with a T = 3 quasi-equivalent symmetry and the two genomic RNAs. Provirions are not infectious unless they undergo an autocatalytic maturation process, which results in the cleavage of protein α into protein β (38 kDa) and protein γ (5 kDa) (23, 51). sgF3 has been shown to trans activate F2 replication (20). The replication of FHV occurs on the outer mitochondrial membranes (36). An unusual and remarkable feature associated with FHV is its ability to cross the kingdom barrier and to infect a wide variety of cells, including cells of insects (23), mammals (8), yeasts (41), and several species of monocotyledonous and dicotyledonous plants (although the virus replicates only in single cells) (53).
In contrast to FHV, the genome of BMV is divided among three RNAs. Genomic RNAs 1 (B1) and 2 (B2) encode the replicase proteins 1a and 2a, respectively (54). A functional RdRp enzyme of BMV is a complex of proteins 1a and 2a and some host factors (1, 2). BMV replicates on the outer perinuclear endoplasmic reticulum membranes (36, 47). A third genomic RNA, RNA 3 (B3), is dicistronic and encodes a nonstructural movement protein gene and the structural CP. Although the movement protein is directly translated from B3, the CP is translated from a subgenomic RNA 4 (sgB4), synthesized by internal initiation on the progeny minus-strand RNA 3 (38). The 3′ ends of all four BMV RNAs contain a highly conserved sequence with a tRNA-like secondary structure (TLS) (18). This TLS is known to contain sequence elements that are intimately involved in minus-strand initiation by viral RdRp (18) and plays an important role in virus assembly by functioning as a nucleating element of CP subunits (13). Like FHV, BMV is a nonenveloped icosahedral virus with a T = 3 symmetry and is composed of 180 subunits of a single 19.4-kDa protein (34). Neither the BMV CP nor the assembled virion undergoes any maturation.
Regarding genome packaging, in FHV only F1 and F2 are copackaged into a single virion, while sgF3 is not (31, 32). By contrast, in BMV, B1 and B2 are packaged independently into two virions, whereas B3 and sgB4 are copackaged into a third virion (44). However, these three virion populations are physically and morphologically indistinguishable. The mechanism by which the CP regulates this balanced distribution of four BMV RNAs into three individual virions is still unknown. More recent studies using experimental systems that are competent to effectively uncouple replication from packaging revealed that efficient packaging of viral RNA is functionally coupled to replication-dependent transcription and translation in FHV and BMV (5, 58). However, it is not known whether such functional coupling between replication-dependent transcription and translation requires homologous replicase machinery. To address this issue, in this study, we extended the Agrobacterium-mediated transient expression system (agroinfiltration) (6), which facilitates the synchronized delivery and coexpression of multiple transfer DNA (T-DNA)-based plasmids to the same cell, to initiate replication of the FHV genome in plant cells. We also exploited the agroinfiltration system to evaluate the mechanism of replication-coupled packaging by coexpressing FHV and BMV CP under the control of homologous and heterologous replication machinery. The results demonstrate that the packaging specificity exhibited by the CPs of BMV and FHV requires the homologous combination of synchronized replication and translation of CP.
MATERIALS AND METHODS
Construction of FHV T-DNA plasmids for agroinfiltration.
The characteristic features of the binary vector pCass4Rz, which is amenable for DNA-based Agrobacterium-mediated transient expression (agroinfiltration) of viral genomes, were described previously (6). A full-length cDNA sequence corresponding to F1 was amplified by PCR, with the forward primer 5′AAAACTGCAGTTTTCGAAACAAATAAAAC3′ (the PstI site is underlined) and the reverse primer 5′GCGGCGGGATCCACCTCTGCCCTTTCGG3′ (the BamHI site is underlined). The resulting PCR product was first digested with PstI and then treated with T4 DNA polymerase to create blunt-ended products, followed by BamHI digestion. The product was finally subcloned into the StuI/BamHI-digested pCass4Rz vector. Similarly, a full-length cDNA sequence of F2 was amplified in a PCR with the forward primer 5′GGGGTACGTAAACAATTCCAAGTTCCAAAATGG3′ (the SnaBI site is underlined) and the reverse primer 5′CCGTACGTACCTTAGTCTGTTGACTT3′ (the SnaBI site is underlined). The resulting product was digested with SnaBI and subcloned into the StuI-digested pCass4Rz vector. The resulting plasmids of F1 and F2 (Fig. 1A) contain, in sequential order, a double cauliflower mosaic virus 35S promoter (35S), cDNA corresponding to respective full-length FHV RNAs, a tobacco ringspot virus ribozyme (Rz) sequence, and a 35S terminator (T). The presence of the desired cDNA regions was confirmed by sequencing.
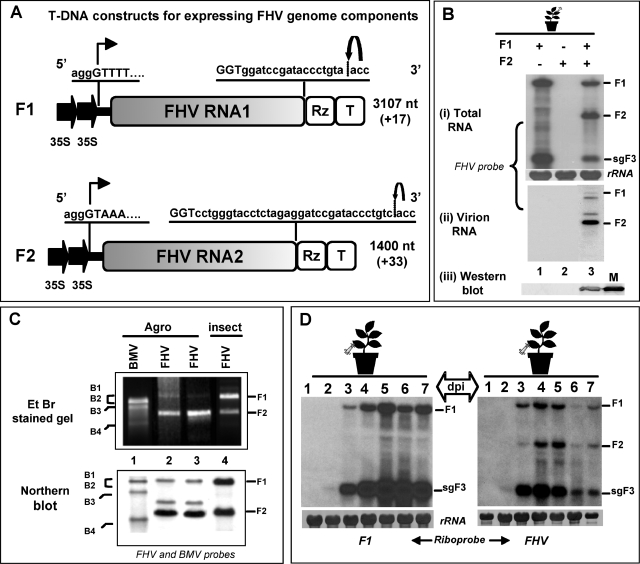
FIG. 1.
(A) Characteristic features of T-DNA constructs of FHV RNA. F1 and F2 constructs contain full-length cDNA copies of FHV genomic RNAs 1 (F1) and 2 (F2), respectively. Single lines and stippled boxes represent noncoding and coding regions, respectively. The position of the double 35S promoters (filled arrows) at the 5′ ends and positions of the ribozyme (Rz) and the 35S terminator (T) at the 3′ end are shown. At the 5′ junction, the nucleotide sequence of the 35S promoter (lowercase letters) and the 5′ sequence of each genomic cDNA (uppercase letters) are shown. A bent arrow indicates the expected transcription start site. At the 3′ end, viral (uppercase letters) and nonviral nucleotide sequences (lowercase letters) left after self-cleavage by the tobacco ringspot virus ribozyme are shown. A bent arrow shows the predicted self-cleavage site. The lengths of wt genomic RNAs and the number of nonviral nucleotides left after self-cleavage by the ribozyme are indicated (values in parentheses). (B) Progeny analysis. Northern blotting analysis of total RNA (i) and virion RNA (ii) and Western blotting analysis of FHV CP (iii) accumulated in N. benthamiana leaves infiltrated with the following agrotransformants: F1 (lane 1), F2 (Lane 2), and F1+F2 (lane 3). For Northern blotting analysis, approximately 5 to 10 μg of total nucleic acid and 0.5 to 1 μg of virion RNA preparations isolated from agroinfiltrated leaves were denatured with formamide/formaldehyde and subjected to 1.2% agarose gel electrophoresis prior to vacuum blotting to a nylon membrane. The blot was hybridized with a mixture of negative-sense riboprobe complementary to F1 and F2. For CP detection, the Western blot was incubated with FHV antiserum; M, CP marker of native FHV virions from Drosophila cells. (C) Virion RNA analysis. Total virion RNA extracted from wt BMV as a marker (lane 1) and wt FHV from two independent agroinfiltration preparations (lanes 2 and 3) and Drosophila cells (lane 4) were subjected to nondenaturing agarose gel electrophoresis, followed by staining with ethidium bromide (EtBr), or to Northern blotting hybridization using the indicated riboprobes. The position of BMV and FHV RNAs are shown. (D) Time course accumulation of FHV RNA. Agrotransformant F1 (left panel) or F1+F2 (right panel) was infiltrated into N. benthamiana leaves and harvested at various days postagroinfiltration, and total RNA was subjected to Northern blotting hybridization with the indicated riboprobes, as described above. The positions of genomic F1, F2, and sgF3 are shown to the right.
T-DNA plasmids for the expression of BMV genomic RNA.
The construction, characteristics, and biological activities of T-DNA plasmids corresponding to BMV B1, B2, and B3 have been described previously (6).
Construction of DI-eGFP agrotransformant.
cDNA clones of the FHV defective interfering RNA 634 (DI-634) and its derivative harboring the open reading frame (ORF) of enhanced green fluorescent protein (DI-eGFP) (15) were amplified in a PCR using the forward primer 5′GGGGTACGTAAACAATTCCAAGTTCCAAAATGG3′ (the SnaBI site is underlined) and the reverse primer 5′CCGTACGTACCTTAGTCTGTTGACTT3′ (the SnaBI site is underlined). The resulting products were digested with SnaBI and independently subcloned into the StuI-digested pCass4Rz vector. The orientation of the inserted cDNAs was verified by sequencing.
Plasmids for riboprobes.
For detecting F1 and sgF3, a 3′ 399-nt sequence complementary to sgF3 was amplified with a PCR with the forward primer 5′CCGTGGACGAAGCTTTACCAATG3′ (the HindIII site is underlined) and the reverse primer 5′AACGGGTGTGGGAATTCCTAAGAGCCA3′ (the EcoRI site is underlined). The resulting product was digested with HindIII and EcoRI and subcloned into the similarly treated pT7/T3 vector (45). For detecting F2, a 385-nt sequence located between nt 403 and 787 of the CP ORF was amplified in a PCR with the forward primer 5′GTTCCTGCTGGTAAGCTTCCTACTAGTGC3′ (the HindIII site is underlined) and the reverse primer 5′TATGTCATTGAATTCAAAGTCAGGCTC3′ (the EcoRI site is underlined). The resulting product was digested with HindIII and EcoRI and finally subcloned into the pT7/T3 vector as described above. For detecting eGFP, a 321-nt fragment was amplified from pEGFP-1 (Clontech Laboratories) in a PCR with the forward primer 5′CAGCGTGTGTCCCGGGAGGGCGAGGG3′ (the SmaI site is underlined) and the reverse primer 5′ACTCCAGCTTGTTAACCAGGATGTTG3′ (the HpaI site is underlined). The resulting fragment was digested with SmaI and HpaI and subcloned into the SmaI-digested pT7/T3 vector. The following terminology was used to represent the riboprobes used in this study. (i) The F1 probe represents a riboprobe complementary to FHV RNA 1; (ii) the F2 probe represents a riboprobe complementary to FHV RNA 2; (iii) the FHV probe represents a mixture of riboprobes complementary to F1 and F2; (iv) the BMV probe represents a riboprobe complementary to a 3′ tRNA-like structure that detects all four BMV RNAs (45); (v) the B1 and B2 probes represent a mixture of riboprobes specific for BMV RNAs 1 and 2 (13); and (vi) the B4 probe represents a riboprobe complementary to BMV sgRNA4 that detects both B3 and B4 (14).
Agroinfiltration and progeny analysis.
Procedures used to grow Agrobacterium cultures and infiltrate Nicotiana benthamiana leaves (4, 6) and Northern (5, 17) and Western blotting analyses (5, 17) were performed as described previously. Virions of BMV (46) and FHV (31) were purified as described previously. Packaging efficiency was quantitated as described previously (13).
Microscopy.
Confocal laser scanning microscopy (CLSM) and transmission electron microscopy (TEM) were performed as described previously (3, 6). For rhodamine treatment, healthy leaves of N. benthamiana were infiltrated with rhodamine 123 (Invitrogen), diluted in distilled water according to the manufacturer's recommendation. For electron microscopy, following agroinfiltration of N. benthamiana leaves with the desired agrotransformants of FHV, leaf tissue was harvested at various days postagroinfiltration (dpa) and excised into small squares with a sterile razor blade. For general observations, the tissue was fixed in 4% formaldehyde and 2.5% glutaraldehyde in 50 mM phosphate buffer, pH 7.2, rinsed in buffer, and postfixed in 1% osmium tetraoxide in the same buffer, followed by dehydration with a graded acetone series and embedding in Spurr's resin. For immunogold labeling of the desired antigens, the excised tissue was fixed in 4% formaldehyde and 2.5% glutaraldehyde in 50 mM phosphate buffer, pH 7.2, followed by dehydration with graded alcohol and embedding in LR white resin (EM Sciences). Tissue sections of approximately 80 nm were mounted onto gold grids and placed in phosphate buffer (PB) containing 0.1% sodium borohydride to inactivate residual aldehyde groups. After sections were washed several times in PB, they were incubated in PB containing 0.05% Triton X-100 to increase reagent penetrability, followed by a 1-h incubation in a blocking solution (phosphate-buffered saline [PBS], pH 7.4, containing 5% preimmune serum, 5% bovine serum albumin, and 0.1% gelatin). Sections were then incubated for 1 h in either anti-protein A (36) or anti-eGFP (Invitrogen) diluted in PBS. After sections were washed three times for 5 min each, they were incubated in F(ab′)2 goat anti-rabbit-gold conjugate (Ultra Small Gold; EM Sciences). After sections were rinsed with distilled water as described above, they were subjected to silver enhancement (using a kit from EM Sciences) for 12 to 15 min and fixed in 2.5% glutaraldehyde in PBS for 15 min. After the grids were rinsed with sterile distilled water, they were poststained with 3% (wt/vol) uranyl acetate and lead citrate for 30 min prior to being examined with TEM (Tecnai 12 model instrument; FEI Co.).
RESULTS
Characteristics of FHV constructs used for agroinfiltration.
The characteristic features of T-DNA plasmids harboring full-length cDNAs of F1 and F2 are shown in Fig. 1A. It has been well documented that the presence of a natural 5′ sequence is essential for the efficient replication and the wild-type (wt) biological activity of several eukaryotic RNA viruses (7, 11). By contrast, the effects of 3′ nonviral nt extensions on the biological activities of viral RNA transcripts varied significantly among the expression systems tested (7, 19, 26). Consequently, the cloning of F1 and F2 cDNAs was designed such that the cauliflower mosaic virus 35S promoter could precisely initiate transcription at the authentic viral 5′ end (Fig. 1A). De novo synthesized RNA transcripts of F1 and F2, respectively, will terminate with +17-nt and +33-nt extensions, beyond the natural 3′ GGUOH sequence due to the presence of self-cleaving ribozyme at the 3′ end (Fig. 1A).
Expression and biological activity of FHV agrotransformants.
The agroinfiltration system has been used successfully for the DNA-based expression of biologically active RNA components of BMV in N. benthamiana (6, 25). To extend this system to FHV, N. benthamiana leaves were infiltrated with Agrobacterium cultures containing T-DNA constructs of either F1 or F2 or F1+F2. At 4 dpa, the total and virion RNA were isolated from the infiltrated leaves, and the preparations were subjected to Northern blotting analysis. The results are shown in Fig. 1B. The FHV replication profile observed for these assays was similar to that observed for insect cells. For example, in FHV, F1 is competent for autonomous replication, since it encodes the required RdRp (8). Thus, N. benthamiana leaves infiltrated with the agrotransformant of F1 resulted in the synthesis of sgF3, which is generated from the 3′ half of F1. This observation validated the fact that de novo-generated F1 RNA is biologically active (Fig. 1B, lane 1). Since the F2 encoding the CP gene cannot replicate without the F1, the autonomous expression of this RNA component did not result in either the replication or translation of CP (Fig. 1B, panels i and iii, lane 2). However, an accumulation of progeny F2 as well as a translation of CP was evident in leaves coinfiltrated with F1+F2 (Fig. 1B, panels i and iii, lane 3).
FHV virions of the expected size and morphology were purified from agroinfiltrated leaves (data not shown). A hallmark of FHV virions assembled in insect cells is the presence of F1 and F2 in equimolar ratios (31). Northern blotting analysis of virion RNA revealed that F1 was packaged less than F2 (Fig. 1B, panel iii, lane 3). In addition to F1 and F2, two additional RNA species were consistently detected by riboprobes specific for FHV RNA (Fig. 1B, panel ii, lane 3). Although the origin of these bands is currently obscure, we speculate that these RNAs might represent DI RNAs. These observations were further confirmed by comparing FHV virion RNA profiles obtained from agroinfiltrated plants with those of Drosophila cells (kindly provided by Show-Wei Ding) by using nondenaturing agarose gel electrophoresis and Northern blotting hybridization (Fig. 1C). Despite efficient replication and accumulation of F1 in F1+F2-infiltrated leaves (Fig. 1B), the reasons that virions package less F1 than F2 are not known. One likely explanation is that the packaging of putative DI RNAs could have competed and inhibited F1 packaging. Nevertheless, FHV virions purified from agroinfiltrated leaves are infectious to Drosophila cells (data not shown), suggesting that the amount of F1 that was packaged was sufficient to initiate infection. Unlike BMV, where the sgB4 is also copackaged with B3 into virions (44), a feature that defines the packaging specificity of FHV is the absence of sgF3 in mature virions. Thus, in the present study, any packaging phenotype that was different from that of the wt was considered nonspecific.
To analyze the temporal accumulation of FHV progeny RNA, N. benthamiana leaves were infiltrated with either F1 or F1+F2, harvested at 1 to 7 dpa, and total RNA preparations were subjected to Northern blotting hybridization (Fig. 1D). In the absence of F2, trace amounts of sgF3 were seen at 2 dpa, and thereafter, high levels of synthesis and accumulation of F1 and sgF3 were detected at 5 and 6 dpa (Fig. 1D). In the presence of F2, the synthesis and accumulation of sgF3 were decreased (Fig. 1D, compare lanes 4 to 7 in both blots), confirming previous observations that sgF3 production was downregulated in the presence of F2 (20). Taken together, the results indicated that FHV RNAs generated from their respective T-DNA plasmids were biologically active as they faithfully mimicked the complete replication behavior that was similarly observed with insect cells.
Evidence for codelivery and expression of multiple FHV agrotransformants to the same cell using FHV DI-eGFP.
To verify the efficiency with which multiple FHV agrotransformants are delivered to a given plant cell, we engineered an agrotransformant of FHV DI-eGFP, a derivative of DI-634 (derived from F2), to express eGFP marker protein in the presence of F1. The characteristic features of DI-634 (15) and DI-634-eGFP are shown schematically in Fig. 2A. The biological activities of DI-634 and DI-eGFP agrotransformants were confirmed by Northern blotting analysis (Fig. 2B). The size of DI-634 is 634 nt (Fig. 2A) and can easily be distinguished from either F1 or F2 (Fig. 2B, lane 2). By contrast, the insertion of eGFP into DI-634 resulted in a sequence size that is identical to that of F2 (Fig. 2B, lane 1) and is therefore indistinguishable when it is coexpressed with F1 and F2 (Fig. 2B, lanes 4 and 5). However, the expression of eGFP, when coinfiltrated with F1, either with or without F2, was evident in a Northern blotting hybridized with a riboprobe specific for eGFP (Fig. 2B, bottom panel).
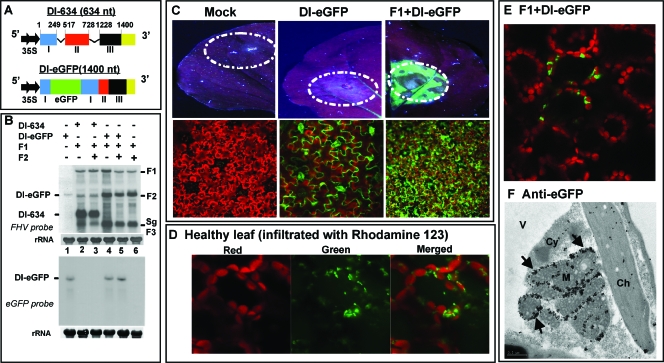
FIG. 2.
(A) Characteristic features of agrotransformants of FHV DI-RNA constructs. The three retained regions, I (nt 1 to 249), II (nt 517 to 728), and III (nt 1228 to 1400), of F2 RNA constituting the DI-634 construct are shown in different colors. Deleted regions are shown with thin lines. In the DI-eGFP construct, the location of eGFP is shown in green. cDNA regions corresponding to DI-634 and DI-eGFP were amplified by PCR and subcloned into the pCass4Rz vector amenable for agroinfiltration. Filled arrows at the 5′ end indicate the position of a double 35S promoter, and at the 3′ end, a yellow box represents the 35S terminator (T). The sizes of DI-634 and DI-eGFP are shown in parentheses. (B) Replication characteristics of DI-634 and DI-eGFP agrotransformants. N. benthamiana leaves were infiltrated with the indicated mixtures of agrotransformants, and total RNAs were subjected to Northern blotting hybridization as described in the legend to Fig. 1. The blots were hybridized with the indicated riboprobes. The positions of DI-634 and DI-eGFP are shown to the left, and the positions of FHV RNAs are shown to the right. (C) Visualization of eGFP. Top panels show macroscopic images of N. benthamiana leaves infiltrated with the indicated cultures of agrotransformants. The infiltrated leaf harvested at 5 dpa was scanned through a Typhoon 9410 model imager equipped with a 670 BP 30-633 nm (red laser) emission filter and a 520 BP-488 nm (blue laser) emission filter. Dotted circles indicate infiltration patches. Bottom panels show CLSM images of subcellular localization of eGFP. Note that the mock infiltrated leaf (left panels) emitted reddish autofluorescence; in the leaf infiltrated with DI-eGFP alone (middle panels), the transiently expressed green fluorescence was confined to epidermal cells, whereas in the leaf infiltrated with F1+DI-eGFP (right panels), green fluorescence was seen in mesophyll cells. (D) Distribution of fluorescent mitochondria in leaves stained with rhodamine 123. Healthy N. benthamiana leaves were infiltrated with the appropriately diluted rhodamine 123 and subjected to CLSM. Representative confocal images show the distribution of chlorophyll (left panel, red), mitochondria (middle panel, green), and merged signals (right panel). (E) Distribution of eGFP in the presence of FHV replication. Representative confocal image shows the distribution of eGFP in N. benthamina leaves infiltrated with F1+DI-eGFP. Note that the distribution of fluorescent mitochondria in panel D (merged image) and that of eGFP in panel E are identical (see text for details). (F) Immunogold EM localization of anti-eGFP. Representative EM image shows the localization of eGFP in N. benthamiana leaves infiltrated with F1+DI-eGFP. Arrows, locations of gold particles on the outer mitochondrial membrane; Ch, chloroplast; Cy, cytoplasm; M, mitochondria; V, vacuole. Bar = 0.5 μm.
Next, to verify the expression of eGFP, N. benthamiana leaves were infiltrated with a mixture of inoculum containing F1+DI-eGFP. Uninfiltrated leaves and leaves infiltrated with DI-eGFP alone served as controls. Leaves were harvested at 4 dpa and subjected to macroscopic and microscopic analyses. Results are summarized in Fig. 2C. Since mRNAs of DI-eGFP are competent for the autonomous translation of eGFP, macroscopic examination of leaves infiltrated with DI-eGFP alone displayed weak green fluorescence (Fig. 2C, middle panels). By contrast, leaves infiltrated with F1+DI-eGFP displayed strong green fluorescence (Fig. 2C, right panel). CLSM examination revealed a distinct pattern of green fluorescence distribution in leaves infiltrated with DI-eGFP and F1+DI-eGFP. For example, the distribution of green fluorescence in leaves infiltrated with DI-eGFP alone was seen throughout epidermal cells (Fig. 2C, bottom middle panel). In these samples, green fluorescence was not seen in any other cells. By contrast, in leaves infiltrated with F1+DI-eGFP, the distribution of green fluorescence was confined mostly to mesophyll cells (Fig. 2C, bottom right panel). No such fluorescence was seen in epidermal cells (data not shown), and this was perhaps masked by the strong signals emitted from the eGFP expressed from replicating DI-RNA in the underlying mesophyll cells. This differential subcellular localization of DI-eGFP can be attributed to the retargeting of transcription and translation of DI-eGFP by FHV replicase. To address this issue further, the following experiment was performed.
The positively charged fluorescent dye rhodamine 123 has been shown to specifically stain the mitochondria in living plant tissue, producing a green fluorescence (63). Thus, a healthy N. benthamiana leaf was infiltrated with rhodamine 123 and subjected to CLSM. As expected, the red autofluorescence produced by chlorophyll could be clearly distinguished from the green fluorescence of mitochondria (Fig. 2D). These mitochondria frequently accumulated in the border areas between chloroplasts. Next, to identify the subcellular localization of DI-eGFP expressed in the presence of FHV replication, N. benthamiana leaves were infiltrated with F1+DI-eGFP and subjected to CLSM. As shown in Fig. 2E, green fluorescence emitted by eGFP was confined to the border areas between chloroplasts, similar to that observed for the rhodamine 123-treated leaves. To further confirm these observations and to precisely identify the subcellular localization of DI-eGFP expressed in the presence of F1, infiltrated leaf tissue was subjected to immunogold EM analysis using anti-eGFP. Results are shown in Fig. 2F and Table 1. In contrast to the control samples (Table 1), more than 90% of the gold particles were clustered around mitochondria in F1+DI-eGFP-infiltrated leaf material. Since FHV replicase protein also localizes on the outer mitochondrial membranes (36) (also see below), these observations suggested that FHV replicase retargets coexpressed DI-eGFP mRNA to mitochondria, resulting in its transcription and translation of eGFP. Collectively, these data also suggest that agroinfiltration was competent to codeliver multiple plasmids to the same cell.
TABLE 1.
Immunogold EM analysis of anti-eGFP and anti-protein Aa
| Sample infiltrate | No. of gold particles per cm2
|
|||
|---|---|---|---|---|
| Anti-eGFP
|
Anti-protein A
|
|||
| Mitochondria | Cytoplasm | Mitochondria | Cytoplasm | |
| F1+DI-eGFP | 61 | 3 | ||
| F1 | 57 | 4 | ||
| Healthy | 0 | 1 | 0 | 3 |
The number of gold particles per cm2 represents the average of three independently processed samples. The preparation of leaf samples for immunogold localization of anti-eGFP and anti-protein A is described in Materials and Methods.
FHV RdRp localizes to outer membranes of mitochondria.
Immunogold localization studies using FHV-infected Drosophila cells revealed that protein A, the RdRp enzyme required for the replication of FHV, was localized on outer mitochondrial membranes (36). Subsequent studies also showed that FHV replication was not obligatorily associated with specific intracellular membranes, since retargeting FHV protein A to endoplasmic reticulum did not impair replication (37). To verify which of the intracellular membranes of plant cells are involved in the replication of FHV, subcellular localization assays of FHV RdRp in N. benthamiana leaves infiltrated with the F1 agrotransformant were performed using immunogold EM with protein A antiserum (Fig. 3). Preimmune serum did not show any gold labeling (Fig. 3B). However, when samples were immunolabeled with protein A serum, a clustering of gold particles (∼90%) around mitochondria in cells agroinfiltrated with F1 T-DNA was clearly evident but not with healthy leaf material (Table 1; Fig. 3C). Consistent with observations for the subcellular localization of protein A in FHV-infected Drosophila cells, higher magnification microscopic views particularly revealed clustering of gold particles around the double-walled outer mitochondrial membrane (Fig. 3D). These results confirmed that, analogous to virus activity in Drosophila cells, FHV replication in plant cells also occurs on the outer mitochondrial membranes.
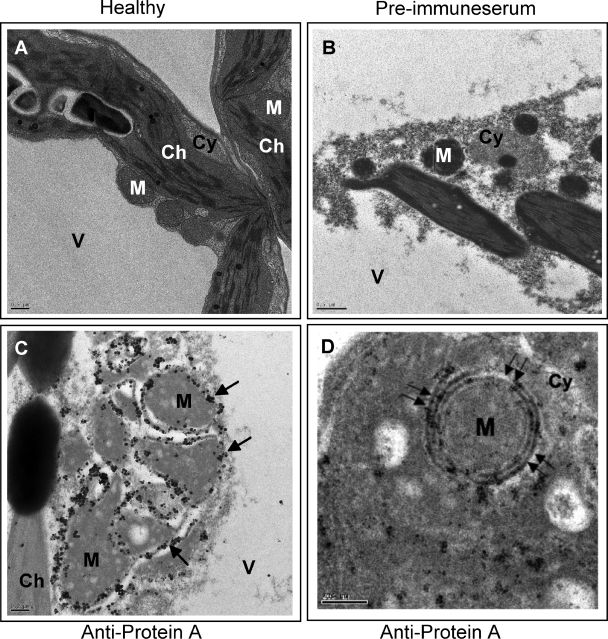
FIG. 3.
Immunogold EM localization of FHV protein A. (A) EM of a section of healthy N. benthamiana leaf showing vacuole (V), chloroplast (Ch), cytoplasm (Cy), and mitochondria (M). (B) EM of a section of F1-infiltrated N. benthamiana leaf treated with preimmune serum. (C) EM showing localization of FHV protein A on mitochondria of N. benthamiana infiltrated with the F1 agrotransformant. Arrows show the location of gold particles on outer mitochondrial membranes. (D) EM (at a higher magnification) reveals localization of gold particles to outer double-walled mitochondrial membrane (double arrows). Scale bars = 0.5 μm (panels A and B), 0.2 μm (panel C), and 200 nm (panel D).
Expression and packaging phenotypes of FHV and BMV agrotransformants in N. benthamiana.
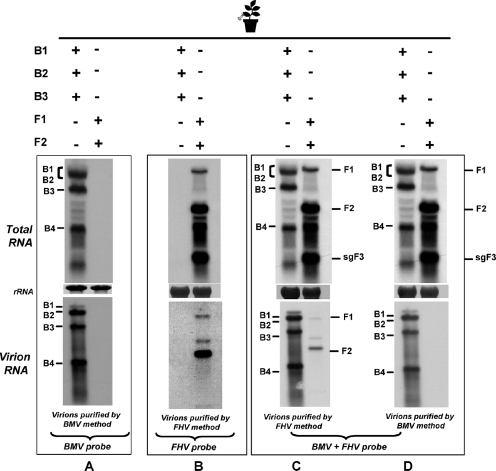
N. benthamiana leaves were infiltrated with the desired mixture of BMV or FHV agrotransformants. At 4 dpa, leaf samples were divided into three lots. One lot was used to extract total RNA, while the second and third lots were used for virion (and packaged RNA) isolation of either FHV or BMV. Three sets of identical Northern blots containing total and virion RNA preparations were produced. One set was hybridized with riboprobes complementary to the highly conserved 3′ TLS region of BMV that detects the positive strands of all four BMV progeny RNAs (6). A second set was hybridized with a mixture containing riboprobes of F1 and F2, designed to detect all three FHV RNAs. A third set of blots was hybridized with a mixture containing BMV and FHV riboprobes. The results are summarized in the legend to Fig. 4.
FIG. 4.
Expression and packaging profiles for BMV and FHV agrotransformants in N. benthamiana. Northern blotting analyses of total (top panels A to D) and virion (bottom panels A to D) RNA recovered from N. benthamiana leaves infiltrated with the indicated sets of inocula. Multiple blots were produced and hybridized with the indicated riboprobes. Hybridization conditions are as described in the legend to Fig. 1. The positions of FHV and BMV RNA are shown. The method used to purify virions is shown below each panel.
The specificity of riboprobes complementary to BMV and FHV is exemplified by the detection of the respective RNAs in total or virion RNA preparations obtained from leaves infiltrated independently with either BMV or FHV agrotransformants (Fig. 4A and B). Figures 4C and D show that FHV virions could be purified only by the homologous purification method, while BMV virions could be isolated by either procedure (Fig. 4C and D, bottom panels). To verify whether CPs of BMV and FHV would trans encapsidate heterologous RNA, the following additional experiments were performed.
Independent expression of BMV CP or FHV CP from heterologous replication results in nonspecific packaging.
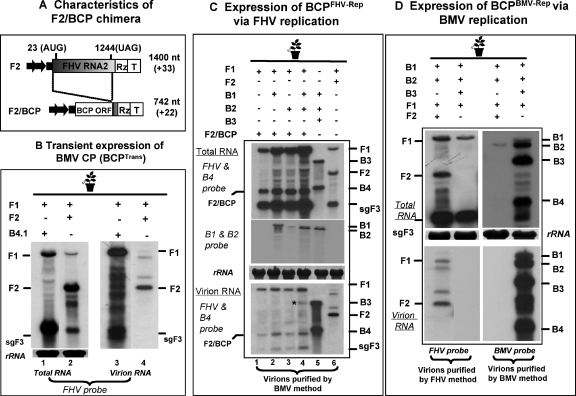
First, to test the packaging specificity of BMV CP, we sought to express CP subunits by using the following three different forms. (i) Agroinfiltration of a previously constructed B4.1 agrotransformant (6) would result in the transient expression of BMV CP (BCPTrans); (ii) when an F2 chimera (F2/BCP) (Fig. 5A) was coinfiltrated with F1, it would result in the expression of BMV CP from FHV-directed replication (i.e., BCPFHV-Rep); and (iii) the coexpression of wt B3 with wt B1 and B2 would result in the expression of BMV CP via homologous replication (i.e., BCPBMV-Rep). The encapsidation competence of FHV progeny RNA by BCPTrans is shown in Fig. 5B. In contrast to the data shown in the previous experiment, where BMV CP was found to specifically package homologous RNAs in leaves infiltrated with both BMV and FHV (Fig. 4D, bottom panel), the efficient packaging of F1 and sgF3 by BCPTrans was evident (Fig. 5B, compare lanes 1 and 3). Since wt FHV virions do not package sgF3, the packaging phenotype observed for the BCPTrans was considered nonspecific, as reported previously (5, 6).
FIG. 5.
Packaging phenotype of BMV CP synthesized from FHV replication. (A) Schematic representation of a T-DNA construct of an F2 chimera harboring a BMV CP ORF (BCP). In wt F2, the positions of the start (AUG) and stop (UAG) codons of the FHV CP ORF are indicated. In the F2/BCP chimera, FHV CP was precisely replaced with that of BCP. The lengths of the wt F2 and F2/BCP and the number of nonviral nucleotides left after self-cleavage by the ribozyme (shown in parentheses) are indicated. The positions of the cauliflower mosaic virus 35S promoter, the ribozyme (Rz), and the 35S terminator (T) are identical to those of the T-DNA cassettes of wt FHV genomic RNAs shown in Fig. 1A. (B) The packaging phenotype of FHV progeny by BMV CP expressed in trans (BCPTrans). N. benthamiana leaves were infiltrated with the indicated mixture of agrotransformants. Agrotransformant B4.1 was coinfiltrated with F1 to provide BCP in trans. Plants infiltrated with F1+F2 served as positive controls. Total and virion RNA profiles were analyzed by Northern blotting hybridization as described in the legend to Fig. 1. The positions of FHV RNA are shown. (C and D) Packaging phenotypes of BCP expressed via either (C) FHV replication (BCPFHV-Rep) or (D) BMV replication (BCPBMV-Rep). N. benthamiana leaves were infiltrated with the indicated mixture of agrotransformants. Total (top and middle panels) and virion RNA (bottom panel) profiles were analyzed by Northern blotting hybridization with the indicated riboprobes as described in the legend to Fig. 1. The positions of FHV and BMV RNA are shown at the right. The asterisk indicates a band of unknown origin (see text for details).
Recently, we reported that the BMV CP expressed from replication-derived mRNA exhibits packaging specificity (5). To verify this, we examined the packaging specificity of BMV CP expressed from FHV replication machinery (i.e., BCPFHV-Rep). The results of these experiments and those of the control experiments involving BMV CP expression from homologous replication (i.e., BCPBMV-Rep) are summarized in the legends to Fig. 5C and D, respectively. FHV RdRp directed the replication of F2/BCP, produced progeny RNAs of the expected size (Fig. 5C, top panel), and resulted in the efficient translation of BMV CP and assembly of virions with morphological features indistinguishable from those of the wt (data not shown). BCPFHV-Rep packaged F1, F2/BCP, and sgF3 progeny (Fig. 5C, bottom panel) with 40% to 50% efficiency.
Results obtained from the above-described experiments suggested that the packaging phenotype exhibited by BCPFHV-Rep is nonspecific, whereas that of BCPBMV-Rep is specific (Fig. 5C and D, bottom panels). These results also imply that replication-derived transcription per se is not sufficient to confer packaging specificity. It is likely that the expression of homologous replicase would enhance the packaging specificity of CP expressed from a heterologous derived transcription. The rationale behind performing the following experiment was to verify whether trans complementation of BMV replicase would enhance the packaging of BMV CP translated under FHV replication. Therefore, inocula containing F1+F2/BCP were complemented with either B1+B2 (which provides functional BMV replicase), B1 (which provides replicase protein 1a), or B2 (which provides replicase protein 2a). The last two served as negative controls, since the assembly of functional replicase requires coexpression of both the BMV 1a and the 2a replicase proteins (28). Total and virion RNA preparations isolated from infiltrated leaves were subjected to Northern blotting hybridization with a set of desired riboprobes. Results are shown in Fig. 5C (lanes 2 to 4). Compared to wt BMV control infections (e.g., Fig. 5C, lane 5), complementation with either B1 (Fig. 5C, lane 2) or B2 (Fig. 5C, lane 3) or B1+B2 (Fig. 5C, lane 4) did not alter the packaging phenotype of BCPFHV-Rep (Fig. 5C, bottom panel, lanes 2 to 4). The presence of complemented B1 and B2 in total RNA preparations was confirmed by Northern blotting hybridization (Fig. 5C, middle panel). Note that in Fig. 5C (bottom panel, lane 4 marked with an asterisk), a band comigrating with genomic B3 was detected consistently. The origin of this band is currently unknown, and we conclude that it is not genomic B3, based on the following additional findings: (i) hybridization with a riboprobe complementary to the movement protein ORF (which is specific for B3) failed to detect this RNA (data not shown); and (ii) had this been genomic B3, it would have replicated and accumulated in total RNA preparations (Fig. 5C, top panel, lane 4).
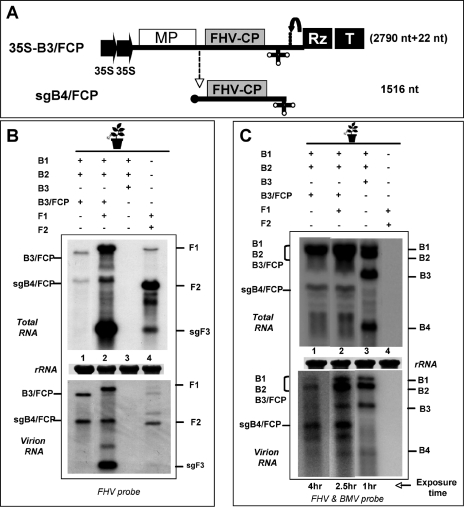
Finally, to verify whether FHV CP, like BMV CP, expressed via heterologous replication would also exhibit a nonspecific packaging phenotype, a B3 chimera in which the FHV CP ORF was substituted for that of BMV CP was constructed (Fig. 6A). As a result of these cloning manipulations, the size of the resulting B3/FCP was increased by 673 nt (the size of wt B3 is 2,117 nt). Following replication of the B3/FCP chimera by BMV RdRp, the size of the corresponding sgB4/FCP would be 1,516 nt, compared to 887 nt for wt B4. When the expression of B3/FCP was complemented with BMV RdRp by coinfiltrating with wt B1 and B2 agrotransformants, the replication of B3/FCP was evident by the synthesis and accumulation of a detectable level of sgB4/FCP (Fig. 6B, top panel, lane 1).
FIG. 6.
Expression and packaging phenotypes of FHV CP (FCP) via BMV replication. (A) Schematic representation of a T-DNA construct of BMV RNA3 (B3) and its subgenomic RNA (sgB4/FCP) harboring the FCP ORF. The length of the B3 chimera and its sgB4 and the number of nonviral nucleotides left after self-cleavage by the ribozyme (shown in parentheses) are indicated. (B and C) Northern blotting analysis of progeny RNA. N. benthamiana leaves were infiltrated with the indicated mixture of agrotransformants. Total and virion RNAs were subjected to multiple Northern blotting analyses and hybridized with the indicated riboprobes as described in the legend to Fig. 1. The positions of progeny RNA of wt FHV, wt BMV, B3/FCP, and sgB4/FCP are shown. The band at the B2 position is of a higher intensity since B2 (2,864 nt) and B3/FCP (2,790 nt) comigrate, and in panel C, lanes 1, 2, and 3 were exposed for 4 h, 3 h, and 1 h, respectively, to reveal bands of interest.
Northern blotting analysis of the FHV virions assembled in the presence of BMV RdRp exhibited a nonspecific packaging phenotype. For example, FCP subunits synthesized via BMV replication packaged not only B3/FCP and its sgB4/FCP (Fig. 6B, bottom panel, lane 1) but also BMV genomic RNAs 1 and 2 (Fig. 6C, bottom panel, lane 1). To verify whether complementation with FHV replicase would alter this nonspecific packaging phenotype, F1 was coinfiltrated with B1+B2+B3/FCP. Since BMV RdRp can replicate only B1 and B2 and the B3/FCP chimera, detection of the progeny F1, sgF3, B3/FCP, and B4/FCP (Fig. 6B and C, lane 2) suggested that all four transformants (i.e., B1+B2+B3/FCP+F1) were concurrently delivered to and expressed in a single cell. Detection of both the BMV and the FHV progeny in virions suggested that complementation with FHV replicase did not alter the nonspecific packaging phenotype of FHV CP expressed from BMV replication-dependent transcription (Fig. 6B and C, lane 2). Taken together, the data indicate that packaging specificity is coupled to the translation of BMV CP and FHV CP to respective homologous replication-dependent transcription (see Discussion).
DISCUSSION
The rationale for this study emerged from observations that the packaging specificities in BMV and FHV infections require a replication-dependent transcription and translation of CP subunits, a mechanism commonly referred to as replication-coupled packaging (5, 58). Two hypotheses were proposed to justify the mechanism: (i) CP subunits are synthesized close to the cellular compartment where replication occurs, permitting immediate access to interact with and encapsidate progeny RNA (5, 58); and (ii) viral replicase functioning as a chaperone induces a conformational change to CP subunits that selectively packages viral RNA (12). Although these studies implicate replicase and/or replication as the integral part of genome packaging, how replicase and/or replication confers the packaging specificity is not clear. In this study, we exploited shared traits (e.g., the ability to replicate in plant cells) and distinguishable traits (e.g., differential intracellular replication sites) inherent in FHV and BMV, in conjunction with an agroinfiltration system that facilitates the synchronized delivery and expression of multiple plasmids to a single plant cell (5, 35) to decipher the mechanism of replication-coupled packaging. According to our present findings, BMV CP and FHV CP expressed via heterologous replication-dependent transcription and translation failed to confer packaging specificity (Fig. 5 and 6). Thus, we conclude that transcription of CP mRNA from homologous replication, followed by its translation, must be synchronized to confer packaging specificity.
During packaging, viral nucleic acids must be distinguished from other cellular RNA molecules present in the compartment where assembly takes place. Undoubtedly, one of the major factors that influences selectivity in packaging viral genomes from a large pool of cellular RNAs is the specific interaction between the CP and the RNA (44). Viral CPs with the arginine-rich RNA binding motif (ARM) (44, 56) and sequences or structures unique to viral nucleic acids, often termed origin-of-assembly sequences or packaging signals (9, 21, 44), have been shown to play a crucial role in selective packaging. However, since the presence of either the ARM or the packaging signal alone does not guarantee packaging specificity (44), other factors or events integral to the virus infection cycle must also be involved. Analogous to findings with FHV and BMV, the experimental evidence from a diverse group of positive-strand RNA viruses such as poliovirus (40), Kunjin virus (29), and Venezuelan equine encephalitis virus (60) has also established that selective packaging is functionally coupled to replication. In all these viral systems, efficient packaging of viral progeny RNA requires the translation of CP from replication-derived mRNA.
What does the link between RNA replication and the translation of CP mean for packaging?
In eukaryotic viruses, the regulation of viral translation plays an important role in replication and encapsidation. In some plant (e.g., BMV) and animal (e.g., Sindbis virus) viruses, CP is expressed via subgenomic mRNA (sg-mRNA), whose synthesis is replication contingent (38, 55). More recent studies of the mechanism of Sindbis virus sg-mRNA translation into CP suggested that only replication-derived sg-mRNA is translated efficiently (48). A similar mechanism involving a coupling between the translation and the transcription was also reported for positive-strand (24, 39) and negative-strand RNA viruses (61). Interestingly, in FHV, CP translation is linked to F2 replication. Similarly, in BMV, the translation of CP is coupled to the synthesis of sgRNA from minus-strand B3 progeny. Autonomous transfection of plant protoplasts with either virion-purified (30) or in vitro-generated capped transcripts of sgRNA failed to translate (A. L. N. Rao, unpublished data). Complementation with functional BMV replicase could not restore the messenger activity of sgB4 in transfected protoplasts (A. L. N. Rao, unpublished data). This suggests that the viral RNA replication pathway and the associated viral RNA complexes are required to recruit translation factors to sgRNA. RNA replication complexes of positive-strand RNA viruses often contain proteins that are involved as host factors in translation. For example, RNA polymerase complexes of Qβ, TMV, and BMV have been shown to contain ribosomal proteins and translational elongation factors (10, 43, 57). Thus, similar to alphaviruses, the translation of BMV and FHV CP is also coupled to replication-dependent transcription and provides a specific replicase-CP interaction. This scenario perhaps explains why BMV CP or FHV CP synthesized under heterologous replication failed to exhibit packaging specificity (Fig. 5 and 6). In this context, the results of this study pertaining to subcellular localization of DI-eGFP (Fig. 2E, F) need to be discussed. Compared to results with autonomous expression, the coexpression of F1 resulted in retargeting DI-eGFP to outer mitochondrial membranes, a response identical to that of viral replicase (Fig. 2F and 3D). Extrapolating these observations for the wt FHV, it is reasonable to speculate that the translation of CP that is dependent on F2 replication also occurs on or near replication sites, as proposed previously (58). Thus, this commonly shared site of replication and translation, as discussed below, allows replicase to interact with CP to induce a conformation that specifically packages viral RNA.
Does viral replicase interact with CP to enhance packaging specificity?
Several experimental findings argue in favor of this hypothesis. First, in BMV only the CP expressed from homologous replication-derived mRNA exhibited packaging specificity (5, 58, 59). Second, Willits et al. (62) reported that a cowpea chlorotic mottle bromovirus CP variant defective in β-hexamer formation assembled in vitro into a heterogeneous population of empty icosahedral virions without RNA and aberrant assembly products in the presence of RNA. However, the in planta expression of Δβ-hexamer via replication-dependent transcription and translation resulted in the assembly of RNA containing virions indistinguishable from the wt particles. Third, we reported that a BMV variant CP competent to assemble in vivo failed to assemble in vitro (12). Collectively, the data suggested that the packaging phenotypes exhibited by CP expressed in the presence and the absence of replication are clearly distinct. Since dimerization of CP subunits plays an important role in the assembly pathway of icosahedral virions (64), we propose that viral replicase, functioning as a chaperone, interacts (perhaps transiently) with CP subunits to induce a conformational change that confers packaging specificity.
In FHV, it is well established that replicase is sequestered in membrane spherules on the outer mitochondrial membrane (36). These spherules are connected to the cytoplasm by a narrow neck from which newly synthesized RNAs exit into the cytoplasm for translation. However, there is no evidence that CP translation and virus assembly occur inside the spherules. The fact that the FHV CP that translated from replication-derived RNA 2 exhibited packaging specificity (58, 59) suggests that the translation of FHV CP occurs outside the spherules and could be coupled to transcription, similar to that observed recently for Sindbis virus (48). But the question is, how does this interaction between FHV replicase and CP occur? We propose that in FHV, like in BMV (as discussed above), the translation of CP is coupled to replication-dependent transcription. While docking the translational apparatus to initiate translation, viral replicase interacts with newly synthesized polypeptides and induces a conformational change. This follows the assembly of dimers with optimal conformation that recognizes only specific viral sequences (e.g., packaging signals), conferring packaging specificity. Furthermore, our experimental evidence obtained from this study (Fig. 5C and 6B and C) suggested that the expression of either BMV CP or FHV CP via replication-dependent transcription per se would not suffice to confer packaging specificity. Thus, we propose that packaging specificity in positive-strand RNA viruses such as BMV and FHV is conferred by two uniquely coordinated sequential events: homologous replication-derived transcription coupled to translation, followed by replication-coupled packaging. Further experiments are required to elucidate the existence of a physical interaction between replicase and CP subunits to unravel this unique packaging mechanism shared by many viruses pathogenic to humans, animals, and plants.
Acknowledgments
We thank Shou-Wei Ding for the generous gift of pCass4 plasmid and FHV cDNA clones, Paul Ahlquist for protein A antiserum, Anette Schneemann for FHV CP antiserum, Ranjit Dasgupta for cDNA clones of DI-634 and DI-eGFP, Roghiyh Aliyari (S.-W. Ding's laboratory) for providing native FHV virions and RNA from Drosophila cells, David Carter of the Center for Plant Cell Biology for confocal microscopy analysis, and Theo Dreher and James Ng for editorial comments.
Research in this laboratory was supported by National Institutes of Health grant GM 064465-01A2. F.R. is a trainee from the NSF- CEPCEB Research Experiences for Undergraduates (REU) internship program.
Footnotes
Published ahead of print on 21 November 2007.
REFERENCES
- 1.Ahlquist, P., A. O. Noueiry, W. M. Lee, D. B. Kushner, and B. T. Dye. 2003. Host factors in positive-strand RNA virus genome replication. J. Virol. 778181-8186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahlquist, P., M. Schwartz, J. Chen, D. Kushner, L. Hao, and B. T. Dye. 2005. Viral and host determinants of RNA virus vector replication and expression. Vaccine 231784-1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Annamalai, P., S. Apte, S. Wilkens, and A. L. Rao. 2005. Deletion of highly conserved arginine-rich RNA binding motif in cowpea chlorotic mottle virus capsid protein results in virion structural alterations and RNA packaging constraints. J. Virol. 793277-3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Annamalai, P., and A. L. Rao. 2006. Unit 16B.2: delivery and expression of functional viral genomes in planta by agroinfiltration. Curr. Protocols in Microbiol. 12.1-2.15. [DOI] [PubMed] [Google Scholar]
- 5.Annamalai, P., and A. L. Rao. 2006. Packaging of brome mosaic virus subgenomic RNA is functionally coupled to replication-dependent transcription and translation of coat protein. J. Virol. 8010096-10108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Annamalai, P., and A. L. Rao. 2005. Replication-independent expression of genome components and capsid protein of brome mosaic virus in planta: a functional role for viral replicase in RNA packaging. Virology 33896-111. [DOI] [PubMed] [Google Scholar]
- 7.Ball, L. A. 1994. Replication of the genomic RNA of a positive-strand RNA animal virus from negative-sense transcripts. Proc. Natl. Acad. Sci. USA 9112443-12447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ball, L. A. 1995. Requirements for the self-directed replication of flock house virus RNA 1. J. Virol. 692722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berkowitz, R., J. Fisher, and S. P. Goff. 1996. RNA packaging. Curr. Top. Microbiol. Immunol. 214177-218. [DOI] [PubMed] [Google Scholar]
- 10.Blumenthal, T., and G. G. Carmichael. 1979. RNA replication: function and structure of Qbeta-replicase. Annu. Rev. Biochem. 48525-548. [DOI] [PubMed] [Google Scholar]
- 11.Boyer, J. C., and A. L. Haenni. 1994. Infectious transcripts and cDNA clones of RNA viruses. Virology 198415-426. [DOI] [PubMed] [Google Scholar]
- 12.Calhoun, S. L., J. A. Speir, and A. L. Rao. 2007. In vivo particle polymorphism results from deletion of a N-terminal peptide molecular switch in brome mosaic virus capsid protein. Virology 364407-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Choi, Y. G., T. W. Dreher, and A. L. Rao. 2002. tRNA elements mediate the assembly of an icosahedral RNA virus. Proc. Natl. Acad. Sci. USA 99655-660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Choi, Y. G., and A. L. Rao. 2003. Packaging of brome mosaic virus RNA3 is mediated through a bipartite signal. J. Virol. 779750-9757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dasgupta, R., L. L. Cheng, L. C. Bartholomay, and B. M. Christensen. 2003. Flock house virus replicates and expresses green fluorescent protein in mosquitoes. J. Gen. Virol. 841789-1797. [DOI] [PubMed] [Google Scholar]
- 16.Dasmahapatra, B., R. Dasgupta, A. Ghosh, and P. Kaesberg. 1985. Structure of the black beetle virus genome and its functional implications. J. Mol. Biol. 182183-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dong, X. F., P. Natarajan, M. Tihova, J. E. Johnson, and A. Schneemann. 1998. Particle polymorphism caused by deletion of a peptide molecular switch in a quasiequivalent icosahedral virus. J. Virol. 726024-6033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dreher, T. W. 1999. Functions of the 3′-untranslated regions of positive strand RNA viral genomes. Annu. Rev. Phytopathol. 37151-174. [DOI] [PubMed] [Google Scholar]
- 19.Dreher, T. W., A. L. Rao, and T. C. Hall. 1989. Replication in vivo of mutant brome mosaic virus RNAs defective in aminoacylation. J. Mol. Biol. 206425-438. [DOI] [PubMed] [Google Scholar]
- 20.Eckerle, L. D., C. G. Albarino, and L. A. Ball. 2003. Flock house virus subgenomic RNA3 is replicated and its replication correlates with transactivation of RNA2. Virology 31795-108. [DOI] [PubMed] [Google Scholar]
- 21.Fosmire, J. A., K. Hwang, and S. Makino. 1992. Identification and characterization of a coronavirus packaging signal. J. Virol. 663522-3530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Friesen, P. D., and R. R. Rueckert. 1982. Black beetle virus: messenger for protein B is a subgenomic viral RNA. J. Virol. 42986-995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gallagher, T. M., and R. R. Rueckert. 1988. Assembly-dependent maturation cleavage in provirions of a small icosahedral insect ribovirus. J. Virol. 623399-3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gamarnik, A. V., and R. Andino. 1998. Switch from translation to RNA replication in a positive-stranded RNA virus. Genes Dev. 122293-2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gopinath, K., B. Dragnea, and C. Kao. 2005. Interaction between brome mosaic virus proteins and RNAs: effects on RNA replication, protein expression, and RNA stability. J. Virol. 7914222-14234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ishikawa, M., M. Janda, M. A. Krol, and P. Ahlquist. 1997. In vivo DNA expression of functional brome mosaic virus RNA replicons in Saccharomyces cerevisiae. J. Virol. 717781-7790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johnson, K. L., and L. A. Ball. 1997. Replication of flock house virus RNAs from primary transcripts made in cells by RNA polymerase II. J. Virol. 713323-3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kao, C. C., and P. Ahlquist. 1992. Identification of the domains required for direct interaction of the helicase-like and polymerase-like RNA replication proteins of brome mosaic virus. J. Virol. 667293-7302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Khromykh, A. A., A. N. Varnavski, P. L. Sedlak, and E. G. Westaway. 2001. Coupling between replication and packaging of flavivirus RNA: evidence derived from the use of DNA-based full-length cDNA clones of Kunjin virus. J. Virol. 754633-4640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kiberstis, P., L. S. Loesch-Fries, and T. C. Hall. 1981. Viral protein synthesis in barley protoplasts inoculated with native and fractionated brome mosaic virus RNA. Virology 112804-808. [DOI] [PubMed] [Google Scholar]
- 31.Krishna, N. K., D. Marshall, and A. Schneemann. 2003. Analysis of RNA packaging in wild-type and mosaic protein capsids of flock house virus using recombinant baculovirus vectors. Virology 30510-24. [DOI] [PubMed] [Google Scholar]
- 32.Krishna, N. K., and A. Schneemann. 1999. Formation of an RNA heterodimer upon heating of nodavirus particles. J. Virol. 731699-1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li, H., W. X. Li, and S. W. Ding. 2002. Induction and suppression of RNA silencing by an animal virus. Science 2961319-1321. [DOI] [PubMed] [Google Scholar]
- 34.Lucas, R. W., S. B. Larson, and A. McPherson. 2002. The crystallographic structure of brome mosaic virus. J. Mol. Biol. 31795-108. [DOI] [PubMed] [Google Scholar]
- 35.Marillonnet, S., A. Giritch, M. Gils, R. Kandzia, V. Klimyuk, and Y. Gleba. 2004. In planta engineering of viral RNA replicons: efficient assembly by recombination of DNA modules delivered by Agrobacterium. Proc. Natl. Acad. Sci. USA 1016852-6857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miller, D. J., M. D. Schwartz, and P. Ahlquist. 2001. Flock house virus RNA replicates on outer mitochondrial membranes in Drosophila cells. J. Virol. 7511664-11676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miller, D. J., M. D. Schwartz, B. T. Dye, and P. Ahlquist. 2003. Engineered retargeting of viral RNA replication complexes to an alternative intracellular membrane. J. Virol. 7712193-12202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miller, W. A., T. W. Dreher, and T. C. Hall. 1985. Synthesis of brome mosaic virus subgenomic RNA in vitro by internal initiation on (−)-sense genomic RNA. Nature 31368-70. [DOI] [PubMed] [Google Scholar]
- 39.Mizumoto, H., H. -O. Iwakawa, M. Kaido, K. Mise, and T. Okuno. 2006. Cap-independent translation mechanism of Red clover necrotic mosaic virus RNA2 differs from that of RNA1 and is linked to RNA replication. J. Virol. 803781-3791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nugent, C. I., K. L. Johnson, P. Sarnow, and K. Kirkegaard. 1999. Functional coupling between replication and packaging of poliovirus replicon RNA. J. Virol. 73427-435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Price, B. D., P. Ahlquist, and L. A. Ball. 2002. DNA-directed expression of an animal virus RNA for replication-dependent colony formation in Saccharomyces cerevisiae. J. Virol. 761610-1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Price, B. D., M. Roeder, and P. Ahlquist. 2000. DNA-directed expression of functional flock house virus RNA1 derivatives in Saccharomyces cerevisiae, heterologous gene expression, and selective effects on subgenomic mRNA synthesis. J. Virol. 7411724-11733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Quadt, R., C. C. Kao, K. S. Browning, R. P. Hershberger, and P. Ahlquist. 1993. Characterization of a host protein associated with brome mosaic virus RNA-dependent RNA polymerase. Proc. Natl. Acad. Sci. USA 901498-1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rao, A. L. 2006. Genome packaging by spherical plant RNA viruses. Annu. Rev. Phytopathol. 4461-87. [DOI] [PubMed] [Google Scholar]
- 45.Rao, A. L., T. W. Dreher, L. E. Marsh, and T. C. Hall. 1989. Telomeric function of the tRNA-like structure of brome mosaic virus RNA. Proc. Natl. Acad. Sci. USA 865335-5339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rao, A. L., and G. L. Grantham. 1995. Biological significance of the seven amino-terminal basic residues of brome mosaic virus coat protein. Virology 21142-52. [DOI] [PubMed] [Google Scholar]
- 47.Restrepo-Hartwig, M. A., and P. Ahlquist. 1996. Brome mosaic virus helicase- and polymerase-like proteins colocalize on the endoplasmic reticulum at sites of viral RNA synthesis. J. Virol. 708908-8916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sanz, M. A., A. Castello, and L. Carrasco. 2007. Viral translation is coupled to transcription in Sindbis virus-infected cells. J. Virol. 817061-7068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schneemann, A. 2006. The structural and functional role of RNA in icosahedral virus assembly. Annu. Rev. Microbiol. 6051-67. [DOI] [PubMed] [Google Scholar]
- 50.Schneemann, A., V. Reddy, and J. E. Johnson. 1998. The structure and function of nodavirus particles: a paradigm for understanding chemical biology. Adv. Virus Res. 50381-446. [DOI] [PubMed] [Google Scholar]
- 51.Schneemann, A., W. Zhong, T. M. Gallagher, and R. R. Rueckert. 1992. Maturation cleavage required for infectivity of a nodavirus. J. Virol. 666728-6734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Scotti, P. D., S. Dearing, and D. W. Mossop. 1983. Flock house virus: a nodavirus isolated from Costelytra zealandica (White) (Coleoptera: Scarabaeidae). Arch. Virol. 75181-189. [DOI] [PubMed] [Google Scholar]
- 53.Selling, B. H., R. F. Allison, and P. Kaesberg. 1990. Genomic RNA of an insect virus directs synthesis of infectious virions in plants. Proc. Natl. Acad. Sci. USA 87434-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sivakumaran, K., M. Hema, and C. C. Kao. 2003. Brome mosaic virus RNA syntheses in vitro and in barley protoplasts. J. Virol. 775703-5711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Strauss, J. H., and E. G. Strauss. 1994. The alphaviruses: gene expression, replication, and evolution. Microbiol. Rev. 58491-562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tan, R., and A. D. Frankel. 1995. Structural variety of arginine-rich RNA-binding peptides. Proc. Natl. Acad. Sci. USA 925282-5286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Taylor, D. N., and J. P. Carr. 2000. The GCD10 subunit of yeast eIF-3 binds the methyltransferase-like domain of the 126 and 183 kDa replicase proteins of tobacco mosaic virus in the yeast two-hybrid system. J. Gen. Virol. 811587-1591. [DOI] [PubMed] [Google Scholar]
- 58.Venter, P. A., N. K. Krishna, and A. Schneemann. 2005. Capsid protein synthesis from replicating RNA directs specific packaging of the genome of a multipartite, positive-strand RNA virus. J. Virol. 796239-6248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Venter, P. A., and A. Schneemann. 2007. Assembly of two independent populations of flock house virus particles with distinct RNA packaging characteristics in the same cell. J. Virol. 81613-619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Volkova, E., R. Gorchakov, and I. Frolov. 2006. The efficient packaging of Venezuelan equine encephalitis virus-specific RNAs into viral particles is determined by nsP1-3 synthesis. Virology 344315-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Whitlow, Z. W., J. H. Connor, and D. S. Lyles. 2006. Preferential translation of vesicular stomatitis virus mRNAs is conferred by transcription from the viral genome. J. Virol. 8011733-11742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Willits, D., X. Zhao, N. Olson, T. S. Baker, A. Zlotnick, J. E. Johnson, T. Douglas, and M. J. Young. 2003. Effects of the cowpea chlorotic mottle bromovirus beta-hexamer structure on virion assembly. Virology 306280-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wu, F.-S. 1987. Localization of mitochondria in plant cells by vital staining with rhodamine 123. Planta 171346-357. [DOI] [PubMed] [Google Scholar]
- 64.Zlotnick, A., R. Aldrich, J. M. Johnson, P. Ceres, and M. J. Young. 2000. Mechanism of capsid assembly for an icosahedral plant virus. Virology 277450-456. [DOI] [PubMed] [Google Scholar]