Abstract
Modified vaccinia virus Ankara (MVA) is a highly attenuated vaccinia virus that is under consideration as an alternative to the conventional smallpox vaccine Dryvax. MVA was attenuated by extensive passage of vaccinia virus Ankara in chicken embryo fibroblasts. Several immunomodulatory genes and genes that influence host range are deleted or mutated, and replication is aborted in the late stage of infection in most nonavian cells. The effect of these mutations on immunogenicity is not well understood. Since the structural genes appear to be intact in MVA, it is hypothesized that critical targets for antibody neutralization have been retained. To test this, we probed microarrays of the Western Reserve (WR) proteome with sera from humans and macaques after MVA and Dryvax vaccination. As most protein sequences of MVA are 97 to 99% identical to those of other vaccinia virus strains, extensive binding cross-reactivity is expected, except for those deleted or truncated. Despite different hosts and immunization regimens, the MVA and Dryvax antibody profiles were broadly similar, with antibodies against membrane and core proteins being the best conserved. The responses to nonstructural proteins were less well conserved, although these are not expected to influence virus neutralization. The broadest antibody response was obtained for hyperimmune rabbits with WR, which is pathogenic in rabbits. These data indicate that, despite the mutations and deletions in MVA, its overall immunogenicity is broadly comparable to that of Dryvax, particularly at the level of antibodies to membrane proteins. The work supports other information suggesting that MVA may be a useful alternative to Dryvax.
The eradiation of smallpox by use of vaccinia virus was one of the major accomplishments of vaccination. However, the potential threat of smallpox (variola virus) or monkeypox viruses being used as a biological weapon may again require mass vaccination of the general public, which is largely vaccinia virus naïve. Many of the laboratory and vaccine strains available today are derived from the prototype vaccinia virus strain deposited at the New York City Board of Health in 1874 and include the Dryvax (Wyeth) strain, which was widely used in the Americas and West Africa during the smallpox eradication campaign. The production of Dryvax was discontinued in 1982, and current stocks are over 25 years old. Production methods in use then (i.e., propagation on calf skin) are less acceptable today, owing to the potential for contamination with adventitious agents. Moreover the vaccine is associated with a significant risk of adverse reactions. For example, data collected during the eradication campaign revealed the risk of complications to be 188 per million vaccinations, with death occurring at a rate of 1 to 5 per million (15). Generalized vaccinia was the most commonly observed side effect, with more-serious reactions (eczema vaccinatum, progressive vaccinia, and neurological/cardiac complications) responsible for 4 to 7% of all adverse reactions. Conventional nonattenuated vaccines are now considered unsuitable for a significant proportion of the population, including those individuals and their families that are immunocompromised and those individuals who have atopic dermatitis (eczema) or other skin conditions or are pregnant. There is therefore considerable interest in developing safer alternatives to Dryvax which are equally immunogenic but lack the pathogenicity.
The highly attenuated vaccine strain modified vaccinia virus Ankara (MVA) is under consideration as an alternative to Dryvax. MVA was developed towards the end of the eradication campaign and so has not been evaluated in areas of smallpox endemicity. Since it is no longer possible to evaluate the efficacy of new-generation smallpox vaccines in humans, estimations are being made from animal models using related orthopoxviruses. The MVA prototype was developed by Anton Mayr in Germany through a process of 516 serial passages of the chorioallantois vaccinia virus Ankara strain of the vaccinia virus on chicken embryo fibroblasts (CEF) (18). As a result of adapting to avian cells in vitro, several genes required for immune escape and host range were mutated or deleted (six regions totaling ∼31 kb) near the termini of the genome (3, 22). This causes a block in MVA morphogenesis in most nonavian cells, resulting in reduced cytopathic effect or plaque formation (5) and causing replication to be aborted at the late stage of infection (5, 7). The consequence is severe attenuation of MVA in mammalian hosts in vivo. Despite these gene mutations and deletions, MVA has retained its ability to protect animals against orthopoxvirus challenge nearly as effectively as nonattenuated strains (4, 11, 13, 20, 21, 26, 27, 32, 37). Moreover, the immunogenicity of MVA is thought to be equivalent to that of conventional smallpox vaccines (13, 21, 27). MVA also displays reduced virulence in animals (1, 31, 37), and clinical trials in West Germany in the 1970s demonstrated it has an excellent safety profile in humans (19). MVA is therefore considered more suitable for immunocompromised individuals, children, or those with skin conditions such as atopic dermatitis, for whom conventional vaccines are contraindicated. MVA is also under consideration as a delivery vehicle for other vaccines (6, 23, 33, 34) and for immunization prior to Dryvax administration to reduce its reactogenicity (13, 25). Molecular studies have shown that the structural genes are intact in MVA (3, 22), suggesting that critical targets for antibody-mediated neutralization have been retained. However, the effect of the mutations and deletions in MVA on the antibody response is not well understood. To address this, we have used vaccinia virus proteome microarrays (9) to profile antibody specificities in sera from rabbits, macaques, and humans inoculated with MVA and compared these with profiles induced by the conventional vaccine Dryvax in humans and macaques and the pathogenic strain WR in rabbits.
MATERIALS AND METHODS
Viruses.
MVA is a clonal, host-restricted vaccinia virus that was developed by >500 serial passages of the Ankara strain of vaccinia virus on CEF. Two MVA preparations were used in this study. Each was derived by plaque purification of seed stocks from A. Mayr (University of Munich) and propagated in CEF. MVA-BN (Imvamune from Bavarian Nordic A/S, Kvistgård, Denmark) (36) was used for clinical studies, and sucrose gradient-purified MVA 1974/NIH clone 1 (37) was used to immunize macaques and rabbits. Dryvax (Wyeth) was derived from the New York City Board of Health (NYCBOH) strain and is a replicative, nonclonal heterogeneous population of vaccinia virus. Lyophilized Dryvax was obtained from the CDC and reconstituted according to the manufacturer's instructions. Vaccinia virus strain Western Reserve (WR) (ATCC VR-1354) is a clonal and fully replicative laboratory strain derived from Wyeth NYCBOH by repeated passage on cells including mouse brain and cultured cells. WR is neurotropic in mice and more virulent than the Wyeth strain. Stocks of WR were prepared in serum-free medium in rabbit kidney fibroblasts and purified by sedimentation through a sucrose cushion (14).
Sera.
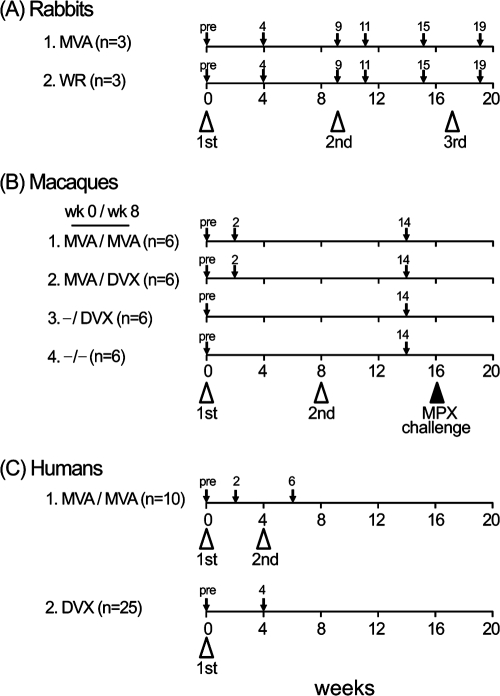
Hyperimmune rabbit sera (Fig. 1A) were generated in two groups of three animals inoculated with MVA and WR by the intramuscular (i.m.) and intradermal routes, respectively, followed by intravenous boosting. New Zealand White rabbits (Harlan) (13 weeks old and weighing between 2.75 and 3.0 kg) were inoculated with 108 PFU of vaccinia virus strain WR intradermally at each of four sites on the shaved back (100 μl/site) or inoculated with 5 × 108 PFU of MVA i.m. at two sites in each hind leg (100 μl/site). Booster intravenous inoculations of 109 PFU of each virus were given at weeks 9 and 17. Sera were collected before inoculation (preimmune sera) and at weeks 4, 9, 11, 15, and 19. Cynomolgus macaque sera (Fig. 1B) were obtained from four groups of six animals immunized and boosted on week 8 as previously described (13): group 1 (“MVA/MVA”) animals were immunized and boosted i.m. with MVA, group 2 (“MVA/DVX”) animals were immunized i.m. with MVA followed by a percutaneous boost with Dryvax, group 3 (“−/DVX”) animals were immunized by percutaneous immunization with Dryvax alone at week 8, and group 4 animals were unimmunized. Sera from prebleed, week 2, and week 14 animals were analyzed for this study. All groups were challenged with monkeypox virus on week 16. Unimmunized animals became gravely ill or died, whereas vaccinated animals were healthy and asymptomatic, except for transient skin lesions in the MVA/MVA group (13). Human sera (Fig. 1C) were obtained from a randomized, partially blinded placebo controlled trial of Imvamune conducted at the NIAID-sponsored Saint Louis University Vaccine and Treatment Evaluation Unit (15a). Ninety subjects were equally randomized into six arms as follows. Seventy-five subjects received two doses of Imvamune (2 × 107, 5 × 107, or 1 × 108 50% tissue culture infective dose [TCID50]) or saline placebo at weeks 0 and 4, followed by 1 × 108 PFU/ml of Dryvax or saline placebo by scarification at month 4 after the first vaccination. Fifteen subjects also received two doses of saline administered subcutaneously followed by Dryvax. Blood was drawn for plaque reduction neutralizing antibody response at 2 weeks after the first and second vaccinations and 4 weeks after the third vaccination. The mean (± standard deviation [SD]) age was 24.8 (± 3.8) years. The sera used in this study were from 10 vaccinia virus naïve-individuals that were inoculated and boosted with 1 × 108 TCID50 of MVA; sera were collected prevaccination and 2 weeks after the second dose of vaccine. Human Dryvax sera were collected from 25 laboratory staff members before inoculation and 4 weeks after a single dose of Dryvax as described previously (10). The mean (± SD) age of the subjects was 33.3 (± 8.8) years. Thirteen subjects were vaccinia virus naïve at the time of vaccination, and 12 were receiving a boost.
FIG. 1.
Immunization and bleeding schedules. Vaccinia virus inoculations are designated by open arrowheads, and blood draws for serum are designated by small arrows. (A) Rabbits. Group 1 was primed i.m. and then boosted twice intravenously with MVA, and group 2 was primed intradermally with WR and boosted twice intravenously. (B) Macaques (13). Group 1 was immunized and boosted i.m. with MVA, group 2 was immunized i.m. with MVA followed by a percutaneous boost with Dryvax (DVX), group 3 was immunized by percutaneous immunization with Dryvax alone, and group 4 was unimmunized. All groups were challenged with monkeypox (MPX) on week 16. (C) Humans. Group 1 was inoculated and boosted with 1 × 108 TCID50 of MVA (Imvamune), and group 2 was immunized by single intradermal inoculation with Dryvax (10). pre, preimmunization.
Virus neutralization assays.
Neutralization titers of rabbit and monkey sera were determined by incubation of twofold serial dilutions of sera with recombinant vaccinia virus intracellular mature virions (MVs) that expresses green fluorescent protein (GFP) and then quantifying infected cells by flow cytometry, as described previously (12). Dilutions required for 50% neutralization relative to no-serum neutralization (50% inhibitory concentration values) were calculated with PRISM software (GraphPad, San Diego, CA). Titers of MVA-neutralizing activity in human Imvamune sera were determined by overnight incubation of twofold serial dilutions of heat-inactivated sera with MVA (30 to 50 PFU), followed by incubation with BSC-40 cell monolayers for 2 days, as described previously (24). Plaques were counted using a dissecting microscope and the 60% plaque reduction neutralization test titer was determined by interpolation by use of a linear regression function in Microsoft Excel.
Measurement of serum antibody by enzyme-linked immunosorbent assay (ELISA).
Procedures for measurement of serum antibody responses by use of purified WR virus-coated plates and specific vaccinia virus protein-coated plates have been previously reported (13).
Proteome microarrays.
Arrays displaying the prototype vaccinia virus strain WR proteome were produced as described previously (10). Briefly, vaccinia virus WR genomic DNA was used as a template for PCRs to amplify individual open reading frames (ORFs), which were then cloned into a T7 expression vector by use of homologous recombination. All proteins were expressed from purified plasmids in Escherichia coli-based coupled in vitro transcription/translation reaction (RTS 100 kits; Roche), except for L1, which was expressed in RTS disulfide kits (Roche) according to the manufacturer's instructions and designated “L1ss” in the figures. Detection of antibodies to L1 by microarray was found to be dependent on expression of the L1 protein in conditions that permitted disulfide bond formation (see Fig. S1 in the supplemental material). Control reactions that lacked template DNA or empty expression vector were also set up. Reactions were printed without further purification onto nitrocellulose-coated FAST slides (Whatman) by use of an Omni Grid 100 microarray printer (Gene Machines). For probing, sera were used at 1/50 or 1/100 dilution in protein array blocking buffer (Whatman) plus 10% E. coli lysate to block antibodies to E. coli. Bound hyperimmune rabbit antibodies were visualized with Cy3-conjugated goat anti-rabbit immunoglobulin G (IgG) (Jackson ImmunoResearch) diluted 1/200 in blocking buffer. For human and macaque sera, the directly conjugated antibody was found to give low signals, and experiments were repeated using a biotinylated anti-human IgG (Jackson) followed by streptavidin-PBXL3 conjugate (Martek Biosciences), both used at 1/200 in blocking buffer. After being washed five times, slides were air dried under brief centrifugation and examined in a ScanArray ExpressHT microarray scanner (PerkinElmer). Fluorescence intensities were quantified by using ProScanArray Express software (PerkinElmer). Data handling and statistical analyses were performed as described in the figure legends and table footnotes.
RESULTS
Production of hyperimmune MVA and WR sera in rabbits.
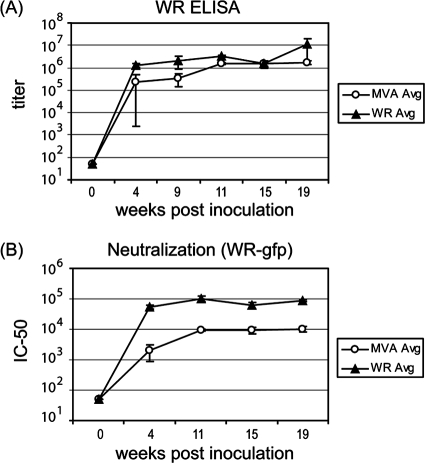
Hyperimmune sera were generated in rabbits against MVA and the replication-competent strain WR as laboratory reagents and used in this study to partially verify the arrays against standard ELISAs. In rabbits, MVA infections were asymptomatic, whereas WR was pathogenic and caused a transient loss of appetite (days 4 to 7), lethargy (days 5 to 6), and lesions at the injection site. Virus-specific antibody titers were monitored by ELISA of whole vaccinia virus and by virus neutralization measured by flow cytometry with vaccinia virus expressing GFP. Robust immune responses occurred to both MVA and WR inoculation, although WR induced higher antibody titers and ∼10-fold-higher virus-neutralizing titers (Fig. 2A and B, respectively). ELISAs were also performed with the following purified membrane proteins expressed in insect cells: L1 and A27, as representatives of MVs, and B5 and A33, as representatives of extracellular mature virions (EVs). Both hyperimmune MVA and WR sera showed strong responses to all four proteins (Fig. 3A), with the data essentially mirroring that obtained by ELISAs against whole vaccinia virus. Responses to L1, B5, and A33 reached similar maximal titers in MVA- and WR-inoculated rabbits, although the rise to plateau was more rapid in the latter. The titers to A27 were lower after MVA inoculation, which may contribute to the lower neutralizing activity of MVA hyperimmue sera (Fig. 2B). The differences in immunogenicity between MVA and WR viruses are probably due to the fact that while WR is replication competent and pathogenic in mammalian hosts, MVA is not.
FIG. 2.
Development of vaccinia virus antibodies in hyperimmunized rabbits. Two groups of rabbits were inoculated with MVA or WR to generate hyperimmune sera, as shown in Fig. 1A. (A) Serum antibody titers of individual rabbits inoculated with MVA or WR were determined by whole-virus ELISA. Shown are average titers (± SD) of three rabbits in each group. (B) Neutralization titers were determined by incubation of twofold serial dilutions of sera with a recombinant vaccinia virus that expresses enhanced GFP and then quantifying infected cells by flow cytometry. Shown are average (Avg) neutralization titers (± SD) of three rabbits in each group. IC50, 50% inhibitory concentration.
FIG. 3.
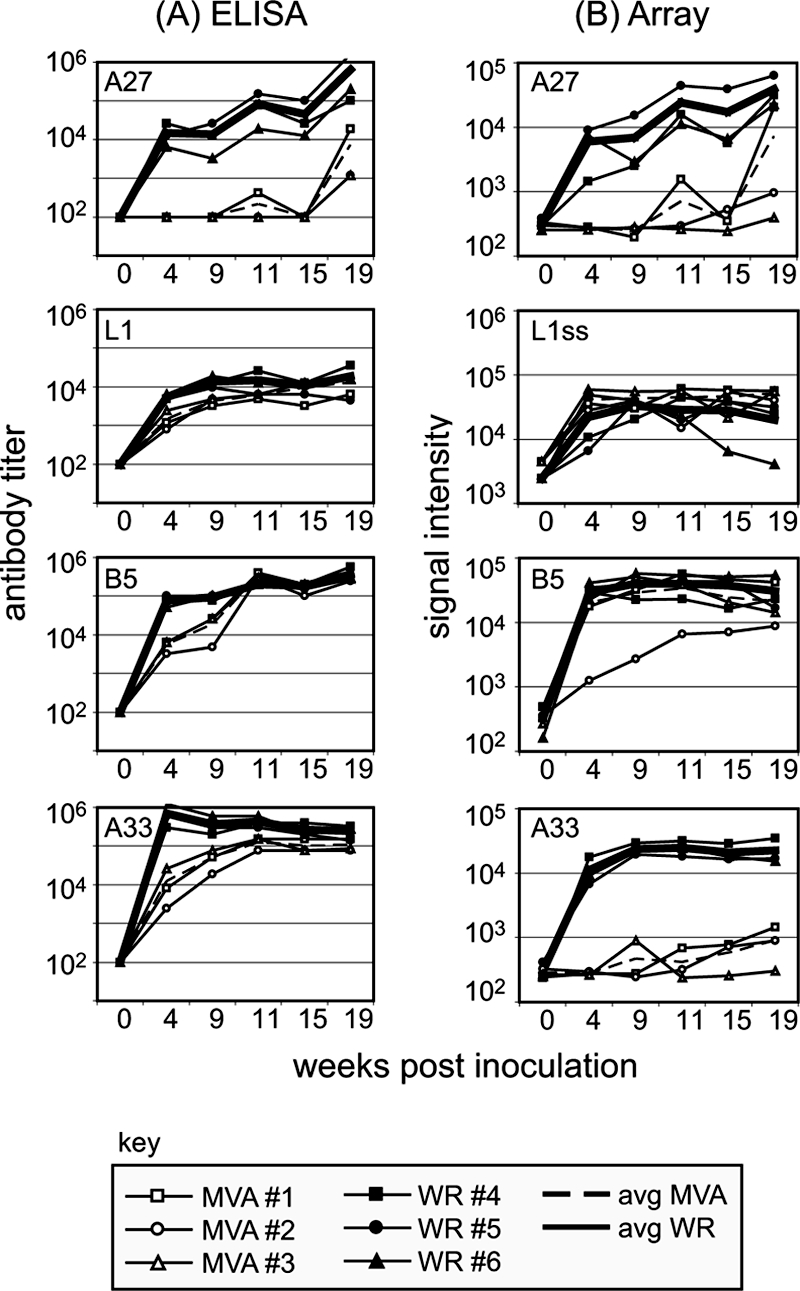
Comparable data obtained by ELISA and by protein microarray for four signature membrane proteins. (A) ELISAs of rabbit hyperimmune WR and MVA sera by use of plates coated with baculovirus-expressed vaccinia virus proteins. MV membrane proteins were represented by L1 and A27 and EV proteins by B5 and A33. (B) Corresponding SIs revealed by proteome microarrays probed with the same hyperimmune rabbit sera as used for the ELISAs in panel A. L1ss, L1 expressed in RTS disulfide kits (see text for details); avg, average.
We then used the hyperimmune rabbit sera to probe proteome microarrays displaying 210 different vaccinia virus WR proteins. Shown in Fig. 3B are the array signal intensities (SIs) for A27, L1, B5, and A33 plotted alongside the corresponding ELISA data for comparison. Array data for A27, L1, and B5 corresponded well to that obtained by ELISA, providing verification of the array platform. The response to A27 matched particularly closely, with both assays reporting a low titer in MVA sera relative to that in WR sera. The array format also readily detected antibodies to A33, but only in WR-hyperimmune serum. Since antibodies from MVA-inoculated rabbits were detected by ELISA using A33 secreted from insect cells, we assume that these sera recognized native protein predominantly, whereas WR sera recognized both native insect cell and nonnative E. coli-expressed protein.
Hyperimmune rabbit MVA sera recognize mainly late virion proteins, whereas WR sera also recognize early proteins.
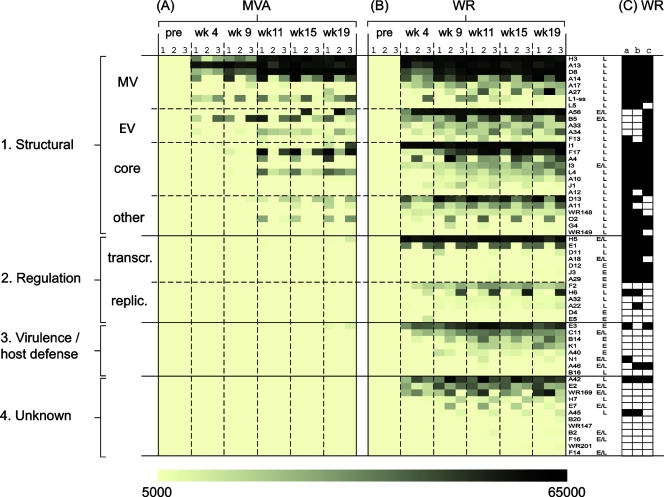
We next analyzed the antibody responses at the level of the vaccinia virus proteome (Fig. 4). Responses to individual proteins were very consistent between individual rabbits, particularly for the strongly immunoreactive antigens. In hyperimmune MVA sera (Fig. 4A), 18 different antigens were recognized in total, of which 13 were recognized by at least two of the three animals in each group. The MVA antibody profile was focused overwhelmingly on virion proteins (summarized in Table 1). Of the 13 commonly recognized antigens, 7 were membrane proteins and 4 were core proteins. Longitudinal sampling revealed the responses to envelope proteins appeared first and prior to the first boost, whereas the appearance of antibodies to the core proteins was not seen until after the first boost.
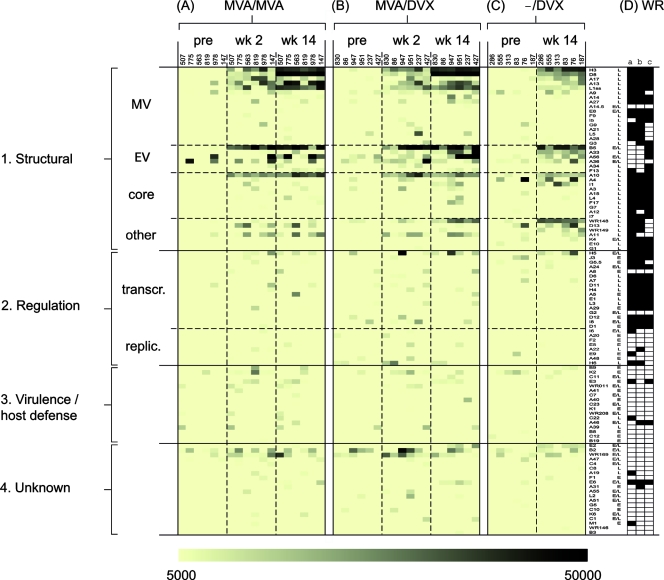
FIG. 4.
Rabbit hyperimmune MVA sera predominantly contain antibodies to late virion proteins, whereas WR is pathogenic in rabbits and also induces antibodies to early proteins. Hyperimmune rabbit sera generated against MVA (A) and WR (B) according to the schedules shown in Fig. 1A were used to probe WR proteome microarrays. The “no-DNA” control signals were subtracted from the SI for each protein and assigned a shade of color according to the strength of the signal, shown at the bottom of the figure. Antigens that were uniformly seronegative (i.e., SI of <5,000 in all six animals) have been omitted for clarity. The antigens have been classified into main groups 1 to 4 as follows. Group 1 consists of structural proteins, which have been subclassified into membrane proteins on intracellular MV and EV, core proteins, and other virion-associated late proteins. Group 2 consists of regulation proteins, subclassified into “transcr.” (transcription, translation) and “replic.” (DNA synthesis and genome replication). Group 3 consists of host range, virulence, and host defense proteins (virokines, cytokine receptors, and modulators of apoptosis, etc.). Group 4 consists of proteins of unknown function. Promoter designations (from www.poxvirus.org): L, late; E/L, early/late; E, early. (C) Virion proteins determined by mass spectroscopy studies of WR virions are indicated by the filled cells; data in columns a to c are from references 8, 28, and 38, respectively.
TABLE 1.
The commonly recognized antigens are predominantly structural proteinsa
| Animal | Protein of indicated type recognized by:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| MVA sera
|
Dryvax sera
|
WR sera
|
|||||||
| Membrane | Core | Other | Membrane | Core | Other | Membrane | Core | Other | |
| Rabbit | D8*b, H3*, A13*, A14*, B5*, L1*, A34* | F17*, L4*, A4, I1 | O2, D13 | D8*, H3*, A56*, A13*, B5*, A14*, A34*, A33*, A27*, A17*, L1*, F13*, L5 | I1*, F17*, I3*, A4*, L4*, A10*, J1* | H5*, E3*, D13*, A42*, C11*, E2*, WR169*, E1, A11*, F2*, B14*, H6, K1*, WR148*, H7*, A40*, A32*, A22*, A45*, J3 | |||
| Macaque | D8*, H3*, B5*, L1*, A13*, A56, A17, F13, A14 | A10*, L4 | A11, D13, H5 | B5*, H3*, D8*, A33*, A17*, A56, A13, L1 | A10*, I1 | WR148*, H5, E2, WR149 | |||
| Humanc | H3*, D8*, L1, B5, A14, A13, A34 | A10* | H3*, A13, B5, D8, A17, A33 | A10* | WR148*, H5, A11, D13, E2, B2 | ||||
Listed here are all the antigens recognized by 50% or more of the rabbits, macaques, and humans sampled in this study and ranked in each cell in descending order of average SI. Antigens were scored as positive if the SI was >5,000 after subtraction of the corresponding SI obtained with preimmune serum.
Antigens marked with asterisks were recognized by 100% of the individuals in the group.
The 25 humans sampled after Dryvax vaccination consisted of 13 individuals undergoing primary responses and 12 undergoing secondary responses (10).
By comparison, the responses to WR evolved more rapidly, and near-maximal signals were attained prior to the first boost (Fig. 4B). WR also engendered a much broader antibody profile than MVA. Fifty antigens were recognized overall (∼22% of the proteome), of which 40 were recognized by two or more animals. Moreover, all of the antigens in the MVA profile were also present in the WR profile. The additional antigens in the latter were mostly nonstructural/early proteins. Several of these are present in virions (8, 28, 38), while others, particularly those involved in host defense, are absent from virions (Fig. 4C).
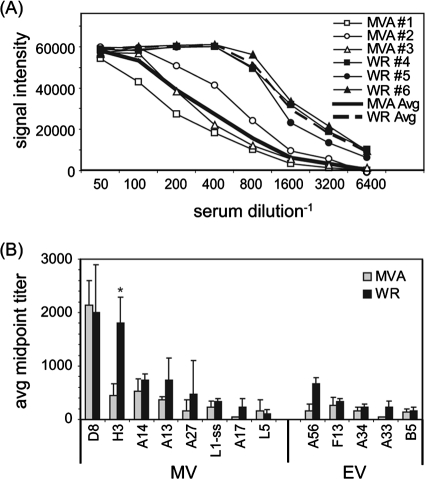
Antibodies to vaccinia virus EV and MV membrane proteins are known to mediate virus neutralization and protection against infection (2). Therefore, we determined antibody titers to all the vaccinia virus proteins by probing arrays with serially diluted hyperimmune MVA and WR sera. Titration data for the MV membrane protein H3 are shown in Fig. 5A as a representative example. Titers for the top immunoreactive membrane proteins (seven MV and five EV proteins) are shown in Fig. 5B. Titers were generally lower in the MVA sera, although only H3 antibodies were significantly lower (P < 0.05). Overall, the reduced titers of antibodies to MV membrane proteins in MVA sera, as determined both by ELISA (Fig. 2A) and array (Fig. 5B), are consistent with reduced MV-neutralizing activity (Fig. 2B).
FIG. 5.
Titers to some anti-MV antibodies are lower for hyperimmune MVA sera than for hyperimmune WR sera. (A) Sera from rabbits taken at the final time point (Fig. 1A) were serially diluted and used to probe vaccinia virus protein microarrays. Shown are titration curves for H3; average “no-DNA” control signals were subtracted from all SIs. (B) Average antibody titers (+ 1 SD) against MV and EV membrane proteins only. Titers were determined from titration plots by interpolating from the inflection point. *, Significant difference between MVA and WR responses by two-tailed, paired t test (P < 0.05). L1ss, L1 expressed in RTS disulfide kits (see text for details); avg, average.
The data set of immunoreactive viral proteins also allowed us to identify properties of viral proteins that were associated with immunogenicity (Table 2). We found that membrane and core proteins, proteins with late or early/late temporal expression, and proteins with transmembrane domains were overrepresented in the immunoreactive antigen set relative to the whole proteome. These predictors are strongest in MVA profiles, since the antibody profile to MVA is more heavily skewed toward structural proteins. In contrast, early proteins were underrepresented relative to the whole proteome, and there was negligible influence of molecular weight, isoelectric point, or the presence of a signal sequence on immunogenicity.
TABLE 2.
Properties of commonly recognized vaccinia virus antigens
| Categoryd | No. of vaccinia virus ORFs (%)e | No. of antigens recognized by indicated group (%) and fold enrichmenta
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MVAb
|
WR/DVXc
|
||||||||||||
| Rabbit (n = 3)
|
Macaque (n = 6)
|
Human (n = 10)
|
Rabbit (WR) (n = 3)
|
Macaque (−/DVX) (n = 6)
|
Human (DVX) (n = 25)
|
||||||||
| Antigen | Fold enrichment | Antigen | Fold enrichment | Antigen | Fold enrichment | Antigen | Fold enrichment | Antigen | Fold enrichment | Antigen | Fold enrichment | ||
| Total | 179 | 13 | 14 | 8 | 40 | 14 | 13 | ||||||
| Late promoter | 63 (35) | 9 (69) | 2.0 | 9 (64) | 1.8 | 5 (63) | 1.8 | 19 (48) | 1.4 | 7 (50) | 1.4 | 6 (46) | 1.3 |
| Early/late promoter | 63 (35) | 4 (31) | 0.9 | 5 (36) | 1.2 | 3 (38) | 1.1 | 15 (38) | 1.1 | 7 (50) | 1.4 | 6 (46) | 1.3 |
| Early promoter | 53 (30) | 0 (0) | 0 | 0 (0) | 0 | 0 (0) | 0 | 6 (15) | 0.5 | 0 (0) | 0 | 1 (8) | 0.3 |
| Membrane/core | 45 (25) | 11 (85) | 3.4 | 11 (79) | 3.2 | 8 (100) | 4.0 | 20 (50) | 2.0 | 10 (71) | 2.8 | 7 (54) | 2.2 |
| TM | 40 (22) | 7 (54) | 2.2 | 9 (64) | 2.9 | 7 (88) | 4.0 | 15 (38) | 1.7 | 8 (57) | 2.6 | 7 (54) | 2.5 |
| Signal S | 28 (16) | 3 (23) | 1.4 | 5 (36) | 2.3 | 3 (38) | 2.3 | 7 (18) | 1.1 | 5 (36) | 2.3 | 4 (31) | 1.9 |
| Avg pI | 6.99 | 7.24 | 6.32 | 7.46 | 6.96 | 6.25 | 5.96 | ||||||
| Avg Mr | 33,860 | 27,181 | 35,963 | 34,553 | 31,022 | 43,564 | 44,363 | ||||||
Values are the numbers of antigens recognized by 50% or more of the vaccinated animal or human sera in each group, with each expressed as a percentage of the total number of antigens in parentheses. “Fold enrichment” is a ratio obtained by dividing this percentage by the percentage of each category of the whole genome. In this analysis, an antigen was scored positive for recognition if the SI was >5,000 after subtraction of the corresponding SI obtained with preimmune serum.
Rabbit sera from week 12 (group 1 in Fig. 1A); macaque sera from week 14 (group 1 in Fig. 1B); human sera from week 16 (group 1 in Fig. 1C).
DVX, Dryvax. Rabbit sera from week 12 (group 2 in Fig. 1A); macaque sera from week 14 (group 3 in Fig. 1B); human primary plus secondary responses from week 4 post-Dryvax (group 2 in Fig. 1C).
Promoter designations and protein functions obtained from the Poxvirus Bioinformatics Resource Center (www.poxvirus.org). TM, transmembrane domain; Signal S, signal sequence.
Arrays fabricated with proteins from WR; pseudogenes and noncoding ORFs are excluded from this list.
MVA and Dryvax antibody profiles in macaques are broadly similar to each other.
Hyperimmune rabbit sera have been particularly useful here for the verification of arrays against existing immunoassay platforms. However, from a vaccine perspective, the responses by humans and nonhuman primates are more informative. The macaque is of particular importance since it is a close model of the human immune response and because smallpox vaccines can no longer be evaluated in human populations with endemic infections. Therefore, we probed arrays with sera from macaques inoculated with MVA and boosted with either MVA or Dryvax or else given Dryvax alone (13). Each of these regimens was shown to be equally protective in macaques against a lethal monkeypox challenge. Macaque profiles, shown in Fig. 6, showed interindividual heterogeneity greater than that seen for rabbits. In the MVA/MVA group (Fig. 6A), the strongest and most frequent positive signals were against structural proteins, particularly to the membrane proteins H3, D8, L1, and B5 and to core protein A10. The total number of commonly recognized antigens was 14, of which 8 were also commonly recognized by rabbits (Table 1). Robust responses were also seen to virion proteins A11 and D13, the latter being a highly expressed protein detectable in MVs (8, 28).
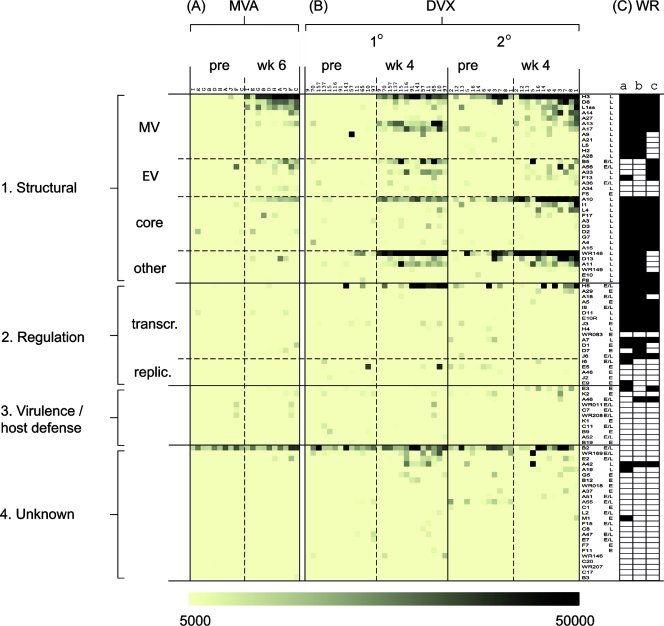
FIG. 6.
Antibody profiling of macaques inoculated with MVA or Dryvax shows both profiles are dominated by antibodies to structural proteins. Antibody profiles of cynomolgus macaques pre- and postimmunization with MVA/MVA (n = 6) (A), MVA/DVX (n = 6) (B), or −/DVX (n = 6) (C) according to the schedules shown in Fig. 1B. Note that week 14 of the Dryvax-alone experiment (C) corresponds to week 6 postvaccination (Fig. 1B). Data representation is as described for Fig. 4. (D) Virion proteins determined by mass spectroscopy studies of WR virions are indicated by the filled cells; data in columns a to c are from references 8, 28, and 38, respectively.
The response to Dryvax alone (Fig. 6C) was broadly similar to that seen for the MVA/MVA group, with the correlation coefficients being slightly higher for membrane proteins, or membrane and core proteins, than for other proteins (Table 3). The most striking difference between macaque MVA/MVA and Dryvax antibody profiles was the response to the WR148 A-type inclusion protein homolog. This protein is immunodominant in the Dryvax profile but absent from the MVA profile owing to a deletion of this region of the MVA genome. Other differences were antibodies to A33, I1, E2, and WR149, which were found only in the response to Dryvax (the lack of A33 response in MVA sera being a possible conformation issue, as discussed earlier). As seen previously, strong correlates of antigenicity in responses to both MVA and Dryvax were membrane/core proteins, late temporal expression, and/or the presence of transmembrane domain(s) (Table 2).
TABLE 3.
Correlations between MVA and Dryvax antibody profiles are strongest for viral membrane and membrane/core proteins
| Vaccination group |
R2 fora:
|
|||
|---|---|---|---|---|
| Membrane proteins | Membrane/core proteins | Nonmembrane/core proteins | All proteins | |
| Macaque | 0.691 | 0.684 | 0.354 | 0.614 |
| Human | 0.671 | 0.625 | 0.265 | 0.428 |
The values shown are correlation coefficients (R2) obtained from scatter plots comparing array SIs of MVA and Dryvax profiles obtained from vaccinated macaques and humans. The “no DNA” control background was subtracted from the SIs for each group and averages plotted.
Macaques inoculated with MVA followed by Dryvax show signature WR148 antibody.
It has been proposed that MVA could be used to prevaccinate individuals prior to receiving Dryvax to reduce its reactogenicity (13, 25). However, there are concerns that preexisting immunity to MVA may block the infectivity of Dryvax when given as a boost. We found that profiles from macaques given MVA and boosted with Dryvax were indistinguishable from the Dryvax-alone profiles (Fig. 6B). In particular, the MVA/DVX profile included modest titers to the WR148 protein, which are not produced in response to MVA alone. This observation confirms that prior immunization with MVA does not inhibit the immunogenicity of Dryvax given subsequently as a boost.
MVA and Dryvax antibody profiles in humans are also broadly similar to each other.
We next profiled human responses to MVA for comparison with the protective Dryvax response. All human subjects inoculated with Imvamune were confirmed for the development of MVA-neutralizing antibodies, with postvaccination titers ranging from 1/45 to 1/4,569 (Table 4). The sera were then used to probe vaccinia virus arrays and the profiles compared to human Dryvax profiles obtained previously (10) (Fig. 7 and Table 1). Human MVA profiles were dominated by responses to structural antigens, particularly the membrane proteins H3, D8, L1, and B5 and the core protein A10, consistent with macaque profiles. The response to Dryvax was also broadly similar (Fig. 7B), with the greatest concordance again being antibodies against membrane and core proteins (Table 3). As seen for macaques, a major difference between human MVA and Dryvax profiles was the response to WR148, which was lacking from the MVA response. Responses to D13, A11, and H5 were also stronger in the Dryvax profiles. All of these proteins are present in WR MVs (8, 28) and are probably present in Dryvax virions. As we reported previously (10), a strong signal to the B2 protein (function unknown) was seen in naïve as well as postvaccination human sera. This is due either to cross-reactive antibody or to the nonspecific capture of human IgG by the vaccinia virus B2 protein. As before, membrane/core proteins, late temporal expression, and the presence of transmembrane domain(s) are correlated with immunogenicity (Table 2). Differences between human primary and secondary responses to Dryvax have been described previously (10).
TABLE 4.
Titers of MVA-neutralizing antibodies in human sera on week 6 after MVA (Imvamune) vaccination on weeks 0 and 4a
| Subject | Titer for indicated serum
|
|
|---|---|---|
| Preimmunization | Postimmunization | |
| A | <4 | 963 |
| B | <4 | 128 |
| C | <4 | 472 |
| D | <4 | 553 |
| E | <4 | 45 |
| F | <4 | 1,089 |
| G | <4 | 2,253 |
| H | <4 | 4,569 |
| I | <4 | 208 |
| J | <4 | 768 |
Figure 1C shows further detail.
FIG. 7.
Antibody profiling of humans inoculated with MVA or Dryvax (DVX) shows both profiles are dominated by antibodies to structural proteins. Antibody profiles for humans pre- and postvaccination with MVA (A) and WR (B) according to the schedule shown in Fig. 1C. Data representation is as described for Fig. 4. For Dryvax responses, primary (n = 13) and secondary (n = 12) infections are shown. (C) Virion proteins determined by mass spectroscopy studies of WR virions are indicated by the filled cells; data in columns a to c are from references 8, 28, and 38, respectively. The B2 antigen was consistently recognized by vaccinia virus-naïve human IgG, although this was nonspecific and independent of vaccination.
Macaques and humans share similar antibody profiles to both MVA and Dryvax.
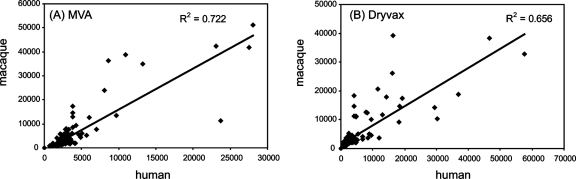
Finally, we aligned the human and macaque profiles. In Fig. 8, the average SIs for each vaccinated group are shown. Overall, human and macaque profiles were very similar. Immunodominant membrane proteins in Dryvax shared in human and macaque responses were the MV proteins H3, A13, D8, A17, and L1 and the EV proteins B5, A33, and A56. Similarly, immunodominant membrane proteins in MVA shared by human and macaque profiles were H3, A13, D8, and L1 (MV) and B5 (EV). When all SIs on the chip were compared, the correlation coefficients (R2) were 0.656 and 0.722 for Dryvax and MVA, respectively (Fig. 9). The concordance of human and macaque signals was highest for membrane proteins, with R2 for Dryvax and MVA profiles of 0.696 and 0.827, respectively (data not shown). By comparison, the concordance of MVA profiles in humans and rabbits was lower (R2 = 0.357) but slightly higher between rabbits and macaques (R2 = 0.437) (data not shown). These differences may be a consequence of phylogenetic relatedness between these different host species.
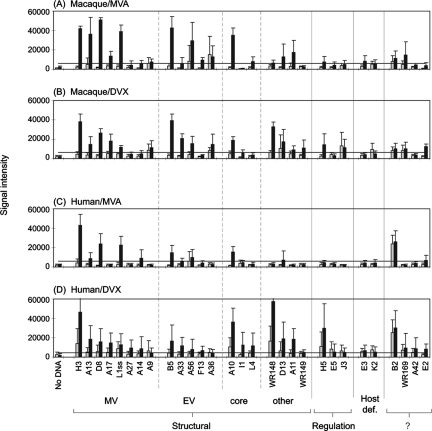
FIG. 8.
Summary of antibody profiles for humans and macaques. Humans and macaques were inoculated with Dryvax (DVX) or MVA according to the schedules shown in Fig. 1. Bars represent average SIs (+ SD) of the top-ranking antigens for all four cohorts combined: gray bars, prevaccination; black bars, postvaccination. (A) Macaque responses, pre- and 14 weeks post-MVA (“MVA/MVA” in Fig. 1B; n = 6). (B) Macaque responses pre- and 6 weeks post-Dryvax (“−/DVX” in Fig. 1B; n = 6). (C) Human responses pre- and 6 weeks post-MVA (n = 10). (D) Human responses pre- and 4 weeks post-Dryvax (n = 25). This last panel consisted of 13 individuals undergoing primary responses and 12 individuals after boosting. Positive signals in prevaccination signals in the human/Dryvax group (e.g., H3, A10, and WR148) are due to antibodies still detectable in the sera of previously vaccinated individuals (n = 12); a cutoff, represented by the horizontal bar, was set as the average signal (+ 10 SD) of “no-DNA” control spots with postvaccination sera.
FIG. 9.
Human and macaque antibody profiles show good correlation. Data points are the “no-DNA” control background subtracted from the average SIs on protein arrays. (A) MVA responses. Human sera (n = 10) at 6 weeks postvaccination versus macaque sera (n = 6) at week 14 postvaccination (“MVA/MVA” in Fig. 1B). (B) Dryvax responses. Human sera (n = 25) at 4 weeks postvaccination versus macaque sera at 6 weeks post vaccination (“−/DVX” week 14 in Fig. 1). R2 equals the square of the Pearson product moment correlation coefficient.
DISCUSSION
In this study we have applied a method developed in our laboratory to produce protein microarray chips from microorganisms on a whole-proteome scale (9). An array displaying the prototypic vaccinia virus strain WR has been used to screen the sera of mammalian hosts infected with vaccinia virus strains MVA, Dryvax, and WR. Since most protein sequences of MVA are 97 to 99% identical to those of other vaccinia virus strains (3), extensive binding cross-reactivity is expected. The first aim of the study was to describe antibody signatures that correlated with protection against orthopoxvirus challenge. Since macaques are protected from monkeypox challenge by both MVA and Dryvax, protective antibodies are likely to be among those conserved between both profiles. The second aim was to quantify the correlation between macaque and human profiles in response to both vaccines. Since humans are protected against smallpox by Dryvax, the relatedness of Dryvax antibody signatures in macaques and humans will help gauge the macaque as a model for human infection.
The first main finding of the study is that MVA and Dryvax profiles were very similar to each other, in both human and macaque responses. Precise alignments of MVA and Dryvax profiles in humans and macaques are complicated by their heterogeneity, although antibodies to membrane proteins were particularly well conserved between the vaccines. Importantly, antibodies to both MV and EV forms of the virus were found. MVs are robust, enveloped virions that may mediate transmission between individual hosts, whereas EVs are double-enveloped particles that are thought to mediate the intercellular spread of infection within an individual host (29, 30). Given the general acceptance that a replacement smallpox vaccine must elicit antibodies that neutralize both MV and EV forms of infectious particles in order to be successful (16, 35), it is encouraging that we observed antibodies to EV and MV membrane proteins in MVA profiles. The responses to some nonmembrane proteins were less well conserved, although these antibodies would not be expected to influence virus neutralization. The high concordance of MVA and Dryvax profiles in macaques seen here is consistent with previously published ELISA data showing antibody binding and neutralizing titers were equivalent or higher in the MVA/MVA and MVA/DVX groups than those induced by a single dose of Dryvax (13). The most significant difference we saw was a lack of a response in MVA profiles to the immunodominant A-type inclusion protein homolog WR148, owing to its deletion from MVA. This confirms an earlier Western blot comparison of vaccinia virus Ig and MVA sera (17) and suggests that this antigen may provide a useful diagnostic tool.
Given the ability of MVA to synthesize both early and late proteins, it is not surprising that the Dryvax and MVA profiles are so similar. Indeed, abundant late structural components can account for the majority of antigens recognized in both profiles. By comparison, WR profiles in hyperimmune rabbits were characterized by a significant expansion in the response against nonstructural/early proteins relative to the MVA profile and included several against proteins not found in virions. The difference is likely to be related to the increased pathology associated with WR infection in rabbits compared to MVA. One possibility is that WR infection in the rabbit leads to significant necrotic cell death, allowing intracellular viral antigens to become accessible to B cells. Alternatively, WR infection may lead to a more “systemic” infection in rabbits and substantially higher amounts of all viral antigens in comparison to the inoculation of replication-defective MVA.
Despite this expanded WR profile relative to what was seen for MVA, the responses to membrane proteins were broadly similar. Titers were lower in the MVA response, although the significance of this in terms of the protection afforded by MVA in vivo is not clear.
The second main finding of the study was a high concordance between vaccination signatures from macaques protected against lethal orthopox challenge and the corresponding human signatures. Overall, these data support the notion that the macaque is a close model for the human antibody response and that, by extension, MVA should provide protection against lethal orthopoxvirus challenge in humans. Since MVA was developed towards the end of the eradication campaign, it was not used in areas of smallpox endemicity, and no efficacy data for the prevention of smallpox were obtained. This is also a problem for other pathogens of importance in biodefense that cannot be tested in humans. To address this, the U.S. Food and Drug Administration (FDA) instituted the “two-animal rule” specifically for vaccines and other agents that need to be licensed against diseases of low or no incidence in the population. A vaccine for smallpox, such as MVA, would be required under this rule to show protection in two animal species expected to react with a response predictive for humans.
This study has also helped determine whether MVA has utility to reduce the reactogenicity of conventional vaccines. A concern is that MVA may negate the effect of Dryvax altogether by virtue of engendering prior immunity. We found that immunization of macaques with MVA followed by Dryvax engendered a profile indistinguishable from that achieved with Dryvax alone. In particular, the presence of antibodies to WR148 in the MVA/DVX profile signified that the Dryvax boost engendered an immune response. Previous studies in these macaques showed antibody titers attained after two doses of MVA or one dose of MVA followed by one dose of Dryvax were higher than those after a single dose of Dryvax (13). In a recent study in which results for humans who were given two or more doses of MVA followed by Dryvax challenge were compared to results for Dryvax challenge alone, it was noted that although neutralizing antibody titers were similar, individuals receiving a prior MVA inoculation had elevated titers of antibodies to EV proteins B5 and A33 (25). Together, the data indicate the MVA prime-boost regimen does not diminish the immunogenicity engendered by Dryvax and in some cases may enhance it.
In summary, we have profiled the humoral response to MVA and compared it with the currently licensed vaccine Dryvax and the pathogenic strain WR. The data reveal that despite the deletion of several genes from the MVA genome, there has been little impact on the profile of antibodies to membrane proteins. Differences between profiles depend on whether MVA is compared to Dryvax or WR, but in each case, the major differences are found among the responses to nonmembrane proteins.
Supplementary Material
Acknowledgments
We thank G. Cohen and R. Eisenberg for gifts of purified L1, B5, A27, and A33 membrane proteins expressed in insect cells for the rabbit ELISA studies; Cangene Corporation (Winnipeg, Canada) for the gift of vaccinia virus Ig; and Joe Miller, Jens Wrammert, and Rafi Ahmed (Emory University) for human Dryvax sera. We are grateful to Elliot J. Lefkowitz, Curtis Hendrickson, and Chunlin Wang of the Department of Microbiology, University of Alabama at Birmingham, and the NIH/NIAID Viral Bioinformatics Resource Center (HHSN266200400036C) for sharing their vaccinia virus WR and vaccinia virus Copenhagen promoter analysis. We thank Jeff Americo, Jennifer Vogt, and Norman Cooper for excellent technical assistance.
This work was supported in part by NIH/NIAID grants U01AI056464, AI058365, and 1U01AI061363 (P. L. Felgner, principal investigator [PI]) and contract N01-AI-25464 (R. B. Belshe, Saint Louis University, PI). The study was also supported in part by the Division of Intramural Research, NIAID, NIH.
We declare that we have no competing financial interests.
Footnotes
Published ahead of print on 31 October 2007.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Abdalrhman, I., I. Gurt, and E. Katz. 2006. Protection induced in mice against a lethal orthopox virus by the Lister strain of vaccinia virus and modified vaccinia virus Ankara (MVA). Vaccine 244152-4160. [DOI] [PubMed] [Google Scholar]
- 2.Amanna, I. J., M. K. Slifka, and S. Crotty. 2006. Immunity and immunological memory following smallpox vaccination. Immunol. Rev. 211320-337. [DOI] [PubMed] [Google Scholar]
- 3.Antoine, G., F. Scheiflinger, F. Dorner, and F. G. Falkner. 1998. The complete genomic sequence of the modified vaccinia Ankara strain: comparison with other orthopoxviruses. Virology 244365-396. [DOI] [PubMed] [Google Scholar]
- 4.Belyakov, I. M., P. Earl, A. Dzutsev, V. A. Kuznetsov, M. Lemon, L. S. Wyatt, J. T. Snyder, J. D. Ahlers, G. Franchini, B. Moss, and J. A. Berzofsky. 2003. Shared modes of protection against poxvirus infection by attenuated and conventional smallpox vaccine viruses. Proc. Natl. Acad. Sci. USA 1009458-9463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carroll, M. W., and B. Moss. 1997. Host range and cytopathogenicity of the highly attenuated MVA strain of vaccinia virus: propagation and generation of recombinant viruses in a nonhuman mammalian cell line. Virology 238198-211. [DOI] [PubMed] [Google Scholar]
- 6.Carroll, M. W., W. W. Overwijk, R. S. Chamberlain, S. A. Rosenberg, B. Moss, and N. P. Restifo. 1997. Highly attenuated modified vaccinia virus Ankara (MVA) as an effective recombinant vector: a murine tumor model. Vaccine 15387-394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chahroudi, A., D. A. Garber, P. Reeves, L. Liu, D. Kalman, and M. B. Feinberg. 2006. Differences and similarities in viral life cycle progression and host cell physiology after infection of human dendritic cells with modified vaccinia virus Ankara and vaccinia virus. J. Virol. 808469-8481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chung, C. S., C. H. Chen, M. Y. Ho, C. Y. Huang, C. L. Liao, and W. Chang. 2006. Vaccinia virus proteome: identification of proteins in vaccinia virus intracellular mature virion particles. J. Virol. 802127-2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davies, D. H., X. Liang, J. E. Hernandez, A. Randall, S. Hirst, Y. Mu, K. M. Romero, T. T. Nguyen, M. Kalantari-Dehaghi, S. Crotty, P. Baldi, L. P. Villarreal, and P. L. Felgner. 2005. Profiling the humoral immune response to infection by using proteome microarrays: high-throughput vaccine and diagnostic antigen discovery. Proc. Natl. Acad. Sci. USA 102547-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davies, D. H., D. M. Molina, J. Wrammert, J. Miller, S. Hirst, Y. Mu, J. Pablo, B. Unal, R. Nakajima-Sasaki, X. Liang, S. Crotty, K. L. Karem, I. K. Damon, R. Ahmed, L. Villarreal, and P. L. Felgner. 2007. Proteome-wide analysis of the serological response to vaccinia and smallpox. Proteomics 71678-1686. [DOI] [PubMed] [Google Scholar]
- 11.Drexler, I., C. Staib, W. Kastenmuller, S. Stevanovic, B. Schmidt, F. A. Lemonnier, H. G. Rammensee, D. H. Busch, H. Bernhard, V. Erfle, and G. Sutter. 2003. Identification of vaccinia virus epitope-specific HLA-A*0201-restricted T cells and comparative analysis of smallpox vaccines. Proc. Natl. Acad. Sci. USA 100217-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Earl, P. L., J. L. Americo, and B. Moss. 2003. Development and use of a vaccinia virus neutralization assay based on flow cytometric detection of green fluorescent protein. J. Virol. 7710684-10688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Earl, P. L., J. L. Americo, L. S. Wyatt, L. A. Eller, J. C. Whitbeck, G. H. Cohen, R. J. Eisenberg, C. J. Hartmann, D. L. Jackson, D. A. Kulesh, M. J. Martinez, D. M. Miller, E. M. Mucker, J. D. Shamblin, S. H. Zwiers, J. W. Huggins, P. B. Jahrling, and B. Moss. 2004. Immunogenicity of a highly attenuated MVA smallpox vaccine and protection against monkeypox. Nature 428182-185. [DOI] [PubMed] [Google Scholar]
- 14.Earl, P. L., B. Moss, L. S. Wyatt, and M. W. Carroll. 1998. Generation of recombinant vaccinia viruses, vol. 2. Greene Publishing Associates & Wiley Interscience, New York, NY.
- 15.Feery, B. J. 1977. Adverse reactions after smallpox vaccination. Med. J. Aust. 2(6)180-183. [DOI] [PubMed] [Google Scholar]
- 15a.Frey, S. E., F. K. Newman, J. S. Kennedy, V. Sobek, F. A. Ennis, H. Hill, L. K. Yan, P. Chaplin, J. Vollmar, B. R. Chaitman, and R. B. Belshe. Clinical and immunologic responses to multiple doses of Imvamune (modified vaccinia Ankara) followed by Dryvax challenge. Vaccine, in press. [DOI] [PMC free article] [PubMed]
- 16.Hooper, J. W., D. M. Custer, and E. Thompson. 2003. Four-gene-combination DNA vaccine protects mice against a lethal vaccinia virus challenge and elicits appropriate antibody responses in nonhuman primates. Virology 306181-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jones-Trower, A., A. Garcia, C. A. Meseda, Y. He, C. Weiss, A. Kumar, J. P. Weir, and M. Merchlinsky. 2005. Identification and preliminary characterization of vaccinia virus (Dryvax) antigens recognized by vaccinia immune globulin. Virology 343128-140. [DOI] [PubMed] [Google Scholar]
- 18.Mayr, A., V. Hochstein-Mintzel, and H. Stickl. 1975. Abstammung, Eigenschaften und Verwendung des attenuierten Vaccinia-Stammes MVA. Infection 36-14. [Google Scholar]
- 19.Mayr, A., H. Stickl, H. K. Muller, K. Danner, and H. Singer. 1978. The smallpox vaccination strain MVA: marker, genetic structure, experience gained with the parenteral vaccination and behavior in organisms with a debilitated defence mechanism. Zentralbl. Bakteriol. B 167375-390. [PubMed] [Google Scholar]
- 20.McCurdy, L. H., J. A. Rutigliano, T. R. Johnson, M. Chen, and B. S. Graham. 2004. Modified vaccinia virus Ankara immunization protects against lethal challenge with recombinant vaccinia virus expressing murine interleukin-4. J. Virol. 7812471-12479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meseda, C. A., A. D. Garcia, A. Kumar, A. E. Mayer, J. Manischewitz, L. R. King, H. Golding, M. Merchlinsky, and J. P. Weir. 2005. Enhanced immunogenicity and protective effect conferred by vaccination with combinations of modified vaccinia virus Ankara and licensed smallpox vaccine Dryvax in a mouse model. Virology 339164-175. [DOI] [PubMed] [Google Scholar]
- 22.Meyer, H., G. Sutter, and A. Mayr. 1991. Mapping of deletions in the genome of the highly attenuated vaccinia virus MVA and their influence on virulence. J Gen. Virol. 721031-1038. [DOI] [PubMed] [Google Scholar]
- 23.Moss, B. 1996. Genetically engineered poxviruses for recombinant gene expression, vaccination, and safety. Proc. Natl. Acad. Sci. USA 9311341-11348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Newman, F. K., S. E. Frey, T. P. Blevins, M. Mandava, A. Bonifacio, Jr., L. Yan, and R. B. Belshe. 2003. Improved assay to detect neutralizing antibody following vaccination with diluted or undiluted vaccinia (Dryvax) vaccine. J. Clin. Microbiol. 413154-3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parrino, J., L. H. McCurdy, B. D. Larkin, I. J. Gordon, S. E. Rucker, M. E. Enama, R. A. Koup, M. Roederer, R. T. Bailer, Z. Moodie, L. Gu, L. Yan, and B. S. Graham. 2007. Safety, immunogenicity and efficacy of modified vaccinia Ankara (MVA) against Dryvax challenge in vaccinia-naive and vaccinia-immune individuals. Vaccine 251513-1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Phelps, A., A. J. Gates, M. Hillier, L. Eastaugh, and D. O. Ulaeto. 2005. Comparative efficacy of replicating smallpox vaccine strains in a murine challenge model. Vaccine 233500-3507. [DOI] [PubMed] [Google Scholar]
- 27.Phelps, A. L., A. J. Gates, M. Hillier, L. Eastaugh, and D. O. Ulaeto. 2007. Comparative efficacy of modified vaccinia Ankara (MVA) as a potential replacement smallpox vaccine. Vaccine 2534-42. [DOI] [PubMed] [Google Scholar]
- 28.Resch, W., K. K. Hixson, R. J. Moore, M. S. Lipton, and B. Moss. 2007. Protein composition of the vaccinia virus mature virion. Virology 358233-247. [DOI] [PubMed] [Google Scholar]
- 29.Smith, G. L., and M. Law. 2004. The exit of vaccinia virus from infected cells. Virus Res. 106189-197. [DOI] [PubMed] [Google Scholar]
- 30.Smith, G. L., and A. Vanderplasschen. 1998. Extracellular enveloped vaccinia virus. Entry, egress, and evasion. Adv. Exp. Med. Biol. 440395-414. [PubMed] [Google Scholar]
- 31.Stittelaar, K. J., T. Kuiken, R. L. de Swart, G. van Amerongen, H. W. Vos, H. G. Niesters, P. van Schalkwijk, T. van der Kwast, L. S. Wyatt, B. Moss, and A. D. Osterhaus. 2001. Safety of modified vaccinia virus Ankara (MVA) in immune-suppressed macaques. Vaccine 193700-3709. [DOI] [PubMed] [Google Scholar]
- 32.Stittelaar, K. J., G. van Amerongen, I. Kondova, T. Kuiken, R. F. van Lavieren, F. H. Pistoor, H. G. Niesters, G. van Doornum, B. A. van der Zeijst, L. Mateo, P. J. Chaplin, and A. D. Osterhaus. 2005. Modified vaccinia virus Ankara protects macaques against respiratory challenge with monkeypox virus. J. Virol. 797845-7851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sutter, G., and B. Moss. 1992. Nonreplicating vaccinia vector efficiently expresses recombinant genes. Proc. Natl. Acad. Sci. USA 8910847-10851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sutter, G., L. S. Wyatt, P. L. Foley, J. R. Bennink, and B. Moss. 1994. A recombinant vector derived from the host range-restricted and highly attenuated MVA strain of vaccinia virus stimulates protective immunity in mice to influenza virus. Vaccine 121032-1040. [DOI] [PubMed] [Google Scholar]
- 35.Viner, K. M., and S. N. Isaacs. 2005. Activity of vaccinia virus-neutralizing antibody in the sera of smallpox vaccinees. Microbes Infect. 7579-583. [DOI] [PubMed] [Google Scholar]
- 36.Vollmar, J., N. Arndtz, K. M. Eckl, T. Thomsen, B. Petzold, L. Mateo, B. Schlereth, A. Handley, L. King, V. Hulsemann, M. Tzatzaris, K. Merkl, N. Wulff, and P. Chaplin. 2006. Safety and immunogenicity of IMVAMUNE, a promising candidate as a third generation smallpox vaccine. Vaccine 242065-2070. [DOI] [PubMed] [Google Scholar]
- 37.Wyatt, L. S., P. L. Earl, L. A. Eller, and B. Moss. 2004. Highly attenuated smallpox vaccine protects mice with and without immune deficiencies against pathogenic vaccinia virus challenge. Proc. Natl. Acad. Sci. USA 1014590-4595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yoder, J. D., T. S. Chen, C. R. Gagnier, S. Vemulapalli, C. S. Maier, and D. E. Hruby. 2006. Pox proteomics: mass spectrometry analysis and identification of Vaccinia virion proteins. Virol. J. 310. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.