Abstract
The future of genetic interventions in humans critically depends on the selectivity and efficiency of gene transfer to target tissues. The viral gene vectors explored to date cannot selectively transduce the desired targets. While substantial progress has been made in developing targeting strategies for adenovirus (Ad) vectors, future advances in this direction are severely limited by the shortage of naturally existing molecules available for use as targeting ligands. This shortage is due to fundamental and irresolvable differences at the level of both posttranslational modifications and intracellular trafficking between the Ad structural proteins and those natural proteins that are involved in interactions with the cell surface and could otherwise be considered as potential targeting ligands. We hypothesized that this problem could be resolved by altering the natural tropism of Ad vector through incorporation into its capsid of a rationally designed protein ligand, an affibody, whose structural, functional, and biosynthetic properties make it compatible with the Ad assembly process. We tested this hypothesis by redesigning the receptor-binding Ad protein, the fiber, using affibodies specific for human epidermal growth factor receptor type 2 (Her2), a major molecular marker of human tumors. The biosynthesis and folding of these fiber chimeras were fully compatible with Ad virion formation, and the resultant viral vectors were capable of selective delivery of a dual-function transgene to Her2-expressing cancer cells. By establishing the feasibility of this affibody-based approach to Ad vector targeting, the present study lays the foundation for further development of Ad vector technology toward its clinical use.
Human adenovirus serotype 5 (Ad5) has long been explored as a prototype vector for genetic immunization and therapy (13). However, the success of this work has been hampered by Ad5's inability to express transgenes in a target-specific manner and by the fact that Ad5 infection of some types of human cells, both normal and malignant, is inefficient. Therefore, to improve both the selectivity and the infectivity of Ad5 vectors in target tissues or tumors, the mechanism of cell attachment that has been evolutionarily developed by the virus has to be altered (5, 18).
Ad virion consists of the genome-containing core enclosed within an icosahedral capsid, the faces of which are formed mainly by the hexon protein and the vertices of which are occupied by the penton capsomers. The components of the penton—the penton base protein and the fiber—mediate virus attachment to the cell surface and virus internalization. The fiber protein is a homotrimer that has a distinct three-domain structure (7) (Fig. 1A). Its amino-terminal tail anchors the fiber within the penton base, the carboxy-terminal knob binds to the primary cell receptor (11), and the central shaft extends the knob away from the capsid and toward the receptor. It is the binding of the fiber knob to the Ad5 receptor, the coxsackievirus and Ad receptor (CAR), that anchors the virion on the cell surface and makes the infection possible (3, 29). While the secondary interaction between Ad5 and the cell, which involves the penton base and cellular integrins (31, 32), is crucial for virus internalization, it cannot happen unless the virus is attached to CAR first. Importantly, CAR expression is not associated with any known disease, and thus CAR is a poor target for therapeutic gene vectors. Therefore, to be useful, Ad5 vectors have to be directed to disease-linked cell surface molecules by modification of their natural tropism.
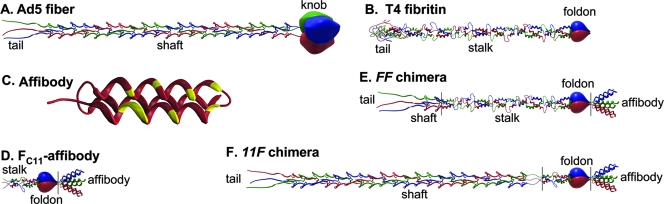
FIG. 1.
Design of Her2-targeted Ad fiber protein. (A) The Ad5 fiber is a homotrimer whose tail anchors it within the Ad virion. The globular knob domain initiates the trimerization of individual subunits by aligning them and also makes viral infection possible by binding to Ad5 natural receptor, CAR. The fibrous shaft domain consists of 21 pseudorepeats that form a characteristic β-spiral structure. (B) Similar to the Ad5 fiber, the fibritin is a homotrimer that has the tail, the stalk, and the foldon domains. The stalk connects the globular tail and foldon and consists of 13 α-helical coiled coils connected with loops. The carboxy-terminal foldon is a very small (30 amino acids) and stable trimerization motif. (C) The affibody is a 58-amino-acid-long artificially designed ligand derived from Staphylococcus protein A. An array of 13 amino acids, which is engrafted into two of the three α-helices of an affibody by combinatorial randomization, forms a target-binding site (shown with yellow). (D) The Fc11-affibody protein is designed as a replacement for the Ad5 fiber knob domain. It consists of the foldon and the two preceding coiled coils of the fibritin's stalk. Within this molecule, the foldon holds the three subunits of the chimera together, while the affibody binds to the target receptor. The FF-type (E) and 11F-type (F) fiber-fibritin-affibody chimeras are designed as targeted equivalents of the Ad5 fiber. They are similar in that each contains three structural elements—the affibody ligand (or the parental dZ scaffold) and the fragments of both the fiber and the fibritin. The FF-type chimera incorporates the tail and the first two pseudorepeats of the Ad5 fiber shaft fused with the fragment of fibritin truncated at the sixth coiled coil of the stalk. The 11F-type chimera resembles the structure of the fiber more closely, as it contains two complete domains of the fiber, the tail and the shaft. The flexible linker sequence, which connects the shaft and knob domains in the wt fiber, links the β-spiral shaft to the α-helical segment of the fibritin, thus facilitating this structural transition within the 11F protein. Vertical black lines in panels D, E, and F indicate the fusion points between individual components of the chimeric proteins.
Ad5 receptor specificity can be modified by inserting target receptor-specific ligands into the Ad fiber knob. However, advancement in this direction is severely limited by the shortage of naturally existing molecules suitable for use as ligands. The repertoire of candidate protein ligands is narrowed not only by their size and tertiary structure but also, more importantly, by incompatibility between the ligands and the Ad fiber with respect to biosynthesis and intracellular trafficking. Because Ad particles are assembled in the nucleus of the infected cell, all virion proteins have to localize to the nucleus. Thus, none of them can undergo posttranslational modifications, which take place in the endoplasmic reticulum or Golgi apparatus. Consequently, such modifications as the formation of disulfide bonds, which occur in the endoplasmic reticulum, are unavailable to both the Ad fiber and to any ligand that is fused with it. These limitations make such seemingly attractive ligand candidates as antibodies, growth factors, and many others useless for Ad targeting (20). Attempts to resolve this conundrum by engrafting into the fiber knob the peptide ligands identified by phage display had very limited success because of the low affinities of peptides and frequent loss of their function upon fusion with the Ad fiber (34).
Here, we report a solution to the problem of biosynthetic incompatibility between targeting ligands and Ad capsid components. Key to the success of this work is the use of affibodies, a novel type of artificial protein ligand. The affibodies are one of the types of emerging designed proteins that are derived from existing protein scaffolds using combinatorial randomization of amino acids in preselected positions (4). In particular, affibodies are derived from a three-helix bundle domain Z (dZ) of Staphylococcus protein A, in which 13 of 58 amino acids are randomized to create a target-binding site (23) (Fig. 1). The biosynthesis of affibodies, their small size, their high affinity to and selectivity for targets, and their simple structure, which does not require any posttranslational modifications, together have provided a strong rationale for using affibodies as ligands for Ad targeting.
In the study reported here, we altered the natural tropism of Ad5 by redesigning the fiber of the virus to include an affibody to human epidermal growth factor receptor type 2 (Her2), a major molecular marker of human cancers (21), which is now used by the modified vector as a surrogate receptor. This work establishes the feasibility of employing artificially designed proteins for therapeutic vector targeting, and our findings suggest that affibodies may be the ligands of choice for Ad. Furthermore, this study shows that affibody-based Ad5 targeting yields fully functional vector prototypes, which could be further developed into therapeutic agents for Her2-expressing human tumors.
MATERIALS AND METHODS
Cells and antibodies.
293, 293T, and SK-BR-3 cells (all from American Type Culture Collection, Manassas, VA), 293A cells (Invitrogen, Carlsbad, CA), MDA-MB-231 cells (provided by Janet Price, The University of Texas M. D. Anderson Cancer Center), and 293/F28 cells (1) were maintained as previously described (2). 293/Her2 cells, a Her2-expressing derivative of 293A cells, were made by stable transfection of 293A cells with a Her2 cDNA-containing plasmid derived from pIRES.neo (Clontech, Mountain View, CA). The hybridoma line 5E1, expressing anti-fibritin stalk monoclonal antibody (MAb), was generated using recombinant Fc11 protein (below). Anti-fiber tail MAb 4D2 (12) was kindly provided by Jeff Engler (University of Alabama at Birmingham).
Expression and purification of recombinant proteins.
All recombinant proteins produced in Escherichia coli BL21(DE3) were His6 tagged at their amino termini and were expressed using pET20b (EMD Biosciences, La Jolla, CA). The affibody-coding genes were assembled with oligonucleotides using published amino acid sequences (33). The affibody ligands were then genetically fused to the carboxy-terminal fragment of the phage T4 fibritin protein comprising the foldon domain and two preceding α-helical coiled coils, the Fc11. All proteins were purified by affinity chromatography under native conditions on HisTrap HP columns (GE Healthcare, Piscataway, NJ) and then subjected to gel filtration on a Superdex 75 300/10 GL column (GE Healthcare).
Design and transient expression of the Ad5 fiber-replacing protein chimeras.
The genes encoding the affibody-containing or dZ-containing fiber-fibritin chimeras were assembled in the plasmid pDV67 (30). These proteins were expressed by transient transfection of 293T cells using Lipofectamine (Invitrogen). Details of all DNA manipulation experiments and DNA sequences of all described constructs are available upon request.
Generation of Ad genomes and rescue, propagation, and purification of Ad vectors.
The genomes of all Ad vectors were assembled in the Ad rescue plasmid pVK500C by two sequential DNA recombinations in E. coli BJ5183 (6) using the reporter genes and the fiber genes cloned in plasmids pShuttle (Clontech) and pZ3.1, respectively. The plasmid pZ3.1 is a derivative of pZeRo2Kan (Invitrogen) into which a fragment of Ad5 genome that includes the fiber gene and the adjacent genomic sequences was cloned. The genome of the “tester” Ad, Ad5LucF0, lacks the fiber gene and has its E1 region replaced with a cytomegalovirus promoter-driven gene cassette that contains the firefly luciferase gene. The genomes of the fiber-modified Ad vectors all contained the same reporter cassette as the tester Ad genome but with a dual reporter TL gene, which encoded a genetic fusion of the herpes simplex virus thymidine kinase and firefly luciferase.
As described previously (17), to rescue the Ad vectors, the genomes of these fiber-deleted or fiber-modified viruses were released from plasmid backbones by PacI restriction endonuclease and used to transfect the wild-type (wt) Ad5 fiber-expressing 293/F28 cells. These cells were further used for intermediate rounds of propagation of the fiber-modified vectors, whereas the final amplification of these viruses was done by infecting 293 cells, which ensured that no wt fiber was present in these preparations. The amplification of the fiber-deleted tester vector, Ad5LucF0, was done entirely in 293/F28 cells, thus yielding a vector equipped with wt fibers. All Ad vectors used in the study were purified by equilibrium centrifugation in CsCl gradients. Particle titers of Ad preparations were determined using light absorbance at 260 nm and by measuring total protein concentration. trans-complementation of the fiber-deleted tester vector, Ad5LucF0, with the transiently expressed fiber chimeras was done exactly as described by Jakubczak et al. (14).
Western blotting.
The detection of fibers in lysates of transfected 293T cells and in purified viruses was done by Western blotting essentially as previously described (1). The modifications to the protocol were that the secondary antibodies were fluorescently labeled with RDye800 (LI-COR, Lincoln, NB) and that the membrane was scanned in a near-infrared fluorescent laser scanner (Odyssey; LI-COR).
To obtain accurate quantitative data on fiber expression and/or incorporation into Ad virions, the samples were loaded on the gels in triplicate, and after the scanning, the images of individual bands were processed using Odyssey software (LI-COR). Statistical analysis of the intensities of these bands was done using analysis of variance and the Tukey procedure to adjust for multiple comparisons.
Enzyme-linked immunosorbent assay.
Detection of binding of Fc11-derived proteins to a recombinant Her2 protein, ErbB2/Fc (BD Biosciences, Palo Alto, CA), was done according to the established protocol (8). The ErbB2/Fc protein was absorbed in the wells of an enzyme-linked immunosorbent assay plate (500 ng/well) and probed with either Fc11 or its affibody-modified derivative (50 ng/well). Bound proteins were detected with the anti-fibritin stalk MAb 5E1 followed by the secondary antibody-horseradish peroxidase conjugate (DAKO, Glostrup, Denmark). The plates were scanned at 490 nm in a multimodality reader (DTX 880; Beckman-Coulter, Fullerton, CA).
Flow cytometry.
Detection of binding of the targeted Fc11-derived proteins to cells was done according to a previously published method (1). Briefly, the cells were incubated with purified recombinant proteins (100 ng/ml), and the cell-bound proteins were detected with the fluorescently labeled Penta-His MAb (Qiagen, Valencia, CA). Similarly, binding of the wt fiber and fiber chimeras expressed in 293T cells was tested using the lysates of transfected cells. In this instance, the quantities of the expressed fibers in the lysates were normalized using quantitative Western blotting data, and the bound proteins were detected by anti-fiber tail MAb 4D2 followed by fluorescently labeled secondary antibody. When the competing free affibody was used, it was added to cells 1 h before the addition of the fiber-containing cell lysates.
Gene transfer.
Transduction of cells was done as previously reported (1). Briefly, cells grown in monolayer culture in 24-well plates were rinsed with a serum-free medium, and then a fresh medium either with or without a competitor protein (the knob or the affibody) was added. Next, the viral vectors were added at the desired multiplicity of infection, and incubation was continued for 30 min. The virus-containing medium was removed, and 24 h later, the cells were lysed using reporter lysis buffer (Promega, Madison, WI). The activity of luciferase in the lysate was measured using the luciferase assay system (Promega) and a tube luminometer (Sirius; Berthold, Pforzheim, Germany).
Ganciclovir-mediated cell killing.
Cells seeded in 96-well plates were infected with Ad vectors at a multiplicity of infection of 100 viral particles per cell, and 8 h later, ganciclovir was added to the wells to a final concentration of 0.25 mM. The number of viable cells per well was determined 6 days later using a nonradioactive cell proliferation assay (Promega).
Nucleotide sequence accession numbers.
The GenBank accession numbers for the sequences that encode the protein chimeras constructed in this study are EU219996 (FF.dZ), EU223251 (FF.Her2:7), EU223252 (FF.Her2:4), EU223253 (11F.Her2:7), EU223254 (11F.Her2:4), and EU223255 (11F.dZ).
RESULTS
Design and functional validation of affibody-containing equivalent of the Ad5 fiber knob domain.
As the first step toward the design of a Her2-targeted Ad, we sought to make a structural and functional equivalent of the fiber knob domain by fusing the Her2-specific affibodies Zher2:7 and Zher2:4 (33) to the carboxy-terminal fragment of the phage T4 fibritin protein (Fig. 1). The Zher2:7 and Zher2:4 proteins were chosen as targeting ligands for Ad because they were the best characterized of all Her2-specific affibody species available at the time this work started. This ligand presentation approach was chosen on the basis of our previous successes with using fibritin-based chimeras for Ad5 vector targeting (1, 16). The resultant proteins, termed Fc11.Her2:7 and Fc11.Her2:4, comprised two α-helical coiled coils and the trimerization foldon domain of the fibritin (Fc11) connected to the affibody ligands with a flexible peptide linker (Fig. 1D). In these proteins, the two functions that are performed by the fiber knob in an Ad particle—trimerization and receptor binding—were disengaged by being assigned to the foldon and the affibody, respectively. The proteins were produced in bacteria, and their ability to self-assemble into homotrimers, which is required for association of the fiber with the penton base, was confirmed (Fig. 2A).
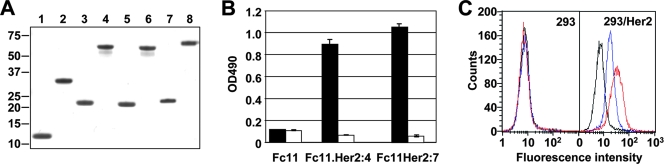
FIG. 2.
Molecular properties of the Fc11-affibody chimeras. (A) Sodium dodecyl sulfate-polyacrylamide gel electrophoresis of purified recombinant proteins. The Fc11 backbone (lanes 1 and 2), its affibody-modified derivatives Fc11.Her2:7 (lanes 3 and 4) and Fc11.Her2:4 (lanes 5 and 6), and Ad5 fiber knob for comparison (lanes 7 and 8) purified from bacterial cells were loaded on the gel with or without prior denaturation by boiling (odd-numbered and even-numbered lanes, respectively). While the proteins in the denatured samples are monomeric polypeptides, the proteins in the nondenatured samples are trimers. The positions and molecular masses (in kDa) of protein standards are shown on the left. (B) Binding of affibody-containing proteins to recombinant Her2. Purified ErbB2/Fc protein immobilized on an enzyme-linked immunosorbent assay plate (500 ng per well) was probed with the Fc11 backbone, Fc11.Her2:7, or Fc11.Her2:4 (black bars) followed by anti-His5 MAb and secondary antibody-horseradish peroxidase conjugate. The white bars indicate background binding to casein. Error bars indicate the standard deviations obtained in triplicate samples. OD490, optical density at 490 nm. (C) Target receptor-selective attachment of the affibody-containing proteins to Her2-positive cells. Fc11 (black), Fc11.Her2:7 (red), and Fc11.Her2:4 (blue) were used to probe the surface of Her2-negative 293 cells and Her2-expressing 293/Her2 cells.
Because the target-binding site of the affibody was now adjacent to the intermolecular fusion point, the function of this site could be compromised by the immediate proximity of the foldon. To rule this out, the functional integrity of the ligand was verified by enzyme-linked immunosorbent assay, which confirmed binding of the affibody-containing proteins, but not the Fc11 backbone, to purified recombinant Her2 protein (Fig. 2B). Furthermore, flow cytometry revealed that the affibody-modified proteins selectively attached to the target receptor on the surface of Her2-positive 293/Her2 cells while demonstrating no binding to Her2-negative 293 cells (Fig. 2C). These results showed that the functional duality of the Ad5 fiber knob could be successfully recreated using an unrelated self-trimerizing protein scaffold in combination with an artificially designed protein ligand.
Affibody-targeted knob-deleted fibers are fully functional and compatible with Ad virion assembly.
Next, two types of Her2-targeted derivatives of the Ad5 fiber were designed by replacing the knob domain of the wt Ad5 fiber with the fibritin-affibody constructs described in the preceding section. Chimeras of the FF type were built on the basis of the previously developed backbone (16), which consists of the tail and the two amino-terminal repeats of the fiber shaft fused with the fragment of the phage T4 fibritin protein that includes the coiled coils 6 through 13 of the stalk domain and the carboxy terminal foldon (Fig. 1E). Chimeras of the 11F type were designed to more closely resemble the wt Ad5 fiber. The 11F chimeras contained the tail and the complete shaft of the fiber fused with coiled-coils 12 and 13 and the foldon of the fibritin. Chimeras of both types carried either the carboxy-terminal affibody ligands or dZ connected via a flexible peptide linker (Fig. 1F).
Transient expression of both types of Her2-targeted fibers in human cells showed that despite the significant redesign, these modified proteins were produced as stable polypeptides and at levels comparable with those of the wt Ad5 fiber. For instance, digital processing of the Western blots of these fully denatured proteins showed that the expression of 11F.dZ, 11F.Her2:4, and 11F.Her2:7 was 54%, 54%, and 64% of unmodified Ad5 fiber expression, respectively (data not shown). Most important, the folding patterns of these chimeras proved to be consistent with the formation of the fiber-like trimers (Fig. 3A and data not shown). In a cell-binding study, the chimeras carrying the affibodies, but not the proteins fused with the affibody scaffold (dZ), recognized cell-associated Her2, confirming that the Her2 specificity of these chimeras was due to the affibody ligands (Fig. 3B and data not shown). This finding was further corroborated by efficient dose-dependent inhibition of Her2 binding with a free affibody competitor (Fig. 3C).
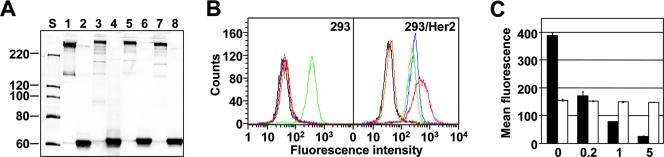
FIG. 3.
Validation of the affibody-modified fiber replacement proteins. (A) Western blot analysis of the transiently expressed proteins in cell lysates containing the wt Ad5 fiber (lanes 1 and 2), 11F.dZ (lanes 3 and 4), 11F.Her2:7 (lanes 5 and 6), or F11.Her2:4 (lanes 7 and 8) in seminative form (odd-numbered lanes) or denatured form (even-numbered lanes). Each line of the gel was loaded with the aliquot of cell lysate containing 8 μg of total soluble protein. S, protein molecular mass standards. (B) Fluorescence-activated cell sorting assay of the transiently expressed fibers: wt Ad5 fiber (green), 11F.dZ (orange), 11F.Her2:4 (blue), and 11F.Her2:7 (red). The black curve represents the background signal obtained with the lysate of mock-transfected cells. (C) Dose-dependent inhibition of cell attachment of the 11F.Her2:7 protein detected by flow cytometry. Mean fluorescence intensity of the 293/Her2 cells probed with either the 11F.Her2:7-containing lysate (black bars) or the wt fiber-containing lysate (white bars). Shown below the graph are the concentrations of the free affibody competitor (in mg/ml) used to inhibit Her2 binding. Error bars indicate the standard deviations obtained in triplicate samples.
In addition, functional equivalency of the chimeras to the wt Ad5 fiber was demonstrated by encapsidating these proteins into the virions of the fiber-deleted tester vector, Ad5LucF0. First, virus lacking the fiber gene in its genome was rescued and propagated in cells stably expressing the wt fiber. Next, cells transiently transfected to produce affibody-modified chimeras were infected with this tester Ad to allow for encapsidation of the targeted fibers into Ad5LucF0 progeny virions. This trans-complementation yielded Ad particles that contained the Her2-targeted chimeras, as confirmed by Western blot analysis of purified virions (Fig. 4).
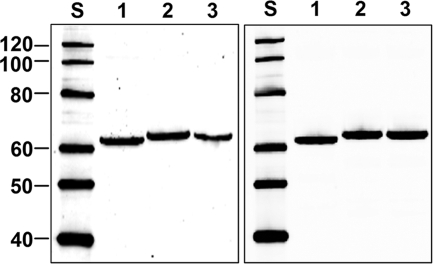
FIG. 4.
Encapsidation of the modified 11F-type fiber chimeras into Ad particles. CsCl-purified Ad virions were obtained either by trans-complementation of the tester vector, Ad5LucF0, or by standard Ad rescue and propagation protocol. All samples were fully denatured before electrophoresis. (Left) Tester Ad trans-complemented with the wt fiber (lane 1), 11F.Her2:7 (lane 2), or 11F.Her2:4 (lane 3) with 3 × 1010 virus particles per lane. (Right) AdTL (lane 1), AdTL.11FHer2:7 (lane 2), and AdTL.11FHer2:4 (lane 3) with 1010 viral particles per lane. The molecular masses (in kDa) of protein standards (lane S) are shown on the left.
In aggregate, these findings proved that the biosynthesis, the folding, and the intracellular trafficking of the affibody-modified fiber chimeras were consistent with the Ad5 virion assembly process.
Incorporation of affibody-targeted fiber chimeras into Ad particles yields complete and fully functional virions.
The genes encoding the affibody-modified fibers were transferred into the E1-deleted Ad5 genome containing an expression cassette with a dual reporter gene termed the TL. The viruses were rescued by transfection of 293/F28 cells, which stably express the wt Ad5 fiber. The rescued mosaic virions were further amplified in 293/F28 cells and then propagated in 293 cells. The yields of the targeted vectors were very similar to that of the control Ad containing the wt Ad5 fiber. For instance, the yields for AdTL.11F.Her2:4 and AdTL.11F.Her2:7 were 7,450 and 9,720 viral particles per infected cell, respectively, while the yield for the control AdTL was 8,140 particles per cell. The efficiency with which the chimeras were encapsidated in Ad particles was assessed by Western blot analysis of highly purified viral preparations. Importantly, this assay showed that the amount of chimeric 11F-type fibers incorporated per modified Ad virion was the same as seen with the wt Ad5 protein (Fig. 4). Incorporation of chimeras of the FF type was similarly efficient (data not shown).
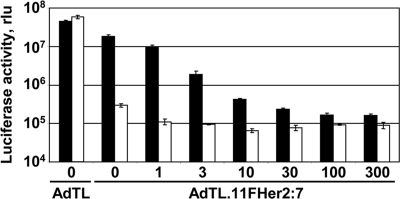
The infectivity of the tropism-modified vectors, AdTL.11F.Her2:4 and AdTL.11F.Her2:7, was tested in 293 and 293/Her2 cells. In line with the cell binding data (above), the retargeted vectors showed much greater infectivity in Her2-positive cells than in control cells. The efficiency of gene delivery to Her2-expressing cells by the best performer, AdTL.11F.Her2:7, was similar to that achieved with the isogenic vector containing the wt fiber (Fig. 5). The concentration-dependent inhibition of this infectivity with free affibody further confirmed the Her2-dependent mode of gene transfer by the targeted Ads (Fig. 5).
FIG. 5.
Affibody-mediated viral gene transfer to Her2-expressing cells. Her2-positive 293/Her2 cells (black bars) or Her2-negative 293 cells (white bars) were transduced with Ad vectors containing either wt fiber (AdTL) or the affibody-targeted fiber chimera AdTL.11F.Her2:7. The efficacy of transduction was gauged by the activity of luciferase expressed by the viruses. Shown below the graph are the concentrations of free affibody (in μg/ml) used to inhibit cell transduction. Error bars indicate the standard deviations obtained in triplicate samples. This competitor protein had no effect on the transduction of either cell line by AdTL (not shown).
Affibody-containing Ad5 vectors use Her2 as a surrogate receptor to transduce and kill human cancer cells.
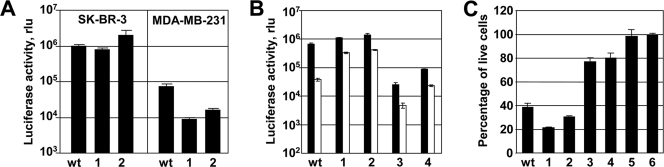
Next, we tested the ability of the affibody-targeted chimeras of the 11F type to direct Ad vectors to human mammary carcinoma cell lines with different Her2 phenotypes. Transduction of these cell lines by vectors encapsidating targeted chimeras of the 11F type correlated with the cells' Her2 expression level. Transduction was efficient in SK-BR-3 cells, which expressed 1 × 106 to 2 × 106 copies of Her2 receptor per cell (28); compared with unmodified Ad5 vector, the AdTL.11F.Her2:4 vector yielded an equal level of transgene expression, and the AdTL.11F.Her2:7 vector was twice as efficient as the control (Fig. 6A). In contrast, Her2-negative MDA-MB-231 cells were infected very poorly by both of the affibody-containing Ad vectors: signals were at a background level. These results further confirmed the key role of Her2 in infection by these tropism-modified viruses.
FIG. 6.
Transduction of human breast cancer cells with the Her2-targeted Ad vectors. The following viruses were used: wt, AdTL; lanes 1, AdTL.11F.Her2:4; lanes 2, AdTL.11F.Her2:7; lanes 3, AdTL.FF.Her2:4; lanes 4, AdTL.FF.Her2:7; lane 5, Ad5Luc1, Ad expressing only luciferase with the wt fiber; and lane 6, no virus. All transductions were done at a multiplicity of infection of 100 virus particles per cell. In all three panels, error bars indicate the standard deviations obtained in triplicate samples. (A) Comparison of gene delivery to Her2-positive SK-BR-3 and Her2-negative MDA-MB-231 cells. Each cell line was infected with either Ad vector containing wt fiber (AdTL) or Ad vector containing a Her2-targeted fiber chimera (AdTL.11F.Her2:4 or AdTL.11F.Her2:7). The efficacy of transduction was measured using the activity of vector-encoded luciferase reporter. (B) Comparison of gene delivery to SK-BR-3 cells with AdTL, AdTL.11F.Her2:4, AdTL.11F.Her2:7, AdTL.FF.Her2:4, and AdTL.FF.Her2:7. Black bars show luciferase expression by unblocked vectors. White bars show the levels of bioluminescence in the cells preincubated with a free competitor (Ad5 fiber knob for AdTL or Her2:4 affibody for targeted Ads, each used at a final concentration of 30 μg/ml). (C) Targeting of Ad with redesigned fiber-fibritin-affibody proteins yields more efficient killing of tumor cells. SK-BR-3 cells transduced with Ad vectors were treated with ganciclovir to engage the drug-converting activity of the TL transgene. The number of viable cells determined using an MTT [3-(4,5-dimethylthiazol-2-yl)2 2,5-diphenyl tetrazolium bromide] assay is shown. rlu, relative light units.
To see whether vector infectivity depended on the type of targeting chimera encapsidated, we did a comparative transduction of SK-BR-3 cells with vectors containing either the FF-type or 11F-type proteins. In this test (Fig. 6B), the 11F-type vectors outperformed their FF-type counterparts by a large margin (16- and 43-fold for the vectors containing the Zher2:7 and Zher2:4 affibodies, respectively). These findings indicated that the 11F-type proteins, by mimicking the native structure of the wt fiber more closely than the FF-type proteins, supported more efficient viral infection. Additional confirmation of the Her2 specificity of the targeted Ads was obtained using free affibody ligand as a competitor (Fig. 6B).
Next, the prodrug-converting activity of the TL transgene carried by the Ad vectors was used to determine whether the designed viruses could serve as prototypes for gene therapeutics. Toward this end, SK-BR-3 cells were transduced by the same panel of vectors and then treated with ganciclovir. In line with the gene transfer data, the 11F-type vectors mediated much more efficient killing of target cells than did the FF-type Ads. The 11F-type vectors also proved to be more efficient than the nontargeted control AdTL (Fig. 6C).
Together, these data proved that affibody-enabled targeting of Ad vectors to Her2 is an efficient strategy for improving both the specificity of Ad-mediated gene delivery to Her2-positive tumor cells and the therapeutic outcomes of this transduction.
DISCUSSION
The goal of this study was to explore the possibility of genetically altering the natural tropism of Ad5 by using artificially designed proteins, the affibodies. This was attempted in order to resolve a fundamental problem that has hampered progress in the development of targeted Ad vectors for in vivo gene delivery—the shortage of appropriate targeting ligands. Since these ligands are to be incorporated into the Ad capsid genetically, they have to satisfy a set of stringent structural, functional, and biosynthetic criteria, which in aggregate effectively eliminate from the list of potential ligand candidates nearly all naturally existing proteins. Therefore, instead of continuing an unpromising search for such molecules in nature, we took an alternative route of employing artificial proteins that have been rationally designed to fit the profile of a targeting ligand for Ad.
In this study, affibody proteins derived from a compact, simply structured, and stable scaffold of Staphylococcus protein A dZ were used to target knob-deleted Ad fiber proteins to Her2. The trimeric structure of these chimeras was facilitated by the inclusion of a trimerization domain of the phage T4 fibritin protein. These modifications yielded fully functional tripartite protein chimeras that proved to be an adequate replacement for the wt Ad5 fiber. Namely, they formed stable fiber-like trimers and consequently incorporated into Ad particles with efficacy of encapsidation equal to that of the wt fiber. The affibody ligands within these targeting proteins retained their Her2 binding specificity and directed the virus to the target receptor. Furthermore, this engineered tropism to Her2 allowed two of the targeted vectors to deliver the transgene to Her2-positive human breast cancer cells with efficiency equal to or greater than that of the virus with unaltered receptor specificity. Thus, this tropism modification improved both the vector's specificity for the target cells and the efficiency of gene delivery.
The following considerations suggest that for Ads, genetic modification of virus tropism with affibodies could be the targeting strategy of choice. First, both of the affibody species we tried (Zher2:7 and Zher2:4) resulted in functional fibers and virions. These findings, taken together with the high level of structural homology among affibodies, suggest that the overall success rate with other affibodies should be high, too. Second, the efficacy of Ad vector targeting and gene delivery may be improved further by using affibody multimers as ligands. Of note, it has been shown that compared to affibody monomers, affibody dimers have higher affinities for Her2 and better tumor localization properties in vivo (26, 27). Alternatively, affibodies with higher affinities for their targets could be used. While the dissociation equilibrium constants (KD) for the Zher2:4 and Zher2:7 affibodies used in this work are 50 nM and 140 nM (33), respectively, recent affinity maturation work resulted in much better Her2 binders, with KDs in the lower picomolar range (24). Third, in addition to their high affinities, the affibodies show excellent target selectivity both in vitro and in vivo as they are able to discriminate between closely related members of the same receptor families (9). Fourth, the repertoire of affibodies with specificities for disease-related targets is growing, and significant commercial interest in this technology is expected to ensure its further growth. Fifth, the fact that some affibodies trigger internalization upon binding to the target (9) provides an opportunity to design targeted Ad vectors that would not require integrin-mediated internalization. For such vectors, integrin expression by the target tissues would not be a limiting factor for efficient gene delivery. Sixth, affibodies may be developed that are specific for molecular markers of diseases for which natural ligands do not exist, allowing Ad targeting of these molecules.
It should be noted that although the use of affibody ligands is an attractive strategy, it does not by itself guarantee that Ad vector tropism will be successfully altered. In this regard, recent attempts to target Ad5 by using an affibody-modified, knob-deleted fiber fused with the neck region peptide of lung surfactant protein D have been unsuccessful (19). In the same study, incorporation of the affibody within the HI loop of the fiber knob resulted in a poor encapsidation of the resultant proteins in Ad virions, which was only 25% of that seen with the wt fiber. Similarly, our affibody-modified chimeras of the FF type were only marginally useful in directing the virus. It was thus the particular combination of the affibody with the improved ligand-presenting molecule, the 11F chimera, that made our strategy a success. It is possible, however, that alternative fiber-derived chimeric backbones, such as the recently developed fusions between the Ad5 fiber and reovirus protein σ1 (22, 25), might be successful in accommodating affibodies as targeting ligands for Ad. Regardless of which ligand-presenting protein is used to replace Ad fiber, targeting Ads to new receptors may affect virus uptake, intracellular trafficking, and nuclear import of viral genomes, and, thus, these potential problems must be considered for each new ligand/receptor pairing. Also, despite our success with the 11F chimera, it is possible that some affibodies may fail to function as ligands when fused to this scaffold.
What makes the combination of the fiber replacement approach with the use of affibodies particularly attractive is that this strategy should be equally efficient when applied to Ad serotypes other than human Ad5. Several of those serotypes are currently being explored as alternative vector prototypes with the goal of overcoming the preexisting anti-Ad5 immunity in humans (10, 15). The high degree of structural similarity between various Ad fibers (7) suggests that replacement of the fiber knob in these alternative vectors with fibritin-affibody fusions is likely to be a successful approach. Targeting of those other Ad fibers by replacing their knobs with the fibritin-affibody fusions will be independent of both the identity of their natural receptors and the identification of their receptor-binding sites; thus, this strategy would be predicted to be rather straightforward.
From the standpoint of basic science research, efficient remodeling of the Ad's cell binding mechanism with affibodies makes the Ad-cell interaction manipulatable and thus allows the study of important biological questions that could not be answered using viruses with native tropism. For instance, affibody-based modification of Ad vector tropism could be used to study the dependence of the overall efficacy and temporal dynamics of the infection process on the nature of the target receptor, the localization of the virus-binding site within the receptor molecule, or its position relative to the cell surface. It is thus envisioned that the work reported here, by establishing that affibodies can be used to successfully manipulate Ad vector tropism, will lead to many new research opportunities in both the basic and applied fields of science.
Acknowledgments
We are grateful to the personnel of the DNA Analysis Facility at M. D. Anderson Cancer Center for their help with DNA sequencing. We are indebted to Stephanie Deming, Diane Hackett, and David Bier for their excellent assistance with preparing the manuscript for publication. We extend our thanks to Marine Kojanyan for her technical help, to Roland Bassett for doing biostatistical analyses, and to Amer Najjar for his help with the flow cytometry experiments. We thank Vladimir Ponomarev (Memorial Sloan-Kettering Cancer Center) for making the gene of the TL reporter available for this work. We gratefully acknowledge Mary Ann Accavitti-Loper (University of Alabama at Birmingham) for generating the anti-fibritin hybridomas.
This work was supported by Susan G. Komen for the Cure research grant BCTR0504193, Public Health Service grant R01 CA116621, and the Commonwealth Foundation for Cancer Research Award (all to V.K.). Additional support was provided by Cancer Center support grant P30 CA 16672.
Footnotes
Published ahead of print on 7 November 2007.
REFERENCES
- 1.Belousova, N., N. Korokhov, V. Krendelshchikova, V. Simonenko, G. Mikheeva, P. L. Triozzi, W. A. Aldrich, P. T. Banerjee, S. D. Gillies, D. T. Curiel, and V. Krasnykh. 2003. Genetically targeted adenovirus vector directed to CD40-expressing cells. J. Virol. 7711367-11377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Belousova, N., V. Krendelchtchikova, D. T. Curiel, and V. Krasnykh. 2002. Modulation of adenovirus vector tropism via incorporation of polypeptide ligands into the fiber protein. J. Virol. 768621-8631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bergelson, J. M., J. A. Cunningham, G. Droguett, E. A. Kurt-Jones, A. Krithivas, J. S. Hong, M. S. Horwitz, R. L. Crowell, and R. W. Finberg. 1997. Isolation of a common receptor for coxsackie B viruses and adenoviruses 2 and 5. Science 2751320-1323. [DOI] [PubMed] [Google Scholar]
- 4.Binz, H. K., P. Amstutz, and A. Pluckthun. 2005. Engineering novel binding proteins from nonimmunoglobulin domains. Nat. Biotechnol. 231257-1268. [DOI] [PubMed] [Google Scholar]
- 5.Campos, S. K., and M. A. Barry. 2007. Current advances and future challenges in adenoviral vector biology and targeting. Curr. Gene Ther. 7189-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chartier, C., E. Degryse, M. Gantzer, A. Dieterle, A. Pavirani, and M. Mehtali. 1996. Efficient generation of recombinant adenovirus vectors by homologous recombination in Escherichia coli. J. Virol. 704805-4810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chroboczek, J., R. W. Ruigrok, and S. Cusack. 1995. Adenovirus fiber. Curr. Top. Microbiol. Immunol. 199163-200. [DOI] [PubMed] [Google Scholar]
- 8.Dmitriev, I., V. Krasnykh, C. R. Miller, M. Wang, E. Kashentseva, G. Mikheeva, N. Belousova, and D. T. Curiel. 1998. An adenovirus vector with genetically modified fibers demonstrates expanded tropism via utilization of a coxsackievirus and adenovirus receptor-independent cell entry mechanism. J. Virol. 729706-9713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Friedman, M., E. Nordberg, I. Hoiden-Guthenberg, H. Brismar, G. P. Adams, F. Y. Nilsson, J. Carlsson, and S. Stahl. 2007. Phage display selection of affibody molecules with specific binding to the extracellular domain of the epidermal growth factor receptor. Protein Eng. Des. Sel. 20189-199. [DOI] [PubMed] [Google Scholar]
- 10.Havenga, M., R. Vogels, D. Zuijdgeest, K. Radosevic, S. Mueller, M. Sieuwerts, F. Weichold, I. Damen, J. Kaspers, A. Lemckert, M. van Meerendonk, R. van der Vlugt, L. Holterman, D. Hone, Y. Skeiky, R. Mintardjo, G. Gillissen, D. Barouch, J. Sadoff, and J. Goudsmit. 2006. Novel replication-incompetent adenoviral B-group vectors: high vector stability and yield in PER.C6 cells. J. Gen. Virol. 872135-2143. [DOI] [PubMed] [Google Scholar]
- 11.Henry, L. J., D. Xia, M. E. Wilke, J. Deisenhofer, and R. D. Gerard. 1994. Characterization of the knob domain of the adenovirus type 5 fiber protein expressed in Escherichia coli. J. Virol. 685239-5246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hong, J. S., and J. A. Engler. 1996. Domains required for assembly of adenovirus type 2 fiber trimers. J. Virol. 707071-7078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Imperiale, M. J., and S. Kochanek. 2004. Adenovirus vectors: biology, design, and production. Curr. Top. Microbiol. Immunol. 273335-357. [DOI] [PubMed] [Google Scholar]
- 14.Jakubczak, J. L., M. L. Rollence, D. A. Stewart, J. D. Jafari, D. J. Von Seggern, G. R. Nemerow, S. C. Stevenson, and P. L. Hallenbeck. 2001. Adenovirus type 5 viral particles pseudotyped with mutagenized fiber proteins show diminished infectivity of coxsackie B-adenovirus receptor-bearing cells. J. Virol. 752972-2981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kobinger, G. P., H. Feldmann, Y. Zhi, G. Schumer, G. Gao, F. Feldmann, S. Jones, and J. M. Wilson. 2006. Chimpanzee adenovirus vaccine protects against Zaire Ebola virus. Virology 346394-401. [DOI] [PubMed] [Google Scholar]
- 16.Krasnykh, V., N. Belousova, N. Korokhov, G. Mikheeva, and D. T. Curiel. 2001. Genetic targeting of an adenovirus vector via replacement of the fiber protein with the phage T4 fibritin. J. Virol. 754176-4183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krasnykh, V., I. Dmitriev, G. Mikheeva, C. R. Miller, N. Belousova, and D. T. Curiel. 1998. Characterization of an adenovirus vector containing a heterologous peptide epitope in the HI loop of the fiber knob. J. Virol. 721844-1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krasnykh, V. N., J. T. Douglas, and V. W. van Beusechem. 2000. Genetic targeting of adenoviral vectors. Mol. Ther. 1391-405. [DOI] [PubMed] [Google Scholar]
- 19.Magnusson, M. K., P. Henning, S. Myhre, M. Wikman, T. G. Uil, M. Friedman, K. M. Andersson, S. S. Hong, R. C. Hoeben, N. A. Habib, S. Stahl, P. Boulanger, and L. Lindholm. 2007. Adenovirus 5 vector genetically re-targeted by an affibody molecule with specificity for tumor antigen HER2/neu. Cancer Gene Ther. 14468-479. [DOI] [PubMed] [Google Scholar]
- 20.Magnusson, M. K., S. See Hong, P. Henning, P. Boulanger, and L. Lindholm. 2002. Genetic retargeting of adenovirus vectors: functionality of targeting ligands and their influence on virus viability. J. Gene Med. 4356-370. [DOI] [PubMed] [Google Scholar]
- 21.Menard, S., S. M. Pupa, M. Campiglio, and E. Tagliabue. 2003. Biologic and therapeutic role of HER2 in cancer. Oncogene 226570-6578. [DOI] [PubMed] [Google Scholar]
- 22.Mercier, G. T., J. A. Campbell, J. D. Chappell, T. Stehle, T. S. Dermody, and M. A. Barry. 2004. A chimeric adenovirus vector encoding reovirus attachment protein sigma1 targets cells expressing junctional adhesion molecule 1. Proc. Natl. Acad. Sci. USA 1016188-6193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nord, K., E. Gunneriusson, J. Ringdahl, S. Stahl, M. Uhlen, and P. A. Nygren. 1997. Binding proteins selected from combinatorial libraries of an alpha-helical bacterial receptor domain. Nat. Biotechnol. 15772-777. [DOI] [PubMed] [Google Scholar]
- 24.Orlova, A., M. Magnusson, T. L. Eriksson, M. Nilsson, B. Larsson, I. Hoiden-Guthenberg, C. Widstrom, J. Carlsson, V. Tolmachev, S. Stahl, and F. Y. Nilsson. 2006. Tumor imaging using a picomolar affinity HER2 binding affibody molecule. Cancer Res. 664339-4348. [DOI] [PubMed] [Google Scholar]
- 25.Schagen, F. H., F. M. Wensveen, J. E. Carette, T. S. Dermody, W. R. Gerritsen, and V. W. van Beusechem. 2006. Genetic targeting of adenovirus vectors using a reovirus sigma1-based attachment protein. Mol. Ther. 13997-1005. [DOI] [PubMed] [Google Scholar]
- 26.Steffen, A. C., A. Orlova, M. Wikman, F. Y. Nilsson, S. Stahl, G. P. Adams, V. Tolmachev, and J. Carlsson. 2006. Affibody-mediated tumour targeting of HER-2 expressing xenografts in mice. Eur. J. Nucl. Med. Mol. Imaging 33631-638. [DOI] [PubMed] [Google Scholar]
- 27.Steffen, A. C., M. Wikman, V. Tolmachev, G. P. Adams, F. Y. Nilsson, S. Stahl, and J. Carlsson. 2005. In vitro characterization of a bivalent anti-HER-2 affibody with potential for radionuclide-based diagnostics. Cancer Biother. Radiopharm. 20239-248. [DOI] [PubMed] [Google Scholar]
- 28.Tang, Y., D. Scollard, P. Chen, J. Wang, C. Holloway, and R. M. Reilly. 2005. Imaging of HER2/neu expression in BT-474 human breast cancer xenografts in athymic mice using [(99m)Tc]-HYNIC-trastuzumab (Herceptin) Fab fragments. Nucl. Med. Commun. 26427-432. [DOI] [PubMed] [Google Scholar]
- 29.Tomko, R. P., R. Xu, and L. Philipson. 1997. HCAR and MCAR: the human and mouse cellular receptors for subgroup C adenoviruses and group B coxsackieviruses. Proc. Natl. Acad. Sci. USA 943352-3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Von Seggern, D. J., S. Huang, S. K. Fleck, S. C. Stevenson, and G. R. Nemerow. 2000. Adenovirus vector pseudotyping in fiber-expressing cell lines: improved transduction of Epstein-Barr virus-transformed B cells. J. Virol. 74354-362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wickham, T. J., E. J. Filardo, D. A. Cheresh, and G. R. Nemerow. 1994. Integrin alpha v beta 5 selectively promotes adenovirus mediated cell membrane permeabilization. J. Cell Biol. 127257-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wickham, T. J., P. Mathias, D. A. Cheresh, and G. R. Nemerow. 1993. Integrins alpha v beta 3 and alpha v beta 5 promote adenovirus internalization but not virus attachment. Cell 73309-319. [DOI] [PubMed] [Google Scholar]
- 33.Wikman, M., A. C. Steffen, E. Gunneriusson, V. Tolmachev, G. P. Adams, J. Carlsson, and S. Stahl. 2004. Selection and characterization of HER2/neu-binding affibody ligands. Protein Eng. Des. Sel. 17455-462. [DOI] [PubMed] [Google Scholar]
- 34.Xia, H., B. Anderson, Q. Mao, and B. L. Davidson. 2000. Recombinant human adenovirus: targeting to the human transferrin receptor improves gene transfer to brain microcapillary endothelium. J. Virol. 7411359-11366. [DOI] [PMC free article] [PubMed] [Google Scholar]