Abstract
The crystal structure of the vesicular stomatitis virus nucleoprotein (N) in complex with RNA reveals extensive and specific intermolecular interactions among the N molecules in the 10-member oligomer. What roles these interactions play in encapsidating RNA was studied by mutagenesis of the N protein. Three N mutants intended for disruption of the intermolecular interactions were designed and coexpressed with the phosphoprotein (P) in an Escherichia coli system previously described (T. J. Green et al., J. Virol. 74:9515-9524, 2000). Mutants N (Δ1-22), N (Δ347-352), and N (320-324, (Ala)5) lost RNA encapsidation and oligomerization but still bound with P. Another mutant, N (Ser290→Trp), was able to form a stable ring-like N oligomer and bind with the P protein but was no longer able to encapsidate RNA. The crystal structure of N (Ser290→Trp) at 2.8 Å resolution showed that this mutant can maintain all the same intermolecular interactions as the wild-type N except for a slight unwinding of the N-terminal lobe. These results suggest that the intermolecular contacts among the N molecules are required for encapsidation of the viral RNA.
All negative-strand RNA viruses contain a ribonucleoprotein (RNP) complex that consists of the viral genome RNA completely enwrapped by the nucleoprotein (N). Vesicular stomatitis virus (VSV) is a nonsegmented negative-strand RNA virus that belongs to the rhabdovirus family. The genome of VSV encodes five proteins: the nucleoprotein (N), the phosphoprotien (P), the matrix protein (M), the glycoprotein (G), and the large subunit of the polymerase (L). The RNP of VSV is packaged in a bullet-shaped virion by M, the protein that condenses the RNP during virion assembly, and G, the surface protein that is embedded in the viral envelope (14, 16, 22, 29). P and L, which are other viral proteins, are packaged through their association with the RNP. After entry into the cytoplasm via membrane fusion mediated by G, the RNP is released from the virion and serves as the active template with which the copackaged polymerase proteins transcribe mRNAs from the five viral genes in the RNP. In the later stage of the virus replication cycle, a positive strand of the viral genome (cRNA) is produced in the form of an RNP. The cRNA-containing RNP then serves as the template for replication that also generates the viral genomic RNA in the form of an RNP ready to be packaged in the virion. Throughout the entire virus replication cycle of a negative-strand RNA virus, the genome-length viral RNA (cRNA or viral genomic RNA) is only present in the form of an RNP that is either serving as a template for RNA synthesis or being packaged in the virion. The assembly of the RNP is therefore a critical step in the replication of negative-strand RNA viruses.
Functions of the N protein require it to have two essential properties. First, the N protein needs to bind a single-strand RNA. Second, the N protein has to polymerize in order to cover the entire length of the viral genome RNA. A number of experiments have attempted to probe these properties. In the case of the VSV N protein, the region responsible for RNA binding was initially searched by comparisons of the amino acid sequences of nucleoproteins from different negative-strand RNA virus families (3). The sequence alignments suggested that the first 350 residues or so may have a common structure that binds RNA. The RNA binding region was experimentally narrowed to residues 298 to 352 of the N protein of rabies virus (RABV), another member of the rhabdovirus family (23). Digestion of the RABV RNP with trypsin removed a C-terminal portion (residues 377 to 450) but left the RNA binding region of the RABV N protein unchanged. By UV-laser cross-linking followed by protease digestion, a fragment comprising residues 298 to 353 was shown to be bound to the probing RNA. In a baculovirus expression system, the RABV N protein formed a ring structure that contained multiple copies of the N protein and a single-strand RNA with random sequences (21). The trypsin digestion of this RABV N protein complex resulted in a similar RNP structure that protects the RNA and shows the same N-N organizational characteristics as seen with the wild-type (wt) RNP. These data suggest that the RNA binding properties of the N protein are localized in a defined region within the full-length protein. However, other data showed that the terminal residues of the N protein also play a role in RNA encapsidation. Deletion or mutation of the last five residues from the C terminus of the VSV N protein completely abolished its ability to encapsidate RNA in an in vitro assembly experiment (6). It appears that the full-length N protein is required to encapsidate the RNA but that some of the C-terminal portion may be removed after the N-RNA complex has been assembled. It is known that the N protein has to be maintained in an RNA binding-competent form before RNA encapsidation can take place. The viral P protein has been shown to play this role during virus replication. An N-P complex, which was the only active form that could carry out N encapsidation of RNA, was isolated from VSV-infected cells (32). Such a complex was also isolated from coexpression of the N and P protein and was shown to be RNA-free (7, 25, 26, 31). Only this N/P complex, not N alone, can support viral RNA replication efficiently (24, 32). The N protein was incorporated into the RNP or formed N oligomers with dissociation of the P protein (31, 32). The N protein is always in a polymeric state when associated with RNA.
The crystal structure of the N protein in complex with a single-strand RNA was recently determined for two members of the rhabdovirus family, VSV (20) and RABV (2). The regions in contact with RNA were clearly mapped to a cavity formed between two structural lobes of the N protein (Fig. 1A). The cavity has a hydrophobic space to accommodate the bases of the RNA, and positively charged residues are placed along the cavity to interact with the phosphate backbone. More importantly, intermolecular interactions can be described in atomic detail among the adjacent N molecules, including the side-by-side interactions and unique contacts by the N-terminal arm and the extended loop in the C-terminal lobe (C-lobe). The contacts are divided into three groups (I, II, and III), and they span four neighboring N molecules in a repeated pattern. In this study, we generated mutations in each unique contact to delineate their roles in RNA encapsidation in a system that coexpresses the N and P proteins in Escherichia coli. The N/P coexpression system is more relevant than a system expressing N alone, which could also produce N-RNA complexes, to study the RNA encapsidation related to viral replication, because the N/P complex is the entity that participates in the RNP assembly. In addition, the mutations were generated in the area of side-by-side interactions between adjacent N molecules and in the RNA binding cavity. The results indicated that the intermolecular interactions rendered by the unique contacts, as well as the side-by-side interactions, are required for RNA encapsidation. At the same time, the N molecules could form an N oligomer structure similar to that seen in the RNP even in the absence of RNA, as illustrated by the Ser290→Trp mutation. In this mutant, a bulky side chain was introduced in the cavity to block RNA binding. Our findings provide insights into how N oligomerization may contribute to the regulation of RNP assembly.
FIG. 1.
(A) A cartoon depicting the intermolecular interactions of the N protein molecules in the N-RNA complex. The two lobes in the reference N molecule (red) are labeled AN and AC, standing for the A N-lobe and the A C-lobe, respectively. The 5′ end of the RNA molecule (darker yellow) is on the left, and the 3′ end is on the right, across all the N molecules. Three other unique N molecules are labeled B (green), C (blue), and D (yellow), respectively, and a fifth N molecule (E) (white) is added to indicate that the intermolecular interactions continue to other N molecules. The key elements involved in the intermolecular interactions are also labeled, including the N-terminal arm, the extended loop, and contacts I, II, and III. (B) Composite of mutations designed on the basis of the N protein. The N- and C-lobes are colored green and yellow, respectively. Mutations are colored red with appropriate labels. The RNA binding cavity formed between the two lobes is shown as a semitransparent surface.
MATERIALS AND METHODS
Plasmid construction.
A QuikChange site-directed mutagenesis kit (Stratagene) was used to create mutant N plasmids pET-N (Δ347-352)-P, pET-N (320-324, (Ala)5)-P, and pET-N (Ser290→Trp)-P from plasmid pET-N/P (19) following the manufacturer's protocol. For plasmid pET-N (Δ1-22)-P, the coding sequence of N(Δ1-22) was amplified by PCR from pET-N/P with NcoI restriction sites introduced at both its 5′ and 3′ ends. The 3′ primer also contained a ribosomal binding site immediately downstream of the N gene stop codon to allow the independent translation of the P protein. The amplicon was digested with NcoI restriction enzyme and purified with a gel extraction kit (Novagen). Plasmid pET-N/P was digested with NcoI and treated with (calf intestinal) alkaline phosphatase. The purified amplicon was then ligated with linearized pET-N/P by use of T4 DNA ligase to create pET-N (Δ1-22)-P.
Protein expression and purification.
The coexpression and copurification of the VSV mutant N proteins with P protein followed the protocols described previously (19). Briefly, each plasmid containing a mutant N protein, and the P protein coding sequence was transformed into E. coli BL21(DE3) strain individually. The cell culture was grown at 37°C in LB broth (EM Science) in the presence of 0.1 g/liter ampicillin until the optical density at 595 nm reached 0.6 to 0.8. Protein expression was induced by adding isopropyl-β-d-thiogalactopyranoside (IPTG) at a final concentration of 1 mM. Cells were allowed to continue growing at 29°C for 8 h and were harvested by centrifugation. The cell pellet was resuspended in 20 mM Tris buffer (pH 7.9) containing 500 mM NaCl, sonicated, and centrifuged at 18,000 rpm for 1 h. The soluble portion was purified by use of a nickel-affinity column according to the Novagen protocol. The sample eluted from the nickel-affinity column was further purified by size exclusion chromatography using a gel filtration column (Superdex 200) and 50 mM Tris buffer (pH 7.5) containing 50 mM NaCl. To dissociate the N (Ser290→Trp) and P proteins, the complex was dialyzed in 100 mM citrate buffer (pH 4.0) containing 250 mM NaCl. The P protein dissociated from the N (Ser290→Trp) protein and precipitated at pH 4.0. After removing the precipitant by centrifugation, the sample of the N (Ser290→Trp) protein was dialyzed in 50 mM Tris buffer (pH 7.5) containing 300 mM NaCl and was further purified by size exclusion chromatography using a Superdex 200 column and 50 mM Tris buffer (pH 7.5) containing 300 mM NaCl.
The molecular weight of protein entities obtained by gel filtration purification was estimated by a standard curve of elution time versus molecular weight. The standard curve was plotted by fitting the molecular weights of standard proteins (Bio-Rad Laboratories) to their elution times using OriginPro 7 software (OriginLab) (R2 = 0.99597) (5).
Gel electrophoresis and immunoblotting.
Samples were boiled for 5 min in a reducing sodium dodecyl sulfate (SDS) sample buffer. The denatured proteins in the sample were separated on a 10% SDS-polyacrylamide gel (SDS-PAGE) containing 0.13% Bis and were stained with Coomassie brilliant blue. For immunoblotting, proteins were electrophoretically transferred from the SDS-PAGE to a nitrocellulose membrane and treated with the polyclonal anti-N antibody or the polyclonal anti-P antibody as the first antibody by use of a Western Breeze chromogenic Western blot immunodetection kit (Invitrogen) following the manufacturer's protocol. Development of the blots was performed with 5-bromo-4-chloro-3-indolylphosphate-nitroblue tetrazolium substrate for alkaline phosphatase.
RNA detection.
The presence or absence of RNA bound to N-mutant/P protein complexes was detected by UV absorbance analysis. UV absorbance scanning was performed with a Beckman DU-600 spectrometer using a wavelength range of 240 nm to 340 nm. RNA extraction was performed following the TRIzol (Invitrogen) reagent protocol. Briefly, 1 mg mutant N and P protein complex (nickel-affinity column purified or gel filtration purified) was used for each extraction. After phase separation by use of chloroform, the RNA in the aqueous phase was precipitated by addition of isopropyl alcohol. The precipitated RNA was washed with 75% ethanol and briefly dried by vacuum evaporation. The RNA was visualized by use of a 10% polyacrylamide-8 M urea gel and stained with ethidium bromide.
Electron microscopy.
The gel filtration purified N (Ser290→Trp)-P and N (320-324, (Ala)5)-P complexes were diluted to a concentration of 0.05 mg/ml. Samples were negatively stained with 2% aqueous uranyl acetate. Micrographs were taken using a conventional transmission electron microscope.
Crystallization and structure determination.
N (Ser290→Trp) crystals were grown by the hanging drop method in 24-well VDX plates (Hampton Research) at 293 K in conditions similar to those previously reported for the wt N-RNA complex (18). A protein solution (1 μl [8.8 mg/ml]) was mixed with 1 μl of reservoir solution consisting of 5.5% (wt/vol) polyethylene glycol 4000 in 100 mM sodium acetate buffer containing 200 mM NaCl (pH 4.5). Prior to data collection, crystals were cryoprotected in a final solution containing 20% (wt/vol) polyethylene glycol 4000-20% glycerol in the reservoir buffer by increasing the protectant concentration stepwise, as previously described (18), and flash frozen in liquid nitrogen.
X-ray diffraction data were collected at a wavelength of 1.0 Å on South East Regional Collaborative Access Team beamline 22-BM hardware at the Advanced Photon Source research facility. Data were collected with an oscillation angle of 0.3° and a crystal-to-detector distance of 300 mm on a MAR 225 charge-coupled device detector. The crystals grew as elongated bars, which allowed for multiple incomplete data wedges to be collected from a single crystal. As significant radiation damage was observed by visual inspection of diffraction images, the crystal was recentered and data collection continued on a fresh area of the crystal. Raw intensity images were processed with the HKL2000 software package (30), and structure factors were calculated with TRUNCATE software (15).
The structure was determined by molecular replacement using the previously determined wt N protein structure (Protein Data Bank accession number 2GIC), from which the RNA coordinates were removed. Rotation and translation functions were calculated with MolRep software (35) as part of the CCP4 package. Rigid-body refinement of the individual domains and extensions was carried out with REFMAC5 software (28). In-place model rebuilding was carried out with the Phenix software package (1) followed by additional manual model building and real-space refinement with COOT software (13). Refinement of the final model was initially performed with CNS software (4) followed by additional refinement, including translation/libration/screw refinement with REFMAC5 software. No outliers were found in the Ramachandran plot. Refinement statistics are shown (see Table 3).
TABLE 3.
Crystallographic refinement
| Characteristic | Value(s) |
|---|---|
| Space group | P21212 |
| Unit cell a, b, c (Å) | 166.4, 236.6, 74.7 |
| Resolution (Å) | 29.97-2.80 |
| No. of reflections (no. unique/total no. observed) | 52,030/240,608 |
| Completeness (2.90-2.80 Å) | 74.3 (44.9) |
| I/σ (2.90-2.80 Å) | 16.8 (4.8) |
| Rmerge (2.90-2.80 Å) | 0.088 (0.172) |
| Rcryst | 0.243 |
| Rfree | 0.285 |
| Model | |
| No. of atoms | 16,675 |
| No. of residues | 2,105 |
| RMSDa | |
| Bonds (Å) | 0.006 |
| Angles (°) | 0.953 |
RMSD, root mean square deviation.
Analytical ultracentrifugation.
A Beckman XL-A analytical ultracentrifuge was used for sedimentation velocity measurements for the gel filtration-purified N (320-324, (Ala)5)-P complex. The protein sample (400 μl) was centrifuged using a concentration of 1 mg/ml at 4°C and a four-cell An-60 Ti rotor, with 400 μl of sample buffer (50 mM Tris, 50 mM NaCl [pH 7.5]) filling the reference sector as the control. Data were collected at 40,000 rpm every 3 min for 12.5 h at a wavelength of 230 nm. The density (1.00172) and viscosity (1.0224E−2) of the buffer at 4°C were calculated by use of the Sednterp software program. The partial specific volume of the N (320-324, (Ala)5)-P complex (0.7294) was also calculated using the density and viscosity values given above. The complex molecular mass (113 kDa) was calculated by use of the SEDFIT software program (33) and the continuous C(s) distribution model.
Protein structure accession number.
The coordinates of this structure has been deposited in the Protein Data Bank with an accession number of 2QVJ.
RESULTS
In the crystal structure of the N-RNA complex, a network of protein interactions was observed across four neighboring molecules (Fig. 1A, Table 1). One N molecule (for instance, the red molecule labeled A in the figure) may be selected as the reference point. Contact I represents the interaction of the N-terminal arm (residues 2 to 22) with the outer surface of the C-lobe in the preceding N molecule B. Contact II represents the interaction of the extended loop (residues 340 to 375) in the C-lobe with the following N molecule C. Contact III represents the interaction of the N-terminal arm with the extended loop in the N molecule D preceded by two N molecules. The pattern of interactions in contact III may be considered to be that the N-terminal arm of the N molecule A locks on the extended loop of the N molecule D on the outer surface of the C-lobe in the N molecule B. Details of the designed mutations to the N protein presented here are summarized in Fig. 1B.
TABLE 1.
Residues related to intermolecular contacts
| Interaction location | Interaction in: |
|
|---|---|---|
| Residue 1 | Residue 2 | |
| Contact I | ||
| H bond | O of Ser2 | Side chain of Glu243 |
| Salt bridge | Side chain of Arg7 | Side chain of Asp256 |
| H bond | —NH2 of Lys17 | O of Met262 |
| Contact II | ||
| H bond | Side chain of Ser340 | Side chain of Lys388 |
| H bond | —NH— of Asp343 | Side chain of Ser326 |
| Hydrophobic | Side chain of leu344 | Surface pocket |
| H bond | —NH— of Ala345 | O of Leu250 |
| Hydrophobic | Side chain of Phe348 | Surface pocket |
| Hydrophobic | Side chain of Lys354 | Side chain of Leu379 |
| Contact III | ||
| H bond | —NH— of Lys6 | O of Cys349 |
| H bond | O of Lys6 | —NH— of Cys349 |
| H bond | —NH— of Ile8 | O of Gln347 |
Deletion of the N-terminal arm.
The N-terminal arm makes two hydrogen bonds and one salt bridge with the C-lobe BC through contact I and three hydrogen bonds with the extended loop in molecule C through contact III (Fig. 2A). It was observed that the orientation of the N-terminal lobes (N-lobe) in the oligomeric N-RNA complex is dependent on the crystal contact. The five independent molecules in the crystallographic asymmetric unit have a root mean square deviation of up to 1.1 Å between the Cα coordinates in two N lobes when their C-lobes are superimposed. The side-by-side interaction between the neighboring N-lobes involves half the buried surface area of that between the neighboring C-lobes. This suggests that the N-lobe requires forces in addition to the side-by-side interactions to stabilize its association and orientation within the oligomer such as those of contacts I and III. Deletion of the N-terminal arm eliminates these interactions and destabilizes the N lobe.
FIG. 2.
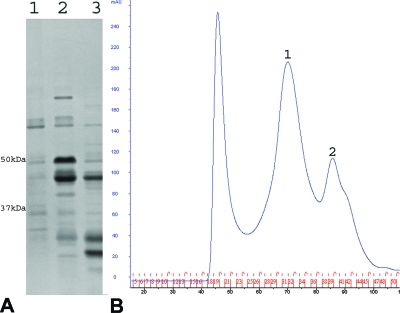
(A) Cartoon illustration of the contacts between N protein molecules. The bulk of each N molecule is shown as a solid surface. Interactions for contacts I and III as described in Table 1 are shown as dashed lines (cyan) with residues labeled. Residues involved in hydrogen bonds and salt bridges are shown as ball and stick models. Coloring of the N molecules follows the colors used as described for Fig. 1. The cartoon illustrations in this and the following figures were generated with PyMOL software (8). (B) Coexpression and copurification of N (Δ1-22) with the His-tagged P protein. Lanes 1 to 8, SDS-PAGE stained with Coomassie blue. Lane 1, molecular mass marker; lane 2, the cell lysate supernatant after induction; lane 3, the cell lysate pellet after induction; lane 4, the N (Δ1-22)-P protein complex copurified by use of a nickel-affinity column. Lanes 5 to 8, purification of the N (Δ1-22)-P complex by size exclusion chromatography on a S200 column, showing the protein contents in the void volume peak (lane 5), peak 1 (lane 6), peak 2 (lane 7), and peak 3 (lane 8) (peak numbering refers to the peaks in panel C). Polyclonal anti-N antibody (lane N) and polyclonal anti-P antibody (lane P) were used to confirm the expression and purification of N (Δ1-22)-P (peak 1) by Western blot analysis. (C) Size exclusion chromatography profile of the N (Δ1-22)-P protein complex after nickel-affinity purification. Peaks are labeled sequentially. The peak before peak 1 corresponds to the void volume.
A mutant, N (Δ1-22), was engineered by use of a plasmid that was used to coexpress N and P protein in E. coli (19). The recombinant proteins were overexpressed in E. coli and were shown by means of SDS-PAGE to have their expected sizes (Fig. 2B). The mutant N protein was able to be copurified with the His-tagged P protein (his6P) by use of a nickel-affinity column, which suggested that the P binding site on the N protein remains structurally intact, since only an N-his6P complex could bind the nickel-affinity column. Further purification of the N-his6P complex by size exclusion chromatography on an S200 column resulted in a number of peaks (Fig. 2C). The complex in peak 1 contains the mutant N and P proteins (Fig. 2C) and its size corresponds to that of the wt N-P-RNA complex, while peaks 2 and 3 in this purification had not been observed previously when the wt complex was purified on the S200 column. The identities of the mutant N and P proteins were confirmed by Western blot analysis (Fig. 2B). Peak 3 contains the N protein only, as a result of separation performed with the P protein during purification by size exclusion chromatography. The estimated size of the complex in peak 2 is about 261 kDa, which implies that this complex might contain multiple copies of mutant N and/or P molecules. A scan of UV absorbance shown by the protein complex represented by peak 1 or other two peaks revealed that the ratio of the absorbance at 260 nm to that at 280 nm was 0.5845 or similar (not shown), respectively, indicating the absence of RNA from all of the complexes on the basis of comparison to the ratio of 0.9899 for the wt complex (Table 2). All the complexes were unstable, since they disintegrated into entities with much smaller sizes when stored for 2 weeks at 4°C. Therefore, the exact number of mutant N or P molecules in each complex could not be determined accurately, but it is likely that there were 10 copies each of N and P in the peak 1 complex based on its size being similar to that seen with the wt N-P-RNA complex. Peak 3 contains only the mutant N protein in a monomeric form with an estimated molecular mass of 55 kDa (Fig. 2B).
TABLE 2.
Ratio of UV absorbance at 260 nm (UV260) to that at UV280a
| Protein complex | UV260/UV280 ratio |
|---|---|
| wt N-P | 0.99 |
| N (Δ1-22)-P | 0.58 |
| N (Δ347-352)-P | 0.56 |
| N (320-324, (Ala)5)-P | 0.61 |
| N (Ser290→Trp)-P | 0.57 |
The UV absorbance scan for the wt N-P complex that contained RNA gave values of 0.62 for UV260 and 0.62 for UV280, resulting in a UV260/UV280 ratio of 0.99. The UV absorbance scan for the N (Ser290→Trp)-P complex gave values of 0.18 for UV260 and 0.31 for UV280, resulting in a UV260/UV280 ratio of 0.57.
Disruption of the interactions by the extended loop.
The extended loop in the C-lobe contributes to interactions involving both contact II and contact III, in similarity to the results seen with the N-terminal arm, which contributes to interactions involving contacts I and III (Fig. 2A). These interactions appear to be as extensive as those contributed by the N-terminal arm, including three hydrogen bonds and three hydrophobic interactions in contact II in addition to interactions through contact III. Mutations in the extended loop were therefore expected to have effects on both contact II and contact III.
Before selection of the mutation sites, a survey of the residues involved in contacts II and III was conducted using the crystal structure. It was noticed that none of the residues is conserved among rhabdoviruses and that interactions of each residue could contribute only limited binding energy because they are located on the protein surface and not fully buried. It was reasoned that the structural stability maintained by contacts II and III is the result of the collective interactions by all the residues involved. Mutation of one single residue may not be sufficient to disrupt the contacts. In order to adequately disrupt the contacts made by the extended loop, a deletion mutant, N (Δ347-352), was engineered by use of the same parental plasmid as that used in generating the previous mutant. Only six residues at the tip of the extended loop were deleted rather than the entire extended loop. This mutation was mainly intended for use in examination of the contributions of contacts II and III to the oligomerization of the N protein molecules and RNA encapsidation. Larger deletions in the extended loop may have other consequences, such as changes in the tertiary structure of the C-lobe that may potentially affect N oligomerization and RNA encapsidation, unrelated to the intermolecular interactions rendered by the extended loop. The limited deletion directly removed the hydrophobic interactions by the side chain of Phe348 and all of the hydrogen bond potential related to contact III. However, it can also be expected that the conformational change of the extended loop as the result of this deletion may abolish some, if not all, of the other contact II interactions. With this mutant, the recombinant mutant N and P proteins were expressed with expected sizes in E. coli (Fig. 3A), and the mutant N retained the P binding site, as confirmed by copurification with his6P on the nickel-affinity column. Further purification on the S200 size exclusion column showed that the mutant N-P complex with an estimated size of 318 kDa was much smaller than the wt N-P-RNA complex (Fig. 3B). The ratio of UV absorbance at 260 nm to that at 280 nm showed that no RNA was bound in the complex (Table 2). No RNA was detected in this sample by phenol extraction either (data not shown). This complex was unstable during storage at 4°C as well. There was another peak found in the size exclusion purification that had an estimated size of 81 kDa. The sample in this peak contained the mutant N protein and fragments of the P protein but no RNA, according to UV absorbance scan results (data not shown). This suggests that the deletion in the extended loop diminished not only N oligomerization and RNA encapsidation but also, to some extent, the integrity of the P binding site in the N protein. When the P protein was not associated with the N protein, it was degraded, as previously observed for purified recombinant P protein expressed in E. coli (9).
FIG. 3.
(A) SDS-PAGE stained with Coomassie blue, showing the coexpression and copurification of N (Δ347-352) with the His-tagged P protein. Lane 1, the void volume peak; lane 2, peak 1; lane 3, peak 2 (peak numbering refers to the peaks in panel B). (B) Size exclusion chromatography profile of the N (Δ347-352)-P protein complex after nickel-affinity purification. Peaks are labeled sequentially. The peak before peak 1 corresponds to the void volume.
Disruption of the side-by-side interactions.
Extensive side-by-side interactions were observed between the neighboring N molecules in the structure of the N-RNA complex. The total calculated buried area between two neighboring N molecules was 1,954 Å2. Two-thirds of that area was contributed by the contacts between two neighboring C-lobes. Disruption of the side-by-side interactions, especially at the contact between two neighboring C-lobes, would be expected to destabilize N oligomerization. We designed a mutation, N (320-324, (Ala)5), that was aimed at removing a stretch of interactions between two neighboring N molecules. In addition to reducing the hydrophobic interactions involving the side chain of Tyr324 by mutating it to an alanine, the mutation of other four residues (Asp, Asp, Ile, and Glu) to alanines eliminated five hydrogen bonds with the neighboring N molecule (Fig. 4A).
FIG. 4.
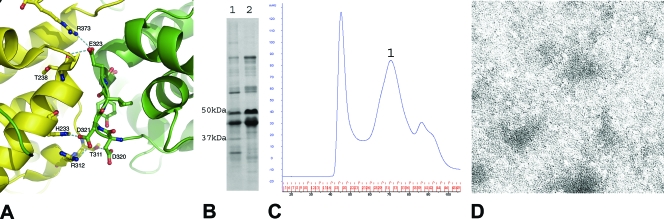
(A) Cartoon illustration for a stretch of residues (residues 320 to 324) involved in side-by-side interactions. These residues form five hydrogen bonds with the neighboring N molecule in the oligomeric N-RNA complex, including those between the side chain of Asp320 and the side chain of Thr311; the side chain of Asp321 and the side chain of His233, as well as that of Arg312; the side chain of Asp323 and the carbonyl of Thr238; and the side chain of Thr325 and the side chain of Arg309. The side chain of Tyr309 fits into a hydrophobic pocket on the surface of the C-lobe of the neighboring N molecule. (B) SDS-PAGE stained with Coomassie blue, showing the coexpression and copurification of N (320-324, (Ala)5) with the His-tagged P protein. Lane 1, void volume peak; lane 2, peak 1 (peak numbering refers to the peaks in panel C). (C) The size exclusion chromatography profile of the N (320-324, (Ala)5)-P protein complex after nickel-affinity purification. Peaks are labeled sequentially. The peak before peak 1 corresponds to the void volume. (D) Conventional transmission electron micrograph of the sample represented by peak 1. The sample was prepared at a concentration of 0.05 mg/ml protein and was stained with 2% aqueous uranyl acetate. The micrograph was taken at a magnification of ×50,000.
This mutant N protein was also able to be coexpressed and copurified with his6P (Fig. 4B), suggesting the preservation of a functional P binding site. In the S200 size exclusion purification, a stable complex of the mutant N and P proteins was isolated with an estimated size of 318 kDa (Fig. 4C), in similarity to the results seen with the complex formed by the mutant N (Δ347-352) (peak 1). No RNA was detected in this complex, according to UV absorbance ratio results (Table 2) and phenol extraction results (data not shown). Electron micrographs taken with this sample did not show any regular complex (Fig. 4D). To further investigate the composition of this complex, the sample was subjected to analytical ultracentrifugation studies. Based on sedimentation velocity measurements, an estimated molecular mass of 113 kDa was calculated using SEDFIT and the continuous C(s) distribution model for the protein complex, suggesting that the complex was formed by one N molecule (47.4 kDa) and two P molecules (30.2 kDa per P molecule). The presence of two P molecules is consistent with the idea that the P protein always functions as a stable dimer (10). The N (320-324, (Ala)5)-P complex appears to have a composition similar to that of the N0/P complex purified from insect cell coexpression of the RABV N and P proteins (27). This suggests that the N (320-324, (Ala)5) protein and the P dimer may form a complex in which the N protein is stabilized in the conformation prior to that in the RNP assembly. The discrepancy in the molecular mass estimated in the ultracentrifugation analysis compared to that estimated in the gel filtration analysis may be the result of distorted migration by this complicated complex in the gel filtration column.
Blocking RNA binding.
The 90-base RNA found in the structure of the N-P-RNA complex was tightly associated with the N proteins. All attempts to release or destroy the RNA failed, including treatment using various RNA nucleases or pH 11.0 (19). The issue was therefore raised of whether the RNA is required to stabilize the N oligomer, since it seems to be an inseparable part of the complex. The RNA was bound in a cavity located between the N- and C-lobes (Fig. 5A). The residue that is closest to the bound RNA is Ser290, with its side chain next to the 3′ backbone phosphate group of nucleotide 3. Our design was to replace residue Ser290 with a tryptophan in mutant N (Ser290→Trp), which would introduce a bulky side chain in the cavity to prevent RNA from coming into close contact with the N protein.
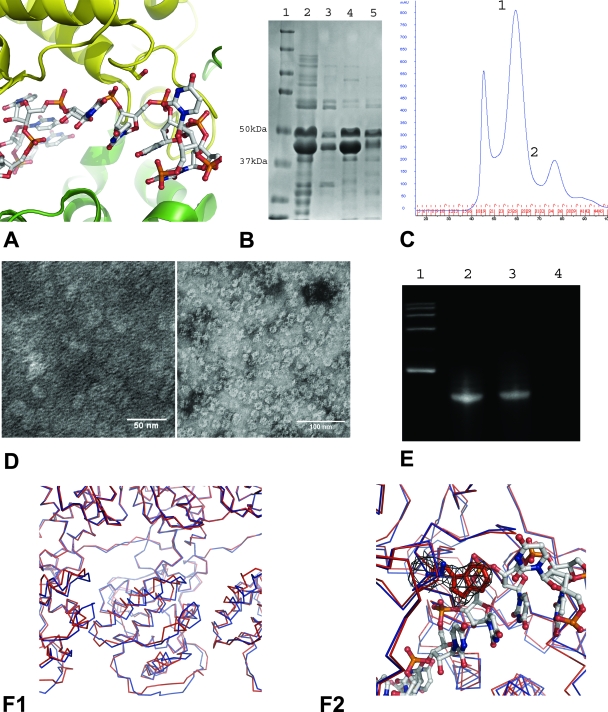
FIG. 5.
(A) Cartoon illustration of the RNA binding cavity between the N-lobe (green) and the C-lobe (yellow). In the N-RNA complex, the RNA (ball-and-stick model) is bound in close proximity of Ser290. Ser290 was mutated to the much bulkier Trp in order to block RNA binding by steric hindrance. (B) SDS-PAGE stained with Coomassie blue, showing the coexpression and copurification of N (Ser290→Trp) with the His-tagged P protein. Lane 1, molecular mass marker; lane 2, the N (Ser290→Trp)-P protein complex copurified by use of a nickel-affinity column; lane 3, the void volume peak; lane 4, peak 1; lane 5, peak 2 (peak numbering refers to the peaks in panel C). (C) Size exclusion chromatography profile of the N (Ser290→Trp)-P protein complex after nickel-affinity purification. Peaks are labeled sequentially. The peak before peak 1 corresponds to the void volume. (D) A conventional transmission electron micrograph of the sample represented by peak 1 in comparison with that of the wt N-P protein complex used as the control (peak 2). The samples were prepared at a concentration of 0.05 mg/ml protein and were stained with 2% aqueous uranyl acetate. The wt N-P complex micrograph was taken at a magnification of ×150,000 (left panel). The N (Ser290→Trp)-P protein complex micrograph was taken at the magnification of ×80,000 (right panel). (E) Phenol extraction from the N (Ser290→Trp)-P RNA complex in comparison with results seen with the wt N-RNA and the N-P-RNA complexes used as controls. Lane 1, RNA molecular mass marker; lane 2, RNA extractant from the wt N-RNA complex; lane 3, RNA extractant from the wt N-P-RNA complex; lane 4, RNA extractant from the N (Ser290→Trp)-P RNA complex. (F1) Structural overlay of wild-type N (blue) protein and N (Ser290→Trp) (red) protein. One complete N-lobe in the center flanked by partial lobes to the right and left is shown. The superposition shows a slight unwinding of the N-lobe with respect to the center of the 10-member ring. (F2) The overlay with the wild-type N-RNA complex shows how the Trp mutation blocks RNA from accessing the RNA binding cavity. Electron density (2Fo − Fc) from the structure determination of the N (Ser290→Trp) mutant is shown for the side chain of Trp290 at a level of 1.1 σ. The side chains for Ser290 and Trp290 as well as the RNA are shown as ball and stick models. One complete N-lobe in the center flanked by partial lobes to the right and left is shown. The superposition shows a slight unwinding of the N-lobe with respect to the center of the 10-member ring. (F2) The overlay with the wild-type N-RNA complex shows how the Trp mutation blocks RNA from accessing the RNA binding cavity. Electron density (2Fo − Fc) from the structure determination of the N (Ser290→Trp) mutant is shown for the side chain of Trp290 at a level of 1.1 σ. The side chains for Ser290 and Trp290 as well as the RNA are shown as ball and stick models.
The recombinant mutant N protein was coexpressed and copurified with his6P (Fig. 5B). The complex purified by the S200 size exclusion column has a size similar to that of the wt N-P-RNA complex (Fig. 5C). Electron micrographs showed the same ring-like structures indistinguishable from the structures seen with the wt complex (Fig. 5D). However, the ratio of UV absorbance at 260 nm to that at 280 nm indicated the absence of RNA, and phenol extraction showed no RNA in the new mutant N-P complex compared with the results determined using the wt complex as the control (Fig. 5E). This indicated that mutant N (Ser290→Trp) can still form a stable ring-like oligomer with 10 subunits and bind with the P protein when coexpressed with P even in the absence of encapsidated RNA.
After the P protein was removed by dialysis of the new mutant N-P complex at pH 4.5 as described previously (19), the mutant N oligomer retained its stability in solution and was crystallized under the same conditions as those used with the wt N-RNA complex (18). X-ray diffraction data were collected with these crystals under cryogenic conditions, and the structure was solved by molecular replacement and averaging using the coordinates of the oligomeric N protein without including the RNA molecule. The crystallographic statistics are presented in Table 3. The overall structure of the N (Ser290→Trp) protein is very similar to that of the wt N protein, as seen in the N-RNA complex results (20). There is a slight unwinding of the N-lobe relative to the center of the 10-member ring, which results in a small opening of the RNA binding cavity after an N-lobe movement of 2.2 Å (Fig. 5F1). The results with respect to electron density for the tryptophan side chain are clear, showing that it blocks access of RNA to the RNA binding cavity (Fig. 5F2). In addition, the structure of the N (Ser290→Trp) protein has better quality at higher resolution (2.8 Å) than that seen with the wt N-RNA complex. For instance, residues 175 to 180 are shown to form a better-resolved part of an α-helix.
DISCUSSION
The RNP is assembled during replication in which the viral genomic RNA is synthesized concomitant with its encapsidation by the N protein. The N protein forms a copolymer with the RNA in a fixed ratio of nine nucleotides per N molecule. The template for RNA synthesis by the viral polymerase is the N-enwrapped genomic RNA (negative or positive strand) and not the naked RNA. This intimate association between the N protein and viral RNA indicates the structural and functional interdependence of the two identities. The crystal structure of an oligomeric N-RNA complex mapped the RNA binding site in a cavity of the N protein (2, 20). More importantly, the extensive intermolecular interactions of the N molecules in the complex emphasize the importance of these interactions in formation of a stable N-RNA complex.
In this study, we designed a number of mutations for disruption of the intermolecular interactions of the N protein found in the N-RNA complex (Fig. 1B). The results showed as a general principle that stabilization of the N oligomerization requires not only the intermolecular interactions but also the encapsidation of RNA. Having positively charged residues in the RNA binding cavity alone is insufficient for assembly of a stable N-RNA complex. The interactions between the neighboring C-lobes appear to offer the most stabilization forces in the N-RNA complex, as evidenced by a mutant with a deletion in the extended loop and a mutant with five alanine substitutions at the side-by-side interface. The deletion of the N-terminal arm may not have diminished the N oligomerization completely but certainly subverted the ability of the N protein to encapsidate RNA. This is consistent with the observation that the N-lobe seems to be more flexible than the C-lobe, as shown by the different orientations of the N-lobes in the crystallographic asymmetric unit (20). Without the interactions between the N-terminal arm and the neighboring N molecules, the N-lobe is unlikely to maintain the orientation required for the formation of the RNA binding cavity together with the C-lobe to bind the RNA as tightly as is seen with the wt N-RNA complex. This property of the RNP structure may be essential for serving as a template for RNA synthesis by the viral polymerase. The N-lobe was able to be temporarily and locally dissociated from the body of the RNP template to give the polymerase access to the RNA sequence, while the C-lobe remained associated to maintain the integrity of the template. After the RNA synthesis, the N-lobe was able to recapture the template RNA to restore the RNP structure.
The P protein is a cofactor of the viral polymerase, and it dictates the specific attachment of the L protein to the RNP template (11). The binding site for P has been mapped to the C-lobe of the N protein (12, 17, 19, 34). In all the mutants generated in this study, the P binding site was not affected, as demonstrated by the copurification of the mutant N proteins with P, in similarity to the results seen with respect to the copurification of the wt N with P. However, we observed that mutant N (Δ347-352) protein free of P was present during size exclusion chromatography after copurification with P by use of a nickel-affinity column. This observation may suggest that the association of mutant N (Δ347-352) with P was weakened by the deletion of the extended loop in the C-lobe. It also implies that the binding site for P may be near, if not at, the extended loop.
In mutant N (Ser290→Trp), on the other hand, the intermolecular interactions appear to be the same as in the wt N-RNA complex, whereas RNA binding was completely blocked by the substitution of a bulky residue (tryptophan) in the RNA binding cavity. This observation implies that the N protein alone contains all the information for the assembly of a RNP-like structure that has all the proper intermolecular interactions among the N protein molecules. The cage formed by such an N protein assembly can package the viral RNA that fits in the volume encapsulated by the N proteins. The structural comparison of mutant N (Ser290→Trp) with that of the wt N in the N-RNA complex revealed a slight rotation of the N-lobe which resulted in some opening of the N-lobe. These observations would suggest that the interactions between the RNA and the N protein in the N-RNA complex may further tighten the complex, but the main forces that stabilize the overall complex structure to completely encapsidate RNA are from the intermolecular interactions of the N protein. The RNA molecule in the N-RNA complex is primarily a subject completely protected by the assembly of the N protein. Mutant N (Ser290→Trp) may be an interesting candidate for studying the role of the N protein in transcription or replication, because it retains a complete wt surface of the N protein that is capable of interacting with the L and P proteins. It may be used to address the issue of whether RNA encapsidation is required for the polymerase activities. When a system becomes available for preparing adequate active templates, mutant N (Ser290→Trp) may be studied together with the L and P proteins for effects on transcription and/or replication.
The critical role of the intermolecular interactions in the formation of a stable N-RNA complex may be a mechanism to regulate the RNP assembly. When the N protein molecule is first synthesized by the ribosome, it has to remain RNA-free in order to encapsidate the RNA during the assembly of RNP. The individual N protein molecule could not bind any RNA molecule very tightly, despite the presence of a RNA binding cavity, when the N protein molecule was not at the proper site to encapsidate the viral RNA. However, the P protein has been suggested to act as a chaperone to keep the newly synthesized N protein molecules from binding random RNA molecules by forming a P-N complex (32). Since the N protein alone would not bind the RNA as tightly as a monomeric protein, as shown here, the P protein in the N-P complex should have additional functions to keep N from binding RNA. We have postulated that the P protein in the P-N complex may direct the N protein molecule to the site of viral replication by formation of a tetramer with the P protein that is associated with the L protein in the viral polymerase, a hypothesis based on the crystal structure of the oligomerization domain of the P protein (10). Once at the appropriate location, the N protein molecule could be added to the nascent RNA via extensive intermolecular interactions with the other N molecules that have been recruited in the growing RNP. When the N protein becomes associated with the RNP, it is almost irreversibly associated. This tight intermolecular interaction may be a regulation step to ensure that the N protein molecule is assembled correctly and efficiently with the viral RNA only at the site of replication. Interactions of the N protein with random RNA or other proteins may be weak and are likely reversible if an RNP-like structure is not formed.
Acknowledgments
We thank Peter Prevelige and Chi-Yu Fu for their assistance in ultracentrifugation analyses. We also thank Shu-zhen Wang and Weiming Mao for their assistance in analyses of protein molecular mass with OriginPro 7 software. We thank the staff of the electron microscope center in Mississippi State University for their assistance in taking the electron micrographs. We thank the staff of the South East Regional Collaborative Access Team (SER-CAT) at the Advanced Photon Source (APS) research facility, Argonne National Laboratory, for their assistance in data collection.
Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under contract W-31-109-Eng-38. SER-CAT supporting institutions may be found listed at www.ser-cat.org/members.html. The work is supported in part by National Institutes of Health grant AI050066.
Footnotes
Published ahead of print on 14 November 2007.
REFERENCES
- 1.Adams, P. D., K. Gopal, R. W. Grosse-Kunstleve, L. W. Hung, T. R. Ioerger, A. J. McCoy, N. W. Moriarty, R. K. Pai, R. J. Read, T. D. Romo, J. C. Sacchettini, N. K. Sauter, L. C. Storoni, and T. C. Terwilliger. 2004. Recent developments in the PHENIX software for automated crystallographic structure determination. J. Synchrotron Radiat. 11:53-55. [DOI] [PubMed] [Google Scholar]
- 2.Albertini, A. A., A. K. Wernimont, T. Muziol, R. B. Ravelli, C. R. Clapier, G. Schoehn, W. Weissenhorn, and R. W. Ruigrok. 2006. Crystal structure of the rabies virus nucleoprotein-RNA complex. Science 313:360-363. [DOI] [PubMed] [Google Scholar]
- 3.Barr, J., P. Chambers, C. R. Pringle, and A. J. Easton. 1991. Sequence of the major nucleocapsid protein gene of pneumonia virus of mice: sequence comparisons suggest structural homology between nucleocapsid proteins of pneumoviruses, paramyxoviruses, rhabdoviruses and filoviruses. J. Gen. Virol. 72:677-685. [DOI] [PubMed] [Google Scholar]
- 4.Brünger, A. T., P. D. Adams, G. M. Clore, W. L. DeLano, P. Gros, R. W. Grosse-Kunstleve, J. S. Jiang, J. Kuszewski, M. Nilges, and N. S. Pannu. 1998. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D 54:905-921. [DOI] [PubMed] [Google Scholar]
- 5.Bruns, C. K., and R. R. Kopito. 2007. Impaired post-translational folding of familial ALS-linked Cu, Zn superoxide dismutase mutants. EMBO J. 26:855-866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Das, T., B. K. Chakrabarti, D. Chattopadhyay, and A. K. Banerjee. 1999. Carboxy-terminal five amino acids of the nucleocapsid protein of vesicular stomatitis virus are required for encapsidation and replication of genome RNA. Virology 259:219-227. [DOI] [PubMed] [Google Scholar]
- 7.Davis, N. L., H. Arnheiter, and G. W. Wertz. 1986. Vesicular stomatitis virus N and NS proteins form multiple complexes. J. Virol. 59:751-754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Delano, W. L. 2002. The PyMOL user's manual. Delano Scientific, San Carlos, CA.
- 9.Ding, H., T. J. Green, and M. Luo. 2004. Crystallization and preliminary X-ray analysis of a proteinase-K-resistant domain within the phosphoprotein of vesicular stomatitis virus (Indiana). Acta Crystallogr. D 60:2087-2090. [DOI] [PubMed] [Google Scholar]
- 10.Ding, H., T. J. Green, S. Lu, and M. Luo. 2006. Crystal structure of the oligomerization domain of the phosphoprotein of vesicular stomatitis virus. J. Virol. 80:2808-2814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Emerson, S. U., and R. R. Wagner. 1972. Dissociation and reconstitution of the transcriptase and template activities of vesicular stomatitis B and T virions. J. Virol. 10:297-309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Emerson, S. U., and M. Schubert. 1987. Location of the binding domains for the RNA polymerase L and the ribonucleocapsid template within different halves of the NS phosphoprotein of vesicular stomatitis virus. Proc. Natl. Acad. Sci. USA 84:5655-5659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Emsley, P., and K. Cowtan. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60:2126-2132. [DOI] [PubMed] [Google Scholar]
- 14.Flood, E. A., and D. S. Lyles. 1999. Assembly of nucleocapsids with cytosolic and membrane-derived matrix proteins of vesicular stomatitis virus. Virology 261:295-308. [DOI] [PubMed] [Google Scholar]
- 15.French, S., and K. Wilson. 1978. On the treatment of negative intensity observations. Acta Crystallogr. A 34:517-525. [Google Scholar]
- 16.Gaudin, Y., J. Sturgis, M. Doumith, A. Barge, B. Robert, and R. W. Ruigrok. 1997. Conformational flexibility and polymerization of vesicular stomatitis virus matrix protein. J. Mol. Biol. 274:816-825. [DOI] [PubMed] [Google Scholar]
- 17.Gill, D. S., D. Chattopadhyay, and A. K. Banerjee. 1986. Identification of a domain within the phosphoprotein of vesicular stomatitis virus that is essential for transcription in vitro. Proc. Natl. Acad. Sci. USA 83:8873-8877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Green, T. J., and M. Luo. 2006. Resolution improvement of X-ray diffraction data of crystals of a vesicular stomatitis virus nucleocapsid protein oligomer complexed with RNA. Acta Crystallogr. D 62:498-504. [DOI] [PubMed] [Google Scholar]
- 19.Green, T. J., S. Macpherson, S. Qiu, J. Lebowitz, G. W. Wertz, and M. Luo. 2000. Study of the assembly of vesicular stomatitis virus N protein: the role of the P protein. J. Virol. 74:9515-9524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Green, T. J., X. Zhang, G. W. Wertz, and M. Luo. 2006. Structure of the vesicular stomatitis virus nucleoprotein-RNA complex. Science 313:357-360. [DOI] [PubMed] [Google Scholar]
- 21.Iseni, F., A. Barge, F. Baudin, D. Blondel, and R. W. Ruigrok. 1998. Characterization of rabies virus nucleocapsids and recombinant nucleocapsid-like structures. J. Gen. Virol. 79:2909-2919. [DOI] [PubMed] [Google Scholar]
- 22.Jayakar, H. R., and M. A. Whitt. 2002. Identification of two additional translation products from the matrix (M) gene that contribute to vesicular stomatitis virus cytopathology. J. Virol. 76:8011-8018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kouznetzoff, A., M. Buckle, and N. Tordo. 1998. Identification of a region of the rabies virus N protein involved in direct binding to the viral RNA. J. Gen. Virol. 79:1005-1013. [DOI] [PubMed] [Google Scholar]
- 24.La Ferla, F. M., and R. W. Peluso. 1989. The 1:1 N-NS protein complex of vesicular stomatitis virus is essential for efficient genome replication. J. Virol. 63:3852-3857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Masters, P. S., and A. K. Banerjee. 1988. Resolution of multiple complexes of phosphoprotein NS with nucleocapsid protein N of vesicular stomatitis virus. J. Virol. 62:2651-2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Masters, P. S., and A. K. Banerjee. 1988. Complex formation with vesicular stomatitis virus phosphoprotein NS prevents binding of nucleocapsid protein N to nonspecific RNA. J. Virol. 62:2658-2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mavrakis, M., F. Iseni, C. Mazza, G. Schoehn, C. Ebel, M. Gentzel, T. Franz, and R. W. Ruigrok. 2003. Isolation and characterisation of the rabies virus N0-P complex produced in insect cells. Virology 305:406-414. [DOI] [PubMed] [Google Scholar]
- 28.Murshudov, G. N., A. A. Vagin, and E. J. Dodson. 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D 53:240-255. [DOI] [PubMed] [Google Scholar]
- 29.Ogden, J. R., R. Pal, and R. R. Wagner. 1986. Mapping regions of the matrix protein of vesicular stomatitis virus which bind to ribonucleocapsids, liposomes, and monoclonal antibodies. J. Virol. 58:860-868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Otwinowski, Z., and W. Minor. 1997. Processing of X-ray diffraction data collected in oscillation mode, p. 307-326. In C. W. Carter and R. M. Sweet (ed.), Methods in enzymology. Academic Press, New York, NY. [DOI] [PubMed]
- 31.Peluso, R. W. 1988. Kinetic, quantitative, and functional analysis of multiple forms of the vesicular stomatitis virus nucleocapsid protein in infected cells. J. Virol. 62:2799-2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peluso, R. W., and S. A. Moyer. 1988. Viral proteins required for the in vitro replication of vesicular stomatitis virus defective interfering particle genome RNA. Virology 162:369-376. [DOI] [PubMed] [Google Scholar]
- 33.Schuck, P. 2000. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys. J. 78:1606-1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Takacs, A. M., T. Das, and A. K. Banerjee. 1993. Mapping of interacting domains between the nucleocapsid protein and the phosphoprotein of vesicular stomatitis virus by using a two-hybrid system. Proc. Natl. Acad. Sci. USA 90:10375-10379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vagin, A., and A. Teplyakov. 1997. MOLREP: an automated program for molecular replacement. J. Appl. Crystallogr. 30:1022-1025. [Google Scholar]