Abstract
The Us5 gene of herpes simplex virus (HSV) encodes glycoprotein J (gJ). The only previously reported function of gJ was its ability to inhibit apoptosis. However, the mechanism by which gJ prevents apoptosis is not understood, and it is not known whether gJ mediates additional cellular effects. In this study, we evaluated the expression, localization, and cellular effects of Us5/gJ. Us5 was first expressed 4 h after infection. gJ was detectable at 6 h and was expressed in glycosylated and unglycosylated forms. Us5 was regulated as a late gene, with partial dependency on DNA replication for expression. Us5 expression was delayed in the absence of ICP22; furthermore, expression of Us5 in trans protected cells from apoptosis induced by an HSV mutant with deletion of ICP27, suggesting that the antiapoptotic effects of ICP22 and ICP27 are mediated in part through effects on gJ expression. Within HSV-infected or Us5-transfected cells, gJ was distributed widely, especially to the endoplasmic reticulum, trans-Golgi network, and early endosomes. gJ interacted with FoF1 ATP synthase subunit 6 by a yeast two-hybrid screen and had strong antiapoptotic effects, which were mediated by protein rather than mRNA. Antiapoptotic activity required the extracellular and transmembrane domains of gJ, but not the intracellular domain. Consistent with inhibition of FoF1 ATP synthase function, Us5 was required for HSV-induced reactive oxygen species (ROS) formation, and gJ was sufficient to induce ROS in Us5-transfected cells. Thus, HSV gJ is a multifunctional protein, modulating other cellular processes in addition to inhibition of apoptosis.
Herpes simplex virus (HSV) has at least 84 genes, which are expressed in a tightly regulated fashion during the viral replication cycle (41). The immediate-early (IE, or α) genes are involved in transactivation and regulation of viral gene synthesis (with the notable exception of Us12, which mediates immune evasion by downregulation of major histocompatibility complex class I). The early (E, or β) genes are mainly involved in nucleotide metabolism and viral DNA replication. The late (L, or γ) genes of HSV mainly encode structural proteins, either facilitating the assembly of newly synthesized virions or incorporated into the virion itself. Among the late gene products are the HSV glycoproteins. Most of the HSV glycoproteins have been well characterized in terms of expression class, cellular localization, and function. However, the glycoprotein encoded by the Us5 gene, glycoprotein J (gJ), has been much less carefully studied. The only reported function of gJ is its ability to inhibit apoptosis (3, 27, 28, 47). The mechanism by which gJ prevents apoptosis is unclear, and it is not known whether gJ might mediate additional effects in HSV-infected cells.
The Us5 open reading frame was originally identified by McGeoch et al. as a result of their sequencing of the unique short region of HSV (35). The predicted glycoprotein was subsequently designated gJ (40). However, it was not until 1998 that Us5 was demonstrated to encode a glycoprotein (17). Shortly thereafter, our group demonstrated that viruses with deletion of Us5 were defective in the ability to prevent UV- or anti-fas-induced apoptosis (28). In a different system, Zhou et al. demonstrated that expression of gJ or gD in trans was able to prevent apoptosis induced by mutant virus with deletion of gD (47). We have subsequently shown that gJ contributes to the protection of infected cells from apoptosis induced by cytotoxic T lymphocyte killing mechanisms (3, 27) and that gJ expressed in isolation is sufficient to mediate the protective effect (27).
In this study, we investigate the cellular localization and targets of gJ and its effects on cellular function. We report that Us5 is regulated as a γ1, or leaky late, gene. gJ is produced in small amounts during viral infection and localizes to membranes in multiple compartments throughout the cell. By the yeast two-hybrid assay, gJ interacts with subunit 6 of FoF1 ATP synthase. Consistent with expectations for a molecule disrupting ATP synthase activity, gJ is sufficient to induce reactive oxygen species (ROS) formation in transfected cells and is necessary for ROS generation in HSV-infected cells. These findings demonstrate that gJ is a multifunctional protein in HSV-infected cells and is likely to play an important role in modification of the infected host cell to maximize viral replication.
MATERIALS AND METHODS
Cells.
Jurkat cells (American Type Culture Collection [ATCC], Manassas, VA) were cultured in RPMI supplemented with 10% fetal calf serum (FCS). RAW264.7, HEp-2, and Vero cells (ATCC) were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% FCS. Human primary fibroblasts (HF) were isolated from foreskin. Vero 2.2, generously provided by J. A. Blaho (Mount Sinai School of Medicine, New York) and originally obtained from S. Silverstein (Columbia University, New York), is a derivative Vero cell line expressing ICP27 under its own promoter (43) and was maintained in DMEM supplemented with 10% FCS and G418 (Geneticin; 400 μg/ml). Cultured cell lines were screened regularly for mycoplasma by the Biologics Production Facility at the Fred Hutchinson Cancer Research Center.
Viruses.
Strains of HSV type 1 (HSV-1) were grown in Vero cells, and titers were determined using standard plaque assays. HSV-1 E115 and 17+ are wild-type laboratory strains. HSV-1 strain F, a wild-type strain, and R325, an ICP22 mutant virus, were kind gifts from J. Blaho and were originally obtained from B. Roizman (University of Chicago, Chicago, IL). RAS116, a derivative of F that has a deletion of Us5, and RAS137, the repaired virus, were constructed by A. Sears and H. Lee (28). vBSΔ27 is an ICP27-null derivative of KOS1.1 and must be propagated on the ICP27-complementing cell line Vero 2.2 (44). Both vBSΔ27 and KOS1.1 were generously provided by J. Blaho and originally obtained from S. Silverstein.
Infections.
Jurkat cells were mock infected or infected at 37°C with HSV (10 PFU/cell unless otherwise noted) in low-volume (<0.5 ml) RPMI 1640 with 10% FCS for 1 h, after which medium was added to bring the cells to ideal culture density (0.5 × 106 to 2 × 106 cells/ml). For adherent cells (Vero, HEp-2, HF, or RAW264.7), confluent monolayers were infected (10 PFU/cell unless otherwise noted) at 37°C in low-volume (<1 ml) DMEM with 10% FCS for 1 h with rocking, after which the medium with inoculum was removed and fresh medium was added to normal culture volume. The cells were maintained in 5% CO2 at 37°C until the time of analysis.
Construction and isolation of recombinant viruses.
Recombinant viruses were generated by cotransfection of subconfluent monolayers of Vero cells with 20 μg of linearized plasmid and HSV-1 genomic DNA (capsid preparation) as previously described (37). The transfection was done using a modified CaCl2 transfection protocol (19). When 100% cytopathic effect was observed, the cell monolayers were frozen and thawed once, harvested, and sonicated to obtain a virus stock. The recombinant virus was isolated by plaque assay on Vero cells and selected on the basis of green fluorescent protein (GFP) expression. The isolated recombinant virus was then plaque purified three times. For the generation of the recombinant viruses expressing FLAG-tagged gJ, FUs5FLAG (C-terminally FLAG-tagged gJ), and FUs5-3FLAG (C-terminally three-FLAG-tagged gJ), FΔUs5 virus was used in combination with either pBSKSUs5FLAG or pBSKSUs53FLAG. FΔUs5, which carries a deletion of Us5 sequences and an insertion of GFP sequences at the Us5 locus, was generated using HSV-1(F) genomic DNA and the plasmid pBSKS3.9. The Us5 sequences and the presence of the tag in all recombinant viruses were confirmed by sequencing.
Plasmids.
The 4.8-kb HindIII N fragment from the HSV-1(F) genome (nucleotides 133466 to 138349, containing Us5) was cloned into pBluescript KS(−) between the SacI and PstI sites. First, pBluescript KS(−) was digested with SacI and PstI, followed by fill in with T4 DNA polymerase and finally ligation with the 4.8-kb HindIII N fragment using T4 DNA ligase. The resulting plasmid, pBSKS3, was used to generate pBSKS3.9 (containing a deletion in Us5) by insertion of the GFP coding sequences driven by the cytomegalovirus (CMV) promoter. GFP was inserted as an EcoRV-SacI fragment isolated from pUC21.6 into the NruI and SacI sites of pBSKS3. pUC21.6 was generated by cloning an AseI-SspI fragment containing the enhanced-GFP-encoding gene driven by the CMV promoter from pEGFP-N3 (Clontech) into pUC21 (a gift from J. Vieira, University of Washington) NdeI and HincII sites. For the generation of pBSKSUs5FLAG, pBSKS3.52 was made by deletion of the EcoRV-BamHI region of pBSKS3 by digestion, fill in, and religation (pBSKS3.5), followed by removal of the remaining XhoI site by digestion, fill in, and religation (pBSKS3.52). Then, a new XhoI site was introduced into the Us5 3′ region (pBSKS3.52XhoI) using a QuickChange II site-directed mutagenesis kit (Stratagene) with the following complementary primers: 5′ GGACCCAGTTCTCGAGTAATTTCCC 3′ and 5′ GGGAAATTACTCGAGAACTGGGTCC 3′. The manufacturer's protocol was modified as reported by Wang and Malcolm (46). Next, the 3′ region of Us5 contained in the SacI-XhoI fragment from pBSKS3.52XhoI was introduced into pCMV-Tag4a (Stratagene) SacI and XhoI restriction sites preceding FLAG sequences (pCMVFLAG3′Us5). Finally, the FLAG-tagged 3′ region of Us5 from pCMVFLAG3′Us5 obtained by PCR using the primers 5′ CTGCCCACTTGGCAGTACATCAAGTGTATC 3′ and 5′ TTAAGGTACGTCGACCTACTTATCGTCGTC 3′ and digested with SacI and SalI was used to replaced the 3′ region of Us5 between the SacI and XhoI sites in pBSKS3.52XhoI. To generate pBSKSUs5-3FLAG, first, the NruI-SacI fragment from pBSKS3.52XhoI was introduced into the EcoRV and XhoI restriction sites of pCMV3Tag3a (Stratagene) to create pCMVUs5-3FLAG. The Us5-3FLAG sequences were introduced into the pBSKS3.52XhoI in place of the Us5 sequences. To achieve this replacement, the Us5-3FLAG sequences were amplified from pCMVUs5-3FLAG by PCR using the primers 5′ CTGCCCACTTGGCAGTACATCAAGTGTATC 3′ and 5′ TTAAGGTACGTCGACCTATTTATCGTCATC 3′, followed by digestion with SacI and SalI. The SacI-SalI PCR fragment containing Us5-3FLAG sequences was introduced into the SacI and XhoI sites of pBSKS3.52XhoI, generating pBSKSUs5-3FLAG.
pCMV-mycUs5 and pCMV-HAUs5 were constructed by introducing into the SalI and EcoRI sites of the pCMV-c-myc and pCMV-HA vectors an EcoRI-SalI DNA fragment containing the Us5 sequences from HSV-1 strain 17+ that had been previously cloned into pGBKT7-Us5. pCMV-myc-mUs5 was generated by site-directed mutagenesis in pCMV-mycUs5 using a QuickChange II site-directed mutagenesis kit (Stratagene) according to the manufacturer's directions. The start codon of Us5 was changed from ATG to ACAC using the following complementary primers: 5′ GTTCTGTGTGTCACGACACTCTCTGCGCGCAG 3′ and 5′ CTGCGCGCAGGAGTGTCGTGACACACAGAAC 3′.
The various plasmids for the expression of the c-myc-tagged truncated forms of gJ were generated by PCR of the Us5 sequences encoding the truncated gJ proteins using pCMV-c-mycUs5 as a DNA template, followed by cloning of the PCR products into EcoRI and XhoI restriction sites of the pCMV-c-myc vector. The following primers were used to generate the different PCR products; DS-US5-1UP (5′ ATGGAGGCCCGAATTCGCGTT 3′), DS-US5-74LO (5′ TCTATCTCGAGTTAACGCAGGAGCTCAAGCAGACA 3′), DS-US5-52LO (5′ TGTATCTCGAGTTAGACGGCAAAGCCCCCCAGG 3′), DS-US5-25UP (5′ GATTTGAATTCGACCTGCGGCCAACACAAC), DS-US5-50UP (5′ GATTTGAATTCTTGCCGTCCCCCTCGTAGT 3′), and DS-US5-92LO (5′ TGTATCTCGAGTTATACGACAACTGGGTCCATGTAG 3′). The following primer pairs were used to clone the coding sequences of the truncated form of gJ: DS-US5-1UP and DS-US5-74LO for gJ(1-74), DS-US5-1UP and DS-US5-52LO for gJ(1-52), DS-US5-25UP and DS-US5-92LO for gJ(25-92), DS-US5-25UP and DS-US5-74LO for gJ(25-74), and DS-US5-50UP and DS-US5-92LO for gJ(50-92). The Us5 sequences and the presence of the tag in all of the plasmids generated were confirmed by sequencing.
Real-time RT-PCR.
Total RNA was isolated from Vero cells infected at a multiplicity of infection (MOI) of 5 for the indicated times, using the Qiagen RNeasy kit. After DNase I treatment for 10 min at 65°C, RNA templates were reversed transcribed in the presence of oligo(dT) and random decamers using a RETROscript kit according to the manufacturer's recommendations (Ambion). Controls without reverse transcriptase (RT) were performed in parallel. cDNAs obtained from reverse transcription reactions (without RT and with RT) were amplified by real-time PCR using Brilliant SYBR green QPCR master mix (Stratagene) with primers Us5bR (5′ GCCGGATCCCCGACATCACCCACGCGGAGAAG 3′) and Us5treF (5′ GGTGAATTCGCGTTCTGTGTGTCACGATGTCTCT 3′) at 150 nM each, under the following conditions: 95°C for 10 min and 40 cycles of 95°C for 30 s and 1 min at 68°C. After amplification, automated melting curve analysis was performed (95°C for 15 s, ramping to 60°C for 1 min, and 95°C for 15 s). The fluorescence data were collected during the extension step at 68°C, and the fluorescence curve, along with the cycle threshold values, was obtained using System Detection Software (SDS 1.3) for the ABI7500 real-time PCR system (Applied Biosystem). For visualization, PCR products were resolved on a 1.5% agarose gel and stained with ethidium bromide. The cycle threshold values for all controls (without RT) were above 38, and no specific PCR product was detected (data not shown).
Yeast two-hybrid screen.
Two-hybrid screening was performed using the Clontech (Palo Alto, CA) Matchmaker Gal4 Two-Hybrid System 3, according to the manufacturer's directions. Full-length gJ was cloned into pGBKT7 and used as bait to probe a cDNA library constructed from Jurkat cells (Clontech) in pACT2. Screening was performed under the highest-stringency conditions (synthetic dropout medium without Ade, His, Leu, and Trp and with 5-bromo-4-chloro-3-indoyl-α-d-galactopyranoside (X-α-Gal); for details, see the manufacturer's instructions). A total of 1.35 × 106 independent cDNA clones were screened, resulting in approximately 200 yeast colonies, which grew under the highest-stringency conditions and thus were candidates for expression of gJ-interacting proteins. Thirty-five of these were sequenced, revealing 26 unique clones appearing only once, a single clone appearing twice, and one clone, constituting the C-terminal portion of FoF1 ATP synthase subunit 6, appearing seven times.
Transient transfection.
Cells were transiently transfected with Lipofectamine 2000 (Invitrogen) according to the manufacturer's recommendations and incubated for 24 to 48 h prior to analysis.
Regulated expression of Us5/gJ.
In the Jurkat cell line derivative Us5 ON/OFF (27), Us5 expression was either allowed or repressed with 0.5 μg/ml doxycycline. For generation of the EcR-293-gJ-expressing cell line (Ecdysone-inducible mammalian expression system; Invitrogen), Us5 sequences were cloned into the pIND/V5-HisA vector and transfected into EcR-293 cells, and a stable cell line was isolated under geneticin (600 μg/ml) and zeocin (400 μg/ml) selection. To induce expression of gJ, ponasterone A (PonA) (5 μM) was added to the medium for 20 h.
Apoptosis induction and detection.
Apoptosis was induced with 1 μM staurosporine (STS) or 100 ng/ml of anti-fas (clone CH-11; MBL) for the indicated time. To measure activated caspase 3, cells were fixed with 2% paraformaldehyde for 20 min at room temperature, permeabilized with 0.2% Tween for 15 min at 37°C, and then incubated with fluorescein isothiocyanate-conjugated rabbit anti-active caspase 3 (Pharmingen), which specifically recognizes the large subunit of cleaved, activated caspase 3 but does not bind inactive procaspase 3. The stained cells were analyzed for green (FL1) fluorescence, using a Becton Dickinson FACSCalibur flow cytometer (488-nm argon laser excitation source). Caspase 3 activity was determined using the ApoAlert caspase 3 assay (Clontech). Visualization of apoptotic cells was performed by incubating the cells in phosphate-buffered saline (PBS) containing ethidium bromide (2 μg/ml) and acridine orange (1 μg/ml) and counting a minimum of 200 cells using an epifluorescence microscope. Apoptotic cells showed either green fragmented nuclei (early apoptotic) or red fragmented nuclei (late apoptotic), while intact cells had unfragmented green nuclei.
Cell fractionation.
HEp-2 cells were grown overnight in a 75-cm2 dish and infected at an MOI of 5 for 9 h. They were then washed twice with PBS, scraped into 2 ml of ice-cold lysis buffer (25 mM MES [morpholineethanesulfonic acid], 150 mM NaCl, pH 6.5, with 0.5% Triton X-100), incubated for 20 min on ice, Dounce homogenized (10 strokes), and cleared by centrifugation for 5 min at 2,000 rpm at 4°C. After collection of the supernatant, an equal volume of 80% sucrose solution was added to adjust the cell lysate to 40% sucrose; 1 ml of this mixture was overlaid sequentially with 3.5 ml of 30% sucrose and then 0.5 ml of 5% sucrose. After centrifugation of the sample at 240,000 × g for 18 h at 4°C in a Beckman SW55Ti rotor, 12 equal fractions were collected from the sucrose gradient. Protein from each fraction was precipitated in 10% trichloroacetic acid for 10 min at −20°C and centrifuged at 14,000 rpm for 5 min at 4°C. The pellets were then washed with 90% acetone solution, resuspended in 1× sample loading buffer, boiled, separated onto a 4 to 12% Bis-Tris gel, transferred onto nitrocellulose membrane, and immunoblotted.
Antibodies and immunofluorescence reagents.
The following primary antibodies were used: anti-hemagglutinin (HA) and anti-c-myc (1:1,000; Clontech), anti-flotillin and anti-insulin receptor β (both 1:250; BD Transduction Laboratories), anti-caspase 3 (1:1,000; BD Transduction Laboratories), anti-caspase 9 (Ab-2; Oncogene), anti-actin and anti-gB clone 10B7 (1:1,000; Virusys), anti-gC clone 3G9 (1:1,000; Virusys), anti-FLAG clone M2 (1:1,000; Sigma), anti-TGN46 (1:1,000; Serotec), anti-LAMP1 (1:200; Affinity Bioreagents), anti-Rab5 and anti-calnexin (both 1:100; Stressgen), anti-giantin (1:500; Covance), anti-cytochrome c (1:1,000; PharMingen), anti-oligomycin sensitivity-conferring protein (OSCP) (1:1,000; Molecular Probes), and anti-COX IV (1:1,000; Cell Signaling). Secondary antibodies used for immunoblotting were horseradish peroxidase-conjugated goat anti-mouse immunoglobulin G (IgG) or horseradish peroxidase-conjugated goat anti-rabbit IgG (1:1,000; Cell Signaling). For indirect immunofluorescence, the following secondary antibodies were used at a dilution of 1:1,000: Cy3-conjugated donkey anti-sheep IgG, Cy3-conjugated goat anti-mouse, fluorescein isothiocyanate-conjugated goat anti-rabbit (Jackson Immunoresearch), and Alexa 488-conjugated goat anti-mouse (Molecular Probes). Mitochondria were stained by incubation of the cells at 37°C with MitoTracker red (Molecular Probes) added to the culture medium at a final concentration of 0.5 μM prior to fixation.
Immunoblotting.
Cells were harvested and lysed in RIPA buffer. Protein concentrations were determined using a bicinchoninic acid protein assay (Pierce), and equal amounts of protein were separated in 4 to 12% Bis-Tris gel with MES running buffer (Invitrogen) and transferred onto a nitrocellulose membrane (Invitrogen). The membranes were blocked for 1 h at room temperature with 5% milk in PBS prior to being immunoblotted with primary antibodies, followed by horseradish peroxidase-conjugated secondary antibodies. Reactive bands were visualized on autoradiographic film (Kodak) using the Supersignal West Pico chemiluminescent substrate (Pierce).
Immunofluorescence microscopy.
HEp-2 cells were grown overnight to 70 to 80% confluence on glass coverslips in six-well plates and then transfected or infected with HSV-1 for the indicated time. The cells were then washed, fixed in 2% methanol-free formaldehyde (Polysciences, Inc.) for 20 min at room temperature, and permeabilized in acetone (Sigma) at −20°C for 4 min. For indirect-immunofluorescence staining, the cells were blocked in PBS with 5% bovine serum albumin (or 5% normal donkey serum for anti-TGN46) overnight at 4°C prior to incubation with primary antibodies for 1 h, followed by incubation with secondary antibodies for 45 min at room temperature. Finally, the cells were mounted on slides in 0.1% Mowiol (anti-fade agent; Sigma) and 2.5% 1,4-diazabicylo[2.2.2]-octane (antioxidant; Sigma) used as an antibleaching agent and sealed with clear nail polish. The slides were left at least overnight at 4°C before being viewed using the DeltaVision microscopy system.
Cellular ATP levels.
For HSV-infected cells and tetracycline-regulated Us5/gJ tranfectants, cellular ATP levels were determined using the Calbiochem ATP assay kit. The cells were diluted to 105/ml, and 0.2 ml cell suspension was mixed with 0.2 ml releasing reagent in a cuvette as described in the manufacturer's instructions. Luciferase reagent (0.1 ml) was injected into the cuvette, and ATP levels were determined using a Monolight luminometer (Analytical Luminescence Laboratory, San Diego, CA).
For oligomycin-treated cells, ATP levels were measured in either normal or glucose-free medium, using the Roche ATP bioluminescence assay kit. The cells were treated with 2.5 μM oligomycin at 37°C for 60 min, and cellular ATP was extracted by suspending 0.1 ml of the cell suspension (5 × 105 cells/ml) in 0.05 ml dilution buffer and 0.05 ml of cell lysis reagent, followed by incubation at room temperature for 5 min. ATP levels were measured in an Auto Lumat LB953 luminometer (Berthold Technologies, Wildbad, Germany), using 0.1 ml culture extract injected with 0.1 ml luciferase reagent.
Measuring whole-cell ROS with H2DCFDA.
HSV-infected or control Jurkat cells were washed in PBS and pelleted at 1,200 rpm for 5 min. The cells were resuspended in 500 μl PBS containing 0.5 μΜ2′,7′-dichlorofluorescein diacetate (H2DCFDA) (Molecular Probes). The cells were incubated in the dark at room temperature for 15 min and analyzed immediately by flow cytometry. RAW264.7 cells were allowed to adhere in 12-well plates for 1 h, incubated in 0.5 μΜ H2DCFDA, washed with PBS, and infected at an MOI of 5 for 6 h prior to collection and analysis by flow cytometry on an LSR cytometer (BD Biosciences).
RESULTS
Expression and regulation of gJ.
The only previously reported cellular function of gJ was its ability to inhibit apoptosis (3, 27, 28, 47). In many cell types, HSV induces an antiapoptotic state, starting 4 to 6 h after infection (1). We therefore evaluated the kinetics of Us5/gJ expression during HSV infection. In infected Vero cells, Us5 mRNA could first be detected 4 h after infection and increased over 24 h (Fig. 1A). We next evaluated the expression of gJ. In HSV-infected cells, gJ could not be detected using antisera raised either to a cocktail of three predicted immunogenic peptides from gJ or to a Us5-maltose binding protein fusion (kindly provided by B. Roizman [9]; data not shown). Either antiserum could easily detect overexpressed gJ in transfected cells (data not shown), suggesting that gJ may be produced at relatively low levels during HSV infection. We therefore constructed a recombinant virus containing a C-terminal epitope-tagged gJ (FUs5FLAG). Upon probing infected cell lysates with anti-FLAG antibody, protein could first be detected at 6 h postinfection and reached near-maximal levels by 8 h postinfection (Fig. 1B). Thus, the expression kinetics of Us5/gJ correlate well with the so-called apoptosis “prevention window” during HSV infection.
FIG. 1.
Expression of gJ in HSV-infected cells. (A) Detection by real-time RT-PCR of Us5 mRNA at the indicated hours (top) postinfection in Vero cells infected at an MOI of 5 with HSV-1(F). The numbers within the bars indicate the cycle thresholds of amplicon detection. Amplification beyond 38 cycles did not yield specific product, as determined by melting curve analysis (data not shown). (Inset) Gel electrophoresis detection of Us5 mRNA amplified by RT-PCR. (B) Immunoblot for gJ using anti-FLAG monoclonal antibody (MAb) in HEp-2 cells infected with FUs5-FLAG at an MOI of 5 for the indicated times (top, in hours). (C and D) Immunodetection of gJ (C) and gB (D) in cell lysates obtained from monolayers of different cell lines infected with FUs5-FLAG at an MOI of 5 for 10 h in the presence (+) or absence (−) of tunicamycin (10 μg/ml). Tunicamycin was added from 3 h postinfection for the duration of the infection. Anti-FLAG and anti-gB MAbs were used to detect gJ and gB, respectively. Exposure times were as follows: 5 min (C) and 10 seconds (D). MW, molecular weight.
Previously, two forms of gJ (10 and 17 kDa) were described in insect Sf9 cells infected with recombinant baculovirus containing the Us5 gene, and the two forms were ascribed to differential glycosylation (17). Similarly, in FUs5FLAG-infected mammalian cells, two forms of gJ are detected, migrating at approximately 12 kDa and 18 kDa. The slower-migrating 18-kDa form appears to result from glycosylation of the faster-migrating 12-kDa product, since addition of the glycosylation inhibitor tunicamycin during infection increased the relative abundance of the 12-kDa product and depleted the 18-kDa product (Fig. 1C). Interestingly, the lower-molecular-mass form was not observed in infected HEp-2 cells or HF unless they were treated with tunicamycin. In untreated Vero cells, both forms could be observed, although the relative abundance of the lower-molecular-mass form was increased by tunicamycin treatment. Additionally, gJ appeared to be expressed at low levels in infected cells, since detection by immunoblotting using anti-FLAG antibody required extended exposure times. This was in contrast to other HSV glycoproteins, such as gB (Fig. 1D) or gC (not shown), which could be easily detected in the same cell lysates with very brief exposure times. While the use of different detection antibodies precludes direct comparison, the simplest interpretation of these results is that gJ is produced at low levels relative to the other glycoproteins in HSV-infected cells.
The other glycoproteins of HSV are regulated as late, or γ, genes; that is, their synthesis correlates with the onset of viral DNA replication (41). The γ genes can be further subdivided into γ1 genes, whose expression precedes viral DNA replication but increases once viral DNA replication commences, and γ2 genes, which are transcribed at significant levels only after viral DNA synthesis (41). We therefore evaluated the expression class of gJ in cell lines infected with wild-type virus or virus containing epitope-tagged gJ. In Vero cells, treatment with the DNA synthesis inhibitor acyclovir substantially reduced the expression of Us5 mRNA (Fig. 2A). The effect was even more pronounced in HF, in which Us5 mRNA was not detected in the presence of acyclovir. Similar results were seen at the protein level. Both Vero and HEp-2 cells showed substantially less gJ in the presence of another DNA synthesis inhibitor, phosphonoacetic acid (PAA), than untreated control cells (Fig. 2B). gJ expression was more sensitive to PAA than was expression of gB but less sensitive than expression of gC. Thus, Us5 appears to be regulated as a γ1 gene, showing partial dependence on HSV DNA replication for its expression.
FIG. 2.

Dependence of Us5/gJ expression on viral DNA replication and ICP22 expression. (A) Detection of Us5 mRNA by RT-PCR in Vero or HF infected at an MOI of 10 with HSV-1(F) or R325, an ICP22 mutant virus, in the presence (+) or absence (−) of 50 μM acyclovir (Acy). (B) Immunodetection of gJ, gB (γ1), and gC (γ2) in Vero and HEp-2 cells mock infected or infected with FUs5-FLAG at an MOI of 5 for 17 h in the presence (+) or absence (−) of 300 μg/ml PAA. Anti-FLAG, anti-gB, and anti-gC monoclonal antibodies were used to detect gJ, gB, and gC, respectively. (C) Detection of Us5 mRNA by RT-PCR in Vero cells infected with R325 at an MOI of 10 for the indicated times (hpi, hours postinfection).
A number of HSV genes have been implicated as playing indirect roles in apoptosis; that is, they do not exert antiapoptotic activity themselves but presumably instead regulate the expression of other proteins that have direct antiapoptotic activity. Among these is ICP22. Deletion of ICP22 has been reported to cause a modest decrease in the antiapoptotic function of the virus (4). Mutation of the C-terminal portion of ICP22 leads to reduced expression of a subset of HSV γ proteins (39). We therefore asked whether expression of Us5 was dependent upon ICP22. At 16 h after infection of Vero cells or HF, virus with deletion of ICP22 produced lower levels of Us5 mRNA than wild-type virus (Fig. 2A). To determine whether this might result from a delay in Us5 expression, we performed a time course experiment similar to that in Fig. 1. We found that virus with deletion of ICP22 expressed Us5 mRNA with delayed kinetics, so that Us5 mRNA did not begin to accumulate until approximately 16 h postinfection (Fig. 2C). Thus, the expression of Us5 is delayed in the absence of ICP22, but Us5 is not absolutely dependent upon ICP22 for its expression.
Cellular localization of gJ.
HSV glycoproteins typically are found in abundance on the cell surface and can localize to other cell membranes, as well (45). We had previously shown that gJ can inhibit mitochondrion-dependent apoptosis. Therefore, we investigated whether gJ is localized to the mitochondria. HSV-1-infected HEp-2 cell lysates were fractionated on a discontinuous sucrose gradient, and the resulting fractions were analyzed by immunoblotting. When cell lysates of HEp-2 cells infected with FUs5-FLAG were analyzed using anti-FLAG antibody, gJ was detected in two series of fractions that were distinct from the fractions containing the mitochondria, which were identified with antibodies recognizing cytochrome c, OSCP, and Cox IV (Fig. 3A). These data suggest that gJ is not predominantly localized to the mitochondria, but instead, to another membrane organelle(s).
FIG. 3.
Cellular localization of gJ. (A) Subcellular fractionation. HEp-2 cells infected with FUs5-3FLAG virus were solubilized, depleted of nuclei, and subjected to sucrose gradient fractionation. The 12 fractions (1 to 12, from right to left) were resolved on sodium dodecyl sulfate-polyacrylamide gel electrophoresis and analyzed by Western blotting with the indicated antibodies. P, pellet. (B) Intracellular localization of gJ in HEp-2 cells infected with FUs5-3FLAG at 6, 10, and 24 h. Cells infected at an MOI of 1 for the indicated times were fixed, permeabilized, and stained with anti-FLAG monoclonal antibodies (MAbs) to visualize gJ (red) and specific organelle markers (green) that recognize the ER (Calnexin), trans-Golgi network (TGN46), early endosomes (Rab5), mitochondria (MitoTracker), cis-medial Golgi (giantin), and lysosomes (LAMP1). The merged images of the signals are presented with highlights shown in white depicting colocalization of the two signals and were obtained using the Image J colocalization highlighter function. Cells infected with wild-type virus showed no red fluorescence (data not shown). (C) Intracellular localization of transiently expressed gJ in HEp-2 cells. Cells transfected with pCMVUS5-3FLAG or pCMV-mycUs5 for 48 h were fixed, permeabilized, and stained with anti-FLAG MAbs to visualize gJ (red) and specific organelle markers (green) as in panel B. The merged images of the signals are presented with highlights shown in white depicting colocalization of the two signals and were obtained using the Image J colocalization highlighter function.
To determine more precisely the cellular distribution of gJ, we used an immunolocalization approach. Using the DeltaVision microscopy system, we found that epitope-tagged gJ was widely distributed within HSV-infected cells. A majority of intracellular gJ appeared to localize to the endoplasmic reticulum (ER), trans-Golgi network, and early endosomes (Fig. 3B). A smaller fraction of gJ could be detected in the mitochondria, Golgi apparatus, and lysosomes. Thus, gJ is distributed widely to membrane compartments within infected cells. Furthermore, a similar distribution of gJ was observed in cells transfected with plasmids expressing N- or C-terminally tagged gJ (Fig. 3B), suggesting that the cellular distribution of gJ is not dependent upon the activities of other viral gene products.
Binding partners for gJ.
We hypothesized that gJ might exert its antiapoptotic function via a protein-protein interaction. To identify cellular proteins interacting with gJ, we performed yeast two-hybrid screening using gJ as bait to probe a cDNA library constructed from Jurkat cells (Clontech, Palo Alto, CA). Our screen was performed using the Clontech Matchmaker Gal4 Two-Hybrid System 3, under the highest-stringency conditions (synthetic dropout medium without Ade, His, Leu, and Trp and with X-α-Gal). A total of 1.35 × 106 independent cDNA clones were screened, resulting in approximately 200 yeast colonies, which grew under the highest-stringency conditions and thus were candidates for expression of gJ-interacting proteins. Thirty-five of these were sequenced, revealing 26 unique clones appearing only once, a single clone (snapin) appearing twice, and one clone, constituting the C-terminal portion of FoF1 ATP synthase subunit 6, appearing seven times. The seven FoF1 ATP synthase subunit 6 clones found in our two-hybrid screen were independent, each with a different start point in the subunit 6 sequence. The shortest binding clone expressed the C-terminal 43 amino acids of FoF1 ATP synthase subunit 6. Using the same two-hybrid system, we retransfected the fragment of FoF1 ATP synthase subunit 6, along with Us5/gJ, into yeast, again resulting in growth under high-stringency selection (Fig. 4A). Growth was observed only when both gJ and FoF1 ATP synthase subunit 6 were cotransfected (Fig. 4A, wedge 2), confirming the specificity of the interaction of these proteins in the two-hybrid system.
FIG. 4.
Binding partners for gJ. (A) Yeast two-hybrid screening. The indicated combinations of two-hybrid constructs were transformed into AH109 yeast and grown for 5 days on appropriate low-stringency selection media. Growing colonies were then replated onto high-stringency media (synthetic dropout without Trp, Leu, Ade, and His), grown for 3 days, and photographed. Groups: 1, pGBKT7-Us5; 2, pGBKT7-Us5 plus pACT-FoF1 subunit 6; 3, pACT2-FoF1 subunit 6; 4, pGBKT7-Us5 plus pGADT7 (∼pACT2); 5, no DNA; 6, pGBKT7 plus pACT2-FoF1 subunit 6. (B) Schematic representation of gJ predicted domains. There is a predicted sequence signal with a putative cleavage site between the two alanines (A) at positions 21 and 22, as well as an N-glycosylation site on the asparagine (N) at position 30. (C) Schematic representation of the truncated gJ proteins made to determine the domains required for the interaction between gJ and FoF1 ATP synthase subunit 6. Y2H, yeast two hybrid.
In an attempt to independently confirm the interaction of gJ with FoF1 ATP synthase subunit 6, we constructed epitope-tagged versions of each using the plasmids pCMV-myc and pCMV-HA (Clontech). N-terminal c-myc and HA-tagged versions of each protein were made. Both gJ and the FoF1 ATP synthase subunit 6 fragment could be readily detected in transiently transfected HEp-2 cells by Western blotting (not shown), with both the 12- and 18-kDa forms of gJ present. However, we were unable to reproducibly coimmunoprecipitate gJ and FoF1 ATP synthase subunit 6, suggesting that the binding affinities for these two proteins are too low to consistently detect their interaction in cell lysates.
To define more precisely the minimal binding fragment of gJ, we made a series of mutants of gJ lacking the signal sequence, intracellular domain, transmembrane domain, or extracellular domain (Fig. 4B) and evaluated their abilities to interact with the FoF1 ATP synthase subunit 6 fragment in the two-hybrid system. We found that the full-length gJ and the truncation containing amino acids 1 through 74 were able to interact with the FoF1 ATP synthase subunit 6 fragment (Fig. 4C), suggesting that the intracellular domain of gJ was not required for the interaction. The other mutant forms of gJ did not interact with FoF1 ATP synthase subunit 6, suggesting that the signal sequence, extracellular domain, and transmembrane domain are all required for the interaction in the two-hybrid system.
Antiapoptotic effect of gJ.
We previously described Jurkat T cells transfected with a tetracycline-inducible gJ expression vector, which we used to demonstrate that expression of gJ protects cells from apoptosis mediated by UV, fas, or perforin plus granzyme B (27). However, recent work suggests that HSV infection has both pro- and antiapoptotic effects. In T cells, the balance of these effects ultimately leads to apoptosis (22, 25, 26, 38). Similarly, in the macrophage cell line RAW264.7, infection with wild-type virus strain F induced a modest but reproducible degree of apoptosis (Fig. 5A). Deletion of Us5 (strain RAS116) rendered the virus much more strongly proapoptotic, while the repaired strain, RAS137, induced wild-type levels of apoptosis, confirming the function of gJ in limiting apoptosis in immune cells. In contrast to the results in T cells and macrophages, in nonimmune cells, such as epithelial cells and fibroblasts, the HSV antiapoptotic mechanisms predominate, and infection of such cells does not normally lead to apoptosis (18). We therefore considered the possibility that gJ might function differently in nonimmune cell types. To determine whether gJ was sufficient to protect nonimmune cells from apoptosis, we transfected EcR-293 cells with gJ under the control of an ecdysone-inducible promoter. In agreement with our previous findings in Jurkat cells, induction of gJ expression with PonA was sufficient to protect EcR-293 cells from fas-induced apoptosis (Fig. 5B).
FIG. 5.
Antiapoptotic effects of gJ. (A) Activated caspase 3 was measured by flow cytometry in RAW264.7 cells either mock infected or infected for 6 h with RAS116 (Us5 deletion), RAS137 (repair of RAS116), or the wild-type strain F. Shown are means ± standard deviations (SD) of triplicate determinations from one representative of four independent experiments. *, P < 0.01 versus mock infection; **, P < 0.001 versus mock infection. (B) Inhibition of fas-induced caspase 3 activation in EcR-293 cells expressing gJ. Activated caspase 3 was measured by flow cytometry in control cells (EcR-293), control cells under PonA induction (EcR-293/Pon A), EcR-293 cells transfected with Us5 but without PonA induction (EcR-293-gJ), or EcR-293 cells transfected with Us5 and expressing gJ under PonA induction (EcR-293-gJ/Pon A) either left untreated (white bars; no tx) or treated with anti-fas (100 ng/ml) for 16 h (black bars; anti-fas). PonA (5 μM) was used to induce gJ expression. The values are the averages of eight or nine independent experiments (mean ± SD). *, P < 0.001. (C and D) Protection of Jurkat cells from HSV-1-induced apoptosis. (C) The percentage of apoptotic cells was evaluated as described in Materials and Methods for Jurkat cells mock infected or infected with ICP27 deletion virus (vBSΔ27) and wild-type parental virus (KOS1.1) at an MOI of 5 for 24 h when gJ expression was induced (black bars; Us5 ON) or repressed (white bars; Us5 OFF). *, P < 0.05. (D) Immunoblotting for caspase 3 and 9 cleavage in cell lysates obtained after infection at an MOI of 5 for 24 h of Jurkat cells expressing gJ (Us5 ON) or Jurkat cells with gJ expression repressed (Us5 OFF). The anti-caspase 3 or 9 antibodies detect full-length caspase 3 or 9, and the absence of signal reflects the cleavage of those proteins. Actin was used as a loading control.
The immediate-early protein ICP27 has been shown to be required for the antiapoptotic effects of HSV (2, 4). Infection with virus strains having deletion of the gene encoding ICP27 (vBSΔ27) strongly induces apoptosis in many cell types (2). However, rather than a direct effect, ICP27 is thought to act by promoting the expression of other antiapoptotic proteins (1). We therefore asked whether expression of gJ in trans might complement the proapoptotic defect in cells infected by virus with ICP27 deleted. We found that expression of gJ protected vBSΔ27-infected Jurkat T cells from morphological changes of apoptosis, such as nuclear fragmentation (Fig. 5C). Similarly, Jurkat T cells infected with vBSΔ27 showed almost complete cleavage of caspases 3 and 9 (seen as the absence of bands detectable with antibody to full-length caspase 3 or 9 [Fig. 5D]) compared to mock-infected cells or control cells infected with the parental strain, KOS1.1. However, vBSΔ27-infected cells expressing gJ had levels of caspase 3 and 9 cleavage similar to those of mock-infected cells or control cells infected with the parental strain, KOS1.1 (Fig. 5D). These results suggest that the proapoptotic effect of infection by vBSΔ27 in T cells results, at least in part, from the reduction of gJ expression.
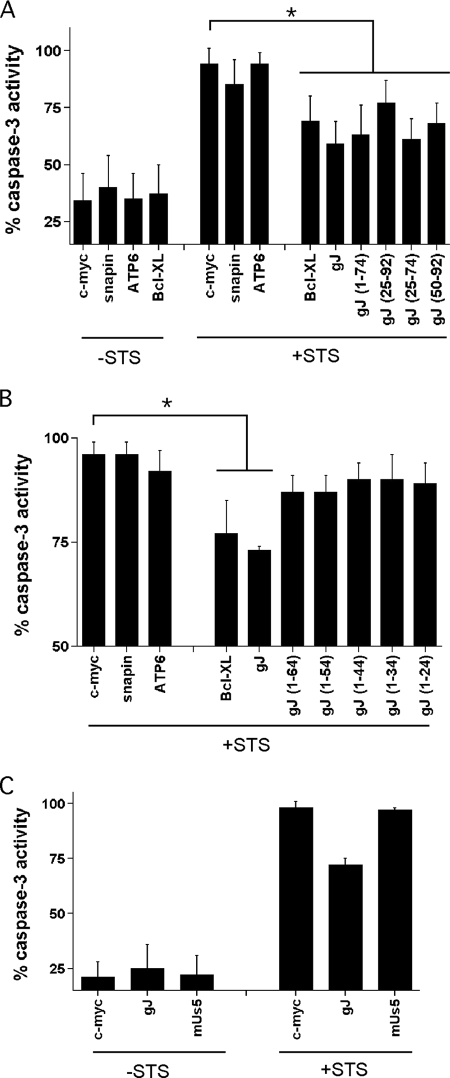
To define the minimal domains of gJ required for the inhibition of apoptosis, we used the mutant forms of gJ shown in Fig. 4 for transient-transfection experiments in human HEp-2 cells. HEp-2 cells are easily induced into apoptosis by STS, and the induction of apoptosis by STS can be blocked by HSV (4). In our hands, transient transfection of HEp-2 cells had an efficiency of 30 to 50%, and we therefore expected that even a highly effective inhibitor of apoptosis would have only a partial protective effect on a population consisting of a mixture of transfected and untransfected cells. Our expectation was confirmed by transient transfection of HEp-2 cells with the strong apoptosis inhibitor Bcl-XL, which showed a partial (but highly reproducible) decrease in STS-induced apoptosis compared to an empty-vector control (Fig. 6A). To control for the possibility of nonspecific effects, we also transfected HEp-2 cells with genes encoding snapin or the FoF1 ATP synthase subunit 6 fragment. Consistent with our expectation, neither the FoF1 ATP synthase subunit 6 fragment nor snapin showed any antiapoptotic activity (Fig. 6A). We then tested myc-tagged full-length gJ and myc-tagged mutant forms of gJ, as shown in Fig. 4. Consistent with our previous results (27), expression of full-length gJ was antiapoptotic, and in fact, in these experiments, it showed somewhat stronger inhibition than Bcl-XL, although the difference between gJ and Bcl-XL did not reach statistical significance (Fig. 6A). These findings also confirm that the N-terminal myc tag did not interfere with the antiapoptotic function of gJ. Mutants 1-74 and 25-74 showed inhibition of apoptosis nearly as strong as full-length gJ, suggesting that the intracellular domain is not required for the full antiapoptotic effect. In addition, mutants 25-92 and 50-92 also had a weaker but reproducible antiapoptotic effect.
FIG. 6.
gJ domains required for its antiapoptotic function. Caspase 3 activity was measured in lysates from transiently transfected HEp-2 cells as described in Materials and Methods. (A) HEp-2 cells were transfected with either the empty vector (c-myc) or the plasmid carrying either the myc-Us5 gene (gJ), the myc-snapin gene, the myc-FoF1 ATP synthase subunit 6 fragment (ATP6), Bcl-XL, or the different truncated forms of the myc-tagged Us5 gene. At 48 h, the cells were either left untreated (−STS) or treated with 1 μM STS for 6 h prior (+STS) to making cell lysates and measuring caspase 3 activity. The bars represent the mean relative caspase 3 activities for three to seven independent determinations (mean ± standard deviation [SD]). *, P < 0.05. (B) HEp-2 cells were transfected with the indicated vectors (as described for panel A) and treated with STS, and caspase 3 activity was determined as for panel A. The bars represent the mean relative caspase 3 activities for six independent determinations (mean ± SD). *, P < 0.01. (C) HEp-2 cells were transfected with empty vector (c-myc), the plasmid carrying the myc-Us5 gene (gJ), or the plasmid carrying a mutant form of the Us5 gene (mUs5) unable to produce gJ. The cells were left untreated or treated with STS, and caspase 3 activity was measured as for panel A. The bars represent the mean relative caspase 3 activities for six independent determinations (mean ± SD).
Since all the gJ truncations able to inhibit apoptosis shared the transmembrane domain, we asked whether the transmembrane domain was essential for antiapoptotic function. Unlike mutant 1-74, mutants missing part or all of the transmembrane domain (amino acids 50 to 70) were unable to effectively inhibit apoptosis (Fig. 6B). However, mutants missing part or all of the transmembrane domain also did not produce detectable levels of protein (data not shown), so it was not possible to determine whether the failure to block apoptosis reflects a direct role for the transmembrane domain in the antiapoptotic effect or instead reflects an indirect role for the transmembrane domain, perhaps by modification of mRNA or protein stability.
Recently, it has been reported that the latency-associated transcript of HSV mediates protection of neurons from apoptosis by encoding a microRNA that downregulates transforming growth factor β1 and SMAD3 (21). To determine whether the protective effect of Us5 expression might be mediated by RNA or instead required the production of protein (gJ), we constructed a mutant form of Us5 (mUs5) with deletion of the translational start site and addition of a frameshift mutation immediately following the mutated start site. HEp-2 cells transfected with this construct synthesized Us5 mRNA, but not gJ (not shown). We then tested HEp-2 cells transfected with the wild-type or mUs5 and evaluated protection from STS-induced apoptosis. While wild-type gJ again showed protection from apoptosis in these experiments (Fig. 6C), the mutant form of Us5 gave no protection, confirming that gJ, rather than the Us5 mRNA, is the mediator of antiapoptotic function.
Effects on ATP synthesis and ROS production.
FoF1 ATP synthase is one component of the respiratory chain responsible for oxidative phosphorylation in the cell. We therefore investigated the effects of HSV and gJ on ATP levels in infected cells. Although one report in the literature suggests that infection of HEp-2 with HSV results in a decrease in ATP levels (36), we did not observe this in Jurkat cells, and instead, ATP levels were stable after HSV infection (Fig. 7). This is perhaps not surprising, given that certain events in HSV replication, such as DNA packaging and capsid maturation, are ATP-dependent processes (11). Furthermore, we saw no difference in ATP levels after infection with wild-type, Us5 deletion (RAS116), or rescue (RAS137) virus. Similarly, no difference in ATP levels after infection with these viruses was observed whether evaluated in the presence of substrate for glycolysis (normal media) (Fig. 7A) or glucose-free media (Fig. 7B). As a complementary approach, we evaluated the effect of exogenously expressed Us5/gJ on cellular ATP levels (Fig. 7C). Again, we saw no difference between cells with Us5/gJ expression induced and those with Us5/gJ expression repressed. These results were in contrast to those with the FoF1 ATP synthase inhibitor oligomycin, which is thought to inhibit apoptosis by depleting cellular ATP stores. Oligomycin had no effect on ATP levels in the presence of glucose (Fig. 7D) but strongly depressed ATP levels in the absence of substrate for glycolysis (Fig. 7E). We interpret these experiments as demonstrating that the inhibition of apoptosis by gJ is not dependent on suppression of cellular ATP levels.
FIG. 7.
gJ and HSV do not lead to ATP depletion. (A and B) Jurkat cells were infected with the indicated HSV strains in glucose-containing medium (A) or glucose-free medium (B) for 16 h, and the ATP contents were determined using the Roche ATP assay kit. Shown are means plus standard deviations (SD) of six replicate determinations representative of three independent experiments. (C) Jurkat cells stably transfected with the tetracycline-regulated Us5/gJ construct with Us5/gJ expression induced or repressed were cultured in glucose-containing medium and evaluated for ATP content. Shown are means plus SD of triplicate determinations representative of three independent experiments. (D and E) Jurkat cells were treated with the indicated concentrations of oligomycin for 1 h in glucose-containing medium (D) or glucose-free medium (E), and ATP contents were determined. Shown are means plus SD of triplicate determinations representative of three independent experiments. In several instances, the error bars are too small to be seen. *, P < 0.02.
A second possible effect of inhibition of complexes within the respiratory chain is leakage of electrons into the mitochondrial matrix or cytoplasm. We therefore asked whether HSV infection in general, and gJ in particular, might lead to an increase in oxidative stress in infected cells. To test this hypothesis, we infected Jurkat cells with wild-type HSV, Us5 deletion virus, or rescue virus for 5 h and measured ROS using 2′,7′-dichlorofluorescein diacetate. Infection with wild-type HSV led to a marked increase in ROS production in Jurkat cells (Fig. 8A). In contrast, infection with the gJ deletion virus caused little increase in ROS production compared to uninfected cells. The rescue virus expressing gJ regained the ability to induce ROS. In the macrophage cell line RAW, infection with wild-type or rescue virus did not lead to ROS levels above those of uninfected cells (Fig. 8B). However, ROS levels were significantly higher in cells infected with wild-type or rescue virus compared to cells infected with the gJ deletion virus (Fig. 8B). This suggests that HSV infection may simultaneously induce and inhibit ROS production and that the balance of these effects differs between cell types. To determine whether gJ was sufficient to cause the induction of ROS, we used the inducible tet-off Jurkat system. In agreement with our results using deletion and rescue virus, induction of gJ expression led to an increase in ROS (Fig. 8C). Taken together, these results demonstrate that gJ is the protein responsible for HSV induction of ROS in infected cells and that gJ itself is sufficient to induce ROS.
FIG. 8.
Effect of gJ expression on the production of ROS. ROS production was measured using 2′,7′-dichlorofluorescein diacetate (DCF) in Jurkat cells infected at an MOI of 10 for 5 h (A) or RAW264.7 cells mock infected or infected with Us5 deletion (RAS116), Us5 rescue (RAS137), or wild-type parental (F) virus at an MOI of 5 for 6 h (B). The bars represent the averages of three independent experiments (mean ± standard deviation). *, P < 0.02; **, P < 0.001. (C) Jurkat cells expressing gJ. Us5 off, Jurkat cells with gJ expression repressed; Us5 on, Jurkat cells with gJ expression induced for 48 h.
DISCUSSION
In this paper, we describe the expression, localization, and cellular effects of the protein product of the HSV Us5 gene, gJ. Us5/gJ has been much less studied than the other glycoproteins of HSV. This likely results from two factors. First, unlike some of the other HSV glycoproteins, gJ is not involved in viral binding or entry and, as such, is not required for viral replication in vitro. Second, gJ is synthesized at much lower levels than other glycoproteins, such as gB or gC. Taken together, these characteristics may have led to a presumption that gJ is likely to be less important to the biology of HSV. Indeed, no function was ascribed to gJ until its antiapoptotic properties were described (28). However, we consider it unlikely that Us5/gJ would have been retained during viral evolution if it were truly irrelevant to HSV biology.
Although gJ in some ways appears unique, it does share certain characteristics with the other HSV glycoproteins. For example, either gJ or gD can rescue cells from apoptosis induced by HSV with deletion of gD (47). The current study demonstrates that gJ is similar to other HSV glycoproteins in that it is regulated as a late, or γ, gene. The HSV γ genes can be subdivided into leaky late (γ1) genes, which are only partially dependent upon viral DNA replication for their expression, and true late (γ2) genes, which, in contrast, are not expressed in the absence of viral DNA replication. Certain HSV glycoproteins, such as gB, are considered γ1 genes, while others, such as gC, are considered γ2 genes. However, it has become increasingly clear that the strict definitions of γ1 and γ2 in fact represent the extremes of a continuum of late-gene expression. gJ regulation varies somewhat between cell types but appears to meet the definition of a γ1 gene, having significant but not strict dependence upon viral DNA replication for its expression.
The paucity of work concerning gJ is illustrated by the fact that only a single previous publication has shown unequivocally that gJ is actually produced in HSV-infected cells. Ghiasi et al. showed expression of gJ in HSV-infected rabbit skin (RS) cells using antisera raised to baculovirus-expressed gJ (17). In that study, gJ was detected on the surfaces of infected RS cells. Our current study is consistent with these previous findings but extends them to demonstrate that gJ is actually found predominantly on intracellular membranes, especially the ER, trans-Golgi network, and endosomes. Similar intracellular distributions have been described for other HSV glycoproteins, including gB, gD, gE/gI, gK, and gM (5, 6, 14, 15, 30). The abilities of glycoproteins, such as gJ, to localize to membranes throughout the cell open up the possibility of the glycoproteins modulating cellular functions not occurring at the cell surface.
Before this study, the only reported function of gJ was its ability to inhibit apoptosis (3, 27, 28, 47). We reasoned that the ability of Us5 to modulate apoptosis would likely be mediated by a protein, an assumption confirmed by the inability of Us5 mRNA to mediate any effect. In an attempt to identify possible cellular targets for protein-protein interactions, we used the yeast two-hybrid approach. Because it requires the interaction between the bait and prey proteins to occur within the nucleus, the yeast two-hybrid screen can be problematic for those transmembrane proteins that misfold or are unstable when they attempt to localize to the nucleus (12). On the other hand, the two-hybrid approach has been used successfully for many transmembrane proteins, and thus, we considered the two-hybrid system a potentially promising approach to identify binding partners for gJ. By the two-hybrid approach, we were able to identify an interaction between gJ and subunit 6 of FoF1 ATP synthase.
The FoF1 ATP synthase is the primary enzyme responsible for ATP synthesis in prokaryotes and eukaryotes, and it exists as a large complex of about 600 kDa with 16 to 18 subunits in mammals (8). Of possible relevance to the antiapoptotic function of gJ, several groups have implicated FoF1 ATP synthase in the regulation of cell death. Using complementation cloning methods, subunits of FoF1 ATP synthase were identified as critical for Bax-mediated killing of Saccharomyces cerevisiae (34). ATP4 and ATP2 yeast, which lack subunits of the Fo component of ATP synthase, are resistant to Bax-mediated death (20, 33). Similar results were obtained using a complementation approach to identify antisense cDNA molecules that could inhibit glucocorticoid-induced apoptosis of thymocytes (42). While this screen identified subunit 6 of FoF1 ATP synthase multiple times, it did not identify other subunits. The involvement of FoF1 ATP synthase in apoptosis is supported by the finding that STS- or calphostin-induced apoptosis can be inhibited by the macrolide antibiotic oligomycin, which interferes with the function of FoF1 ATP synthase (24, 31).
Although it appears that FoF1 ATP synthase is important for the regulation and execution of apoptosis, the exact role that this molecule plays remains unclear. Apoptosis has certain ATP-dependent steps, such as activation of caspase 9 by the apoptosome (13). Thus, the inhibition of apoptosis by oligomycin might reflect inadequate levels of cytoplasmic ATP for the apoptotic program. In at least some systems, however, ATP generated by glycolysis is sufficient to allow apoptosis, and our work and that of others (31) suggest that oligomycin inhibits apoptosis most effectively in situations where glycolysis is limited. A mechanism for gJ dependent upon depletion of ATP would appear unlikely in our system, since neither HSV nor gJ had an effect on cellular ATP, regardless of the presence or absence of glycolytic substrate. Alternatively (and of possible relevance in the context of HSV infections), a significant amount of FoF1 ATP synthase can be found on the plasma membrane (10), and in keratinocytes, plasma membrane FoF1 ATP synthase has been shown to generate extracellular ATP (7). These authors suggested that locally elevated ATP levels might be required for keratinocyte apoptosis induced by the P2X7 receptor. Thus, gJ might affect ATP levels at certain subcellular sites critical for apoptosis, although our current methodology does not allow us to address this.
Quantitative or qualitative defects in FoF1 ATP synthase function have been reported to result in the production of ROS (23), and thus, we predicted that gJ might be associated with ROS production, as well. Indeed, gJ was necessary for the induction of ROS in HSV-infected cells, since mutant virus with deletion of Us5 did not induce ROS. In addition, gJ was sufficient to induce ROS production, since gJ-expressing cells showed increased ROS. These findings are consistent with other situations of disruption of ATP synthase function, in which the failure to fully utilize the proton gradient across the inner mitochondrial membrane leads to leakage of electrons into the mitochondrial matrix or cytoplasm and ROS production. For example, it has been reported that mutation of subunit 6 of FoF1 ATP synthase in the genetic disorder neurogenic ataxia retinitis pigmentosa leads to increased production of ROS (16). Similarly, inhibition of FoF1 ATP synthase with oligomycin can induce increased ROS (29). However, this interpretation must be reconciled with the lack of a detectable decrease in ATP levels in gJ-expressing cells, since this is another logical outcome of ATP synthase inhibition. One possible explanation is that detectable ROS production is a more sensitive indicator of partial inhibition of ATP synthase function than is the total cellular ATP level.
HSV infection has both pro- and antiapoptotic effects on infected cells. In nonimmune cells, such as epithelial cells and fibroblasts, the antiapoptotic mechanisms predominate, and HSV infection of such cells does not normally lead to apoptosis (18). In contrast, in T cells, the balance of these effects ultimately leads to apoptosis (22, 25, 26, 38). Interestingly, infection of RAW264.7 cells with wild-type virus also induced apoptosis, and the induction of apoptosis was much greater using Us5 deletion virus. We are currently working to understand the basis for the differential apoptotic responses of immune and nonimmune cells to HSV infection. Clearly, gJ can inhibit apoptosis in both immune and nonimmune cells, arguing against the possibility that the antiapoptotic mechanisms of HSV are inoperative in immune cells. Instead, we consider it more likely that HSV has proapoptotic functions that are preferentially effective in immune cells. It is tempting to speculate that the ROS production observed in Jurkat and RAW264.7 cells might be related to their apoptotic phenotypes. However, the effects of ROS on apoptosis are complex, with both pro- and antiapoptotic effects reported (32). The addition of ROS inhibitors to our system had variable effects on HSV-induced apotosis of RAW264.7 cells, depending on the inhibitor used (M. Aubert and K. R. Jerome, unpublished data). Thus, the relationship of ROS production to apoptosis in the context of HSV infection remains unclear.
Taken together, our work demonstrates that gJ is produced in HSV-infected cells and distributed to a variety of membrane compartments. gJ is able to modulate certain cellular processes, specifically, apoptosis and ROS generation. Together with the long-term evolutionary conservation of gJ, these findings suggest that gJ plays an important role in viral manipulation of human cells.
Acknowledgments
This work was supported by NIH grant R01AI47378 to K.R.J.
We thank Adam Geballe, Jeff Vieira, and Michael Lagunoff for helpful discussions.
Footnotes
Published ahead of print on 24 October 2007.
REFERENCES
- 1.Aubert, M., and J. A. Blaho. 2001. Modulation of apoptosis during herpes simplex virus infection in human cells. Microbes Infect. 3859-866. [DOI] [PubMed] [Google Scholar]
- 2.Aubert, M., and J. A. Blaho. 1999. The herpes simplex type 1 regulatory protein ICP27 is required for the prevention of apoptosis in infected human cells. J. Virol. 732803-2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aubert, M., E. M. Krantz, and K. R. Jerome. 2006. Herpes simplex virus genes Us3, Us5, and Us12 differentially regulate cytotoxic T lymphocyte-induced cytotoxicity. Viral Immunol. 19391-408. [DOI] [PubMed] [Google Scholar]
- 4.Aubert, M., J. O'Toole, and J. A. Blaho. 1999. Induction and prevention of apoptosis in human HEp-2 cells by herpes simplex virus type 1. J. Virol. 7310359-10370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baines, J. D., E. Wills, R. J. Jacob, J. Pennington, and B. Roizman. 2007. Glycoprotein M of herpes simplex virus 1 is incorporated into virions during budding at the inner nuclear membrane. J. Virol. 81800-812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beitia Ortiz de Zarate, I., K. Kaelin, and F. Rozenberg. 2004. Effects of mutations in the cytoplasmic domain of herpes simplex virus type 1 glycoprotein B on intracellular transport and infectivity. J. Virol. 781540-1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burrell, H. E., B. Wlodarski, B. J. Foster, K. A. Buckley, G. R. Sharpe, J. M. Quayle, A. W. Simpson, and J. A. Gallagher. 2005. Human keratinocytes release ATP and utilize three mechanisms for nucleotide interconversion at the cell surface. J. Biol. Chem. 28029667-29676. [DOI] [PubMed] [Google Scholar]
- 8.Capaldi, R. A., and R. Aggeler. 2002. Mechanism of the F1F0-type ATP synthase, a biological rotary motor. Trends Biochem. Sci. 27154-160. [DOI] [PubMed] [Google Scholar]
- 9.Chang, Y. E., L. Menotti, F. Filatov, G. Campadelli-Fiume, and B. Roizman. 1998. UL27.5 is a novel γ2 gene antisense to the herpes simplex virus 1 gene encoding glycoprotein B. J. Virol. 726056-6064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chi, S. L., and S. V. Pizzo. 2006. Cell surface F1Fo ATP synthase: a new paradigm? Ann. Med. 38429-438. [DOI] [PubMed] [Google Scholar]
- 11.Dasgupta, A., and D. W. Wilson. 1999. ATP depletion blocks herpes simplex virus DNA packaging and capsid maturation. J. Virol. 732006-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Drees, B. L. 1999. Progress and variations in two-hybrid and three-hybrid technologies. Curr. Opin. Chem. Biol. 364-70. [DOI] [PubMed] [Google Scholar]
- 13.Eguchi, Y., A. Srinivasan, K. J. Tomaselli, S. Shimizu, and Y. Tsujimoto. 1999. ATP-dependent steps in apoptotic signal transduction. Cancer Res. 592174-2181. [PubMed] [Google Scholar]
- 14.Farnsworth, A., and D. C. Johnson. 2006. Herpes simplex virus gE/gI must accumulate in the trans-Golgi network at early times and then redistribute to cell junctions to promote cell-cell spread. J. Virol. 803167-3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Foster, T. P., J. M. Melancon, T. L. Olivier, and K. G. Kousoulas. 2004. Herpes simplex virus type 1 glycoprotein K and the UL20 protein are interdependent for intracellular trafficking and trans-Golgi network localization. J. Virol. 7813262-13277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Geromel, V., N. Kadhom, I. Cebalos-Picot, O. Ouari, A. Polidori, A. Munnich, A. Rotig, and P. Rustin. 2001. Superoxide-induced massive apoptosis in cultured skin fibroblasts harboring the neurogenic ataxia retinitis pigmentosa (NARP) mutation in the ATPase-6 gene of the mitochondrial DNA. Hum. Mol. Genet. 101221-1228. [DOI] [PubMed] [Google Scholar]
- 17.Ghiasi, H., A. B. Nesburn, S. Cai, and S. L. Wechsler. 1998. The Us5 open reading frame of herpes simplex virus type 1 does encode a glycoprotein (gJ). Intervirology 4191-97. [DOI] [PubMed] [Google Scholar]
- 18.Goodkin, M. L., E. R. Morton, and J. A. Blaho. 2004. Herpes simplex virus infection and apoptosis. Int. Rev. Immunol. 23141-172. [DOI] [PubMed] [Google Scholar]
- 19.Graham, F. L., and A. J. van der Eb. 1973. Transformation of rat cells by DNA of human adenovirus 5. Virology 54536-539. [DOI] [PubMed] [Google Scholar]
- 20.Gross, A., K. Pilcher, E. Blachly-Dyson, E. Basso, J. Jockel, M. C. Bassik, S. J. Korsmeyer, and M. Forte. 2000. Biochemical and genetic analysis of the mitochondrial response of yeast to BAX and BCL-XL. Mol. Cell. Biol. 203125-3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gupta, A., J. J. Gartner, P. Sethupathy, A. G. Hatzigeorgiou, and N. W. Fraser. 2006. Anti-apoptotic function of a microRNA encoded by the HSV-1 latency-associated transcript. Nature 44282-85. [DOI] [PubMed] [Google Scholar]
- 22.Han, J. Y., D. D. Sloan, M. Aubert, S. A. Miller, C. H. Dang, and K. R. Jerome. 2006. Apoptosis and antigen receptor function in T and B cells following exposure to herpes simplex virus. Virology 359253-263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Houstek, J., A. Pickova, A. Vojtiskova, T. Mracek, P. Pecina, and P. Jesina. 2006. Mitochondrial diseases and genetic defects of ATP synthase. Biochim. Biophys. Acta 17571400-1405. [DOI] [PubMed] [Google Scholar]
- 24.Ikemoto, H., E. Tani, I. Ozaki, H. Kitagawa, and N. Arita. 2000. Calphostin C-mediated translocation and integration of bax into mitochondria induces cytochrome c release before mitochondrial dysfunction. Cell Death Differ. 7511-520. [DOI] [PubMed] [Google Scholar]
- 25.Ito, M., W. Koide, M. Watanabe, H. Kamiya, and M. Sakuri. 1997. Apoptosis of cord blood T lymphocytes by herpes simplex virus type 1. J. Gen. Virol. 781971-1975. [DOI] [PubMed] [Google Scholar]
- 26.Ito, M., M. Watanabe, H. Kamiya, and M. Sakuri. 1997. Herpes simplex virus type 1 induces apoptosis in peripheral blood T lymphocytes. J. Infect. Dis. 1751220-1224. [DOI] [PubMed] [Google Scholar]
- 27.Jerome, K. R., Z. Chen, R. Lang, M. R. Torres, J. Hofmeister, S. Smith, R. Fox, C. J. Froelich, and L. Corey. 2001. HSV and glycoprotein J inhibit caspase activation and apoptosis induced by granzyme B or fas. J. Immunol. 1673928-3935. [DOI] [PubMed] [Google Scholar]
- 28.Jerome, K. R., R. Fox, Z. Chen, A. E. Sears, H.-Y. Lee, and L. Corey. 1999. Herpes simplex virus inhibits apoptosis through the action of two genes, US5 and US3. J. Virol. 738950-8957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kirkland, R. A., and J. L. Franklin. 2007. Bax affects production of reactive oxygen by the mitochondria of non-apoptotic neurons. Exp. Neurol. 204458-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krummenacher, C., I. Baribaud, R. J. Eisenberg, and G. H. Cohen. 2003. Cellular localization of nectin-1 and glycoprotein D during herpes simplex virus infection. J. Virol. 778985-8999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leist, M., B. Single, A. F. Castoldi, S. Kuhnle, and P. Nicotera. 1997. Intracellular adenoside triphosphate (ATP) concentration: a switch in the decision between life and death. J. Exp. Med. 1851481-1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martindale, J. L., and N. J. Holbrook. 2002. Cellular response to oxidative stress: signaling for suicide and survival. J. Cell Physiol. 1921-15. [DOI] [PubMed] [Google Scholar]
- 33.Matsuyama, S., J. Llopis, Q. L. Deveraux, R. Y. Tsien, and J. C. Reed. 2000. Changes in intramitochondrial and cytosolic pH: early events that modulate caspase activation during apoptosis. Nat. Cell Biol. 2318-325. [DOI] [PubMed] [Google Scholar]
- 34.Matsuyama, S., Q. Xu, J. Velours, and J. C. Reed. 1998. The mitochondrial FoF1-ATPase proton pump is required for function of the proapoptotic protein Bax in yeast and mammalian cells. Mol. Cell 1327-336. [DOI] [PubMed] [Google Scholar]
- 35.McGeoch, D. J., A. Dolan, S. Donald, and F. J. Rixon. 1985. Sequence determination and genetic content of the short unique region in the genome of herpes simplex virus type 1. J. Mol. Biol. 1811-13. [DOI] [PubMed] [Google Scholar]
- 36.Murata, T., F. Goshima, T. Daikoku, K. Inagaki-Ohara, H. Takakuwa, K. Kato, and Y. Nishiyama. 2000. Mitochondrial distribution and function in herpes simplex virus-infected cells. J. Gen. Virol. 81401-406. [DOI] [PubMed] [Google Scholar]
- 37.O'Toole, J. M., M. Aubert, A. Kotsakis, and J. A. Blaho. 2003. Mutation of the protein tyrosine kinase consensus site in the herpes simplex virus 1 α22 gene alters ICP22 posttranslational modification. Virology 305153-167. [DOI] [PubMed] [Google Scholar]
- 38.Pongpanich, A., P. Bhattarakosol, and C. Chirathaworn. 2004. Induction of apoptosis by herpes simplex virus in Jurkat cells is partly through caspase-3, -8 and -9 activation. J. Med. Assoc. Thai. 87(Suppl. 2)S140-S145. [PubMed] [Google Scholar]
- 39.Poon, A. P., W. O. Ogle, and B. Roizman. 2000. Posttranslational processing of infected cell protein 22 mediated by viral protein kinases is sensitive to amino acid substitutions at distant sites and can be cell-type specific. J. Virol. 7411210-11214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roizman, B., and W. Batterson. 1985. Herpesviruses and their replication, p. 497-526. In B. N. Fields (ed.), Virology. Raven Press, New York, NY.
- 41.Roizman, B., and D. M. Knipe. 2001. Herpes simplex viruses and their replication, p. 2399-2459. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed., vol. 2. Lippincott Williams and Wilkins, Philadelphia, PA. [Google Scholar]
- 42.Sato, M., Y. Matsuki, T. Oguma, K. Tsujimoto, E. Takayama, and T. Tadakuma. 2000. Inhibition of glucocorticoid-induced apoptosis by the expression of antisense gene of mitochondrial ATPase subunit 6. FEBS Lett. 47834-38. [DOI] [PubMed] [Google Scholar]
- 43.Sekulovich, R. E., K. Leary, and R. M. Sandri-Goldin. 1988. The herpes simplex virus type 1 alpha protein ICP27 can act as a trans-repressor or a trans-activator in combination with ICP4 and ICP0. J. Virol. 624510-4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Soliman, T. M., R. M. Sandri-Goldin, and S. J. Silverstein. 1997. Shuttling of the herpes simplex virus type 1 regulatory protein ICP27 between the nucleus and cytoplasm mediates the expression of late proteins. J. Virol. 719188-9197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Spear, P. G., J. M. Keller, and B. R. Roizman. 1970. Proteins specified by herpes simplex virus. II. Viral glycoproteins associated with cellular membranes. J. Virol. 5123-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang, W., and B. A. Malcolm. 1999. Two-stage PCR protocol allowing introduction of multiple mutations, deletions and insertions using QuikChange site-directed mutagenesis. BioTechniques 26680-682. [DOI] [PubMed] [Google Scholar]
- 47.Zhou, G., V. Galvan, G. Campadelli-Fiume, and B. Roizman. 2000. Glycoprotein D or J delivered in trans blocks apoptosis in SK-N-SH cells induced by a herpes simplex virus 1 mutant lacking intact genes expressing both glycoproteins. J. Virol. 7411782-11791. [DOI] [PMC free article] [PubMed] [Google Scholar]