Influenza epidemics cause morbidity and mortality worldwide (4). Each year in the United States, more than 200,000 patients are admitted to hospitals because of influenza and there are approximately 36,000 influenza-related deaths (14). In recent years, several subtypes of avian influenza viruses have jumped host species to infect humans. The H5N1 subtype, in particular, has been reported in 328 human cases and has caused 200 human deaths in 12 countries (World Health Organization, http://www.who.int/csr/disease/avian_influenza/country/cases_table_2007_09_10/en/index.html). These viruses have the potential to cause a pandemic in humans. Antiviral drugs and vaccines must be developed to minimize the damage that such a pandemic would bring. To achieve this, it is vital that researchers have free access to viral sequences in a timely fashion, and sequence analysis tools need to be readily available.
Historically, the number of influenza virus sequences in public databases has been far less than those of some well-studied viruses, such as human immunodeficiency virus. The number of complete influenza virus genomes has been even smaller. In addition, many of the sequences were collected in the course of influenza surveillance programs that prioritized antigenically novel isolates. Although collecting antigenically novel isolates is appropriate for surveillance, it results in biased samples of sequenced isolates that are not representative of community cases of influenza (2, 13). Therefore, in 2004, the National Institute of Allergy and Infectious Diseases (NIAID) launched the Influenza Genome Sequencing Project (7), which aims to rapidly sequence influenza viruses from samples collected all over the world. Viral sequences were generated at the J. Craig Venter Institute, annotated at the National Center for Biotechnology Information (NCBI), and deposited in GenBank. In just over 2 years after the initiation of the project, more than 2,000 complete genomes of influenza viruses A and B had been deposited in GenBank. To help the research community to make full use of the wealth of information from such a large amount of data, which will be increasing continuously, the Influenza Virus Resource was created at NCBI in 2004.
OVERVIEW OF THE NCBI INFLUENZA VIRUS RESOURCE
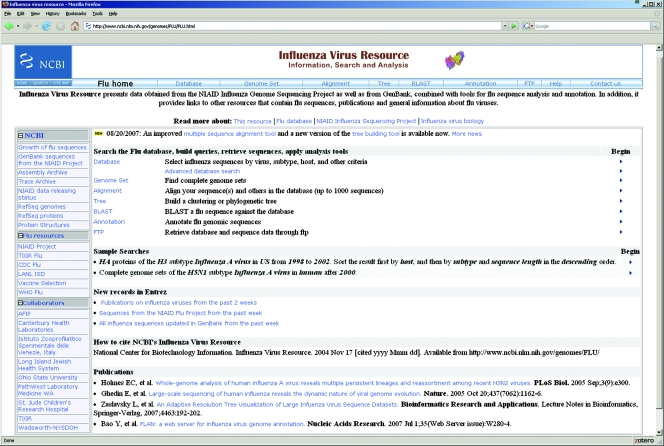
Access to the NCBI Influenza Virus Resource (http://www.ncbi.nlm.nih.gov/genomes/FLU/) is shown in Fig. 1. The main component of the resource is the NCBI Influenza Virus Sequence Database, where users can build queries, retrieve sequences, and find complete genome sets. The resource also provides sequence analysis tools, such as multiple-sequence alignment and clustering of protein sequences based on different metrics, that are integrated with the database. An influenza virus sequence-specific BLAST page and a genome annotation tool for influenza viruses A and B are available. In addition, the resource has links to influenza virus sequences, protein structures, the Trace Archive, publications, and general information about influenza viruses.
FIG. 1.
Home page of the Influenza Virus Resource.
THE NCBI INFLUENZA VIRUS SEQUENCE DATABASE
The NCBI Influenza Virus Sequence Database contains nucleotide sequences of all influenza viruses in the EMBL/DDBJ/GenBank databases, as well as protein sequences and their encoding regions derived from the nucleotide sequences. The influenza database is updated, usually within a day or two, after new sequences become available or older sequence records are updated in GenBank. Information for database fields (subtype, segment, host, country, year, etc.) is extracted automatically from GenBank records and examined by NCBI staff. BLAST searches are performed for all new sequences against the influenza virus sequences in GenBank to verify critical information such as subtype, segment, and year. Information that is not available in GenBank records is obtained from the literature, through direct contact with sequence submitters, or by sequence analysis whenever possible.
Figure 2A shows the basic database query interface (http://www.ncbi.nlm.nih.gov/genomes/FLU/Database/select.cgi?go=1). In querying the database, users may select to search among nucleotide sequences, protein sequences, or coding regions (CDS). Queries may be restricted by using the following additional selectable fields: Virus Species (e.g., Influenzavirus A, Influenzavirus B), Host (e.g., Human, Avian), Subtype (e.g., H3N2, H5), Segment (1 through 8), Country/Region (e.g., Australia, Asia), a range of years (e.g., From year, To year) during which the viruses were isolated, and a range of the lengths of the sequences.
FIG. 2.
(A) Influenza Virus Sequence Database query page. (B) List of sequences retrieved from the query page shown in panel A. (C) Multiple alignment of sequences from those listed in panel B. (D) Graphic view of multiple alignment of sequences from those shown in panel B or C, and parameter selections for tree building. (E) Neighbor-joining tree built from the multiple alignment shown in panel D. Sequences from the United States are marked in green blocks, and sequences from 1987 are highlighted in red.
On the other hand, in the advanced database search tool (http://www.ncbi.nlm.nih.gov/genomes/FLU/Database/multiple.cgi), multiple names can be selected simultaneously for species, host, country/region, segment, and subtype. A list of subtypes separated by commas (e.g., H5N1, H3, N2) can be entered in the boxes after “Only these Subtypes” and/or “All Subtypes except.” The number of sequences found by a query will be displayed after the “Update count” button is clicked.
A string of words or a nucleotide/protein sequence (e.g., New York, AGCGAAAGCAGGGGT, or RSKV) can be added to the “Search by a string” box to be included in the search. Search results can be limited to “Full-length sequences only” (does not apply to protein sequences or CDS sequences of segments that encode more than one protein, i.e., the PB1, MP, and NS segments) by checking the appropriate boxes. For nucleotide sequences, “full-length” is defined as not shorter than the complete coding region.
When the box in front of “Remove identical sequences” is checked, all groups of identical sequences in a data set will be represented by the oldest sequence in the group. By checking the box in front of “Sequences from the FLU project only,” search results can be restricted only to sequences from large-scale influenza virus genome sequencing projects, which usually contain complete genomes, detailed source information, and high-quality annotations. Currently, this includes sequences from the NIAID Influenza Genome Sequencing Project (9), the St. Jude Influenza Genome Project (12), the Centers for Disease Control and Prevention, the Air Force Institute for Operational Health, and the University of Hong Kong. Sequences of recombinant or lab strains (those flagged as “LAB” in the country field) are not included in the search by default, but they may be included by checking the box next to “Include Lab strains.”
After the “Add to Query Builder” button shown in Fig. 2A is clicked, the selected query and the number of resulting sequences will be shown in “Query Builder.” Nucleotide or protein sequences can also be searched by adding the accession number in the box to the left of the “Find sequence by Accession” button. Multiple queries can be built by repeating the above steps. Any combination of queries from the “Query Builder” can be selected to get sequences from the database.
Sequences found by the selected queries will be shown in a separate window (Fig. 2B) once the “Get sequences” button is clicked. The sequence display can be reordered by up to three fields sequentially by selecting one field each from the “Ordered by the following fields” boxes. Sequences of interest can be selected by checking the boxes to the left of the accession numbers. The corresponding protein, coding region, or nucleotide sequences of the selected sequences can be downloaded by selecting the appropriate name in the “Select FASTA sequences to download” drop-down menu. To help users identify the downloaded sequences, the following string is inserted between the GenBank sequence identifier and the sequence title in the FASTA definition lines: /host/segment number(name)/subtype/country/year/month/date/. A list of GenBank accession numbers for selected protein or nucleotide sequences can also be downloaded from the “Select accession list to download” menu.
Further sequence analyses of the selected sequences can be performed by clicking the “Do multiple alignment” or “Build a tree” button. Users' own sequences (of the same sequence type in FASTA format) can be added to the selected sequences for analysis by clicking the “Add your own sequences” button. The number of sequences added cannot be more than 128 kilobytes in file size.
GENOME SET TOOL
The Influenza Virus Genome Set tool (http://www.ncbi.nlm.nih.gov/genomes/FLU/Database/select.cgi?go=4) displays nucleotide sequences obtained from the NCBI Influenza Virus Sequence Database ordered according to the genome segments for each virus. All segments of the same virus are grouped together in the same background color, alternating in light blue and white. Genomes of the same virus isolate that were sequenced in different labs are identified as such in the database and are grouped separately according to the sequence submitters. This tool is a convenient way to check the completeness of genome segments for viruses of interest.
Database searches can be performed similarly to the process described above, and nucleotide sequences can also be searched by adding a complete or partial virus name [e.g., Influenza A virus (A/New York/19/2003(H3N2)), or (New York)] in the box to the left of “Search by a string.” By default, this tool gets only viruses with a complete set of full-length (or nearly full-length) segments. To get all viruses with any number of sequences (full-length or not), check the box before “Show all sequences.” The results are shown in descending order according to the number of segments the viruses have.
MULTIPLE SEQUENCE ALIGNMENT TOOL
Multiple alignments of nucleotide or protein sequences from the NCBI Influenza Virus Sequence Database and/or the user's input file can be obtained by using the MUSCLE program (5). When multiple alignments of a coding region are requested, an alignment of the corresponding protein sequences is performed first, and the alignments of amino acids are mapped back to the coding regions. This not only makes the alignment faster but also leads to a better alignment of coding regions and thus better input for subsequent tree building (see Clustering and Phylogenetic Tool). To start an alignment, the “Alignment” button in the top horizontal bar should be clicked. A database query interface (http://www.ncbi.nlm.nih.gov/genomes/FLU/Database/select.cgi?go=2) similar to the one described in the database section will be opened. Sequences from the same segment of the genome and preferably of similar sizes should be selected for multiple sequence alignment.
The program is set to allow a maximum number of 1,000 sequences to be included in the alignment. For data sets larger than 1,000 sequences, we recommend downloading the sequences using the download tool of the database and running the multiple sequence alignment using a program (e.g., MUSCLE) installed locally.
After sequences of interest are selected from the database and/or added from an input file, users may click the “Do multiple alignment” button in Fig. 2B to get the alignment (Fig. 2C). The consensus sequence is displayed at the top of the alignment; sequences identical to that of the consensus sequence are indicated by dots, and gaps are indicated by dashes. In the coding region alignment, nonsynonymous changes (in triplets) are highlighted in a different background color. The alignment can be navigated horizontally in the following two ways: (i) by typing in the start position in the text box after “Go to position” and clicking “Go” and (ii) by moving the bottom scroll bar that wraps the alignment. The sequence and virus name will be shown in a text box with a yellow background when the cursor is over a sequence. When a sequence in the alignment is clicked, a small menu will pop up. The GenBank record for the sequence can be opened by clicking the accession number in the pop-up menu. The sequence can also be selected to perform BLAST 2 sequencing (click the “BLAST 2 seq.” button after two different sequences are selected from the alignment). When the “Select for anchor” option from the pop-up menu is selected, the consensus sequence will be replaced by the selected sequence. When the anchor sequence is clicked, a small window with options will pop up. The anchor sequence can be reset to the consensus sequence, and the anchor/consensus sequence can be displayed for copying. The multiple sequence alignment file in FASTA format can be downloaded by selecting “Download alignment.” A printer-friendly version of the alignment can be obtained by clicking the “Print-friendly version” button. If desired, a tree can be built from the aligned sequences by clicking the “Build a tree” button.
CLUSTERING AND PHYLOGENETIC TOOL
Phylogenetic/clustering trees can be built for protein sequences and coding regions from the NCBI Influenza Virus Sequence Database and/or the user's input file (Fig. 2E). To accommodate more sequences in a tree, an adaptive approach was used to present an aggregated tree, which can be easily manipulated by users (17).
Clicking the “Tree” button in the top horizontal bar will start the tool. Sequences are acquired from the NCBI Influenza Virus Sequence Database or uploaded by a user as described above. After a data set has been selected, the “Build a tree” button in the database query results page must be clicked to start the process. This will bring up a page with a graphic view of the multiple alignments (Fig. 2D).
Most phylogenetic tree-building algorithms have trouble with partial sequences or a sequence set with various lengths. Many influenza virus sequences are indeed partial segments, and therefore manual editing of the data set is required before an accurate tree can be built. Sometimes a tree derived from a particular region of sequences is desired, and this also requires preprocessing of the sequence set, which is usually done manually. A graphical tool is introduced to allow users to decide which part of the multiple alignment to use for building the tree without manual editing. The blacks and reds in the graphics of multiple sequence alignments represent the presence and absence of amino acid residues at the corresponding positions. The positions for the first and last amino acid of each sequence are shown in the longest sequence of the selected set. A histogram showing the total number of amino acid residues at each position is displayed at the top of the page. The program automatically selects the sequence region to be analyzed so that the majority of the sequences in the set will be included. The sequence region can also be defined by users by first selecting all sequences in the set and then entering the start and end positions in the boxes provided. When the “Select sequences” button is clicked, the region from sequences that have complete coverage between the two positions will be selected and sequences excluded from the selection will be highlighted with a background color in the graphic view. Sequences of interest can be highlighted in the tree, and they can be selected or deselected by checking the boxes to the right of each sequence (Fig. 2D).
A clustering or phylogenetic tree can be built by selecting one of the clustering algorithms and a distance calculating method from the list and clicking the “Next step” button (Fig. 2D).
Phylogenetic and clustering analyses can be performed for data sets of aligned protein sequences or protein coding regions based on pairwise distances between the sequences in the data set. For data sets consisting of aligned protein coding regions, either Felsenstein F84 distance or Hamming distance can be used. For data sets containing aligned protein sequences, users can choose among mPAM distances based on their metric models of amino acid substitution derived from PAM (15), the Kimura formula, the Dayhoff PAM matrix, the Jones-Taylor-Thornton matrix, or the Henikoff/Tillier PMB matrix (8) (data were obtained from the PHYLIP official website at http://evolution.genetics.washington.edu/phylip.html). Trees can be built using a variety of distance methods, such as the neighbor-joining algorithm, average linkage (UPGMA), and complete linkage and single linkage clustering algorithms (6, 8, 10, 11, 16). By default, a tree is built based on the nucleotide distance for the coding region, regardless of what type of sequence (i.e., protein or coding region) was originally obtained from the database.
The visual representation of trees exploits the adaptive resolution technique (17). An aggregated tree with special representation of subscale details is calculated on the client site from the full phylogenetic tree and the amount of available screen space. Metadata, such as distribution over seasons or geographic locations, are aggregated/refined consistently with the tree. Users can interactively request further refinement or aggregation for different parts of the tree.
When the node of a branch on the tree is clicked, the branch is highlighted in orange and a log scale from 1 to the total number of sequences in the branch is shown to the left of the tree. The resolution of the selected branch can be changed by moving along the scale bar. When a leaf contains more than one sequence, the year range and total number of sequences in the leaf will be displayed. Sequences on the tree can be searched by the fields in the database, and the resulting sequences or groups are highlighted in green (Fig. 2E).
GENOME ANNOTATION TOOL
The Influenza Virus Genome Annotation tool (http://www.ncbi.nlm.nih.gov/genomes/FLU/Database/annotation.cgi) is a Web application for user-provided influenza A virus and influenza B virus sequences. It can predict protein sequences encoded by an influenza virus sequence and produce a feature table that can be used for sequence submission to GenBank, as well as a GenBank flat file (1).
A functionality added recently to the genome annotation tool is the ability to detect some of the signature mutations that might confer drug resistance by the virus. Such mutations include V27A and S31N in the M2 protein, H274Y and R292K in the N1 subtype of neuraminidase, and N294S in the N2 subtype of neuraminidase.
FTP
Data in the NCBI Influenza Virus Sequence Database are available through FTP (ftp://ftp.ncbi.nih.gov/genomes/INFLUENZA/). The FTP directory contains the following files that are updated every day: genomeset.dat, a table with supplementary data for the genome set; influenza_na.dat, a table with supplementary nucleotide data; influenza_aa.dat, a table with supplementary protein data; influenza.dat, a table with nucleotide, protein, and coding region IDs; influenza.fna, FASTA nucleotides; influenza.cds, FASTA coding regions; influenza.faa, FASTA proteins; and readme.
Genomeset.dat contains information for sequences of viruses with a complete set of full-length (or nearly full-length) segments. Segments of the same virus are grouped together and separated by an empty line from those of other viruses.
The genomeset.dat, influenza_na.dat, and influenza_aa.dat files are tab-delimited tables that have the following fields: GenBank accession number, Host, Genome segment number, Subtype, Country, Year, Sequence length, Virus name, Age, and Gender. The influenza_na.dat and influenza_aa.dat files have an additional field in the last column to indicate whether a sequence is full length. The influenza.dat file is a tab-delimited table that has the following fields: GenBank accession number for nucleotide; GenBank accession number for protein; and Identifier for protein coding region.
A directory named “updates” contains daily updates for all of the above-listed files in the subdirectories for each date.
A DEMONSTRATION
The main features of the Influenza Virus Resource are demonstrated in the following example, in which the coding regions of the M2 proteins belonging to the H11 subtype of influenza A viruses collected from 1986 to 2004 are retrieved from the database and a multiple-sequence alignment and a phylogenetic tree are built from the sequence set.
From the homepage of the resource (Fig. 1), the link to “Database” is clicked either in the top horizontal bar or on the main section to access the database query interface (Fig. 2A). The radio button in front of “Coding region” is checked to specify the type of sequence to be retrieved. “Influenzavirus A,” “any,” “any,” and “7(MP)” are selected for “Virus Species,” “Host,” “Country/Region,” and “Segment,” respectively. “H11,” “1986,” and “2004” are entered into the text boxes for “Subtype,” “From year,” and “To year,” respectively. The MP segment of influenza A virus encodes two proteins of different sizes. The M2 protein is about 97 amino acids in length (291 nucleotides for the coding region), and they can be defined in the database by entering “291” in both the “Min. length” and “Max. length” text boxes. After the “Add to Query Builder” button is clicked, the selected query and the number of sequences (n = 36, in this example) are shown in the “Query Builder” section.
When the “Get Sequences” button is clicked, the selected sequences can be retrieved and listed in a separate window (Fig. 2B). To sort the sequences first by country and then by year, the corresponding field in the first two drop-down menus under “Ordered by the following fields” is clicked. Then the “Reorder sequences” button is clicked.
A multiple-sequence alignment (Fig. 2C) is obtained by clicking the “Do multiple alignment” button (Fig. 2B).
To build a phylogenetic tree, the “Build a tree” button (Fig. 2B and C) is clicked. This brings up a graphic view of multiple-sequence alignments (not shown), and the “Next step” button is clicked to proceed to the next step (Fig. 2D). The “Neighbor-Joining method” is used to build a tree, so it is selected from the list under “Select Clustering Algorithm,” and the “F84 distance” button is selected from the “Nucleotide Distances for CDS” list. After the “Next step” button is clicked, a tree is created (Fig. 2E). To highlight sequences from 1987 in red on the tree, the “View options” button on top of the tree-displaying page is clicked, “1987” is entered in both the “From year” and “To year” boxes in “Selected time interval,” and the “Make selection” button is clicked. To mark sequences from the United States with green blocks, the “Search the tree” button is clicked, “US” is entered in the country field, and the “Search” button is clicked. To change the resolution of the branch shown in orange, its node is clicked and the cursor is moved along the blue scale bar to the left of the tree to display three leaves in the branch.
SUMMARY
The NCBI Influenza Virus Resource provides an integrated tool for influenza virus sequence retrieval and analysis. Every sequence in the database is curated by an automatic procedure, and NCBI staff make sure that the information presented in the database is complete, accurate, and up to date. The database has open access to all users and has been used as the backbone of several other influenza virus sequence databases, such as the Influenza Virus Database (3), the BioHealthBase BRC (http://www.biohealthbase.org/GSearch/statsAutomation.do?decorator=influenza), the Influenza Virus Genotype Tool (http://www.flugenome.org/index.php), and the Influenza Primer Design Resource (http://www.ipdr.mcw.edu/fludb/search). Sequence analysis tools such as multiple-sequence alignment and clustering of protein sequences are integrated with the database and allow users to quickly modify a data set to optimize the analysis. Using these tools offers a convenient way for preliminary sequence analyses. The influenza virus genome annotation tool makes sequence submission to GenBank much easier and will greatly promote data sharing among the influenza virus research community.
Acknowledgments
We acknowledge Edward Holmes, Yuri Wolf, Scott McDaniel, and numerous users for suggestions on improving the resource. We also acknowledge members of the NIAID Influenza Genome Sequencing Project, which was the initial motivation and driving force for this resource. Those members include Maria Giovanni, Martin Shumway, Steven Salzberg, David Spiro, Elodie Ghedin, Naomi Sengamalay, Claire M. Fraser-Liggett, Kirsten St. George, and Jill Taylor.
This research was supported by the Intramural Research Program of the NIH, National Library of Medicine.
The views expressed in this Commentary do not necessarily reflect the views of the journal or of ASM.
Footnotes
Published ahead of print on 17 October 2007.
REFERENCES
- 1.Bao, Y., P. Bolotov, D. Dernovoy, B. Kiryutin, and T. Tatusova. 2007. FLAN: a web server for influenza virus genome annotation. Nucleic Acids Res. 35W280-W284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bush, R. M., C. B. Smith, N. J. Cox, and W. M. Fitch. 2000. Effects of passage history and sampling bias on phylogenetic reconstruction of human influenza A evolution. Proc. Natl. Acad. Sci. USA 976974-6980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chang, S., J. Zhang, X. Liao, X. Zhu, D. Wang, J. Zhu, T. Feng, B. Zhu, G. F. Gao, J. Wang, H. Yang, J. Yu, and J. Wang. 2007. Influenza Virus Database (IVDB): an integrated information resource and analysis platform for influenza virus research. Nucleic Acids Res. 35D376-D380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cox, N. J., and K. Subbarao. 2000. Global epidemiology of influenza: past and present. Annu. Rev. Med. 51407-421. [DOI] [PubMed] [Google Scholar]
- 5.Edgar, R. C. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 321792-1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Everitt, B. S., S. Landau, and M. Leese. 2001. Cluster analysis, 4th ed. Hodder Arnold, London, United Kingdom.
- 7.Fauci, A. S. 2005. Race against time. Nature 435423-424. [DOI] [PubMed] [Google Scholar]
- 8.Felsenstein, J. 2003. Inferring phylogenies. Sinauer Associates, Sunderland, MA.
- 9.Ghedin, E., N. A. Sengamalay, M. Shumway, J. Zaborsky, T. Feldblyum, V. Subbu, D. J. Spiro, J. Sitz, H. Koo, P. Bolotov, D. Dernovoy, T. Tatusova, Y. Bao, K. St. George, J. Taylor, D. J. Lipman, C. M. Fraser, J. K. Taubenberger, and S. L. Salzberg. 2005. Large-scale sequencing of human influenza reveals the dynamic nature of viral genome evolution. Nature 4371162-1166. [DOI] [PubMed] [Google Scholar]
- 10.Hillis, D. M., C. Moritz, and B. K. Mable. 1996. Molecular systematics, 2nd ed. Sinauer Associates, Sunderland, MA.
- 11.Nei, M., and S. Kumar. 2000. Molecular evolution and phylogenetics. Oxford University Press, New York, NY.
- 12.Obenauer, J. C., J. Denson, P. K. Mehta, X. Su, S. Mukatira, D. B. Finkelstein, X. Xu, J. Wang, J. Ma, Y. Fan, K. M. Rakestraw, R. G. Webster, E. Hoffmann, S. Krauss, J. Zheng, Z. Zhang, and C. W. Naeve. 2006. Large-scale sequence analysis of avian influenza isolates. Science 3111576-1580. [DOI] [PubMed] [Google Scholar]
- 13.Plotkin, J. B., J. Dushoff, and S. A. Levin. 2002. Hemagglutinin sequence clusters and the antigenic evolution of influenza A virus. Proc. Natl. Acad. Sci. USA 996263-6268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thompson, W. W., D. K. Shay, E. Weintraub, L. Brammer, N. Cox, L. J. Anderson, and K. Fukuda. 2003. Mortality associated with influenza and respiratory syncytial virus in the United States. JAMA 289179-186. [DOI] [PubMed] [Google Scholar]
- 15.Xu, W., and D. P. Miranker. 2004. A metric model of amino acid substitution. Bioinformatics 201214-1221. [DOI] [PubMed] [Google Scholar]
- 16.Yang, Z. 2006. Computational molecular evolution. Oxford University Press, New York, NY.
- 17.Zaslavsky, L., Y. Bao, and T. A. Tatusova. 2007. An adaptive resolution tree visualization of large influenza virus sequence datasets, p. 192-202. In I. Mandoiu and A. Zelikovsky (ed.), Bioinformatics research and applications. Springer-Verlag, Heidelberg, Germany.