Abstract
Glycoprotein D (gD) is the receptor binding protein of herpes simplex virus (HSV) and binds to at least two distinct protein receptors, herpesvirus entry mediator (HVEM) and nectin-1. While both receptor binding regions are found within the first 234 amino acids, a crystal structure shows that the C terminus of the gD ectodomain normally occludes the receptor binding sites. Receptor binding must therefore displace the C terminus, and this conformational change is postulated to be required for inducing fusion via gB and gH/gL. When cysteine residues are introduced at positions 37 and 302 of gD, a disulfide bond is formed that stabilizes the C terminus and prevents binding to either receptor. We speculated that if disulfide bonds were engineered further upstream, receptor binding might be separated from the induction of fusion. To test this, we made five additional double cysteine mutants, each potentially introducing a disulfide bond between the ectodomain C terminus and the core of the gD ectodomain. The two mutants predicted to impose the greatest constraint were unable to bind receptors or mediate cell-cell fusion. However, the three mutants with the most flexible C terminus bound well to both HVEM and nectin-1. Two of these mutants were impaired in cell-cell fusion and null-virus complementation. Importantly, a third mutant in this group was nonfunctional in both assays. This mutant clearly separates the role of gD in triggering fusion from its role in receptor binding. Based upon the properties of the panel of mutants we conclude that fusion requires greater flexibility of the gD ectodomain C terminus than does receptor binding.
Herpes simplex virus (HSV) is a human pathogen that infects epithelial cells before spreading to the peripheral nervous system to establish a lifelong latent infection. Herpesviruses are enveloped viruses and must therefore fuse their membrane with a cellular membrane to establish infection.
The first essential step in viral entry is the binding to a cellular receptor. The HSV receptor binding protein gD recognizes at least three distinct receptors: nectin-1, which is a member of the immunoglobulin (Ig) superfamily; herpesvirus entry mediator (HVEM), which is a member of the tumor necrosis factor receptor family; and specific sulfate-modified forms of the glycan heparan sulfate (33). In this report we focused on the interaction of gD with nectin-1 and HVEM.
Once gD binds a receptor, fusion is carried out by three other viral glycoproteins: gB and the heterodimer gH/gL (33, 34). Recent evidence suggests that gD interacts with gH to form a hemifusion intermediate that is resolved into full fusion by gB (35). Initiation of either full fusion or hemifusion may occur via a profusion domain, a region of gD which is not involved in receptor binding but is required for viral membrane fusion (5). The profusion domain is comprised of residues 260 to 285 and was of particular interest because gD truncated at 260 could not rescue infectivity, while gD truncated at 285 could. This led to the hypothesis that residues 260 to 285 contact gB and/or gH/gL (5). However, deletions within residues 255 to 299 did not completely abolish cell-cell fusion (41). These observations suggested that the C terminus of the gD ectodomain (residues 259 to 316) is important for triggering fusion by gB and gH/gL but did not resolve whether a specific region of gD is required (5, 41).
To reveal the structure of the C terminus of the gD ectodomain, a cysteine residue was engineered after amino acid 306 (3, 19), resulting in a 307C-307C disulfide-linked gD dimer molecule and stabilization of the intrinsically flexible C-terminal region which was not solved in previous structures (19). This crystal structure revealed that the C-terminal portion of the ectodomain, hereafter referred to as the gD C terminus, occupies the same space as the N-terminal hairpin loop formed when gD binds HVEM (2, 3, 19). Moreover, the C terminus lies directly on top of residues important for nectin-1 binding (Fig. 1B) (7, 22). We therefore postulated that movement of the C terminus of gD is required for receptor binding, rather than being the result of this event (19). We further hypothesized that this conformational change triggers downstream events.
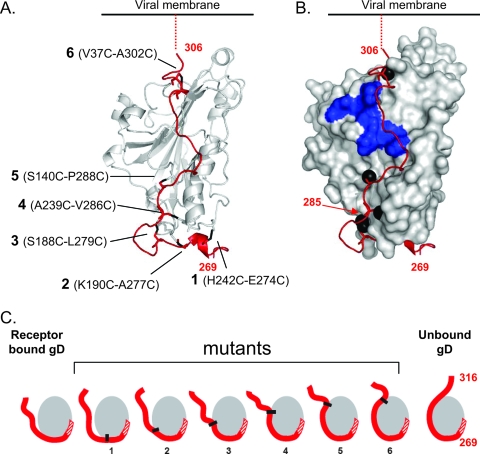
FIG. 1.
Cysteine pairs modeled on gD structure. (A) Model showing each pair of cysteines and the resulting disulfide bonds (black), located on a ribbon diagram of gD (15). (B) The C terminus of the ectodomain (red ribbon) fits into a groove formed by gD (gray space filled). The C terminus blocks access to residues critical for nectin-1 binding (blue-shaded surface). The profusion domain is defined as residues 260 to 285. (C) Model illustrating the hypothetical range of restraint on conformational changes to the ectodomain C terminus (red) imposed by the disulfide bonds (black). The gray ball represents residues 1 to 255. The red hatched region of the C terminus corresponds to the unresolved region of the gD crystal structure, residues 256 to 268.
Indeed, deletion of residues 290 to 299, mutation of residue W294 to alanine, or truncation of gD to residue 285 increases the affinity of gD for receptors (19, 28, 32). The higher affinity for these gD truncations/mutations is due to a higher “on” rate, which is increased over that of wild-type (WT) gD by either completely removing or destabilizing the interaction of the C terminus with the core, the latter being defined here as residues 37 to 259. Furthermore, gD containing two engineered cysteine residues at positions 37 and 302 forms an additional disulfide bond and this protein is unable to bind either HVEM or nectin-1 (19). A crystal structure confirmed that this disulfide bond constrained the entire C terminus by locking the most membrane-proximal portion of the C terminus to the gD core (Fig. 1) (19).
These two sets of mutations represent two extreme situations: increased flexibility of the C terminus or positional constraint of the entire C terminus. Here we made five additional double cysteine mutants each predicted to form a disulfide bond. The positions of these cysteines were chosen to constrain different portions of the ectodomain C terminus (Fig. 1C). In addition, these mutants were designed to determine if we could separate receptor binding by gD from its postulated function in triggering fusion. This type of mutagenesis has proven successful in the past (7, 19), and it has been shown that formation of disulfide bonds is determined by protein folding and not vice versa (1, 11, 12, 16).
Of the five double cysteine mutants that we constructed two mutants were unable to bind either receptor. In contrast, the other three mutants bound both receptors but were impaired or not functional in cell-cell fusion and null-virus complementation assays. The identification of a mutant that binds receptor(s) but cannot trigger fusion is of critical importance in identifying and segregating the steps that occur post-receptor binding.
MATERIALS AND METHODS
Cells and viruses.
293T cells were grown in Dulbecco's minimal essential medium (DMEM) supplemented with 10% fetal calf serum (FCS). CHO-K1 cells were grown in Ham's F-12 medium supplemented with 10% FCS. Vero and VD60 cells (21) were grown in DMEM supplemented with 5% FCS.
The gD-null virus, HSV-1 KOS-gDβ, carries lacZ in place of the coding region for gD under the control of the gD promoter (10). It was propagated and its titers were determined on VD60 cells as described previously (21).
Antibodies.
Rabbit polyclonal antibody (PAb) R8 was raised against HSV type 2 (HSV-2) gD and cross-reacts with HSV-1 gD (17). Anti-gD monoclonal antibodies (MAbs) used in this study include 1D3 (13), which binds residues 11 to 19, as well as DL2 (6), DL11 (6, 25), AP7 (4, 24), and LP2 (24), which recognize discontinuous epitopes.
Construction of mutant gD molecules.
The plasmid pSC390 encodes full-length gD from HSV-1(KOS) in the pcDNA3.1 vector (8). We generated gD mutants using the QuikChange site-directed mutagenesis kit (Stratagene Cloning Systems), as previously described (9). Mutations were made using the pSC390 plasmid as a template, and newly generated plasmids are as follows: pEL721 for H242C-E274C, pEL722 for K190C-A277C, pEL723 for S188C-L279C, pEL724 for A239C-V286C, pEL725 for P288C-S140C, pEL757 for H242C, pEL758 for E274C, pEL759 for K190C, and pEL760 for A277C. The plasmid for the V37C-A302C mutation is pDL485 and was previously described (19).
Immunoprecipitation.
293T cells were transfected with the desired plasmids using GenePorter according to the manufacturer's protocol (Gene Therapy Systems, Inc.). At 24 h posttransfection, cells were lysed in a buffer consisting of 10 mM Tris, pH 8, 150 mM NaCl, 10 mM EDTA, 1% NP-40, 0.5% deoxycholic acid, and 1 mM phenylmethylsulfonyl fluoride. Typically, 1% of the total cell extract (from a six-well plate) was incubated with 10 μg/ml of antibody for 2 h at 4°C. Proteins were precipitated with protein A agarose beads (Gibco BRL) for 2 h at 4°C, washed twice with lysis buffer, separated by electrophoresis on a 10% sodium dodecyl sulfate (SDS)-polyacrylamide gel, and detected by Western blotting with gD PAb R8.
Enzyme-linked immunosorbent assay (ELISA).
293T cells growing in 12-well plates were transfected with the gD plasmids or empty vector, using 2 μg of DNA/well and 10 μl of GenePorter/well for 3 h followed by addition of an equal volume of DMEM containing 20% FCS. Cells were incubated overnight at 37°C and then harvested in extraction buffer (10 mM Tris, pH 8, 150 mM NaCl, 10 mM EDTA, 1% NP-40, 0.5% sodium deoxycholate, 1 mM phenylmethylsulfonyl fluoride).
We used a capture ELISA to normalize the amount of gD in the 293T extracts as previously described (8). Briefly, ELISA plates were coated with 1D3 IgG (overnight at 4°C) and then blocked with PBS-milk (phosphate-buffered saline [PBS] containing 5% nonfat dry milk and 0.2% Tween 20) for 1 h. Dilutions of cell extracts containing gD were made in PBS-milk and added for 1 h at room temperature (RT). Captured gD was detected with PAb R8 IgG followed by goat anti-rabbit antibody coupled to horseradish peroxidase. Plates were rinsed with 20 mM citrate buffer (pH 4.5), 2,2′-azinobis(3-ethylbenzthiazolinesulfonic acid) (ABTS) peroxidase substrate was added, and the absorbance at 405 nm was recorded using a microtiter plate reader. The level of gD in each extract was normalized by dilution in extraction buffer.
To assess receptor binding of the gD mutants, ELISA plates were coated overnight with soluble receptors [5 μg of HVEM(200t)/ml, 10 μg of nectin-1(346t)/ml] (18, 38), blocked with PBS-milk for 1 h, and then incubated for 1 h at RT with normalized cell extracts. Bound gD was detected as described above (7-9). This assay was repeated at least three times for each form of gD.
CELISA.
To determine the amount of gD on cells used in the fusion assay, we used a previously described cell-based ELISA (CELISA) (8, 14, 23). One 96-well plate of CHO-K1 cells was transfected with the same mixture used to transfect the glycoprotein-expressing cells used in the luciferase fusion assay (see below). After 5 h of incubation at 37°C, medium was added to each well and the cells were incubated at 37°C for 18 to 20 h. The medium was removed, and cells were then incubated with PAb R8 serum diluted (1:1,000) in 3% bovine serum albumin-PBS++ (PBS containing 0.9 mM CaCl2 and 0.5 mM MgCl2) for 1 h at RT. Cells were rinsed three times with PBS++ and fixed in 3% paraformaldehyde for CELISA. The cells were again rinsed with PBS++ three times and incubated for 1 h at RT with goat anti-rabbit antibody coupled to horseradish peroxidase. Following another three PBS++ washes, cells were rinsed with 20 mM citrate buffer (pH 4.5). ABTS peroxidase substrate (Moss, Inc.) was added, and the absorbance at 405 nm was recorded using a microtiter plate reader.
Quantitative fusion assay.
To detect cell-cell fusion, we used a luciferase reporter assay (8, 29, 30). Briefly, CHO-K1 cells were grown in 24-well plates and transfected with plasmids encoding T7 RNA polymerase (pCAGT7), gB (pPEP98), gH (pPEP100), and gL (pPEP101) and one of the gD plasmids. To prepare receptor-expressing cells, CHO-K1 cells growing in six-well plates were transfected with a plasmid encoding the firefly luciferase gene under control of the T7 promoter (pT7EMCLuc) and a plasmid encoding either nectin-1 (pBG38) or HVEM (pSC386). After 5 h at 37°C, the transfection mixes were replaced with fresh medium. After 1 h of incubation at 37°C, receptor-expressing cells were trypsinized, added to the glycoprotein-expressing cells, and incubated at 37°C. At 18 to 20 h postcocultivation, cells were washed with PBS, lysed in reporter lysis buffer (luciferase assay system; Promega), and frozen. To measure the extent of fusion, samples were mixed with luciferase substrate (Promega) and immediately assayed for light output using a Synergy 2 system (BioTek). Plasmids pBG38, pT7EMCLuc, pCAGT7, pPEP98, pPEP100, and pPEP101 were gifts of P. Spear (15, 29, 30). This assay was repeated at least three times for each mutant.
Complementation assay.
L cells grown in 12-well plates were transfected with WT (pSC390), vector (pCDNA3.1), or mutant gD plasmids. After 3 h, cells were infected with HSV-1 KOS-gDβ at a multiplicity of infection of 5. The virus contains the lacZ gene in place of the gene for gD. After 1 h at 37°C, the medium was removed and extracellular virus was inactivated by a 1-min exposure to sodium citrate buffer at pH 3.0. Fresh medium was added, and the cells were incubated at 37°C overnight. At 18 h postinfection, cells were subjected to two freeze-thaw cycles.
Titers of complemented virions were determined on both Vero and VD60 cells grown in 24-well plates. VD60 cells contain copies of the entire WT gD gene including its own promoter. Therefore, these cells will express WT gD only upon successful entry of the complemented virus. Expression of WT gD from the infected VD60 cells allows the gD-null virus to propagate and form plaques between cells. After the virus-cell mixture was incubated with cells for 1 h at 37°C, the monolayers were overlaid with DMEM containing 1% methylcellulose and 2% FCS. Between 42 and 72 h cells were fixed with 5% formaldehyde in PBS and stained with 0.1% crystal violet.
RESULTS
Construction of gD cysteine mutants.
In order to determine which gD residues to mutate to cysteine, we located pairs of residues on the C terminus and core of gD that could potentially form disulfide bonds by using the crystal structure of gD 306 (307C) as a guide (19). We took care not to mutate residues known to be involved in receptor binding or gD glycosylation and designed disulfide bonds that would limit the mobility of different proportions of the C terminus (Fig. 1C) (7, 8, 22). This approach yielded five potential cysteine pairs: H242C-E274C, K190C-A277C, S188C-L279C, A239C-V286C, and S140C-P288C (Fig. 1A). Each pair was numbered 1 through 5, respectively. The mutant from our previous study (19), gD A37C-V302C, was included in this study and is numbered 6.
The positions of the potential disulfide bonds are modeled on a crystal structure of gD (Fig. 1A). The C terminus of gD fits into a groove created by the gD core (Fig. 1B). Each of these disulfide bonds should impose different degrees of constraint on the gD C terminus and presumably limit the conformational change(s) required for receptor binding, for triggering later events, or both (Fig. 1C). The disulfide bond on the C terminus of mutant 6 is the most limiting in this regard, and we already know that this bond prevents receptor binding and thereby ablates entry (19). The positions of the other disulfide bonds should impose less constraint on the C terminus and potentially restrict the exposure of different functional regions of gD (Fig. 1C).
Characterization of gD mutants.
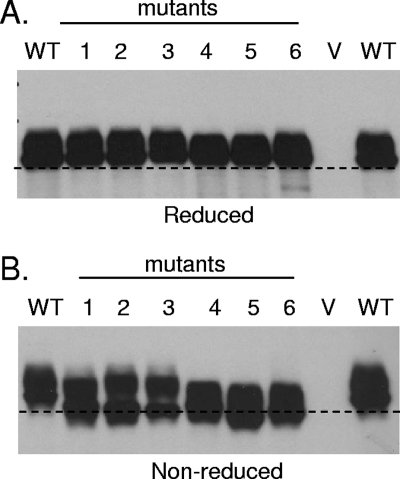
Initially, we asked whether the engineered disulfide bond was formed in each full-length gD mutant. Proteins from lysates of transfected 293T cells were separated by SDS-polyacrylamide gel electrophoresis under denaturing and reducing conditions, and Western blot assays were performed with gD PAb R8. Under these conditions the mutant proteins migrated to the same position as did the WT gD (Fig. 2A). However, when the proteins were electrophoresed under denaturing and nonreducing conditions, which leave disulfide bonds intact, each of the mutant proteins migrated further in the gel than did WT gD (Fig. 2B). This suggests that each mutant protein is more compact, consistent with an additional disulfide bond that constrains the flexible C terminus. The increased migration of mutant 6 was seen in a truncated version of this mutant in our previous study, and X-ray crystallography confirmed the presence of the disulfide bond between residues 37 and 302 (19).
FIG. 2.
Western blot analysis of gD disulfide mutants. The lanes are numbered according to the number designating each mutant, with WT and V denoting WT gD and vector-transfected cell lysates, respectively. (A) Samples were boiled for 5 min in 2% SDS buffer containing 100 mM dithiothreitol and electrophoresed in 10% polyacrylamide gels. (B) Samples were boiled in SDS buffer lacking the reducing agent dithiothreitol. The dotted line corresponds to the front of migration of WT gD. Blots were probed with gD PAb R8.
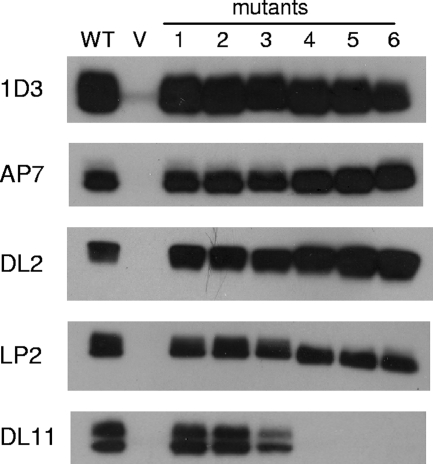
The presence of an additional disulfide bond in any of the gD mutants could have a deleterious effect on the overall conformation of gD. To exclude this possibility, we probed each protein with MAbs to distinct epitopes that are located at different sites on gD (6). First we determined the relative amounts of gD in the cell lysates from transfected 293T cells by capture ELISA with MAb 1D3 (data not shown). This MAb recognizes a linear epitope near the N terminus and therefore was used to normalize the amount of gD in each lysate. Normalized amounts of lysate were immunoprecipitated with each MAb and electrophoresed under denaturing and reducing conditions, and then Western blotting was performed with gD PAb R8 (Fig. 3). The epitope of MAb AP7 consists of portions of the N and C termini of the ectodomain and is a good indicator of proper folding of the N and C termini (4, 24). All six mutants retained this epitope. All of the mutants were also immunoprecipitated by DL2, which recognizes a discontinuous epitope that does not interfere with receptor binding (6). MAbs LP2 and DL11 recognize distinct epitopes near the binding site of nectin-1 (7). All mutants were recognized by LP2.
FIG. 3.
Western blot of immunoprecipitated gD disulfide mutants with MAbs. Protein was immunoprecipitated from lysates of transfected cells by 10 μg/ml of the indicated MAb, gD-MAb complexes were captured with protein A-conjugated Sepharose beads and electrophoresed under denaturing and reducing conditions, and Western blot assays were performed with gD PAb R8. MAb 1D3 recognizes residues 11 to 19 on gD and detects all forms of gD. AP7, DL2, LP2, and DL11 each recognize distinct conformational epitopes. V, vector-transfected cells.
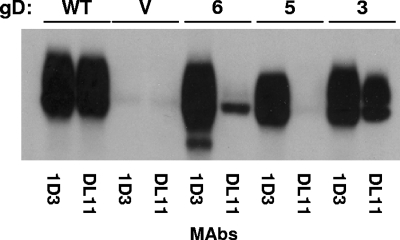
MAb DL11 is highly neutralizing and blocks binding of gD to both receptors (27, 37). As such, DL11 binding is strongly predictive of receptor binding (7). Mutants 1 and 2 reacted as well as WT gD did with DL11, while mutant 3 reacted weakly. Mutants 4, 5, and 6 did not appear to be immunoprecipitated by DL11. Since mutants 1, 2, and 3 reacted with all of the MAbs, we conclude that they were properly folded. Mutants 4, 5, and 6 react with all MAbs except DL11. Since DL11 competes with both HVEM and nectin-1 for receptor binding, the DL11 epitope could be masked by the locked C terminus. The loss of DL11 reactivity suggests that mutants 4, 5, and 6 will not bind receptor (19).
Binding of gD mutants to receptors.
To test each protein for receptor binding, the level of gD in each lysate was again normalized by capture ELISA with MAb 1D3 (data not shown). We normalized the gD lysates so that differences in protein expression would not account for differences in binding. The normalized gD lysates were serially diluted before being added to ELISA plates coated with purified receptor. Lysate from vector-transfected cells did not bind to either receptor, and the minimal absorbance from this lysate has been subtracted from all gDs.
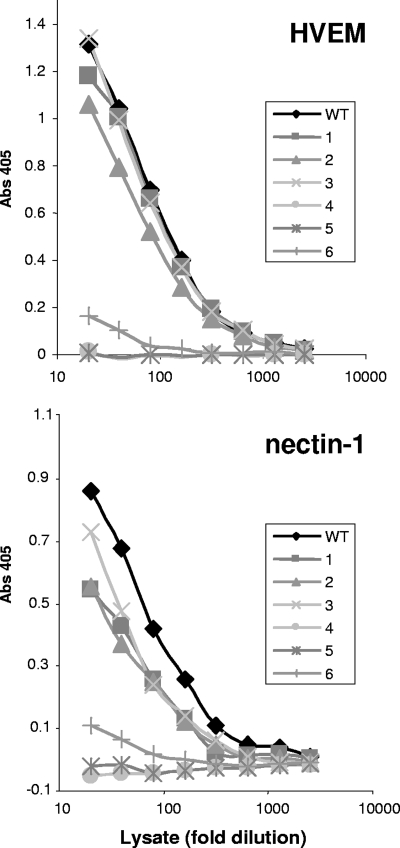
Mutants 1, 2, and 3 bound both HVEM and nectin-1 (Fig. 4), while mutants 4 and 5 did not bind to either receptor. This indicates that the constraints imposed by the disulfide bonds in mutants 4 and 5 completely block receptor binding, while the disulfide bonds in mutants 1, 2, and 3 do not constrain the gD C terminus sufficiently to inhibit receptor binding. Mutant 6 bound weakly to both nectin-1 and HVEM. This result differs from that seen with a soluble truncated version of mutant 6, which failed to bind either receptor (19). One explanation for this minimal binding is that a fraction of mutant 6 might not have formed the disulfide bond in the context of full-length gD.
FIG. 4.
Binding of gD mutant proteins to nectin-1 and HVEM. The amount of gD from transfected 293T cells was first quantitated by capture ELISA with the MAb 1D3. The lysates were then diluted to normalize the amount of gD in each lysate, serially diluted twofold, and incubated with purified receptor adhered to the ELISA plate. gD was then detected with the PAb R8. Binding data are the averages of at least three independent experiments and are representative of the results seen in each individual experiment.
Disulfide bond formation of mutant 6 (V37C-A302C).
In our previous study (19), a soluble truncated form of mutant 6 did not bind HVEM or nectin-1, by either ELISA or optical biosensor analysis. In that study, the ectodomain of gD was expressed in insect cells by a baculovirus and both mass spectrometry analysis and a crystal structure of the protein gD 316t (37C-302C) confirmed the presence of the predicted disulfide bond. In the present study each double mutation was engineered into the gene for full-length gD and the protein was expressed in mammalian cells. We considered the possibility that the proximity of A302C to the transmembrane domain led to inefficient disulfide bonding, enabling a proportion of the protein to bind receptors.
Since the DL11 epitope overlaps the binding site for both receptors, we reevaluated the ability of full-length mutant 6 to be recognized by this MAb (7, 18, 27, 37). The soluble, truncated form of mutant 6 is not recognized by DL11, because the locked C terminus masks the DL11 epitope (19). We reasoned that, if a fraction of the full-length mutant 6 was not disulfide bonded, that form would be recognized by DL11. To address this, we repeated the immunoprecipitation assay with DL11 for mutants 3, 5, and 6 and WT gD (Fig. 5). Once again we used 1D3 as the control MAb and deliberately overexposed the Western blot to detect small amounts of protein. As expected WT gD was immunoprecipitated by DL11, as was mutant 3. In contrast, mutant 5, which did not bind either receptor, was not immunoprecipitated by DL11. Finally, mutant 6 was immunoprecipitated by 1D3, but a small fraction of the total mutant 6 protein was immunoprecipitated by DL11. A similar proportion of mutant 6 was immunoprecipitated by DL11 from transfected CHO-K1 cells, showing that this effect is cell type independent (data not shown). We believe that this small fraction of the total protein that binds DL11 is not disulfide bonded and accounts for the weak binding of mutant 6 to receptors (Fig. 4). As a result of these experiments, it appears that mutants 1 to 5 are completely bonded while mutant 6 is a mixture of bonded and nonbonded protein.
FIG. 5.
Immunoprecipitation of gD mutants with MAb DL11. Immunoprecipitations were performed as described for Fig. 3. The blot was overexposed to show less abundant bands. The reactivity of the mutants at a lower exposure is demonstrated in Fig. 3. V, vector-transfected cells.
Ability of gD mutants to induce cell-cell fusion.
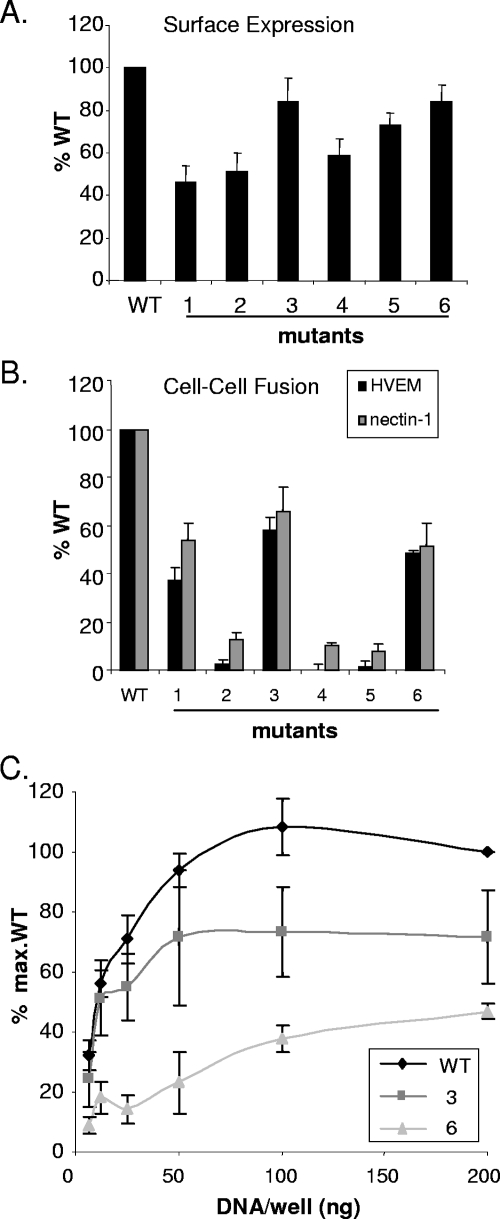
We used a cell-cell fusion assay to determine which cysteine mutants were capable of triggering fusion (30). We first tested whether the cysteine mutants were expressed on the surface of CHO-K1 cells. The level of expression of each mutant protein was measured using a CELISA and is shown as the percentage of that expressed by cells transfected with WT gD (Fig. 6A). Mutants 1, 2, and 4 were expressed at between 40 and 60% of the WT level. Mutants 3, 5, and 6 were expressed at approximately 80% of the amount of WT gD. Cells were transfected with plasmid DNA at a fourfold excess over the amount of DNA needed for maximum fusion to ensure that differences in cell surface expression do not account for differences in fusion activity. Thus, all mutants were expressed on the cell surface and could be directly compared for fusion.
FIG. 6.
Surface expression and cell-cell fusion of gD mutants. CHO-K1 cells were transfected with plasmids expressing a gD mutant and gB, gH, gL, and T7 RNA polymerase. (A) Cell surface expression of gD mutants was measured by PAb R8 binding using a CELISA. (B) A second set of CHO-K1 cells was transfected with plasmids for either HVEM or nectin, as well as a plasmid for the luciferase gene under the control of the T7 RNA polymerase promoter. Glycoprotein-expressing and receptor-expressing cells were mixed, and the resulting luciferase activity was expressed as the percentage of WT gD luciferase activity minus the background of the vector control. The results represent the averages of four independent experiments, with their standard errors shown as error bars. (C) Titration of fusion activity of WT gD and mutants on HVEM-expressing cells. The amount of gD plasmid was titrated from 200 ng/well (as in panel B) to 6.3 ng/well, with constant amounts of plasmids for gB, gH, gL, and T7 RNA polymerase. Cell surface expression of gD mutants was equivalent to or higher than that of WT gD at all points (data not shown).
For the fusion assay (30), one set of CHO-K1 cells were transfected with plasmids for gB, gH, gL, T7 RNA polymerase, and WT or mutant gD. A second set of CHO-K1 cells was transfected with a plasmid encoding luciferase under the control of the T7 promoter, along with plasmids encoding either nectin-1 or HVEM. The receptor-expressing CHO-K1 cells were then seeded onto the glycoprotein-expressing CHO-K1 cells. Fusion was measured by the amount of light generated by luciferase activity with a fluorescent substrate. As a negative control, vector plasmid was transfected with gB, gH, gL, and T7 RNA polymerase and the minimal luciferase activity was subtracted from all other conditions.
First, each mutant gD triggered the same relative amount of fusion on HVEM-expressing cells as it did on nectin-1-expressing cells, indicating that there is no receptor-specific difference (Fig. 6B). Second, the amount of fusion depended on the particular mutant. Thus, mutant 1 induced fusion at approximately 50% and mutant 3 induced fusion at 60% of the WT gD level. Importantly, mutant 2 failed to induce fusion despite its ability to bind to both receptors. This is the first example that we have found of a properly folded, full-length HSV-1 gD that binds receptors but fails to trigger cell fusion. Mutants 4 and 5, which did not bind either receptor, did not trigger fusion. Mutant 6 induced 50% fusion, despite weak binding to both receptors (Fig. 4).
The cells used in the luciferase fusion assay are normally transfected with an excess of DNA to ensure high levels of protein expression. In light of the unexpectedly high level of fusion observed with mutant 6, we wondered whether these conditions enabled the small fraction of unbonded mutant 6 protein (Fig. 5) to cause significant fusion. To test this possibility, we titrated the gD DNA in the cell-cell fusion assay while maintaining the concentrations of the other plasmids at the same level as in Fig. 6B. For WT gD, fusion increased with increasing DNA, reaching a maximum at 50 ng/well DNA (Fig. 6C). For mutant 6 the amount of fusion was significantly reduced at 50 ng/well. Mutant 3 was also titrated for comparison and, like WT gD, reached saturation at 50 ng/well. The amount of gD expressed on the cell surface continued to increase past 50 ng/well, and expression was equivalent throughout the titration for all gDs (data not shown). This continual increase in cell surface expression would allow the small portion of functional gD in mutant 6 to induce abnormally high fusion. Thus, when gD protein expression levels are reduced in the fusion assay, the functional deficit in mutant 6 is obvious and the small amount of fusion can be reasonably explained by the small proportion of non-disulfide-bonded protein.
The important conclusion is therefore that mutants 1 and 3 are partially deficient for cell-cell fusion while mutants 2, 4, 5, and 6 are null.
The effect of cysteine mutations on virus entry.
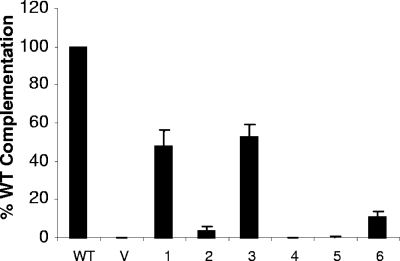
To examine the phenotypes of the gD mutants in the context of a virion, we used a standard complementation assay. L cells were first transfected with the plasmids that encoded WT or mutant gDs and then infected with the genotypically gD-null virus KOS-gDβ (10). The complemented progeny virions were tested for their ability to form plaques on VD60 cells. VD60 cells are Vero cells engineered to express gD under the control of the gD promoter. Thus, these cells express gD only upon virus infection (21). The expression of gD in infected VD60 cells allows plaques to form, and the resulting titers are a measure of the amount of entry of each complemented virus. Of the six gD mutants, virions complemented with mutants 1 and 3 rescued infectivity at 50% of the level of WT-rescued virions (Fig. 7). Mutants 2, 4, and 5 rescued infectivity at less than 5% of WT-rescued virus, and mutant 6 complemented virions at 10% of the WT level.
FIG. 7.
Complementation of a gD-null virus by gD mutants. L cells were transfected to express WT gD or mutants. Transfected cells were then infected with the gD-null virus KOS-gDβ. Cells were freeze-thawed to release intracellular virus, and the virion-containing lysates were used to infect the gD-complementing cell line VD60. The results represent the averages of three independent experiments, with their standard errors shown as error bars. V, vector-transfected cells.
The failure of mutants 2, 4, and 5 to complement virus agrees with their inability to induce cell-cell fusion, as well as the failure of mutants 4 and 5 to bind either receptor (Table 1). Mutant 1 and mutant 3 complemented virions approximately as well as they induced fusion. The complementation data for mutant 6 agree with our previous results (19) and also agree with the data in Fig. 6C for fusion. Of most importance is the phenotype of mutant 2 (K190C-A277C). This protein bound both receptors relatively well but did not function in either cell-cell fusion or virus entry.
TABLE 1.
Properties of double cysteine mutants
| Mutant no. | Mutation | % of WT gD value
|
||
|---|---|---|---|---|
| Receptor bindinga | Fusiona | Complementation | ||
| 1 | H242C-E274C | 96 | 37 | 48 |
| 2 | K190C-A277C | 76 | 3 | 4 |
| 3 | S188C-L279C | 95 | 58 | 53 |
| 4 | A239C-V286C | 0 | 0 | 0 |
| 5 | S140C-P288C | 0 | 1 | 0 |
| 6 | A37C-A302C | 10 | 24b | 11 |
Data are shown for HVEM only; similar results were obtained with nectin-1.
Fusion relative to WT under low-expression conditions (50 ng/well).
Characterization of single cysteine gD mutants.
Mutants 1, 2, and 3 retained the ability to bind receptors but were reduced or nonfunctional in fusion and null-virus complementation assays. The loss of function for these mutants can be explained either as a constraint of conformational changes in gD imposed by the engineered disulfide bond or, alternatively, as the mutation of specific amino acids to cysteine. To address this, we constructed four additional mutants, each containing a single cysteine from each pair of cysteines of mutants 1 and 2: H242C and E274C for mutant 1 and K190C and A277C for mutant 2.
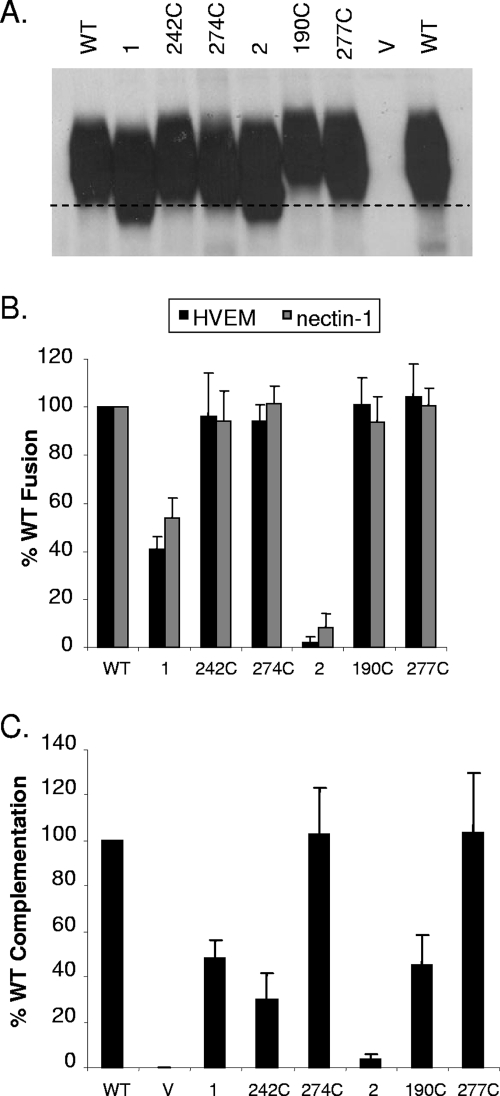
These single cysteine mutants were characterized in the same manner as the double cysteine mutants. In contrast to the latter, the electrophoretic mobility of the single cysteine mutants was similar to that of WT gD under nonreducing conditions (Fig. 8A). Furthermore, all single cysteine mutants bound both receptors better than their respective double mutants and at least as well as WT gD (data not shown).
FIG. 8.
Characterization of gD single cysteine mutants. (A) Western blot of single cysteine mutants. Samples were boiled for 5 min in buffer lacking dithiothreitol. The black dashed line corresponds to migration of WT gD and is used for reference. The blot was probed with gD PAb R8. (B) Cell-cell fusion with single cysteine mutants. Cell-cell fusion was performed as described for Fig. 6B. Fusion activity is the average of five independent experiments, and the error bars represent the standard errors. (C) Virus complementation with single cysteine mutants. Complementation was performed as described for Fig. 7. The results represent the averages of three independent experiments, with their standard errors shown as error bars. V, vector-transfected cells.
Importantly, all single cysteine mutants had WT activity in the cell-cell fusion assay, and gD carrying E274C and 277C complemented the gD-null virus as well as WT (Fig. 8B and C). Surprisingly, changes at residues 190 and 242 reduced the ability of these proteins to complement the gD-null virus, despite WT binding and fusion (Fig. 8B and C). Interestingly, 190C and 242C reside on the core of gD while 274C and 277C are located on the C terminus. Since all four single cysteine mutants function as well as WT gD in binding receptor(s) and inducing cell-cell fusion, it seems unlikely that the deficit in complementation seen with 190C and 242C is due to a direct effect on gD function. Perhaps the location of these free cysteines on the core of gD has an indirect effect in the context of a virion. One possible interpretation is that the cysteines on the core negatively affect gD incorporation into the virus. However, attempts to quantitatively measure gD incorporation into virus were unsuccessful due to the low yield of complemented virions and the high gD background from the infected cells. Nevertheless, we conclude that the engineered disulfide bonds in mutants 1, 2, and 3, rather than the specific mutations, interfere with the post-receptor-binding functions of gD.
DISCUSSION
HSVs differ from most viruses because they employ four different glycoproteins to enter and infect host cells. Since more proteins are involved, the entry processes of herpesviruses are likely to involve more protein-protein interactions than is the case for other viruses. Regardless of the complexity of the entry process, all viruses must engage their target host cells specifically, by binding to a receptor. In HSV the function of receptor binding is carried out by gD. However, because gD does not itself have the ability to fuse viral and cellular membranes (33) and because both gD and its receptors can function as soluble proteins (5, 20, 36, 39), its role is clearly more complex than just to bring the viral and cellular membranes in apposition.
Comparison of the crystal structure of unliganded gD with that of gD bound to HVEM indicates that a conformational change is required in order for receptor binding to occur (3, 19). One aspect of this conformational change must involve the C-terminal portion of the gD ectodomain because residues 288 to 307 occupy nearly the same position as do residues 1 to 16 of the N-terminal HVEM binding loop following receptor engagement. Identification of gD residues involved in nectin-1 binding suggested that the C-terminal portion of the gD ectodomain must also be displaced in order to allow nectin-1 binding (7, 22). Additionally, truncation of gD to residue 285 or destabilization of the interaction between the C terminus and the rest of the gD ectodomain by mutation results in a significant increase in binding affinity for both HVEM and nectin-1 (19, 26, 32, 40). Together, these observations support the hypothesis that the C-terminal portion of the gD ectodomain is a negative regulator of receptor binding.
A separate role for the C terminus of gD as a positive regulator of fusion has also been proposed. This was based on the observation that a soluble form of gD truncated at residue 285 could complement the infectivity of gD-null virions whereas gD truncated at residue 260 could not (5). While 10 residue deletions within the C terminus did not reveal any critical region, chimeras of pseudorabies virus and HSV gD functioned only when the first 285 residues came from HSV (41). In an attempt to reconcile the seemingly disparate observations of this region of gD, it has been proposed that receptor binding initially displaces the C terminus of gD from its native position (against the core of gD). Once displaced, a portion of gD may promote fusion by direct interaction with the downstream effectors, gB and/or gH/gL.
Despite our current understanding of gD structure and function, it remained unclear how much of the C terminus must move in order for receptor binding to occur and how much movement is required for fusion activation. In this study, we used site-directed mutagenesis to generate forms of gD with two additional cysteine residues. The newly introduced cysteine residues were predicted to form disulfide bonds that would lock the C terminus of gD to the core in six different places (Fig. 1). The fact that all mutants formed disulfide bonds to some degree (Fig. 2) validates the crystal structure used to predict the disulfide bonds (19). However, the importance of disulfide bond formation for interpretation of our data is illustrated by mutant 6. A small proportion of this mutant protein did not form a disulfide bond as evidenced by MAb DL11 binding (Fig. 5), and this proportion likely accounted for the minimal binding to receptor, fusion under high expression conditions, and complementation (Fig. 4, 6B, and 7).
Among our other mutant proteins, we observed two distinct phenotypes: (i) those without receptor binding, fusion activity, and virus complementation and (ii) those which retained receptor binding activity but lost fusion activity and were reduced in complementation. Locking the C terminus at gD residue 274, 277, or 279 did not interfere with receptor binding but impaired the ability of the mutants to promote fusion. Receptor binding and fusion activity were both lost when the C terminus was locked at gD residue 286, 288, or 302. The inability of gD to bind receptors when 286 to 306 are fixed fits well with the gD crystal structure, since the residues between 286 and 306 cover amino acids involved in nectin-1 binding and interfere with the formation of the HVEM binding loop (Fig. 1B). Moreover these results are strengthened by the increased binding seen by truncation of the C terminus to residue 285. Since receptor binding is critical for gD function, the loss of receptor binding for mutants 4 and 5 prevents them from promoting fusion or rescuing gD-null virus infectivity.
Our data suggest that two portions of the C terminus must move to allow gD function: residues 286 to 306 for binding and residues at or before residue 279 for fusion. Our cysteine mutagenesis is limited to the resolved regions of gD crystal structures, and residues 256 to 268, due to their flexibility, have not been resolved in any crystal structure to date. Since residues 256 to 268 are flexible, this region could potentially serve as a hinge that moves in response to receptor binding. Limiting the movement of this potential hinge region with disulfide bonds (mutants 1, 2, and 3) would prevent the exposure of residues, either on the C terminus or on the gD core, that may interact with gH/gL or gB. The functional differences between mutant 2 (complete loss of fusion and complementation) and mutants 1 and 3 (partial loss of fusion and complementation) could be due to differences in constraint imposed by the mutants. The constraint imposed by mutants 1 and 3 may lead to a partial exposure of a profusion domain which could result in reduced fusion and complementation. The constraint imposed by mutant 2 completely blocks this region. A disulfide bond further upstream of residue 274 potentially would allow full exposure of a profusion domain and would be completely WT in binding and function.
The mutants in this study forced greater stability between the C terminus and core of gD that interfered with the conformational change(s) required for viral fusion and entry (19). Previously reported deletions or mutations within the C terminus increase the binding of gD to receptors by destabilizing the interaction of the C terminus with the core (19). Importantly, these mutants also complemented virus infectivity poorly (4, 19). In these mutant proteins the C-terminal conformational change likely occurs more easily and possibly pretriggers virions before the virus encounters a host cell.
Our results show that a dynamic interaction between the C terminus and the core of gD is important for fusion and virus infectivity. Too much, or too little, interaction between the C terminus and core negatively impacts the virus. This interaction between the C terminus and core of gD allows the virus to regulate the triggering of the fusion glycoproteins so that fusion occurs only when the virus has encountered the proper host cell and binds a receptor (31).
While our cysteine mutagenesis defines functions for the movement of two different regions of the C terminus, it is unclear if the movement of these two regions is synchronous in WT gD. It is possible that residues 286 to 306 associate only weakly with the core of gD via the interaction with W294 and surrounding residues. We propose that the profusion domain is more tightly associated with the core and that receptor binding is required for stable unwinding of the C terminus and exposure of this domain. Future studies will be directed at testing this and other possibilities using our panel of gD cysteine double mutants.
Acknowledgments
We thank N. W. Fraser, D. C. Johnson, T. Minson, and P. G. Spear for reagents used in this study. We thank H. McGraw for assistance with figures.
This study was supported by Public Health Service grants AI-18289 to G.H.C. and AI-076231 and AI-056045 to R.J.E. C.K. was supported by the University of Pennsylvania Research Foundation.
Footnotes
Published ahead of print on 21 November 2007.
REFERENCES
- 1.Binley, J. M., R. W. Sanders, B. Clas, N. Schuelke, A. Master, Y. Guo, F. Kajumo, D. J. Anselma, P. J. Maddon, W. C. Olson, and J. P. Moore. 2000. A recombinant human immunodeficiency virus type 1 envelope glycoprotein complex stabilized by an intermolecular disulfide bond between the gp120 and gp41 subunits is an antigenic mimic of the trimeric virion-associated structure. J. Virol. 74627-643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carfi, A., H. Gong, H. Lou, S. H. Willis, G. H. Cohen, R. J. Eisenberg, and D. C. Wiley. 2002. Crystallization and preliminary diffraction studies of the ectodomain of the envelope glycoprotein D from herpes simplex virus 1 alone and in complex with the ectodomain of the human receptor HveA. Acta Crystallogr. D Biol. Crystallogr. 58836-838. [DOI] [PubMed] [Google Scholar]
- 3.Carfi, A., S. H. Willis, J. C. Whitbeck, C. Krummenacher, G. H. Cohen, R. J. Eisenberg, and D. C. Wiley. 2001. Herpes simplex virus glycoprotein D bound to the human receptor HveA. Mol. Cell 8169-179. [DOI] [PubMed] [Google Scholar]
- 4.Chiang, H. Y., G. H. Cohen, and R. J. Eisenberg. 1994. Identification of functional regions of herpes simplex virus glycoprotein gD by using linker-insertion mutagenesis. J. Virol. 682529-2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cocchi, F., D. Fusco, L. Menotti, T. Gianni, R. J. Eisenberg, G. H. Cohen, and G. Campadelli-Fiume. 2004. The soluble ectodomain of herpes simplex virus gD contains a membrane-proximal pro-fusion domain and suffices to mediate virus entry. Proc. Natl. Acad. Sci. USA 1017445-7450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cohen, G. H., V. J. Isola, J. Kuhns, P. W. Berman, and R. J. Eisenberg. 1986. Localization of discontinuous epitopes of herpes simplex virus glycoprotein D: use of a nondenaturing (“native” gel) system of polyacrylamide gel electrophoresis coupled with Western blotting. J. Virol. 60157-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Connolly, S. A., D. J. Landsburg, A. Carfi, J. C. Whitbeck, Y. Zuo, D. C. Wiley, G. H. Cohen, and R. J. Eisenberg. 2005. Potential nectin-1 binding site on herpes simplex virus glycoprotein D. J. Virol. 791282-1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Connolly, S. A., D. J. Landsburg, A. Carfi, D. C. Wiley, G. H. Cohen, and R. J. Eisenberg. 2003. Structure-based mutagenesis of herpes simplex virus glycoprotein D defines three critical regions at the gD-HveA/HVEM binding interface. J. Virol. 778127-8140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Connolly, S. A., D. J. Landsburg, A. Carfi, D. C. Wiley, R. J. Eisenberg, and G. H. Cohen. 2002. Structure-based analysis of the herpes simplex virus glycoprotein D binding site present on herpesvirus entry mediator HveA (HVEM). J. Virol. 7610894-10904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dean, H. J., S. S. Terhune, M. T. Shieh, N. Susmarski, and P. G. Spear. 1994. Single amino acid substitutions in gD of herpes simplex virus 1 confer resistance to gD-mediated interference and cause cell-type-dependent alterations in infectivity. Virology 19967-80. [DOI] [PubMed] [Google Scholar]
- 11.Fan, Q. R., E. O. Long, and D. C. Wiley. 2000. A disulfide-linked natural killer cell receptor dimer has higher affinity for HLA-C than wild-type monomer. Eur. J. Immunol. 302692-2697. [DOI] [PubMed] [Google Scholar]
- 12.Farzan, M., H. Choe, E. Desjardins, Y. Sun, J. Kuhn, J. Cao, D. Archambault, P. Kolchinsky, M. Koch, R. Wyatt, and J. Sodroski. 1998. Stabilization of human immunodeficiency virus type 1 envelope glycoprotein trimers by disulfide bonds introduced into the gp41 glycoprotein ectodomain. J. Virol. 727620-7625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Friedman, H. M., G. H. Cohen, R. J. Eisenberg, C. A. Seidel, and D. B. Cines. 1984. Glycoprotein C of herpes simplex virus 1 acts as a receptor for the C3b complement component on infected cells. Nature 309633-635. [DOI] [PubMed] [Google Scholar]
- 14.Geraghty, R. J., C. R. Jogger, and P. G. Spear. 2000. Cellular expression of alphaherpesvirus gD interferes with entry of homologous and heterologous alphaherpesviruses by blocking access to a shared gD receptor. Virology 268147-158. [DOI] [PubMed] [Google Scholar]
- 15.Geraghty, R. J., C. Krummenacher, G. H. Cohen, R. J. Eisenberg, and P. G. Spear. 1998. Entry of alphaherpesviruses mediated by poliovirus receptor-related protein 1 and poliovirus receptor. Science 2801618-1620. [DOI] [PubMed] [Google Scholar]
- 16.Godley, L., J. Pfeifer, D. Steinhauer, B. Ely, G. Shaw, R. Kaufmann, E. Suchanek, C. Pabo, J. J. Skehel, D. C. Wiley, et al. 1992. Introduction of intersubunit disulfide bonds in the membrane-distal region of the influenza hemagglutinin abolishes membrane fusion activity. Cell 68635-645. [DOI] [PubMed] [Google Scholar]
- 17.Isola, V. J., R. J. Eisenberg, G. R. Siebert, C. J. Heilman, W. C. Wilcox, and G. H. Cohen. 1989. Fine mapping of antigenic site II of herpes simplex virus glycoprotein D. J. Virol. 632325-2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krummenacher, C., A. V. Nicola, J. C. Whitbeck, H. Lou, W. Hou, J. D. Lambris, R. J. Geraghty, P. G. Spear, G. H. Cohen, and R. J. Eisenberg. 1998. Herpes simplex virus glycoprotein D can bind to poliovirus receptor-related protein 1 or herpesvirus entry mediator, two structurally unrelated mediators of virus entry. J. Virol. 727064-7074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krummenacher, C., V. M. Supekar, J. C. Whitbeck, E. Lazear, S. A. Connolly, R. J. Eisenberg, G. H. Cohen, D. C. Wiley, and A. Carfi. 2005. Structure of unliganded HSV gD reveals a mechanism for receptor-mediated activation of virus entry. EMBO J. 244144-4153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kwon, H., Q. Bai, H. J. Baek, K. Felmet, E. A. Burton, W. F. Goins, J. B. Cohen, and J. C. Glorioso. 2006. Soluble V domain of nectin-1/HveC enables entry of herpes simplex virus type 1 (HSV-1) into HSV-resistant cells by binding to viral glycoprotein D. J. Virol. 80138-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ligas, M. W., and D. C. Johnson. 1988. A herpes simplex virus mutant in which glycoprotein D sequences are replaced by beta-galactosidase sequences binds to but is unable to penetrate into cells. J. Virol. 621486-1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Manoj, S., C. R. Jogger, D. Myscofski, M. Yoon, and P. G. Spear. 2004. Mutations in herpes simplex virus glycoprotein D that prevent cell entry via nectins and alter cell tropism. Proc. Natl. Acad. Sci. USA 10112414-12421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Milne, R. S., S. A. Connolly, C. Krummenacher, R. J. Eisenberg, and G. H. Cohen. 2001. Porcine HveC, a member of the highly conserved HveC/nectin 1 family, is a functional alphaherpesvirus receptor. Virology 281315-328. [DOI] [PubMed] [Google Scholar]
- 24.Minson, A. C., T. C. Hodgman, P. Digard, D. C. Hancock, S. E. Bell, and E. A. Buckmaster. 1986. An analysis of the biological properties of monoclonal antibodies against glycoprotein D of herpes simplex virus and identification of amino acid substitutions that confer resistance to neutralization. J. Gen. Virol. 671001-1013. [DOI] [PubMed] [Google Scholar]
- 25.Muggeridge, M. I., V. J. Isola, R. A. Byrn, T. J. Tucker, A. C. Minson, J. C. Glorioso, G. H. Cohen, and R. J. Eisenberg. 1988. Antigenic analysis of a major neutralization site of herpes simplex virus glycoprotein D, using deletion mutants and monoclonal antibody-resistant mutants. J. Virol. 623274-3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nicola, A. V., C. Peng, H. Lou, G. H. Cohen, and R. J. Eisenberg. 1997. Antigenic structure of soluble herpes simplex virus (HSV) glycoprotein D correlates with inhibition of HSV infection. J. Virol. 712940-2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nicola, A. V., M. Ponce de Leon, R. Xu, W. Hou, J. C. Whitbeck, C. Krummenacher, R. I. Montgomery, P. G. Spear, R. J. Eisenberg, and G. H. Cohen. 1998. Monoclonal antibodies to distinct sites on herpes simplex virus (HSV) glycoprotein D block HSV binding to HVEM. J. Virol. 723595-3601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nicola, A. V., S. H. Willis, N. N. Naidoo, R. J. Eisenberg, and G. H. Cohen. 1996. Structure-function analysis of soluble forms of herpes simplex virus glycoprotein D. J. Virol. 703815-3822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Okuma, K., M. Nakamura, S. Nakano, Y. Niho, and Y. Matsuura. 1999. Host range of human T-cell leukemia virus type I analyzed by a cell fusion-dependent reporter gene activation assay. Virology 254235-244. [DOI] [PubMed] [Google Scholar]
- 30.Pertel, P. E., A. Fridberg, M. L. Parish, and P. G. Spear. 2001. Cell fusion induced by herpes simplex virus glycoproteins gB, gD, and gH-gL requires a gD receptor but not necessarily heparan sulfate. Virology 279313-324. [DOI] [PubMed] [Google Scholar]
- 31.Rey, F. A. 2006. Molecular gymnastics at the herpesvirus surface. EMBO Rep. 71000-1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rux, A. H., S. H. Willis, A. V. Nicola, W. Hou, C. Peng, H. Lou, G. H. Cohen, and R. J. Eisenberg. 1998. Functional region IV of glycoprotein D from herpes simplex virus modulates glycoprotein binding to the herpesvirus entry mediator. J. Virol. 727091-7098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Spear, P. G., R. J. Eisenberg, and G. H. Cohen. 2000. Three classes of cell surface receptors for alphaherpesvirus entry. Virology 2751-8. [DOI] [PubMed] [Google Scholar]
- 34.Spear, P. G., and R. Longnecker. 2003. Herpesvirus entry: an update. J. Virol. 7710179-10185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Subramanian, R. P., and R. J. Geraghty. 2007. Herpes simplex virus type 1 mediates fusion through a hemifusion intermediate by sequential activity of glycoproteins D, H, L, and B. Proc. Natl. Acad. Sci. USA 1042903-2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tsvitov, M., A. R. Frampton, Jr., W. A. Shah, S. K. Wendell, A. Ozuer, Z. Kapacee, W. F. Goins, J. B. Cohen, and J. C. Glorioso. 2007. Characterization of soluble glycoprotein D-mediated herpes simplex virus type 1 infection. Virology 360477-491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Whitbeck, J. C., M. I. Muggeridge, A. H. Rux, W. Hou, C. Krummenacher, H. Lou, A. van Geelen, R. J. Eisenberg, and G. H. Cohen. 1999. The major neutralizing antigenic site on herpes simplex virus glycoprotein D overlaps a receptor-binding domain. J. Virol. 739879-9890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Whitbeck, J. C., C. Peng, H. Lou, R. Xu, S. H. Willis, M. Ponce de Leon, T. Peng, A. V. Nicola, R. I. Montgomery, M. S. Warner, A. M. Soulika, L. A. Spruce, W. T. Moore, J. D. Lambris, P. G. Spear, G. H. Cohen, and R. J. Eisenberg. 1997. Glycoprotein D of herpes simplex virus (HSV) binds directly to HVEM, a member of the tumor necrosis factor receptor superfamily and a mediator of HSV entry. J. Virol. 716083-6093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Whitbeck, J. C., Y. Zuo, R. S. Milne, G. H. Cohen, and R. J. Eisenberg. 2006. Stable association of herpes simplex virus with target membranes is triggered by low pH in the presence of the gD receptor, HVEM. J. Virol. 803773-3780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Willis, S. H., A. H. Rux, C. Peng, J. C. Whitbeck, A. V. Nicola, H. Lou, W. Hou, L. Salvador, R. J. Eisenberg, and G. H. Cohen. 1998. Examination of the kinetics of herpes simplex virus glycoprotein D binding to the herpesvirus entry mediator, using surface plasmon resonance. J. Virol. 725937-5947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zago, A., C. R. Jogger, and P. G. Spear. 2004. Use of herpes simplex virus and pseudorabies virus chimeric glycoprotein D molecules to identify regions critical for membrane fusion. Proc. Natl. Acad. Sci. USA 10117498-17503. [DOI] [PMC free article] [PubMed] [Google Scholar]