Abstract
Integrase (IN), an essential enzyme of human immunodeficiency virus (HIV), is an attractive antiretroviral drug target. The antiviral activity and resistance profile in vitro of a novel IN inhibitor, elvitegravir (EVG) (also known as JTK-303/GS-9137), currently being developed for the treatment of HIV-1 infection are described. EVG blocked the integration of HIV-1 cDNA through the inhibition of DNA strand transfer. EVG inhibited the replication of HIV-1, including various subtypes and multiple-drug-resistant clinical isolates, and HIV-2 strains with a 50% effective concentration in the subnanomolar to nanomolar range. EVG-resistant variants were selected in two independent inductions, and a total of 8 amino acid substitutions in the catalytic core domain of IN were observed. Among the observed IN mutations, T66I and E92Q substitutions mainly contributed to EVG resistance. These two primary resistance mutations are located in the active site, and other secondary mutations identified are proximal to these primary mutations. The EVG-selected IN mutations, some of which represent novel IN inhibitor resistance mutations, conferred reduced susceptibility to other IN inhibitors, suggesting that a common mechanism is involved in resistance and potential cross-resistance. The replication capacity of EVG-resistant variants was significantly reduced relative to both wild-type virus and other IN inhibitor-resistant variants selected by L-870,810. EVG and L-870,810 both inhibited the replication of murine leukemia virus and simian immunodeficiency virus, suggesting that IN inhibitors bind to a conformationally conserved region of various retroviral IN enzymes and are an ideal drug for a range of retroviral infections.
Three unique and essential HIV enzymes, protease (PR), reverse transcriptase with RNase H (RT), and integrase (IN), appear to be ideal targets for the development of inhibitors of human immunodeficiency virus (HIV) replication. Anti-HIV drugs targeting PR (PR inhibitors [PIs]) and RT (nucleoside/nucleotide RT inhibitors [NRTIs] and nonnucleoside RT inhibitors [NNRTIs]) have been approved for use in the treatment of HIV infection. Combinations of these drugs used in highly active antiretroviral therapy can effectively suppress HIV replication in vivo to undetectable levels and have led to significant declines in HIV-associated mortality (28, 40). However, the emergence of drug-resistant HIV variants can attenuate the efficacy of antiretroviral treatment. Some primary infections also result from the transmission of HIV strains that possess drug-resistant genotypes and phenotypes (9). To suppress these drug-resistant variants, new anti-HIV drugs that block new targets are urgently needed.
IN, a 32-kDa protein resulting from the proteolytic cleavage of the gag-pol precursor, plays an essential role in the integration of proviral DNA into the host genome. As LaFemina et al. previously reported that there is no human homologue of HIV IN (31), it is an attractive target for the development of new antiretroviral therapeutic agents without adverse effects. IN consists of three domains: an N-terminal zinc finger domain and a C-terminal DNA-binding domain flank a central catalytic core domain (CCD) that plays a critical role in its enzymatic activity (13, 14). Following reverse transcription, IN exerts at least two functions: the cleavage of two conserved nucleotides from the 3′ ends of both strands of the viral cDNA (3′ processing) (1) and, subsequently, the ligation of the viral cDNA into the host genome (strand transfer) (14). Gap filling of the interfaces between the viral and host genomic DNA is then completed using the host DNA repair machinery via a mechanism that is not yet fully understood. The completion of integration results in a fully functional provirus, which can then be used to initiate viral DNA transcription.
Several compounds that inhibit IN activity have been described, including diketo acid (DKA) derivatives such as L-731,988 (24) and S-1360 (16), both of which have potent antiviral activity. Crystal structure analysis has indicated that 1-(5-chloroindol-3-yl)-3-hydroxy-3-(2H-tetrazol-5-yl)-prope- none, an S-1360 derivative, binds to the CCD, the putative active site of IN (19). In vitro resistance selection experiments with several IN inhibitors demonstrated that mutations in the CCD of IN play a significant role in the generation of IN inhibitor-resistant viral variants. In vitro selection of HIV-1 in the presence of the DKA IN inhibitors L-731,988 and S-1360 resulted in the emergence of viral variants carrying IN mutations associated with resistance. These mutations, including T66I, S153Y, and M154I, are located in close proximity to the catalytic triad residues (D64, D116, and E152) in the CCD of IN (16, 24). In contrast, L-870,810 (Fig. 1), which has previously demonstrated potent antiviral activity in HIV-1-infected patients in a monotherapy study (33), induced unique IN mutations, including V72I, F121Y, T125K, and V151I, when HIV was selected with the compound in vitro (23). These mutations are also located in the active site of IN, suggesting that a common mechanism may be involved in the acquisition of resistance to IN inhibitors.
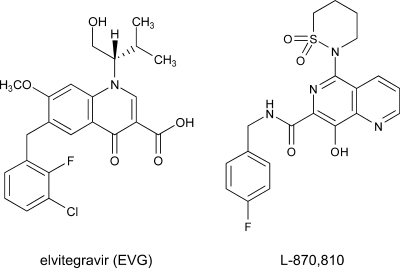
FIG. 1.
Structure of EVG and L-870,810. A dihydroquinoline carboxylic acid derivative, EVG, and a naphthyridine carboxamide derivative, L-870,810 (a representative IN inhibitor), are shown.
Although no IN inhibitors are currently approved for clinical use (41), two IN inhibitors, elvitegravir (EVG) (formerly known as JTK-303/GS-9137, being codeveloped by Gilead Sciences and Japan Tobacco) (Fig. 1) (43, 56) and raltegravir (MK-0518, developed by Merck) (22), are currently being investigated in clinical studies of HIV-1-infected patients. In a phase II study, antiretroviral treatment-experienced patients using 125 mg EVG (boosted with ritonavir) along with an active optimized background regimen showed >2-log10 declines in their viral loads that were durable through week 24 (56).
Here, we describe the antiviral activity, mechanism of action, and resistance profile of EVG in vitro. EVG exerted potent anti-HIV activity against not only wild-type strains but also drug-resistant clinical isolates. Interestingly, EVG also showed antiviral activity against murine leukemia virus (MLV) and simian immunodeficiency virus (SIV). These results imply that IN inhibitors are ideal agents for the treatment of a range of retroviral infections. During the selection of EVG-resistant viral variants, novel IN mutations emerged. Combinations of these mutations conferred resistance to EVG and reduced susceptibility to other IN inhibitors, suggesting that there is a common mechanism underlying the resistance to IN inhibitors. One such mechanism may be conformational changes induced by multiple mutations located in the active site of IN.
MATERIALS AND METHODS
Antiviral agents.
Zidovudine (AZT) and dextran sulfate (DS5000) (average molecular weight, 5,000) were purchased from Sigma (St. Louis, MO). Efavirenz (EFV) (NNRTI) and nelfinavir (NFV) (PI) were used for the control inhibitor. EVG (43), L-731,988 (42), L-870,810 (23), and S-1360 (16) were synthesized as described previously. The structures of EVG and L-870,810 are depicted in Fig. 1.
Cells and viruses.
MT-2 and MT-4 cells were grown in RPMI 1640 medium. 293T cells were grown in Dulbecco's modified Eagle's medium. These media were supplemented with 10% fetal calf serum, 2 mM l-glutamine, 100 U/ml penicillin, and 50 μg/ml streptomycin. HeLa-CD4/CCR5-LTR/β-gal cells (5) were kindly provided by J. Overbaugh through the AIDS Research and Reference Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases (Bethesda, MD), and maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum, 200 μg/ml hygromycin B, 10 μg/ml puromycin, and 200 μg/ml geneticin. Peripheral blood mononuclear cells (PBMC) were obtained from healthy HIV-1-seronegative donors by centrifugation through Ficoll-Hypaque density gradients. PBMC were stimulated with 20 U/ml interleukin-2 (Shionogi, Osaka, Japan) and 0.5 μg/ml phytohemagglutinin (Sigma) for 3 days and then used for assays as described previously (30).
Three laboratory strains, HIV-1IIIB, HIV-2EHO, and HIV-2ROD, were used in this study. Various subtypes of drug-naïve clinical isolates of HIV-1 (four isolates of subtype B and seven isolates of non-B subtypes) were employed. Four drug-resistant clinical isolates of HIV-1, including IVR401, IVR409, IVR411, and IVR415, were kindly provided by S. Oka (AIDS Clinical Center, International Medical Center of Japan, Tokyo, Japan).
Determination of HIV drug susceptibility.
Inhibitory effects of compounds on HIV infection were determined using multinuclear activation of a galactosidase indicator (MAGI) assay, as previously described (37). Inhibitory effects on HIV-1 clinical isolates were measured by p24 production, and cytotoxicity was measured by using a 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) colorimetric assay, as described previously (30). Antiviral activities and cytotoxicities of inhibitors are presented as the concentrations that block viral replication by 50% (50% effective concentration [EC50]) and that suppress the viability of target cells by 50%, respectively.
Quantification of HIV-1 DNA species.
MT-2 cells (5 × 105 cells) were infected with HIV-1IIIB at a multiplicity of infection (MOI) of 0.1 in the absence or presence of various inhibitors. Infected cells were washed after incubation for 2 h at 37°C. At 24 h postinfection, DNA was extracted using DNAzol reagent (Invitrogen, Carlsbad, CA).
Quantification of integrated HIV-1 DNA and the two-long-terminal-repeat (2-LTR) circle was performed by real-time quantitative PCR as described previously (4). To normalize DNA species among inhibitors, β-globin amplification was used as an internal control (51). Reactions were analyzed by using the ABI Prism 7500 sequence detector (PE Applied Biosystems, Foster City, CA), and results were then normalized and expressed as relative HIV-1 DNA species compared to a “no-inhibitor” control.
In vitro strand transfer assay.
An oligonucleotide-based strand transfer assay was performed as previously described (8), with some modifications. Briefly, preprocessed oligonucleotide H-U5V1-2 (5′-ATGTGGAAAATCTCTAGCA-3′), derived from the U5 end of the HIV-1 LTR, was labeled at the 5′ end with [γ-32P]ATP. Radiolabeled H-U5V1-2 was annealed to H-U5V2 (5′-ACTGCTAGAGATTTTCCACAT-3′) and then used for assays. Recombinant HIV-1 IN derived from HIV-1 NL4-3 (wild type) or EVG-selected mutants was prepared using an Escherichia coli expression system. The strand transfer assay was performed with 1 μM IN and 150 nM substrate DNA in 20 mM MOPS (morpholinepropanesulfonic acid) buffer with 30 mM MgCl2 incubated in either the presence or absence of IN inhibitors at 37°C for 60 min. Reaction products were analyzed by electrophoresis on 25% polyacrylamide gels and quantified using a BAS-2500 imaging system (Fuji Photo Film, Tokyo, Japan). The concentration of IN inhibitor that inhibited the production of strand transfer products by 50% (50% inhibitory concentration [IC50]) compared to the control was determined.
Selection of EVG-resistant HIV-1 variants in vitro.
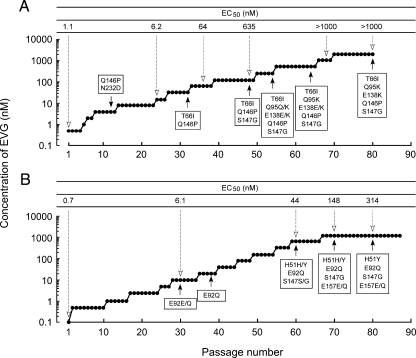
MT-2 cells (2 × 105 cells) were infected with HIV-1IIIB and then cultured in the presence of 0.5 nM (see Fig. 3A) or 0.1 nM (see Fig. 3B) EVG. Cultures were incubated at 37°C until an extensive cytopathic effect (CPE) was observed, and the culture supernatant was then harvested for further passage in fresh MT-2 cells. The concentration of EVG was increased when a significant CPE was observed. At the indicated passages (see Fig. 3A and B), proviral DNA was extracted from infected MT-2 cells and then subjected to PCR, followed by direct population-based sequencing. Susceptibility to EVG at the indicated passages was determined using the MAGI assay (see Fig. 3A) or p24 production (see Fig. 3B).
FIG. 3.
Induction of EVG-resistant HIV-1. Data from MT-2 cells are shown. The initial concentrations of EVG were 0.5 nM (A) and 0.1 nM (B). Results are from two identical but independent experiments. At the indicated passage number (black arrowheads), proviral DNA extracted from infected MT-2 cells was sequenced. Amino acid substitutions are shown. The EC50 values of HIV-1 variants selected by EVG at the indicated passage number (white arrowheads) were determined using MAGI assay (A) or the production of p24 in MT-2 cells (B).
Recombinant HIV-1 clones.
An HIV-1 infectious clone, pNL101 (38), kindly provided by K.-T. Jeang (NIH, Bethesda, MD), was used to generate recombinant HIV-1 clones. Wild-type HIV-1 (HIV-1WT) was constructed by replacing the pol coding region (nucleotide positions 2006 of the ApaI site to 5122 of the NdeI site of pNL101) with HIV-1 strain BH10. The pol coding region contains a silent mutation at nucleotide 4232 (TTTAGA to TCTAGA) resulting in the generation of a unique XbaI site. Recombinant HIV-1 IN infectious clones were generated using a modified pNL101-based vector, pNLRTWT. In brief, mutations were introduced into the XbaI-NdeI region (891 bp) of pSLIntWT, which encodes nucleotides 4232 to 5122 of pNL101, using an oligonucleotide-based site-directed mutagenesis method (54). Next, the XbaI-NdeI fragments were inserted into pBNΔInt, which encodes nucleotides 5122 (NdeI) to 5785 (SalI) of pNL101. Finally, the XbaI-SalI region (1,554 bp) was inserted into pNL101. Each infectious clone was transfected into 293T cells. The following day, MT-2 cells were added, and the supernatants were harvested when an extensive CPE was observed.
Replication kinetics of resistant HIV-1 variants.
MT-2 cells (105 cells) were infected with each virus preparation (500 MAGI units) for 4 h. The infected cells were then washed and cultured in the presence or absence of EVG. The culture supernatants were harvested on day 5 after infection, and p24 levels were quantified using a Retro-Tek HIV-1 p24 antigen enzyme-linked immunosorbent assay (ELISA) (ZeptoMetrix, Buffalo, NY).
Evaluation of antiretroviral activities of IN inhibitors.
The MLV-based retroviral vector pRCV/LIG (15) and plasmid pcDNA-VSVG, encoding the vesicular stomatitis virus envelope glycoprotein (a generous gift from H. Miyoshi, RIKEN Bioresource Center, Tsukuba, Japan), were employed to generate viral particles. These plasmids were cotransfected into an MLV-derived Gag-Pol-expressing packaging cell line, GP293 (Clontech, Palo Alto, CA). After 48 h of transfection, culture supernatants were filtered through a 0.45-μm membrane and stored at −80°C until use.
An HIV-1-based luciferase expression vector, pBC-LIG; pCMVΔ8/9, encoding the HIV-1 viral proteins including IN; and pcDNA-VSVG were transfected into 293T cells to generate pseudotyped HIV-1. The viruses were used to infect 293T cells (105 cells per well in 12-well plates) at an MOI of 0.02 in the absence or presence of inhibitors. After 48 h of transduction, luciferase activity was determined using a luciferase assay system (Promega, Madison, WI) and an LB 9507 luminometer (Berthold, Bad Wildbad, Germany).
An SIV molecular clone, pMA239 (46), containing the full SIVmac239 genome, was a kind gift from E. Ido, Institute for Virus Research, Kyoto University. pMA239 was used to generate viral stocks as previously described (6). Antiviral activities of IN inhibitors against SIVmac239 were determined using the MAGI assay as described above.
Molecular modeling studies.
A three-dimensional model of EVG in complex with HIV-1 IN CCD was prepared by PyMOL software, version 0.97, using previously reported data (44). Amino acid residues involved in resistance to EVG were displayed within this model.
RESULTS
Anti-HIV activities of IN inhibitors.
The antiviral activity of EVG against HIV-1IIIB, HIV-2EHO, and HIV-2ROD was first evaluated by the MAGI assay. EVG showed potent antiviral activity against three laboratory strains of HIV, with EC50 values in the subnanomolar to nanomolar range (Table 1). Next, we evaluated the activity of EVG against wild-type clinical isolates representing various subtypes of HIV-1. EVG suppressed the replication of all HIV-1 subtypes tested, with an antiviral EC50 ranging from 0.10 to 1.26 nM (Table 2). Moreover, EVG suppressed the replication of HIV-1 clinical isolates carrying NRTI, NNRTI, and PI resistance-associated genotypes, as did a control IN inhibitor, the compound L-870,810 (see Table S1 in the supplemental material). The cytotoxicities of these inhibitors were also determined using an MTT colorimetric assay. Mean values for the concentration that suppresses the viability of target cells by 50% for EVG and L-870,810 in PBMC obtained from three independent donors were 4.6 ± 0.5 μM and 2.7 ± 0.6 μM, respectively. Thus, EVG can suppress various HIV strains, including diverse HIV-1 subtypes and clinical isolates carrying multiple mutations associated with resistance to currently approved antiretroviral drugs.
TABLE 1.
Antiviral activities against laboratory HIV strainsa
| Strain | Mean EC50 (nM) ± SD
|
||
|---|---|---|---|
| AZT | EVG | L-870,810 | |
| HIV-1IIIB | 7.1 ± 1.3 | 0.7 ± 0.3 | 6.3 ± 0.3 |
| HIV-2EHO | 22 ± 9.1 | 2.8 ± 0.8 | 11 ± 1.9 |
| HIV-2ROD | 19 ± 4.7 | 1.4 ± 0.7 | 8.6 ± 0.4 |
Antiviral activity was determined using the MAGI assay. Data shown are means and standard deviations obtained from at least three independent experiments.
TABLE 2.
Antiviral activities of EVG against various subtypes of HIV-1a
| Subtype | Isolate | EC50 (nM)
|
|
|---|---|---|---|
| AZT | EVG | ||
| A | RW/92/016 | 7.91 | 0.41 |
| B | 96USHIPS7 | 8.41 | 0.26 |
| BR/92/021 | 2.13 | 0.76 | |
| BR/93/017 | 1.10 | 0.18 | |
| BR/93/022 | 11.7 | 1.13 | |
| C | BR/92/025 | 2.84 | 0.10 |
| D | UG/92/046 | 7.26 | 0.50 |
| E | CMU02 | 9.07 | 1.26 |
| F | BR/93/020 | 25.3 | 0.74 |
| G | JV1083 | 11.1 | 0.35 |
| O | BCF01 | 1.52 | 1.17 |
Antiviral activity was determined using p24 ELISA.
Mechanism of anti-HIV activity of EVG.
First, we performed a “time-of-addition” experiment as described previously (30), with some modifications. MT-4 cells were infected with HIV-1IIIB at an MOI of 0.5. One hour after infection, infected cells were extensively washed, and compounds were added, including an NNRTI (EFV at 100 nM), a PI (NFV at 500 nM), or EVG (100 nM). Amounts of p24 antigen were determined at 31 h postinfection. The antiviral activity of EFV gradually decreased from 6 h postinfection and disappeared at 12 h postinfection, whereas the antiviral activity of EVG decreased from 10 h postinfection and was no longer detected by 12 h postinfection. On the other hand, the PI NFV effectively blocked the infection up to 12 h postinfection and still exerted approximately 20% inhibitory activity up to 24 h postinfection. These results strongly suggest that EVG inhibits the HIV replication at a step that occurs after reverse transcription but before proteolytic cleavage, consistent with the integration step.
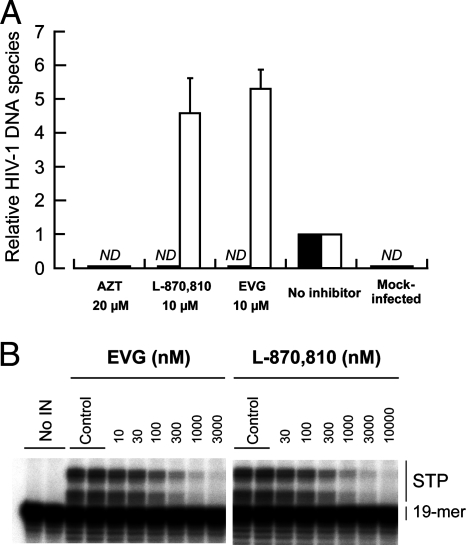
To elucidate the mode of action of EVG on HIV-1 replication, the levels of intracellular HIV-1 DNA species were determined using real-time quantitative PCR (Fig. 2A). MT-2 cells were infected with HIV-1IIIB in the presence or absence of a CD4-gp120 binding inhibitor, DS5000; an NRTI, AZT; an IN inhibitor, L-870,810; and EVG. Unintegrated (2-LTR) and integrated forms of reverse-transcribed HIV-1 genomic DNA were quantified after 24 h of infection and then normalized with β-globin DNA. In the presence of 20 μM AZT, neither 2-LTR nor integrated forms were detected as expected. Similar results were also observed with 20 μM DS5000 (data not shown). In the presence of 10 μM L-870,810, integrated provirus was undetectable, while relative 2-LTR levels increased about 5-fold (4.6-fold ± 1.0-fold). Similar results were observed with 10 μM EVG (2-LTR) (5.3-fold ± 0.5-fold), indicating that EVG exerts anti-HIV activity by blocking the integration step.
FIG. 2.
Mechanism of action of EVG. (A) Quantification of HIV-1 DNA species. MT-2 cells were infected with HIV-1IIIB in the presence or absence of AZT, L-870,810, and EVG. Unintegrated (2-LTR) (white bars) and integrated (black bars) forms of proviral DNA were quantified by real-time PCR and normalized to the β-globin gene after 24 h of infection. The data are represented as means and standard deviations of value relative to that of the no-inhibitor control from three independent experiments. ND means that the signals were not detected even after 40 cycles of amplification. (B) Inhibitory effect of IN inhibitors on strand transfer activity. Gel electrophoresis shows strand transfer products (STP) generated from preprocessed donor DNA substrate (19-mer) covalently bound to acceptor DNA.
To further characterize the mechanism by which EVG inhibits the integration step, the effect of EVG on strand transfer was assessed by characterizing its ability to inhibit the activity of recombinant wild-type HIV-1 IN enzyme in an oligonucleotide-based strand transfer assay (Fig. 2B). EVG and L-870,810 both inhibited the synthesis of strand transfer products with IC50 values of 54 nM and 118 nM, respectively. Taken together, these results indicate that like L-870,810, EVG blocks integration via the inhibition of IN-mediated strand transfer.
Selection of EVG-resistant HIV-1 variants in vitro.
To determine the in vitro resistance profile of EVG, EVG-resistant viral variants were selected using a dose escalation method, and the susceptibilities of the resulting selected variants to EVG (EC50) were determined. Selection of resistant HIV-1IIIB was initiated with 0.5 nM EVG (Fig. 3A). At passage 12 (P-12), where the concentration of EVG was 4 nM, 2 amino acid substitutions, glutamine-to-proline at IN codon 146 (Q146P) and asparagine-to-aspartic acid at IN codon 232 (N232D), were observed (Fig. 3A). An N232D substitution was previously reported to be an IN polymorphism in HIV-1 (34). The EVG EC50 of a P-24 variant containing a Q146P- and N232D-substituted variant was 6.2 nM. At P-32 (32 nM EVG), a T66I IN substitution was newly observed, whereas the N232D substitution had reverted to the baseline sequence. The EVG EC50 against a P-36 variant was 64 nM. An S147G IN substitution was detected at P-48 (128 nM EVG), and the EVG EC50 further increased to 635 nM. In addition, a Q95Q/K IN substitution (mixture of Q and K) and an E138E/K IN substitution were newly identified at P-54 (256 nM EVG). These mixtures, Q95Q/K and E138E/K, fully emerged in the viral pools by P-64 and P-80, respectively. The EVG EC50 at P-68 (1,024 nM EVG) was greater than 1,000 nM.
An independent EVG selection experiment, again using HIV-1IIIB, was performed but began at 0.1 nM EVG (Fig. 3B). An E92E/Q mixture in the IN coding region was first detected at P-30 (10 nM EVG) and was predominantly E92Q by P-38 (20 nM EVG). Additional IN substitutions, H51H/Y and S147S/G, emerged at P-60 (640 nM EVG), and an E157E/Q mixture emerged at P-70 (1,280 nM EVG); the viral pools at the terminal passage P-80 (1,280 nM EVG) had the IN sequence H51Y/E92Q/S147G/E157E/Q (Fig. 3B). The emergence of each of these mutations correlated with an increase in the EVG EC50 of the resulting viral pools (Fig. 3). Other than the N232D polymorphism, all of these mutations are located in the CCD of IN.
Phenotypic analysis of IN recombinant viruses. (i) EVG-selected mutations.
To characterize which mutations are responsible for EVG resistance, infectious HIV-1 clones containing single IN substitutions (H51Y, T66I, E92Q, Q95K, E138K, Q146P, S147G, or E157Q) that were observed to emerge under selection with EVG were generated (Fig. 3 and Table 3). Mutations were classified into two groups based on the level of resistance: mutations that conferred more than 10-fold reduced susceptibility compared to the wild type were defined as primary mutations, and mutations conferring less than 10-fold reduced susceptibility were defined as secondary mutations. T66I and E92Q substitutions conferred significantly reduced susceptibility to EVG (37- and 36-fold reduced, respectively, relative to the wild type), whereas the Q146P and S147G substitutions conferred more moderate reductions in EVG susceptibility (11-fold reduced), indicating that these four IN mutations are primary mutations involved in resistance to EVG. In contrast, H51Y, Q95K, and E157Q substitutions all conferred smaller reductions in EVG susceptibility (each less than 6.3-fold reduced compared to the wild type), suggesting that these substitutions are secondary resistance mutations. Interestingly, the E138K mutation alone conferred no reduction in susceptibility to either EVG or L-870,810. Thus, several distinct mechanisms of resistance may be represented by these different IN mutations.
TABLE 3.
Susceptibilities of HIV-1 IN recombinant molecular clonesa
| Molecular clone(s) | Mean EC50 (nM) ± SD (fold resistance compared to wild type)
|
||||
|---|---|---|---|---|---|
| AZT | EVG | L-870,810 | S-1360 | L-731,988 | |
| HIV-1WT | 32 | 1.1 | 5.8 | 1,239 | 736 |
| EVG mutation (expt 1)b | |||||
| T66Ic | 43 ± 11 (1.3) | 41 ± 14 (37) | 4.7 ± 2.9 (0.8) | 6,403 ± 2,349 (5.2) | 7,234 ± 1,210 (9.8) |
| Q95K | 34 ± 6 (1.1) | 2.9 ± 0.4 (2.6) | 18 ± 2 (3.1) | ND | ND |
| E138K | 33 ± 8 (1.0) | 1.1 ± 0.4 (1.0) | 3.9 ± 0.4 (0.7) | ND | ND |
| Q146P | 26 ± 2 (0.8) | 12 ± 3 (11) | 5.1 ± 0.4 (0.9) | ND | ND |
| S147Gd | 41 ± 5 (1.3) | 12 ± 5 (11) | 23 ± 6 (4.0) | ND | ND |
| T66I/Q146P | 22 ± 2 (0.7) | 131 ± 12 (119) | 18 ± 5 (3.1) | ND | ND |
| T66I/Q146P/S147G | 19 ± 5 (0.6) | 453 ± 62 (412) | 127 ± 37 (22) | ND | ND |
| T66I/Q95K/Q146P/S147G | 31 ± 12 (1.0) | >1,000 | 303 ± 76 (52) | ND | ND |
| T66I/Q95K/E138K/Q146P/S147G | 41 ± 7 (1.3) | >1,000 | 306 ± 76 (53) | >10,000 | >50,000 |
| EVG mutation (expt 2)b | |||||
| H51Y | 34 ± 8 (1.1) | 4.0 ± 0.6 (3.6) | 3.3 ± 0.7 (0.6) | ND | ND |
| E92Q | 32 ± 4 (1.0) | 40 ± 12 (36) | 63 ± 39 (11) | ND | ND |
| E157Q | 34 ± 8 (1.1) | 6.9 ± 1.4 (6.3) | 52 ± 20 (9.0) | ND | ND |
| E92Q/S147G | 39 ± 9 (1.2) | 392 ± 133 (356) | 587 ± 64 (101) | ND | ND |
| H51Y/E92Q/S147G | 54 ± 6 (1.7) | 769 ± 88 (699) | 374 ± 100 (64) | >10,000 | 22,175 ± 1,299 (30) |
| H51Y/E92Q/S147G/E157Q | 21 ± 2 (0.7) | >1,000 | 340 ± 26 (59) | >10,000 | 18,652 ± 4,575 (25) |
| L-870,810 mutation | |||||
| V72I | 17 ± 1 (0.5) | 4.3 ± 1.1 (3.9) | 9.1 ± 2.5 (1.6) | ND | ND |
| L74Mc | 20 ± 3 (0.6) | 3.3 ± 1.1 (3.0) | 4.4 ± 1.7 (0.8) | 1,500 ± 302 (1.2) | 4,471 ± 942 (6.1) |
| F121Y | 15 ± 1 (0.5) | 28 ± 11 (25) | 51 ± 23 (8.8) | ND | ND |
| T125K | 17 ± 3 (0.5) | 2.3 ± 1.1 (2.1) | 9.9 ± 3.7 (1.7) | ND | ND |
| V151I | 21 ± 4 (0.7) | 11 ± 3 (10) | 104 ± 29 (18) | ND | ND |
| G163R | 22 ± 7 (0.7) | 0.8 ± 0.2 (0.7) | 6.5 ± 2.6 (1.1) | ND | ND |
| F121Y/G163R | 36 ± 5 (1.1) | 60 ± 20 (55) | 219 ± 20 (38) | ND | ND |
| F121Y/T125K | 38 ± 12 (1.2) | 195 ± 73 (177) | 393 ± 82 (68) | ND | ND |
| V72I/F121Y/T125K | 33 ± 7 (1.0) | 143 ± 25 (130) | 886 ± 79 (153) | ND | ND |
| V72I/F121Y/T125K/V151I | 64 ± 9 (2.0) | >1,000 | >1,000 | >10,000 | >50,000 |
| DKA mutation | |||||
| T66I/L74M | 46 ± 11 (1.4) | 49 ± 5 (45) | 41 ± 10 (7.1) | >10,000 | 23,043 ± 4,886 (31) |
| T66I/S153Y | 26 ± 8 (0.8) | 285 ± 63 (259) | 29 ± 9 (5.0) | >10,000 | 8,478 ± 1,267 (12) |
Antiviral activity was determined using the MAGI assay. Data shown are means and standard deviations obtained from at least three independent experiments, and resistance (n-fold) of the EC50 of the IN recombinant molecular clone compared to that of parental HIV-1WT is shown in parentheses. ND, not determined.
EVG selection was performed in two independent experiments, and observed mutations are separately represented.
Also observed in the DKA selected mutation.
Observed in two independent EVG-selected experiments.
Multisubstituted clones observed during EVG selection experiments were also generated. HIV-1T66I/Q146P showed high-level resistance to EVG (119-fold reduced susceptibility) (Table 3). Combinations of S147G with T66I/Q146P or E92Q further enhanced resistance, 412- and 356-fold, respectively. The triple mutant HIV-1H51Y/E92Q/S147G showed high-level resistance to EVG (700-fold reduced susceptibility). Interestingly, the addition of the secondary mutation H51Y, which on its own reduced EVG susceptibility only 3.6-fold, substantially enhanced resistance relative to that observed for the double mutant HIV-1E92Q/S147G. HIV-1T66I/Q95K/Q146P/S147G, HIV-1T66I/Q95K/E138K/Q146P/S147G, and HIV-1H51Y/E92Q/S147G/E157Q mutants all showed high-level resistance to EVG, with EC50 values greater than 1,000 nM in all cases. These results indicate that the T66I and E92Q mutations provided the highest change (n-fold) in EVG susceptibility as individual resistance mutations and that the additional substitutions identified further enhance the level of resistance to EVG when combined with these primary mutations.
(ii) L-870,810-selected mutations.
Infectious HIV-1 clones containing mutations (V72I, F121Y, T125K, and V151I) previously shown to be associated with resistance to L-870,810 (23) and two mutations, L74M and G163R, observed in our selection using L-870,810 (data not shown) were generated. Among these variants, HIV-1F121Y and HIV-1V151I demonstrated reduced susceptibility to both L-870,810 and EVG (Table 3). V151I has been observed in some HIV-1 clinical isolates and may be an IN polymorphism (34). Moreover, the effect of V151I on susceptibility to L-870,810 has been controversial (23, 29). This discrepancy might arise from the viral strain or plasmid backbone used, so further experiments to clarify the effect of V151I on IN inhibitor susceptibility are needed. HIV-1F121Y/T125K showed significant resistance to both L-870,810 and EVG (68-fold and 177-fold reduced susceptibility, respectively). HIV-1V72I/F121Y/T125K/V151I showed high-level resistance to both IN inhibitors (EC50 greater than 1,000 nM).
(iii) DKA IN inhibitor-selected mutations.
Highlighting the potential for related mechanisms of IN inhibitor resistance and cross-resistance, the T66I mutation has also been observed to be selected by DKA IN inhibitors such as L-708,906 and S-1360. Additional mutations, L74M and S153Y, in combination with T66I were also observed to be selected by these DKA IN inhibitors (16, 17). L74M also emerged during L-870,810 selection in our studies (data not shown) but conferred no change in susceptibility to L-870,810 when present alone and only low-level resistance (3.0-fold) to EVG (Table 3). The combination of T66I and L74M conferred slightly higher resistance to EVG (45-fold) than did T66I alone but only moderate resistance to L-870,810 (7.1-fold). Another IN mutant, HIV-1T66I/S153Y, observed in L-708,906 selection experiments (24) showed high-level resistance to EVG (260-fold) but low-level resistance to L-870,810 (5.0-fold). These results suggest that the mechanism of EVG resistance may have some similarities to that of DKA IN inhibitors.
Taken together, these results suggest that a variety of IN mutations may be selected by EVG and other IN inhibitors. Most of the IN inhibitor resistance mutations are observed to cluster in the CCD of IN. The resulting mutations and their combinations have the capacity to confer various levels of resistance and potential cross-resistance to EVG and other IN inhibitors. Given their location in the CCD, many of these mutations may act via a common mechanism. The observed development of IN inhibitor resistance mutations resembles that seen for other antiretroviral drugs such as PIs; i.e., multiple mutations are introduced in a stepwise fashion and are required for high-level resistance to the selecting inhibitors (10, 50).
Strand transfer assay.
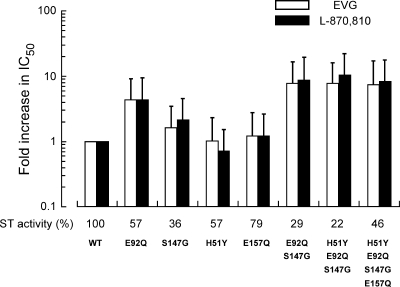
To further characterize the effect of EVG-selected resistance mutations on IN function, the effect of mutations on the enzymatic activity of recombinant IN was evaluated in an in vitro strand transfer assay (Fig. 4). IN enzymes carrying the individual mutations H51Y, S147G, and E157Q had reduced strand transfer activity relative to that of the wild type (57%, 36%, and 79% of wild-type levels, respectively). Strand transfer activities of E92Q, E92Q/S147G, and H51Y/E92Q/S147G IN enzymes decreased with the accumulation of mutations from 57% to 29 and 22% of the wild type, respectively. However, the introduction of E157Q to H51Y/E92Q/S147Q partially restored strand transfer activity to 46% of wild-type activity, suggesting that E157Q may play a role in compensating for the loss of strand transfer activity resulting from the emergence of EVG resistance mutations.
FIG. 4.
Effect of EVG-selected mutations on IN strand transfer activity and on the inhibition of strand transfer by IN inhibitors. The strand transfer activities of recombinant IN enzymes carrying EVG-selected mutations were determined using an oligonucleotide-based strand transfer assay. Strand transfer (ST) activity of IN mutants was compared to that of the wild type (WT); results are shown as percentages of wild-type activity. The effect of IN inhibitors on strand transfer was also determined for wild-type and mutant IN enzymes; results are expressed as the increase (n-fold) in IC50 values of inhibitors relative to those of the wild type.
The effect of EVG-selected mutations on the inhibition of strand transfer by EVG and L-870,810 was also determined (Fig. 4). Recombinant IN enzymes carrying the individual H51Y, S147G, and E157Q substitutions remained susceptible to both EVG and L-870,810 (0.7- to 2.1-fold reduced susceptibility). E92Q IN demonstrated only moderate resistance to both IN inhibitors in the strand transfer assay (4.3-fold reduced for both inhibitors). The combination of E92Q and S147G enhanced resistance to both EVG and L-870,810 (7.6- and 8.5-fold reduced susceptibility, respectively). However, unlike the IN recombinant viruses in the antiviral assay, neither the H51Y/E92Q/S147G nor the H51Y/E92Q/S147G/E157Q IN enzymes showed further enhancement of resistance in the strand transfer assay. This difference in results from the strand transfer assay versus those from the antiviral assay may reflect differences in the recombinant IN enzyme versus the viral IN enzyme in situ. Indeed, structure-activity relationship experiments described in a previous report (43) revealed that antiviral activity and in vitro enzyme inhibition were well correlated. Nevertheless, this biochemical analysis confirmed that the E92Q IN mutation confers significantly reduced susceptibility to EVG at the level of inhibition of strand transfer, consistent with its identification as a primary EVG resistance mutation in the virological analyses.
Replication kinetics of IN inhibitor-resistant variants.
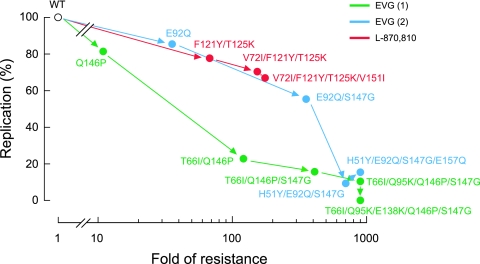
The effects of IN mutations on the replication kinetics of HIV-1 variants were assessed by comparing their levels of p24 production in culture supernatants to that of wild-type virus (Fig. 5). At day 5 postinfection, levels of p24 production by the HIV-1E92Q and HIV-1Q146P variants were 86% and 82% of HIV-1WT levels, respectively. These variants showed high-level (36-fold) or moderate (11-fold) resistance to EVG (Table 3), whereas the replication levels of both were similar to those of the wild type. However, the introduction of additional EVG resistance mutations further decreased p24 production, which is indicative of a decline in the levels of viral replication. In particular, HIV-1T66I/Q146P/S147G, HIV-1T66I/Q95K/Q146P/S147G, HIV-1T66I/Q95K/E138K/Q146P/S147G, HIV-1H51Y/E92Q/S147G/, and HIV-1H51Y/E92Q/S147G/E157Q all showed significantly reduced levels of p24 production (less than 20% of wild-type levels by day 5 in all cases). Thus, there was an inverse correlation between the levels of EVG resistance and the viral replication capacity; that is, as resistance to EVG increased, viral replication decreased. Interestingly, viral variants carrying L-870,810-selected mutations had more moderate reductions in replication capacity, even in the case of the HIV-1V72I/F121Y/T125K/V151I variant that had high-level resistance to both L-870,810 and EVG (68% of wild-type levels). These results indicate that mutations associated with resistance to IN inhibitors can have various effects on viral replication capacity. The reduced replication capacity of EVG-resistant variants was not rescued in the presence of the inhibitor (data not shown), as was observed previously for NFV-resistant variants in the presence of NFV (35). Thus, the reduced replication capacity of IN inhibitor-resistant variants may present a barrier to their emergence in vivo.
FIG. 5.
Replication kinetics of EVG- and L-870,810-resistant viral variants. The replication kinetics of wild-type and IN inhibitor-resistant viral variants were determined by p24 ELISA. The relationship of replication capacity and change (n-fold) in susceptibility (shown in Table 3) is depicted. Variants are plotted according to the observed order of their emergence during selection experiments in vitro. Replication kinetics of EVG-selected mutants derived from the two independent selection experiments (shown in Fig. 3) are plotted in different colors. WT, wild type.
Antiviral effect of IN inhibitors on retroviruses.
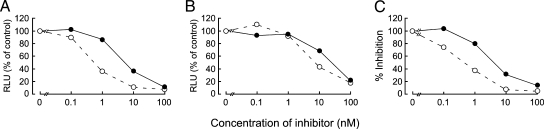
The antiviral activity of EVG against other retroviruses, including MLV and SIV, was assessed. EVG and L-870,810 inhibited the integration of the HIV-based vector used as a positive control for the luciferase assay (EC50 values of 0.8 and 5.0 nM, respectively), as observed in the MAGI assay with HIV-1IIIB (Fig. 6). EVG and L-870,810 suppressed the replication of MLV infection (EC50 values of 5.8 and 22 nM, respectively) as well as that of the primate retrovirus SIV (0.5 and 3.2 nM, respectively), indicating that IN inhibitors have antiviral activity against a broad range of retroviruses.
FIG. 6.
Effect of IN inhibitors on retroviruses. Antiviral activities of EVG (open circles with dashed lines) and L-870,810 (closed circles with solid lines) against HIV-based (A) or MLV-based (B) vectors harboring the luciferase gene were determined by measuring luciferase activity at 48 h posttransduction. Results are expressed as percentages of relative luciferase units (RLU) compared to those of the no-inhibitor control. (C) Anti-SIV activity was determined using the MAGI assay. These results shown are one representative assay from three independent experiments.
DISCUSSION
The data described here show that EVG inhibits HIV replication by specifically blocking the strand transfer reaction mediated by IN, as demonstrated by the intracellular accumulation of 2-LTR DNA products, a signature of nonproductive integration. Furthermore, EVG directly blocked the production of strand transfer products in an in vitro strand transfer assay. Confirming that EVG is a bona fide IN inhibitor, we selected EVG-resistant viral variants in vitro and demonstrated that the resulting viral variants had acquired multiple mutations in the IN coding region and had simultaneously acquired reduced phenotypic susceptibility to EVG. HIV-1 molecular clones carrying the EVG-selected IN mutations had an EVG-resistant phenotype and in many cases also had reduced susceptibility to another IN inhibitor, L-870,810. These data provide formal proof that the observed IN mutations are indeed EVG resistance mutations and that EVG is an IN inhibitor.
Among the IN mutations observed to be selected by EVG, two mutations, T66I and E92Q, appeared to provide the major contribution to EVG resistance. Both of these individual mutations resulted in >30-fold reduced susceptibility to EVG. The T66I mutation conferred cross-resistance to S-1360 and L-731,988 (Table 3) and was also previously observed in an independent EVG selection by Jones et al. (26). The E92Q mutation, when introduced into a recombinant IN enzyme, also reduced the susceptibility of the resulting mutant IN enzyme to EVG, as measured by the reduced EVG inhibition of the in vitro strand transfer assay (Fig. 4). The other IN mutations identified, including H51Y, Q95K, E138K, Q146P, S147G, and E157Q, individually resulted in lower changes (n-fold) in EVG susceptibility (1.0- to 11.0-fold) but, when added to either the T66I or the E92Q mutation, further increased resistance to EVG to various degrees relative to either mutation alone. Interestingly, the accumulation of these EVG-selected IN mutations resulted in a significant attenuation of viral replication kinetics. Thus, the emergence of resistance to IN inhibitors may be associated with reductions in viral fitness, which may provide a barrier to the emergence of these mutations in vivo or be associated with lower viral loads if they do emerge.
Of the three HIV enzymes PR, RT, and IN, the structure and mechanism of IN are the least well understood, and despite extensive efforts, the structure of the complete IN enzyme remains to be determined. Only partial two-domain crystal structures of the IN apoenzyme are available, and no structure showing full-length IN bound to its viral cDNA substrate has been published. During integration in vivo, IN functions in the preintegration complex, which also includes RT and the viral DNA (2, 3). Some limited evidence suggests that RT interacts with the active site of IN (39). IN has also been proposed to function with several cellular factors including IN interactor 1 (Ini1) (27) and lens-epithelium-derived growth factor (LEDGF/p75) (7). In the context of these associated cellular factors, IN may retain a different conformation compared to that of the recombinant enzyme alone. This may be one of the reasons that only moderate EVG resistance was observed in the oligonucleotide-based strand transfer assay compared to a cell-based antiviral assay.
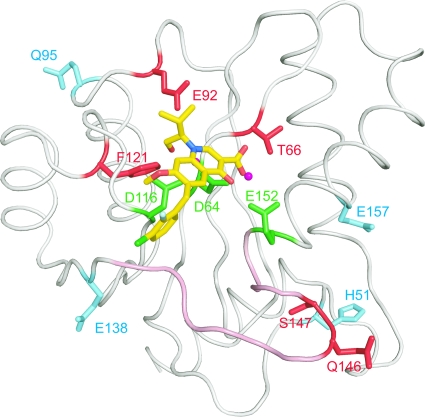
Alignment of several IN CCD structures deposited in the Protein Data Bank indicates that there are two regions with poorly defined or disordered structures, including residues 47 to 56 and 140 to 152 (Fig. 7; see Fig. S1 in the supplemental material). Of these two disordered regions, residues 140 to 152 have been implicated as a flexible loop involved in viral cDNA binding (20, 21, 53). Although the precise structural details are unknown, the flexible loop has been proposed to adopt different conformations in the presence or absence of the viral cDNA (12). Notably, several of the EVG-selected mutations that we observed are located on or adjacent to this proposed flexible loop, including E138K, Q146P, and S147G. The flexible loop is important for the catalytic activity of IN (21, 32), and as shown in Fig. 4, the introduction of mutations in these residues, especially S147G, drastically reduced the catalytic activity of IN. Previously published data also demonstrated that another mutation at codon 147 (S147I) resulted in HIV-1 that was highly replication defective, including effects on viral DNA synthesis (47). Indeed, S147 is highly conserved among various retroviruses (see Fig. S2 in the supplemental material), highlighting the importance of the loop for IN function. It is possible that IN inhibitor resistance mutants may have additional pleiotropic effects on processes in viral replication other than integration; in particular, RT and IN were previously suggested to interact functionally (25).
FIG. 7.
Location of IN mutations associated with resistance to EVG. EVG in complex with the HIV-1 IN CCD is shown along with the catalytic triad residues (D64, D116, and E152) (green) and a magnesium ion (magenta). Amino acid residues conferring resistance to EVG as primary mutations (T66, E92, F121, Q146, and S147) or as secondary mutations (H51, Q95, E138, and E157) are shown in red and cyan, respectively. The flexible loop (residues 140 to 152) is shown in pink.
Recently, an in silico docking simulation of HIV IN with several IN inhibitors including EVG was reported (44). Notably, that author showed that in the best-fit model for EVG docked to IN, the isobutyl substituent on the quionolone moiety of EVG orients directly towards IN residue E92. Interestingly, the hydroxyl component of the isobutyl on the quinolone replaces a water molecule that is coordinated by residue E92 between the two catalytic residues D64 and E152. This docking structure may provide insight into the mechanism of IN inhibition by EVG and provides a starting point for understanding the mechanism of EVG resistance mediated by the E92Q substitution. However, it is uncertain whether this docking simulation represents the precise binding mode of EVG with IN in vivo. Therefore, to accurately assess the binding mode of IN inhibitors with IN, available structural data need to be supplemented by a variety of other approaches. In this study, a virological approach and an enzymatic approach were integrated to characterize the mechanism of action, antiviral activity, and resistance profile of EVG in vitro.
As shown in Fig. 7, primary EVG resistance mutations are located around the catalytic triad of the CCD of IN and are surrounded by the secondary mutations. Among the residues affected by primary mutations, E92 and F121 are located close to EVG on the model and might interact with the IN inhibitor. However, the mechanism by which these mutations interact with the IN inhibitor or with the viral cDNA to mediate resistance is currently unclear. Recently, clinical isolate data from patients experiencing virologic failure in ongoing phase III studies of another IN inhibitor, raltegravir, were reported; E92Q was among the mutations noted to develop in these raltegravir failure patients, usually in combination with another IN mutation, N155H (11, 48). These preliminary clinical data and the data presented here with L-870,810, indicate that the E92Q mutation may be able to mediate resistance and potential cross-resistance to multiple IN inhibitors including EVG and raltegravir. Consistent with the data described here, site-directed mutant HIV carrying the E92Q mutation has been confirmed to show resistance to EVG and to have low-level (approximately sixfold) reduced susceptibility to raltegravir (26).
Several of the IN residues affected by primary mutations observed in EVG-selected variants including T66, E92, and S147 are absolutely conserved among the retroviruses tested (HIV-1, HIV-2, SIV, and MLV) and in retroviruses from multiple mammalian species (see Fig. S2 in the supplemental material). The significant conservation of mammalian retroviral IN CCDs at both the level of sequence homology and structure of the active site was demonstrated by the ability of EVG to inhibit HIV, SIV, and MLV IN activity. This suggests that EVG, and probably other IN inhibitors, binds to a conformationally conserved region of all retroviral INs; the binding of EVG and other IN inhibitors to IN is also likely to involve the catalytic magnesium ion. Taken together, these results suggest that several distinct mechanisms may contribute to IN inhibitor resistance, including conformational changes in the structure of IN that affect the binding of the IN inhibitor, charge effects, steric hindrance, loss of stabilizing binding interactions, or, possibly, alterations in magnesium binding.
A similar reduction in viral replication capacity as a result of drug resistance mutations was previously reported for NRTI resistance mutations (K65R, L74V, and M184V) (45, 55) and for PI resistance mutations (D30N) (49). Mutations that act to compensate for some of the loss of viral replication resulting from drug resistance, for example, GAG processing mutants, have also been described (18, 36, 52). At least one of the EVG secondary mutations, E157Q, may have an analogous role, as it partially restored strand transfer activity that was attenuated by other EVG-selected mutations and also further enhanced resistance to EVG (Fig. 4). Some secondary IN mutations might act to compensate for the altered conformation of IN resulting from the structural effects of primary resistance mutations. The E138K mutation may be such an example, as on its own, it showed no effect on susceptibility to either EVG or L-870,810. The clinical implications of the reduction in fitness resulting from the selection of EVG-resistant mutations are not yet understood.
In conclusion, EVG is a potent inhibitor of the HIV IN enzyme that acts by blocking the strand transfer reaction and is effective not only against HIV but also against other retroviruses. Moreover, the emergence of viral variants that were highly resistant to EVG was associated with significant reductions in viral replication in vitro. These results indicate that EVG should be highly effective for the treatment of HIV-1-infected patients, including those who have had virologic failure of their highly active antiretroviral therapy due to the emergence of HIV-1 drug resistance to approved antiretroviral drugs.
Supplementary Material
Acknowledgments
We thank Shinjiro Hino for technical advice and Mieko Ikeuchi for technical assistance. We appreciate Damian McColl for critically reading and commenting on the manuscript.
This work was supported in part by a grant for the Promotion of AIDS Research from the Ministry of Health and Welfare of Japan (E.K. and M.M.), a grant for Research for Health Science Focusing on Drug Innovation from the Japan Health Science Foundation (E.K. and M.M.), and a grant from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (E.K.). K.S. is supported by the 21st Century COE Program of the Ministry of Education, Culture, Sports, Science, and Technology.
Footnotes
Published ahead of print on 31 October 2007.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Asante-Appiah, E., and A. M. Skalka. 1997. Molecular mechanisms in retrovirus DNA integration. Antivir. Res. 36139-156. [DOI] [PubMed] [Google Scholar]
- 2.Brown, P. O., B. Bowerman, H. E. Varmus, and J. M. Bishop. 1987. Correct integration of retroviral DNA in vitro. Cell 49347-356. [DOI] [PubMed] [Google Scholar]
- 3.Bukrinsky, M. I., N. Sharova, T. L. McDonald, T. Pushkarskaya, W. G. Tarpley, and M. Stevenson. 1993. Association of integrase, matrix, and reverse transcriptase antigens of human immunodeficiency virus type 1 with viral nucleic acids following acute infection. Proc. Natl. Acad. Sci. USA 906125-6129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Butler, S. L., M. S. Hansen, and F. D. Bushman. 2001. A quantitative assay for HIV DNA integration in vivo. Nat. Med. 7631-634. [DOI] [PubMed] [Google Scholar]
- 5.Chackerian, B., E. M. Long, P. A. Luciw, and J. Overbaugh. 1997. Human immunodeficiency virus type 1 coreceptors participate in postentry stages in the virus replication cycle and function in simian immunodeficiency virus infection. J. Virol. 713932-3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chakrabarti, L. A., K. J. Metzner, T. Ivanovic, H. Cheng, J. Louis-Virelizier, R. I. Connor, and C. Cheng-Mayer. 2003. A truncated form of Nef selected during pathogenic reversion of simian immunodeficiency virus SIVmac239Δnef increases viral replication. J. Virol. 771245-1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cherepanov, P., G. Maertens, P. Proost, B. Devreese, J. Van Beeumen, Y. Engelborghs, E. De Clercq, and Z. Debyser. 2003. HIV-1 integrase forms stable tetramers and associates with LEDGF/p75 protein in human cells. J. Biol. Chem. 278372-381. [DOI] [PubMed] [Google Scholar]
- 8.Chow, S. A. 1997. In vitro assays for activities of retroviral integrase. Methods 12306-317. [DOI] [PubMed] [Google Scholar]
- 9.Clavel, F., and A. J. Hance. 2004. HIV drug resistance. N. Engl. J. Med. 3501023-1035. [DOI] [PubMed] [Google Scholar]
- 10.Condra, J. H., W. A. Schleif, O. M. Blahy, L. J. Gabryelski, D. J. Graham, J. C. Quintero, A. Rhodes, H. L. Robbins, E. Roth, M. Shivaprakash, D. Titus, T. Yang, H. Tepplert, K. E. Squires, P. J. Deutsch, and E. A. Emini. 1995. In vivo emergence of HIV-1 variants resistant to multiple protease inhibitors. Nature 374569-571. [DOI] [PubMed] [Google Scholar]
- 11.Cooper, D., J. Gatell, J. Rockstroh, C. Katlama, P. Yeni, A. Lazzarin, J. Chen, R. Isaacs, H. Teppler, and B. Nguyen for the BENCHMRK-1 Study Group. 2007. Abstr. 14th Conf. Retrovir. Opportun. Infect., abstr. 105aLB.
- 12.De Luca, L., G. Vistoli, A. Pedretti, M. L. Barreca, and A. Chimirri. 2005. Molecular dynamics studies of the full-length integrase-DNA complex. Biochem. Biophys. Res. Commun. 3361010-1016. [DOI] [PubMed] [Google Scholar]
- 13.Dyda, F., A. B. Hickman, T. M. Jenkins, A. Engelman, R. Craigie, and D. R. Davies. 1994. Crystal structure of the catalytic domain of HIV-1 integrase: similarity to other polynucleotidyl transferases. Science 2661981-1986. [DOI] [PubMed] [Google Scholar]
- 14.Engelman, A., K. Mizuuchi, and R. Craigie. 1991. HIV-1 DNA integration: mechanism of viral DNA cleavage and DNA strand transfer. Cell 671211-1221. [DOI] [PubMed] [Google Scholar]
- 15.Fan, J., E. Kodama, Y. Koh, M. Nakao, and M. Matsuoka. 2005. Halogenated thymidine analogues restore the expression of silenced genes without demethylation. Cancer Res. 656927-6933. [DOI] [PubMed] [Google Scholar]
- 16.Fikkert, V., A. Hombrouck, B. Van Remoortel, M. De Maeyer, C. Pannecouque, E. De Clercq, Z. Debyser, and M. Witvrouw. 2004. Multiple mutations in human immunodeficiency virus-1 integrase confer resistance to the clinical trial drug S-1360. AIDS 182019-2028. [DOI] [PubMed] [Google Scholar]
- 17.Fikkert, V., B. Van Maele, J. Vercammen, A. Hantson, B. Van Remoortel, M. Michiels, C. Gurnari, C. Pannecouque, M. De Maeyer, Y. Engelborghs, E. De Clercq, Z. Debyser, and M. Witvrouw. 2003. Development of resistance against diketo derivatives of human immunodeficiency virus type 1 by progressive accumulation of integrase mutations. J. Virol. 7711459-11470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gatanaga, H., Y. Suzuki, H. Tsang, K. Yoshimura, M. F. Kavlick, K. Nagashima, R. J. Gorelick, S. Mardy, C. Tang, M. F. Summers, and H. Mitsuya. 2002. Amino acid substitutions in Gag protein at non-cleavage sites are indispensable for the development of a high multitude of HIV-1 resistance against protease inhibitors. J. Biol. Chem. 2775952-5961. [DOI] [PubMed] [Google Scholar]
- 19.Goldgur, Y., R. Craigie, G. H. Cohen, T. Fujiwara, T. Yoshinaga, T. Fujishita, H. Sugimoto, T. Endo, H. Murai, and D. R. Davies. 1999. Structure of the HIV-1 integrase catalytic domain complexed with an inhibitor: a platform for antiviral drug design. Proc. Natl. Acad. Sci. USA 9613040-13043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goldgur, Y., F. Dyda, A. B. Hickman, T. M. Jenkins, R. Craigie, and D. R. Davies. 1998. Three new structures of the core domain of HIV-1 integrase: an active site that binds magnesium. Proc. Natl. Acad. Sci. USA 959150-9154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Greenwald, J., V. Le, S. L. Butler, F. D. Bushman, and S. Choe. 1999. The mobility of an HIV-1 integrase active site loop is correlated with catalytic activity. Biochemistry 388892-8898. [DOI] [PubMed] [Google Scholar]
- 22.Grinsztejn, B., B. Y. Nguyen, C. Katlama, J. M. Gatell, A. Lazzarin, D. Vittecoq, C. J. Gonzalez, J. Chen, C. M. Harvey, and R. D. Isaacs. 2007. Safety and efficacy of the HIV-1 integrase inhibitor raltegravir (MK-0518) in treatment-experienced patients with multidrug-resistant virus: a phase II randomised controlled trial. Lancet 3691261-1269. [DOI] [PubMed] [Google Scholar]
- 23.Hazuda, D. J., N. J. Anthony, R. P. Gomez, S. M. Jolly, J. S. Wai, L. Zhuang, T. E. Fisher, M. Embrey, J. P. Guare, Jr., M. S. Egbertson, J. P. Vacca, J. R. Huff, P. J. Felock, M. V. Witmer, K. A. Stillmock, R. Danovich, J. Grobler, M. D. Miller, A. S. Espeseth, L. Jin, I. W. Chen, J. H. Lin, K. Kassahun, J. D. Ellis, B. K. Wong, W. Xu, P. G. Pearson, W. A. Schleif, R. Cortese, E. Emini, V. Summa, M. K. Holloway, and S. D. Young. 2004. A naphthyridine carboxamide provides evidence for discordant resistance between mechanistically identical inhibitors of HIV-1 integrase. Proc. Natl. Acad. Sci. USA 10111233-11238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hazuda, D. J., P. Felock, M. Witmer, A. Wolfe, K. Stillmock, J. A. Grobler, A. Espeseth, L. Gabryelski, W. Schleif, C. Blau, and M. D. Miller. 2000. Inhibitors of strand transfer that prevent integration and inhibit HIV-1 replication in cells. Science 287646-650. [DOI] [PubMed] [Google Scholar]
- 25.Hehl, E. A., P. Joshi, G. V. Kalpana, and V. R. Prasad. 2004. Interaction between human immunodeficiency virus type 1 reverse transcriptase and integrase proteins. J. Virol. 785056-5067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jones, G., R. Ledford, F. Yu, M. Miller, M. Tsiang, and D. McColl. 2007. Abstr. 14th Conf. Retrovir. Opportun. Infect., abstr. 627.
- 27.Kalpana, G. V., S. Marmon, W. Wang, G. R. Crabtree, and S. P. Goff. 1994. Binding and stimulation of HIV-1 integrase by a human homolog of yeast transcription factor SNF5. Science 2662002-2006. [DOI] [PubMed] [Google Scholar]
- 28.Kaufmann, G. R., and D. A. Cooper. 2000. Antiretroviral therapy of HIV-1 infection: established treatment strategies and new therapeutic options. Curr. Opin. Microbiol. 3508-514. [DOI] [PubMed] [Google Scholar]
- 29.Kehlenbeck, S., U. Betz, A. Birkmann, B. Fast, A. H. Goller, K. Henninger, T. Lowinger, D. Marrero, A. Paessens, D. Paulsen, V. Pevzner, R. Schohe-Loop, H. Tsujishita, R. Welker, J. Kreuter, H. Rubsamen-Waigmann, and F. Dittmer. 2006. Dihydroxythiophenes are novel potent inhibitors of human immunodeficiency virus integrase with a diketo acid-like pharmacophore. J. Virol. 806883-6894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kodama, E. I., S. Kohgo, K. Kitano, H. Machida, H. Gatanaga, S. Shigeta, M. Matsuoka, H. Ohrui, and H. Mitsuya. 2001. 4′-Ethynyl nucleoside analogs: potent inhibitors of multidrug-resistant human immunodeficiency virus variants in vitro. Antimicrob. Agents Chemother. 451539-1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.LaFemina, R. L., C. L. Schneider, H. L. Robbins, P. L. Callahan, K. LeGrow, E. Roth, W. A. Schleif, and E. A. Emini. 1992. Requirement of active human immunodeficiency virus type 1 integrase enzyme for productive infection of human T-lymphoid cells. J. Virol. 667414-7419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee, M. C., J. Deng, J. M. Briggs, and Y. Duan. 2005. Large-scale conformational dynamics of the HIV-1 integrase core domain and its catalytic loop mutants. Biophys. J. 883133-3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Little, S., G. Drusano, R. Schooley, D. Haas, P. Kumar, S. Hammer, D. McMahon, K. Squires, R. Asfour, D. Richman, J. Chen, A. Saah, R. Leavitt, D. Hazuda, B. Y. Nguyen, and the Protocol 004 Study Team. 2005. Abstr. 12th Conf. Retrovir. Opportun. Infect., abstr. 161.
- 34.Los Alamos National Laboratory Theoretical Biology and Biophysics Group T-10. 2001. HIV sequence compendium, 2001. Theoretical Biology and Biophysics Group, Los Alamos National Laboratory, Los Alamos, NM.
- 35.Matsuoka-Aizawa, S., H. Sato, A. Hachiya, K. Tsuchiya, Y. Takebe, H. Gatanaga, S. Kimura, and S. Oka. 2003. Isolation and molecular characterization of a nelfinavir (NFV)-resistant human immunodeficiency virus type 1 that exhibits NFV-dependent enhancement of replication. J. Virol. 77318-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Myint, L., M. Matsuda, Z. Matsuda, Y. Yokomaku, T. Chiba, A. Okano, K. Yamada, and W. Sugiura. 2004. Gag non-cleavage site mutations contribute to full recovery of viral fitness in protease inhibitor-resistant human immunodeficiency virus type 1. Antimicrob. Agents Chemother. 48444-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nameki, D., E. Kodama, M. Ikeuchi, N. Mabuchi, A. Otaka, H. Tamamura, M. Ohno, N. Fujii, and M. Matsuoka. 2005. Mutations conferring resistance to human immunodeficiency virus type 1 fusion inhibitors are restricted by gp41 and Rev-responsive element functions. J. Virol. 79764-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Neuveut, C., and K. T. Jeang. 1996. Recombinant human immunodeficiency virus type 1 genomes with tat unconstrained by overlapping reading frames reveal residues in Tat important for replication in tissue culture. J. Virol. 705572-5581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oz Gleenberg, I., O. Avidan, Y. Goldgur, A. Herschhorn, and A. Hizi. 2005. Peptides derived from the reverse transcriptase of human immunodeficiency virus type 1 as novel inhibitors of the viral integrase. J. Biol. Chem. 28021987-21996. [DOI] [PubMed] [Google Scholar]
- 40.Palella, F. J., Jr., K. M. Delaney, A. C. Moorman, M. O. Loveless, J. Fuhrer, G. A. Satten, D. J. Aschman, S. D. Holmberg, et al. 1998. Declining morbidity and mortality among patients with advanced human immunodeficiency virus infection. N. Engl. J. Med. 338853-860. [DOI] [PubMed] [Google Scholar]
- 41.Pommier, Y., A. A. Johnson, and C. Marchand. 2005. Integrase inhibitors to treat HIV/AIDS. Nat. Rev. Drug Discov. 4236-248. [DOI] [PubMed] [Google Scholar]
- 42.Reinke, R., D. J. Lee, and W. E. Robinson, Jr. 2002. Inhibition of human immunodeficiency virus type 1 isolates by the integrase inhibitor L-731,988, a diketo acid. Antimicrob. Agents Chemother. 463301-3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sato, M., T. Motomura, H. Aramaki, T. Matsuda, M. Yamashita, Y. Ito, H. Kawakami, Y. Matsuzaki, W. Watanabe, K. Yamataka, S. Ikeda, E. Kodama, M. Matsuoka, and H. Shinkai. 2006. Novel HIV-1 integrase inhibitors derived from quinolone antibiotics. J. Med. Chem. 491506-1508. [DOI] [PubMed] [Google Scholar]
- 44.Savarino, A. 2007. In-silico docking of HIV-1 integrase inhibitors reveals a novel drug type acting on an enzyme/DNA reaction intermediate. Retrovirology 421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sharma, P. L., and C. S. Crumpacker. 1997. Attenuated replication of human immunodeficiency virus type 1 with a didanosine-selected reverse transcriptase mutation. J. Virol. 718846-8851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shibata, R., M. Kawamura, H. Sakai, M. Hayami, A. Ishimoto, and A. Adachi. 1991. Generation of a chimeric human and simian immunodeficiency virus infectious to monkey peripheral blood mononuclear cells. J. Virol. 653514-3520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shin, C. G., B. Taddeo, W. A. Haseltine, and C. M. Farnet. 1994. Genetic analysis of the human immunodeficiency virus type 1 integrase protein. J. Virol. 681633-1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Steigbigel, R., P. Kumar, J. Eron, M. Schechter, M. Markowitz, M. Loufty, J. Zhao, R. Isaacs, B. Nguyen, H. Teppler, and the BENCHMRK-2 Study Group. 2007. Abstr. 14th Conf. Retrovir. Opportun. Infect., abstr. 105bLB.
- 49.Sugiura, W., Z. Matsuda, Y. Yokomaku, K. Hertogs, B. Larder, T. Oishi, A. Okano, T. Shiino, M. Tatsumi, M. Matsuda, H. Abumi, N. Takata, S. Shirahata, K. Yamada, H. Yoshikura, and Y. Nagai. 2002. Interference between D30N and L90M in selection and development of protease inhibitor-resistant human immunodeficiency virus type 1. Antimicrob. Agents Chemother. 46708-715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tisdale, M., R. E. Myers, B. Maschera, N. R. Parry, N. M. Oliver, and E. D. Blair. 1995. Cross-resistance analysis of human immunodeficiency virus type 1 variants individually selected for resistance to five different protease inhibitors. Antimicrob. Agents Chemother. 391704-1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Toossi, Z., H. Mayanja-Kizza, J. Baseke, P. Peters, M. Wu, A. Abraha, H. Aung, A. Okwera, C. Hirsch, and E. Arts. 2005. Inhibition of human immunodeficiency virus-1 (HIV-1) by beta-chemokine analogues in mononuclear cells from HIV-1-infected patients with active tuberculosis. Clin. Exp. Immunol. 142327-332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Verheyen, J., E. Litau, T. Sing, M. Daumer, M. Balduin, M. Oette, G. Fatkenheuer, J. K. Rockstroh, U. Schuldenzucker, D. Hoffmann, H. Pfister, and R. Kaiser. 2006. Compensatory mutations at the HIV cleavage sites p7/p1 and p1/p6-gag in therapy-naive and therapy-experienced patients. Antivir. Ther. 11879-887. [PubMed] [Google Scholar]
- 53.Wang, J. Y., H. Ling, W. Yang, and R. Craigie. 2001. Structure of a two-domain fragment of HIV-1 integrase: implications for domain organization in the intact protein. EMBO J. 207333-7343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Weiner, M. P., G. L. Costa, W. Schoettlin, J. Cline, E. Mathur, and J. C. Bauer. 1994. Site-directed mutagenesis of double-stranded DNA by the polymerase chain reaction. Gene 151119-123. [DOI] [PubMed] [Google Scholar]
- 55.White, K. L., N. A. Margot, T. Wrin, C. J. Petropoulos, M. D. Miller, and L. K. Naeger. 2002. Molecular mechanisms of resistance to human immunodeficiency virus type 1 with reverse transcriptase mutations K65R and K65R+M184V and their effects on enzyme function and viral replication capacity. Antimicrob. Agents Chemother. 463437-3446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zolopa, A., M. Mullen, D. Berger, P. Ruane, T. Hawkins, L. Zhong, S. Chuck, J. Enejosa, B. Kearney, and A. Cheng. 2007. Abstr. 14th Confer. Retrovir. Opportun. Infect., abstr. 143LB.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.