Abstract
To better understand the pathogenesis of human herpesvirus 6 (HHV-6), it is important to elucidate the functional aspects of immediate-early (IE) genes at the initial phase of the infection. To study the functional role of the HHV-6B IE gene encoding U95, we generated a U95-Myc fusion protein and screened a pretransformed bone marrow cDNA library for U95-interacting proteins, using yeast-two hybrid analysis. The most frequently appearing U95-interacting protein identified was GRIM-19, which belongs to the family of genes associated with retinoid-interferon mortality and serves as an essential component of the oxidative phosphorylation system. This interaction was verified by both coimmunoprecipitation and confocal microscopic coimmunolocalization. Short-term HHV-6B infection of MT-4 T-lymphocytic cells induced syncytial formation, resulted in decreased mitochondrial membrane potential, and led to progressively pronounced ultrastructural changes, such as mitochondrial swelling, myelin-like figures, and a loss of cristae. Compared to controls, RNA interference against U95 effectively reduced the U95 mRNA copy number and abrogated the loss of mitochondrial membrane potential. Our results indicate that the high affinity between U95 early viral protein and GRIM-19 may be closely linked to the detrimental effect of HHV-6B infection on mitochondria. These findings may explain the alternative cell death mechanism of expiration, as opposed to apoptosis, observed in certain productively HHV-6B-infected cells. The interaction between U95 and GRIM-19 is thus functionally and metabolically significant in HHV-6B-infected cells and may be a means through which HHV-6B modulates cell death signals by interferon and retinoic acid.
The Herpesviridae family includes three subfamilies, i.e., Alphaherpesvirinae, Betaherpesvirinae, and Gammaherpesvirinae. A typical herpesvirus virion consists of a core containing a linear, double-stranded DNA; an icosahedral capsid (approximately 100 to 110 nm in diameter) containing 162 capsomeres, with a hole running down the long axis; an amorphous, sometimes asymmetric material that surrounds the capsid (designated the tegument); and an envelope containing viral glycoprotein spikes on its surface (4). In 1986, a new herpesvirus was isolated from the peripheral blood mononuclear cells of six adults with lymphoproliferative disorders, some of whom were infected with human immunodeficiency virus type 1 (HIV-1). The virus was initially named human B-lymphotropic virus but was subsequently renamed human herpesvirus 6 (HHV-6) and placed within the Betaherpesvirinae, based on its high genetic similarity to human cytomegalovirus (HCMV). HHV-6 infects virtually all children during the early years of life, and like other herpesviruses, establishes latency after primary infection. This infection usually runs a benign course and requires no antiviral treatment. However, in seronegative pregnant women, a primary HHV-6 infection contracted from a child with roseola infantum can rarely lead to spontaneous abortion during the first trimester or produce neurological complications in the newborn following HHV-6 transplacental infection (5). In addition, reactivation of latent HHV-6 can occasionally lead to serious systemic diseases, especially encephalopathy, which may be fatal in immunocompromised or grafted patients (23).
HHV-6 predominantly infects and replicates in CD4+ lymphocytes (39), but it may establish latency in the monocyte/macrophage lineage (22). HHV-6 isolates are further divided into two closely related variants, i.e., A (HHV-6A) and B (HHV-6B), based on their molecular and biological characteristics (1, 34, 44). The complete genome sequences of HHV-6A (strain U1102) and HHV-6B (strains HST and Z29) have been determined (10, 18). The betaherpesviruses have extensive domains of similar genetic organization, i.e., the conserved herpesvirus gene blocks in the unique region of their genome, and they include a number of gene families that are characteristic of this subgroup. These include the US22, G-protein-coupled receptor, and immunoglobulin gene families. The US22 gene family is the most extensive family found in betaherpesviruses but is absent in the alpha- and gammaherpesviruses. HHV-6 carries 11 members of the US22 family (DR1, DR2, DR6, DR7, U2, U3, U7, U8, U16, U25, and U95), which are related to the 12 members of this family found in HCMV (14, 43). Some members of this family are spliced and expressed as immediate-early (IE) proteins (32) and are likely to be transcriptional activators (13, 42). Murine cytomegalovirus IE2 possesses all of these characteristics (7). U95 of HHV-6 has been reported to be expressed at the IE stage of infection (40).
HHV-6 infection of human mononuclear cells induces interferon (IFN), which suppresses HHV-6 replication, and production of IFN suggests that it is an important part of the host response to HHV-6 (21). The combination of IFN-β and all-trans-retinoic acid (RA) induces the death of cells by increasing the intracellular levels of GRIM-19 protein (3). GRIM-19 has also been reported to interact with other herpesvirus proteins, such as viral interferon factor 1 of HHV-8, resulting in deregulation of GRIM-19-induced apoptosis in the presence of IFN-RA and inhibition of IFN-RA-induced cell death (35).
Clues pertaining to the functional aspects of HHV-6B U95 can be derived from the numerous studies on HCMV IE2 due to their similarity at the amino acid level (14, 40). However, there is limited information with respect to the cellular proteins that viral IE proteins interact with during HHV-6B infection. Using yeast two-hybrid analysis, we screened a pretransformed bone marrow cDNA library, using the HHV-6B U95 IE protein as the bait. Here we report that U95 interacts with cellular GRIM-19 protein and that the interaction between U95 and GRIM-19 is functionally important in virus-cell pathogenesis via mitochondrial impairment and may represent a means by which the virus modulates IFN-RA-mediated cell death signals.
MATERIALS AND METHODS
Yeast two-hybrid techniques.
The yeast two-hybrid experiments were performed using a yeast two-hybrid system 2 kit (Clontech, Mountain View, CA) according to the manufacturer's guidelines. The full-length U95 gene of HHV-6B strain HST (GenBank accession no. AB_021506; nucleotides 144230 to 147868) served as the bait and was cloned into the pGBKT7 yeast two-hybrid vector, resulting in an N-terminal in-frame fusion with the GAL4 DNA-binding domain. The insert was sequenced before being transformed into the Saccharomyces cerevisiae host reporter strain PJ69-2A. A Matchmaker pretransformed human bone marrow cDNA library (Clontech) was screened via yeast mating by incubating 500 μl of the pretransformed library (∼25 × 106 CFU) with bait-transformed PJ69-2A. The entire mating culture was plated onto selective medium and incubated at 30°C for 3 to 21 days. The positive control comprising p53 and simian virus 40 large T antigen supplied with the kit was used concurrently in the mating experiments. Colonies were picked, restreaked onto fresh selective medium, and assayed for the LacZ phenotype by using a LacZ colony lift filter assay. Filters were left for 8 h to develop a blue color, which indicated a positive protein-protein interaction. The mating control cross supplied with the library was also used as a positive control for the LacZ colony lift assay.
Activation domain (AD) vector primers FAD (5′-ATGATGAAGATACCCCACCAAACC-3′; sense) and RADO (5′-TTGCGGGGTTTTTCAGTATCTACG-3′; antisense) were designed to amplify AD vector inserts. LacZ-positive colonies were screened via direct colony PCR using the vector primers. Amplified fragments were purified and subjected to cycle sequencing in both directions. BlastN and BLASTP algorithms were used to analyze the resultant sequence data from the clones (38, 45).
Propagation of human MT-4 cell line, transfection, and coimmunoprecipitation assays.
MT-4 cells were cultured in 25-cm2 cell culture flasks with 5 ml of RPMI 1640 medium supplemented with 10% fetal bovine serum at 37°C with 5% CO2. Expression vectors pCMV-HA and pCMV-Myc (Clontech) were used because most of their restriction endonuclease sites were identical to those of the binding domain and AD vectors. The U95 fragment was amplified with PCR primers U95F-SfiI (5′-TGACTAAGCTGTTACTAATATCACTGGCCATTATGGCCATGTCTTCAAATCTGG-3′) and U95R-NotI (5′-TATTAATTGCGGCCGCTTATTTACCTTCCTGAG-3′) and cloned into the pCMV-HA vector at the SfiI and NotI sites. The full-length coding sequence of GRIM-19 (GenBank accession no. NM_015965; nucleotides 260 to 943) was cloned into pCMV-Myc at the SfiI and SalI sites. Both clones were transformed into Escherichia coli strain DH5α, purified using plasmid spin columns (Qiagen, Hilden, Germany), and then transfected, individually or together, into MT-4 cells by using Transfectin reagent (Bio-Rad, Hercules, CA). Empty vectors transfected into MT-4 cells served as negative controls. Expression of the fusion proteins was allowed to occur for 48 h. Total protein was harvested using lysis buffer (1% Triton X-100 and phosphate-buffered saline [PBS], pH 7.4), and the cell lysate was centrifuged at 12,000 rpm for 10 min. The supernatant was pretreated with Sepharose A and A/G beads (Calbiochem, San Diego, CA) at 4°C for 1 h, and the mixture was centrifuged at 5,000 rpm for 5 min to remove the beads. The supernatant was incubated with Sepharose A and A/G beads which were pretreated with anti-hemagglutinin (anti-HA) and/or anti-Myc antibodies at 4°C for 4 h. The mixture was centrifuged at 5,000 rpm for 10 s, the supernatant was discarded, and the beads were washed thrice with PBS. The beads were resuspended with 2× sodium dodecyl sulfate (SDS) sample loading buffer and heated at 99°C for 3 min, and the proteins were subjected to SDS-polyacrylamide gel electrophoresis (10%). Western blot analyses were then carried out using antibodies against the HA or Myc epitope. Negative controls included mixed lysates from cells transfected with plasmid expressing either the HA or Myc tag only (38, 45).
Immunofluorescence confocal microscopy.
MT-4 cells (106 in each 25-cm2 cell culture flask) were grown overnight and then transfected as described above, using the expression plasmid constructs. The cells were incubated at 37°C for 24 h, and transfection was repeated at 48 h. The transfected cells were then washed thrice with PBS and immobilized onto glass slides by use of a Cytospin centrifuge. The immobilized cells were washed twice with cold PBS and incubated with cold methanol at −20°C for 4 min. They were incubated at room temperature for 3 min before being washed thrice with cold PBS. The slides were incubated at room temperature for 1 h with primary anti-Myc monoclonal and/or anti-HA polyclonal antibody. They were then washed five times with cold PBS before being incubated at room temperature for 1 h with secondary fluorescein isothiocyanate (FITC)-tagged goat anti-mouse or rhodamine phalloidin-tagged goat anti-rabbit antibody. The glass slides were washed six times with cold PBS before being mounted on coverslips, using Vectashield mounting medium (Dako, Glostrup, Denmark), and examined by confocal microscopy (38, 45). Mitochondria were labeled with MitoTracker Green FM (Invitrogen, Carlsbad, CA) according to the manufacturer's protocol.
Determination of virus titer by TCID50 and assay for cell viability.
MT-4 cells (105 per well) were propagated in cell culture plates overnight and then infected with HHV-6B strain HST, achieving cytopathic effect in 50% of the cells after 48 h. The 50% tissue culture infective dose (TCID50) was derived from the observation of cytopathic effect, i.e., the induction of syncytium formation, in 50% of the cell population (19). An infecting dose of 105 TCID50 of HHV6-B was selected. After infection for 48 h, cell viability was determined by staining the cells with 0.4% trypan blue and counting them by using a hemocytometer under light microscopy. Viable cells were observed as unstained, translucent bodies, while dead cells were stained blue. No significant cell death or debris was observed for 5 days postinfection, with HHV-6B-infected cells appearing as unstained, large cells with syncytial formation.
shRNA constructs to induce specific antiviral response by RNAi.
The GenBank database was searched for unique sequences within U95 and to exclude homology to known human and other viral genes. Using small interfering RNA software (Ambion, Austin, TX), a 19-mer region of the HHV-6B U95 gene (GenBank accession no. AB_021506; nucleotides 144460 to 144478) was selected as the target for RNA interference (RNAi). U95-specific short hairpin RNA (shRNA) duplex oligonucleotides, consisting of an antisense strand (5′-AGUCGACAGUGAUUCGCGGUU-3′) and a sense strand (5′-CCGCGAAUCACUGUCGACUUU-3′), were designed and chemically synthesized (Sigma-Proligo, Boulder, CO). A scrambled shRNA was designed by random arrangement of the same numbers of A, C, G, and U nucleotides as those in the U95 shRNA construct and confirmed to lack homology to viral and human genes in GenBank. The scrambled-sequence shRNA duplex was synthesized (antisense sequence, 5′-GGGUGACACGUAGUCAGCUUU-3′; and sense sequence, 5′-AGCUGACUACGUGUCACCCUU-3′) to serve as the negative control for all RNAi experiments (41).
Real-time RT-PCR.
Total mRNA samples from uninfected, HHV6B-infected, HHV6B-infected but U95 shRNA-treated, and HHV6B-infected but scrambled shRNA-treated cells were isolated using a total mRNA isolation kit (Qiagen). Genomic DNA contamination was minimized by treating the samples with DNase provided in the kit. Each purified total mRNA was then subjected to reverse transcription-PCR (RT-PCR), using Superscript II reverse transcriptase (Invitrogen) and 100 μM of random hexamers (Promega, Madison, WI) at 42°C for 1 h. The reaction was terminated by incubating the mixture at 95°C for 10 min. The cDNA concentrations were standardized to 120 ng/μl. Real-time RT-PCR analyses were performed in triplicate with a LightCycler instrument (Roche, Mannheim, Germany), using SYBR green with primers flanking a 317-bp fragment (GenBank accession no. AB_021506; nucleotides 145165 to 145481) within the U95 gene (5′-GGCGACACAACTAAGCGCATC-3′ and 5′-CGGAAATGGCCGCATCTACAC-3′) and with primers for the glyceraldehyde-3-phosphate dehydrogenase (G3PDH) housekeeping gene (5′-AGGTGAAGGTCGGAGTCAAC-3′ and 5′-CATGGGTGGAATCATATTGG-3′), which served as a normalization reference. Each 10-μl RT-PCR mix included 1 μl of cDNA, 0.5 μM of each primer, 2.5 to 3 mM MgCl2, and 1 μl of 10× Hot Start reaction mix (Roche). The reaction mixtures were subjected to an initial denaturation of 95°C for 10 min, followed by 35 cycles of 95°C for 10 s, 65°C for 5 s, and 72°C for 10 s, before being subjected to a melting curve analysis of one cycle with a 15-s hold at 65°C. Real-time RT-PCR with each sample, using the two primer sets, was conducted in triplicate to ensure consistency in cycle threshold (CT) values. The mean CT values were used to calculate the relative expression levels, as described previously (25). The normalized CT values for the U95 gene were subjected to Student's t test with a two-tailed distribution to determine significance at the 95% confidence level.
Transmission electron microscopy.
MT-4 cells were propagated for 24 h and were infected with HHV-6B as described above. Uninfected MT-4 cells were included as negative controls. At 0, 1, 4, and 7 h postinfection, both infected and uninfected cells were fixed with 2.5% glutaraldehyde in 0.1 M phosphate buffer (pH 7.4) at room temperature for 1 h. The fixed samples were then postfixed with 2% osmium tetroxide at room temperature for 1 h, dehydrated in a graded alcohol series, and embedded in Epon 812. Ultrathin sections (20 nm thick) were cut with a microtome, stained with uranyl acetate and lead citrate for 20 and 10 min, respectively, and observed using a Philips model 208S electron microscope (26).
Mitochondrial membrane potential assay by flow cytometry.
At 1 h postinfection, the HHV-6-infected cells were transfected with either U95 shRNA or scrambled shRNA (85 nM), using Lipofectamine 2000. They were then washed once with PBS and incubated with MitoProbe DiOC2(3) dye (Invitrogen). Two untreated wells, one with added DiOC2(3) and one without, served as controls.
RESULTS
Yeast two-hybrid screening identifies GRIM-19 as an interactor of U95.
The pGBKT7-U95 plasmid construct was employed as the bait in yeast two-hybrid mating experiments using a pretransformed bone marrow cDNA library. The GRIM-19-interacting clones (with the largest spanning nucleotides 307 to 922; GenBank accession no. NM_015965) exhibited the strongest β-galactosidase activity compared with the positive control (data not shown). GRIM-19 constituted the most frequently appearing positive clones (41 of a total of 210 clones) and was thus selected for further analysis.
U95 interacts with GRIM-19 in human MT-4 cells.
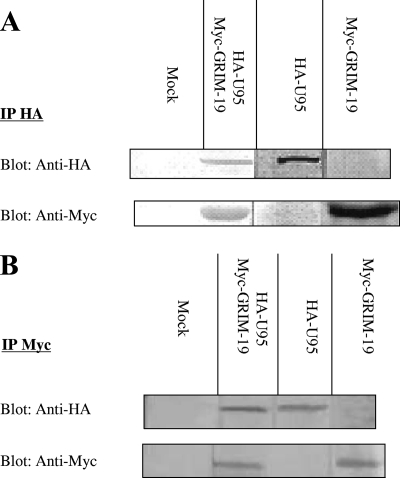
Coimmunoprecipitation experiments were conducted to confirm the interaction between U95 and GRIM-19 in MT-4 cells. Western blot analysis revealed that the U95 protein (∼70 kDa) coimmunoprecipitated with the GRIM-19 protein (∼20 kDa), thereby confirming their interaction in MT-4 cells (Fig. 1). The total cell lysate from MT-4 cells cotransfected with both empty vectors served as the negative control.
FIG. 1.
Coimmunoprecipitation of HHV-6B U95 and human GRIM-19 proteins in MT-4 cells. MT-4 cells were transfected with the pCMV-HA-U95 and/or pCMV-Myc-GRIM-19 construct. Mock controls were mixed lysates from cells transfected with plasmids expressing either the HA or Myc tag only. (A) Immunoprecipitation (IP) of cell lysates with anti-HA polyclonal antibody was performed, except for the Myc-GRIM-19 lysate, which acted as the positive control for Western blotting with anti-Myc. (B) Immunoprecipitation (IP) of cell lysates with anti-Myc monoclonal antibody was performed, except for the HA-U95 lysate, which acted as the positive control for Western blotting with anti-HA. The precipitates were fractionated by Tris-glycine-SDS-polyacrylamide gel electrophoresis (12%) and subjected to Western blot analysis using anti-HA antibody or anti-Myc antibody to detect HA-U95 or Myc-GRIM-19, respectively.
Colocalization of U95, GRIM-19, and mitochondria in MT-4 cells.
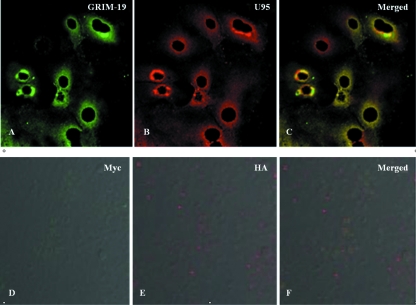
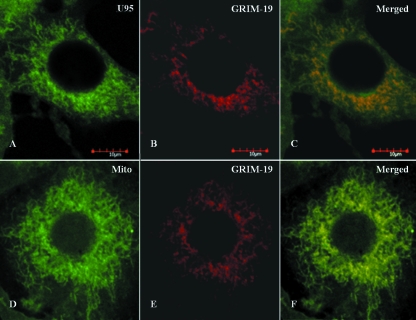
The interaction between U95 and GRIM-19, as well as their intracellular location, was further verified by confocal microscopy. Immunofluorescence staining using specific antibodies revealed strong perinuclear staining and confirmed their colocalization predominantly in the mitochondria (Fig. 2A to C). MT-4 cells transfected with the empty expression vectors served as negative controls (Fig. 2D to F). In cells transfected with the pCMV-HA-U95 construct only, immunofluorescence staining of HA-U95 and endogenous GRIM-19 revealed colocalization, consistent with the results involving dual transfections (Fig. 3). Furthermore, specific staining of mitochondria verified their colocalization with endogenous GRIM-19.
FIG. 2.
Intracellular colocalization of HHV-6B U95 with GRIM-19. MT-4 cells were cotransfected with pCMV-Myc-GRIM-19 and pCMV-HA-U95 constructs, fixed, immunolabeled, and examined by immunofluorescence microscopy. Detection of Myc-GRIM-19 with mouse anti-Myc antibody (A) and of HA-U95 with rabbit anti-HA antibody (B) was performed. The secondary antibodies were FITC-tagged anti-mouse antibody and rhodamine phalloidin-tagged anti-rabbit antibody, respectively. (C) Colocalization was observed by superimposing panels A and B. To serve as negative controls, MT-4 cells were cotransfected with empty pCMV-Myc and pCMV-HA vectors, fixed, immunolabeled, and examined by immunofluorescence microscopy. Reactions with mouse anti-Myc antibody (D) and rabbit anti-HA antibody (E) are shown. The secondary antibodies were FITC-tagged anti-mouse antibody and rhodamine phalloidin-tagged anti-rabbit antibody, respectively. (F) Superimposed image of panels D and E.
FIG. 3.
Colocalization of HHV-6B U95 with endogenous GRIM-19 in MT-4 cells. MT-4 cells were transfected with the pCMV-HA-U95 construct, immobilized onto glass slides by use of a Cytospin centrifuge, fixed, immunolabeled, and examined by immunofluorescence microscopy. (A) Detection of HA-U95 with rabbit anti-HA antibody staining and counterlabeling with FITC-tagged anti-rabbit antibody. (B and E) Detection of endogenous cellular GRIM-19 by mouse monoclonal antibody staining and counterlabeling with rhodamine phalloidin-tagged anti-mouse antibody. (C) Colocalization is displayed by superimposing panels A and B. (D) Mitotracker Green FM dye staining of mitochondria as long, tubular organelles extending from the nucleus. (F) Colocalization is displayed by superimposing panels D and E.
U95 shRNA treatment downregulates the transcription of U95.
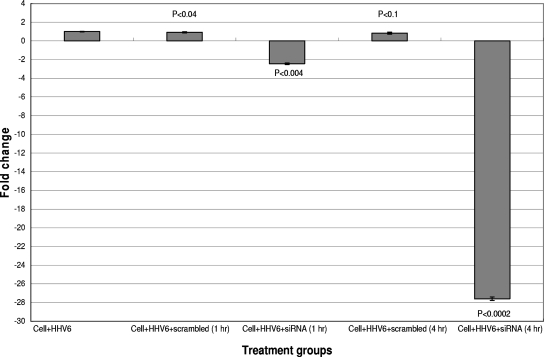
To verify that the U95 shRNA sequence was both specific and effective, real-time RT-PCR experiments were performed on mock-infected and HHV-6B-infected MT-4 cells transfected with shRNA prior to flow cytometry. Figure 4 shows a significant reduction in the U95 transcription level in U95 shRNA-treated samples, especially at 4 h posttreatment, compared to that in untreated HHV-6-infected MT-4 cells. Moreover, less cytopathic effect (i.e., syncytial formation) was observed (data not shown). The G3PDH housekeeping gene served as the reference transcript for RT-PCR and revealed a negligible difference in CT values between infected and mock-infected samples, thereby justifying its use for normalization (data not shown). The shRNA construct with a scrambled sequence served as the negative control.
FIG. 4.
Real-time RT-PCR analyses of relative U95 mRNA expression levels in HHV-6B-infected MT-4 cells following U95 shRNA treatment compared with those in controls. HHV-6B-infected MT-4 cells were transfected with the U95 shRNA construct or with the control scrambled shRNA construct after infection for 1 h. Real-time RT-PCR was performed 1 and 4 h following shRNA transfection. Samples were normalized against the G3PDH housekeeping gene and with untransfected but HHV-6B-infected MT-4 cells at each respective time point as the reference sample (i.e., change of onefold for U95 transcription). Statistical analyses were then performed, with P values of <0.05 denoting significant differences.
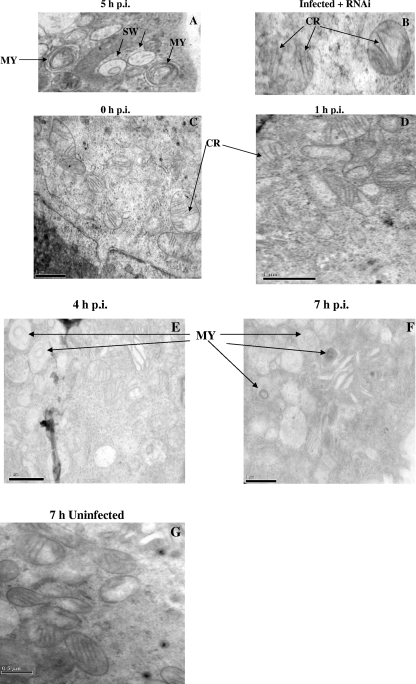
Ultrastructural evidence of the effect of HHV-6B infection on mitochondria.
To understand the implication of the U95-GRIM-19 protein complex on mitochondria, both HHV-6B-infected and HHV-6B-infected but U95 shRNA-treated MT-4 cells were subjected to transmission electron microscopy. The mitochondria of HHV-6B-infected cells exhibited swelling and less-convoluted cristae (Fig. 5A). In contrast, HHV-6B-infected but U95 shRNA-treated MT-4 cells displayed less swelling of the mitochondria, with the majority showing more-convoluted cristae (Fig. 5B). However, no other abnormal phenotypic changes were observed in both populations of mitochondria. To determine the progression and frequency of these structural abnormalities of the mitochondria in HHV-6B-infected cells compared with those in uninfected cells, mitochondrial organelles were monitored by electron microscopy at 0, 1, 4, and 7 h postinfection. Alterations in mitochondrial structures of infected cells were more apparent as the infection progressed (Fig. 5C to G). By using electron microscopy to count the numbers of normal and abnormal mitochondria in 10 random cells per group and obtaining the mean percentages, it was evident that the percentage of abnormal mitochondria steadily increased with the time of infection (Table 1).
FIG. 5.
Transmission electron micrographs of HHV-6B-infected cells without and with U95 shRNA treatment. (A) After 5 h of infection of MT-4 cells with HHV-6B, typical features of mitochondrial swelling (SW), with severe disruption of mitochondrial structures, showing myelin-like figures (MY) and a loss of cristae, were observed. (B) After 1 h of HHV-6B infection, MT-4 cells were transfected with U95 shRNA. At 4 h post-RNAi, swelling of the mitochondria was significantly reduced, and numerous cristae (CR) could be observed clearly. Magnification, ×50,000. At the time of infection of MT-4 cells with HHV-6B (C) and at 1 h postinfection (D), little or no disruption of mitochondrial structures was observed. At 4 h (E) and 7 h (F) postinfection, there were increasingly severe structural abnormalities of mitochondria, including myelin-like figures (MY) and a loss of cristae. (G) At 7 h postinfection, uninfected MT-4 cells generally displayed normal mitochondrial structures. Bars, 1 μm.
TABLE 1.
Frequencies of structural abnormalities of mitochondria in HHV-6B-infected versus uninfected MT-4 cells at 0, 1, 4, and 7 h postinfection
| Time post infection (h) | HHV-6B-infected cells
|
Uninfected cells
|
||||||
|---|---|---|---|---|---|---|---|---|
| No. of mitochondria with normal structure | No. of mitochondria with abnormal structure | Total no. of mitochondria counted | % Abnormal mitochondria | No. of mitochondria with normal structure | No. of mitochondria with abnormal structure | Total no. of mitochondria counted | % Abnormal mitochondria | |
| 0 | 70 | 3 | 73 | 4.11 | 86 | 6 | 92 | 6.52 |
| 1 | 36 | 4 | 40 | 10.00 | 90 | 5 | 95 | 5.26 |
| 4 | 46 | 15 | 61 | 24.59 | 66 | 3 | 69 | 4.35 |
| 7 | 34 | 16 | 50 | 32.00 | 63 | 4 | 67 | 5.97 |
HHV-6B infection and U95 shRNA treatment significantly influence mitochondrial membrane potential.
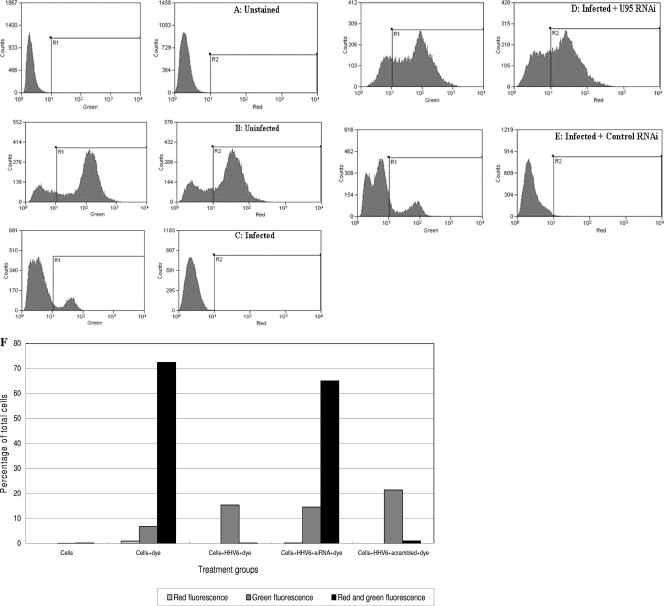
To determine if the interaction between U95 and GRIM-19 has an impact on mitochondrial function, HHV-6B-infected versus uninfected MT-4 cells were assayed for mitochondrial membrane potential activity. The latter is a very good indication of the physiological state of both cells and mitochondria. Unstained cells were included to exclude signals from cells demonstrating autofluorescence (Fig. 6A). Healthy cells were stained with the dye DiOC2(3) and subjected to fluorescence at an excitation wavelength of 488 nm. Signals from 530/30-nm-band-pass and 650-nm-long-pass filters could be detected as green and red fluorescence, respectively (Fig. 6B). However, when MT-4 cells were infected with live HHV-6B, only green fluorescence signals could be detected (Fig. 6C), with the decrease in mitochondrial membrane potential being accompanied by a shift from red to green fluorescence. When HHV6B-infected cells were transfected with U95 shRNA, there was a reversion of the fluorescent signal to a signal comparable to that of healthy cells (Fig. 6D). The negative control experiment involving HHV6B-infected cells transfected with the scrambled shRNA construct could not restore the mitochondrial membrane potential, with a shift to green fluorescence (Fig. 6E). The percentages of cells in different treatment and control groups emitting red, green, or both fluorescence signals are summarized in Fig. 6F.
FIG. 6.
Assessment of mitochondrial membrane potential by MitoProbe DiOC2(3) assay. MT-4 cells were analyzed by flow cytometry using a 488-nm excitation wavelength, with 530/30-nm-band-pass and 650-nm-long-pass detection filters. Cells were resuspended in PBS, and the distribution of fluorescent signals (logarithmic scale) of 20,000 cells was observed. The x axes of the left and right panels depict the intensities of green and red fluorescence, respectively. Each y axis indicates the number of cells, while each area under the curve is equivalent to 20,000 cell events. (A) Unstained MT-4 cells served as a control to exclude autofluorescence. (B) DiOC2(3)-stained uninfected MT-4 cells. (C) DiOC2(3)-stained HHV-6B-infected MT-4 cells showing a reduction in red fluorescence signals. (D) DiOC2(3)-stained HHV-6B-infected MT-4 cells subjected to U95 shRNA treatment at 1 h postinfection. (E) DiOC2(3)-stained HHV-6B-infected MT-4 cells subjected to scrambled shRNA treatment at 1 h postinfection, indicating decreased red fluorescence signals. (F) Effect of HHV-6B RNAi treatment on mitochondrial membrane potential in HHV-6B-infected MT-4 cells. The bar chart illustrates the percentages of cells in different treatment and control groups emitting red, green, or both fluorescence signals by flow cytometry.
DISCUSSION
Many studies have characterized the members of the US22 family, but there are few functional studies pertaining to their interaction with mammalian cellular proteins. Since it is a relatively recently identified member of this family, U95 is known only as an IE protein (40). IE genes are important because they are the first to be transcribed and translated into functional proteins to aid in the downstream processes of viral infection. To address the functional aspects of an IE gene during infection, U95 of HHV-6B was used as bait in a yeast two-hybrid assay to screen for potential interacting human proteins. GRIM-19 was thus identified as a strong interactor of the U95 protein. The U95 protein could immunoprecipitate the GRIM-19 protein from the total cell lysate and vice versa, further demonstrating their strong interaction. To verify this interaction, immunofluorescence assay revealed distinct colocalization in the perinuclear region, predominantly in the mitochondria. Previous studies have demonstrated the localization of GRIM-19 primarily in the mitochondria and have indicated that GRIM-19 is essential for early embryonic development (15). GRIM-19 is a small protein that promotes cell death induced by IFN-β and RA (3). Interestingly, viral interferon regulatory factor 1 of Kaposi's sarcoma-associated HHV-8, human papillomavirus type 16 E6, and simian virus 40 LT antigen also bind GRIM-19, suggesting that this is a target of certain viral proteins (9, 35). Downregulation of GRIM-19 by antisense treatment culminates in a growth advantage to cells in the presence of IFN-RA, and cells that express moderate levels of GRIM-19 are significantly more susceptible to the cytotoxic action of IFN-RA (3, 35). Knockdown of GRIM-19 and NADH dehydrogenase ubiquinone Fe-S protein 3 (the two subunits of the mitochondrial respiratory chain complex I) confers resistance to IFN-RA-induced apoptosis and results in decreased production of reactive oxygen species (16). On the other hand, upregulation of GRIM-19 results in an enhanced caspase-9 activity and apoptotic cell death response to IFN-RA treatment (3). Interestingly, GRIM-19 interacts with endogenous nucleotide oligomerization domain 2, which mediates innate mucosal responses and functions as an antibacterial factor that limits the survival of invasive intracellular bacteria (6).
Cells infected with HHV-6B exhibit the typical appearance of cytopathic effects, becoming large, balloon-type cells or forming syncytia. Cell death almost always results, although it is still unclear whether it occurs by necrosis or apoptosis. Cell death through the apoptotic pathway was indicated by upregulation of the caspase-3 gene during HHV-6 infection (27). The release of cytochrome c from mitochondria can also ultimately lead to activation of caspase-9 and then caspase-3 and thus to apoptosis. While numerous studies indicate that apoptosis is mediated by various viral infections, the precise mechanisms of apoptotic death in virus-infected cells have not been demonstrated clearly. Some studies concluded that apoptosis is rarely observed in virus-infected cells, e.g., HIV-1-producing cells, and that HIV-1 RNA is rarely detected in apoptotic cells (11). Moreover, it is documented that apoptosis occurs predominantly in uninfected CD4+ T cells but not in productively HHV-6-infected cells (17). HHV-6 also prevents monocytes from undergoing spontaneous apoptosis during the initial 72 h of culture (20). Our results indicate that apoptosis is unlikely to be the major event during the early phase of HHV-6B infection of MT-4 cells. Electron micrographs of HHV-6B-infected cells (Fig. 5) revealed swelling of the mitochondria. The other changes in mitochondrial ultrastructure and morphology indicative of mitochondrial degeneration were the loss of cristae and the appearance of myelin-like figures (29, 37). No other structural disruptions were observed in the infected cells, nor were any typical signs of apoptosis, including membrane blebbing, nuclear fragmentation, and chromatin condensation. RNAi against the U95 gene at 1 h postinfection restored the normal phenotypic appearance of the mitochondria without any significant loss of cristae.
In order to ascertain the significance of the U95-GRIM-19 interaction in MT-4 cells, prevention of protein complex formation was achieved by using shRNA treatment targeting U95. Specific and efficacious RNAi targeting against the U95 gene in vitro was confirmed by real-time RT-PCR. U95 shRNA treatment for 4 h resulted in a highly significant reduction of U95 mRNA expression, of 27.6-fold (P < 0.0002), compared to treatment with the control scrambled shRNA sequence.
GRIM-19 is an essential component of the oxidative phosphorylation system. Oxidative phosphorylation activity is closely related to the electrical potential difference across the outer and inner membranes of the mitochondria. This potential is a key indicator of cellular viability, as it reflects the pumping of hydrogen ions across the inner membrane during the process of electron transport, i.e., the driving force behind ATP production. This electrical potential difference can be demonstrated by flow cytometry with the dye DiOC2(3). This membrane potential-sensitive cyanine dye can penetrate the cytosol of eukaryotic cells and accumulates primarily in mitochondria with active membrane potentials, producing bright red and green fluorescence (12, 36). DiOC2(3) staining intensity decreases with decreased mitochondrial membrane potential, accompanied by a shift from red to green fluorescence. The respiratory activity of the mitochondria during HHV-6B infection is a good indication of the physiological state of the cell. The effect of preventing the U95-GRIM-19 interaction via U95 shRNA treatment was documented using flow cytometry experiments with the DiOC2(3) dye. DiOC2(3)-stained uninfected MT-4 cells with active mitochondrial membrane potential exhibited both red and green fluorescence. A similar profile, with emission of both red and green fluorescence, was observed with the HHV-6B-infected cells treated with U95 shRNA. In contrast, HHV-6B-infected cells demonstrated a disruption of the mitochondrial membrane potential, with decreased staining intensity and a shift from red to green fluorescence signals. Scrambled shRNA treatment was unable to inhibit the formation of the U95-GRIM-19 complex, and thus the treated cells displayed a similar fluorescence profile to that of HHV-6B-infected cells. From our data, it is clear that the effect of mitochondrial membrane potential loss due to HHV-6B infection can be abrogated by blocking the formation of the U95-GRIM-19 complex. Previous studies concluded that mitochondrial membrane potential collapse is an early and necessary event which plays an important role in apoptosis of cells (8). Apoptotic mechanisms may involve an alteration in mitochondrial membrane permeability and depolarization of the mitochondrial membrane potential (31). Increased expression of uncoupling protein 2 in HeLa cells leads to a rapid and pronounced fall in mitochondrial membrane potential, culminating in oncosis, which is a form of cell death induced by energy depletion (30). However, we have shown that mitochondrial membrane potential collapse due to HHV-6B infection was not accompanied by overt apoptotic events and that the loss in the membrane potential during HHV-6B infection could be abolished by inhibiting U95 gene transcription. This beneficial effect of U95 downregulation renders U95 a potentially attractive target for novel antiviral design.
Although certain viral homologs of human cellular antiapoptotic genes have been identified within the Betaherpesvirinae, e.g., vbcl-2 and FLIP, none has hitherto been found in HHV-6B (33). Taken together, our results indicate that the U95 protein binds to and may inactivate GRIM-19, thereby interfering with its regulation of cell death in response to IFN-RA. However, GRIM-19 is also an indispensable component of mitochondrial complex I that plays an essential role in mitochondrial function, the integrity of the electron transfer chain, and the oxidative phosphorylation system (15, 16). Our data strongly suggest that U95 is directly responsible for altered mitochondrial structure and function, but it is possible that U95 may regulate the actual agent of mitochondrial dysfunction. Such a hypothesis may be addressed by observing whether mitochondrial structure and function are altered in cells transfected with the U95 gene alone.
Notwithstanding the fact that the specific benefits to the virus resulting from the U95-GRIM-19 protein complex have yet to be elucidated, it is likely that any inactivation of GRIM-19 by HHV-6 will lead to mitochondrial membrane potential loss and ultimately contribute to the cytopathic effect. The febrile illness arising from HHV-6 infection is particularly prevalent in early childhood, and HHV-6 reactivation is a serious problem for immunocompromised patients and organ transplant recipients (23). Furthermore, a putative relationship between HHV-6 infection and multiple sclerosis (MS) has been suggested (28). Biochemical defects in complex I activity may be involved in the pathogenesis of MS (24), and mitochondrial dysfunction plays a key role in progressive axonal loss in MS (2). Future studies on the functional consequences of the interaction between U95 and GRIM-19 are therefore warranted to better comprehend the mechanisms of HHV-6 replication, virulence, reactivation, host pathogenesis, and association with MS.
Acknowledgments
We are grateful to our colleagues for useful discussions. We thank Kelly Lau and Josephine Howe for their excellent technical assistance in electron microscopy.
This study was supported by a research grant from the National University of Singapore.
Footnotes
Published ahead of print on 10 October 2007.
REFERENCES
- 1.Ablashi, D. V., N. Balachandran, S. F. Josephs, C. L. Hung, G. R. Krueger, B. Kramarsky, S. Z. Salahuddin, and R. C. Gallo. 1991. Genomic polymorphism, growth properties, and immunologic variations in human herpesvirus-6 isolates. Virology 184545-552. [DOI] [PubMed] [Google Scholar]
- 2.Andrews, H. E., P. P. Nichols, D. Bates, and D. M. Turnbull. 2005. Mitochondrial dysfunction plays a key role in progressive axonal loss in multiple sclerosis. Med. Hypotheses 64669-677. [DOI] [PubMed] [Google Scholar]
- 3.Angell, J. E., D. J. Lindner, P. S. Shapiro, E. R. Hofmann, and D. V. Kalvakolanu. 2000. Identification of GRIM-19, a novel cell death-regulatory gene induced by the interferon-beta and retinoic acid combination, using a genetic approach. J. Biol. Chem. 27533416-33426. [DOI] [PubMed] [Google Scholar]
- 4.Arvin, A., G. Campadielli-Fiume, E. Mocarski, P. S. Moore, B. Roizman, R. Whitley, and K. Yamanishi (ed.). 2007. Human herpesviruses: biology, therapy, and immunoprophylaxis. Cambridge University Press, Cambridge, United Kingdom. [PubMed]
- 5.Ashshi, A. M., R. J. Cooper, P. E. Klapper, O. Al-Jiffri, and L. Moore. 2000. Detection of human herpes virus 6 DNA in fetal hydrops. Lancet 3551519-1520. [DOI] [PubMed] [Google Scholar]
- 6.Barnich, N., T. Hisamatsu, J. E. Aguirre, R. Xavier, H. C. Reinecker, and D. K. Podolsky. 2005. GRIM-19 interacts with nucleotide oligomerization domain 2 and serves as downstream effector of anti-bacterial function in intestinal epithelial cells. J. Biol. Chem. 28019021-19026. [DOI] [PubMed] [Google Scholar]
- 7.Cardin, R. D., G. B. Abenes, C. A. Stoddart, and E. S. Mocarski. 1995. Murine cytomegalovirus IE2, an activator of gene expression, is dispensable for growth and latency in mice. Virology 209236-241. [DOI] [PubMed] [Google Scholar]
- 8.Charlot, J. F., J. L. Pretet, C. Haughey, and C. Mougin. 2004. Mitochondrial translocation of p53 and mitochondrial membrane potential dissipation are early events in staurosporine-induced apoptosis of wild type and mutated p53 epithelial cells. Apoptosis 9333-343. [DOI] [PubMed] [Google Scholar]
- 9.D'Agostino, D. M., P. Bernardi, L. Chieco-Bianchi, and V. Ciminale. 2005. Mitochondria as functional targets of proteins coded by human tumor viruses. Adv. Cancer Res. 9487-142. [DOI] [PubMed] [Google Scholar]
- 10.Dominguez, G., T. R. Dambaugh, F. R. Stamey, S. Dewhurst, N. Inoue, and P. E. Pellett. 1999. Human herpesvirus 6B genome sequence: coding content and comparison with human herpesvirus 6A. J. Virol. 738040-8052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Finkel, T. H., G. Tudor-Williams, N. K. Banda, M. F. Cotton, T. Curiel, C. Monks, T. W. Baba, R. M. Ruprecht, and A. Kupfer. 1995. Apoptosis occurs predominantly in bystander cells and not in productively infected cells of HIV- and SIV-infected lymph nodes. Nat. Med. 1129-134. [DOI] [PubMed] [Google Scholar]
- 12.Galluzzi, L., N. Zamzami, T. de La Motte Rouge, C. Lemaire, C. Brenner, and G. Kroemer. 2007. Methods for the assessment of mitochondrial membrane permeabilization in apoptosis. Apoptosis 12803-813. [DOI] [PubMed] [Google Scholar]
- 13.Geng, Y. Q., B. Chandran, S. F. Josephs, and C. Wood. 1992. Identification and characterization of a human herpesvirus 6 gene segment that transactivates the human immunodeficiency virus type 1 promoter. J. Virol. 661564-1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gompels, U. A., J. Nicholas, G. Lawrence, M. Jones, B. J. Thomson, M. E. Martin, S. Efstathiou, M. Craxton, and H. A. Macaulay. 1995. The DNA sequence of human herpesvirus-6: structure, coding content, and genome evolution. Virology 20929-51. [DOI] [PubMed] [Google Scholar]
- 15.Huang, G., H. Lu, A. Hao, D. C. Ng, S. Ponniah, K. Guo, C. Lufei, Q. Zeng, and X. Cao. 2004. GRIM-19, a cell death regulatory protein, is essential for assembly and function of mitochondrial complex I. Mol. Cell. Biol. 248447-8456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang, G., Y. Chen, H. Lu, and X. Cao. 2007. Coupling mitochondrial respiratory chain to cell death: an essential role of mitochondrial complex I in the interferon-beta and retinoic acid-induced cancer cell death. Cell Death Differ. 14327-337. [DOI] [PubMed] [Google Scholar]
- 17.Inoue, Y., M. Yasukawa, and S. Fujita. 1997. Induction of T-cell apoptosis by human herpesvirus 6. J. Virol. 713751-3759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Isegawa, Y., T. Mukai, K. Nakano, M. Kagawa, J. Chen, Y. Mori, T. Sunagawa, K. Kawanishi, J. Sashihara, A. Hata, P. Zou, H. Kosuge, and K. Yamanishi. 1999. Comparison of the complete DNA sequences of human herpesvirus 6 variants A and B. J. Virol. 738053-8063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Isegawa, Y., M. Takemoto, K. Yamanishi, A. Ohshima, and N. Sugimoto. 2007. Real-time PCR determination of human herpesvirus 6 antiviral drug susceptibility. J. Virol. Methods 14025-31. [DOI] [PubMed] [Google Scholar]
- 20.Janelle, M. E., and L. Flamand. 2006. Phenotypic alterations and survival of monocytes following infection by human herpesvirus-6. Arch. Virol. 1511603-1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kikuta, H., A. Nakane, H. Lu, Y. Taguchi, T. Minagawa, and S. Matsumoto. 1990. Interferon induction by human herpesvirus 6 in human mononuclear cells. J. Infect. Dis. 16235-38. [DOI] [PubMed] [Google Scholar]
- 22.Kondo, K., T. Kondo, T. Okuno, M. Takahashi, and K. Yamanishi. 1991. Latent human herpesvirus 6 infection of human monocytes/macrophages. J. Gen. Virol. 721401-1408. [DOI] [PubMed] [Google Scholar]
- 23.Krueger, G., and D. V. Ablashi (ed.). 2006. Human herpesvirus-6: general virology, epidemiology and clinical pathology, 2nd ed., vol. 12. Elsevier Science, Oxford, United Kingdom.
- 24.Kumleh, H. H., G. H. Riazi, M. Houshmand, M. H. Sanati, K. Gharagozli, and M. Shafa. 2006. Complex I deficiency in Persian multiple sclerosis patients. J. Neurol. Sci. 24365-69. [DOI] [PubMed] [Google Scholar]
- 25.Leong, W. F., and V. T. Chow. 2006. Transcriptomic and proteomic analyses of rhabdomyosarcoma cells reveal differential cellular gene expression in response to enterovirus 71 infection. Cell. Microbiol. 8565-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lim, K. M., W. S. Yeo, and V. T. Chow. 2004. Antisense abrogation of DENN expression induces apoptosis of leukemia cells in vitro, causes tumor regression in vivo and alters the transcription of genes involved in apoptosis and the cell cycle. Int. J. Cancer 10924-37. [DOI] [PubMed] [Google Scholar]
- 27.Mayne, M., C. Cheadle, S. S. Soldan, C. Cermelli, Y. Yamano, N. Akhyani, J. E. Nagel, D. D. Taub, K. G. Becker, and S. Jacobson. 2001. Gene expression profile of herpesvirus-infected T cells obtained using immunomicroarrays: induction of proinflammatory mechanisms. J. Virol. 7511641-11650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Merelli, E., P. Sola, P. Barozzi, and G. Torelli. 1996. An encephalitic episode in a multiple sclerosis patient with human herpesvirus 6 latent infection. J. Neurol. Sci. 13742-46. [DOI] [PubMed] [Google Scholar]
- 29.Miguet-Alfonsi, C., C. Prunet, S. Monier, G. Bessede, S. Lemaire-Ewing, A. Berthier, F. Menetrier, D. Neel, P. Gambert, and G. Lizard. 2002. Analysis of oxidative processes and of myelin figures formation before and after the loss of mitochondrial transmembrane potential during 7-beta-hydroxycholesterol and 7-ketocholesterol-induced apoptosis: comparison with various pro-apoptotic chemicals. Biochem. Pharmacol. 64527-541. [DOI] [PubMed] [Google Scholar]
- 30.Mills, E. M., D. Xu, M. M. Fergusson, C. A. Combs, Y. Xu, and T. Finkel. 2002. Regulation of cellular oncosis by uncoupling protein 2. J. Biol. Chem. 27727385-27392. [DOI] [PubMed] [Google Scholar]
- 31.Morrison, M. L., K. Williamson, K. Arthur, G. J. Price, P. W. Hamilton, and P. Maxwell. 2005. Phenotypic changes in mitochondrial membrane potential during valinomycin-induced depolarisation and apoptosis. Cell. Oncol. 27231-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nicholas, J., and M. E. Martin. 1994. Nucleotide sequence analysis of a 38.5-kilobase-pair region of the genome of human herpesvirus 6 encoding human cytomegalovirus immediate-early gene homologs and transactivating functions. J. Virol. 68597-610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Raftery, M., A. Muller, and G. Schonrich. 2000. Herpesvirus homologues of cellular genes. Virus Genes 2165-75. [PubMed] [Google Scholar]
- 34.Schirmer, E. C., L. S. Wyatt, K. Yamanishi, W. J. Rodriguez, and N. Frenkel. 1991. Differentiation between two distinct classes of viruses now classified as human herpesvirus 6. Proc. Natl. Acad. Sci. USA 885922-5926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Seo, T., D. Lee, Y. S. Shim, J. E. Angell, N. V. Chidambaram, D. V. Kalvakolanu, and J. Choe. 2002. Viral interferon regulatory factor 1 of Kaposi's sarcoma-associated herpesvirus interacts with a cell death regulator, GRIM19, and inhibits interferon/retinoic acid-induced cell death. J. Virol. 768797-8807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shapiro, H. M. 2000. Membrane potential estimation by flow cytometry. Methods 21271-279. [DOI] [PubMed] [Google Scholar]
- 37.Shishido, S., H. Koga, M. Harada, H. Kumemura, S. Hanada, E. Taniguchi, R. Kumashiro, H. Ohira, Y. Sato, M. Namba, T. Ueno, and M. Sata. 2003. Hydrogen peroxide overproduction in megamitochondria of troglitazone-treated human hepatocytes. Hepatology 37136-147. [DOI] [PubMed] [Google Scholar]
- 38.Sim, D. L., W. M. Yeo, and V. T. Chow. 2002. The novel human HUEL (C4orf1) protein shares homology with the DNA-binding domain of the XPA DNA repair protein and displays nuclear translocation in a cell cycle-dependent manner. Int. J. Biochem. Cell. Biol. 34487-504. [DOI] [PubMed] [Google Scholar]
- 39.Takahashi, K., S. Sonoda, K. Higashi, T. Kondo, H. Takahashi, M. Takahashi, and K. Yamanishi. 1989. Predominant CD4 T-lymphocyte tropism of human herpesvirus 6-related virus. J. Virol. 633161-3163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Takemoto, M., T. Shimamoto, Y. Isegawa, and K. Yamanishi. 2001. The R3 region, one of three major repetitive regions of human herpesvirus 6, is a strong enhancer of immediate-early gene U95. J. Virol. 7510149-10160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tan, E. L., T. M. Tan, V. T. Chow, and C. L. Poh. 2007. Enhanced potency and efficacy of 29-mer shRNAs in inhibition of enterovirus 71. Antivir. Res. 749-15. [DOI] [PubMed] [Google Scholar]
- 42.Thompson, J., S. Choudhury, F. Kashanchi, J. Doniger, Z. Berneman, N. Frenkel, and L. J. Rosenthal. 1994. A transforming fragment within the direct repeat region of human herpesvirus type 6 that transactivates HIV-1. Oncogene 91167-1175. [PubMed] [Google Scholar]
- 43.Thomson, B. J., and R. W. Honess. 1992. The right end of the unique region of the genome of human herpesvirus 6 U1102 contains a candidate immediate early gene enhancer and a homologue of the human cytomegalovirus US22 gene family. J. Gen. Virol. 731649-1660. [DOI] [PubMed] [Google Scholar]
- 44.Yamamoto, T., T. Mukai, K. Kondo, and K. Yamanishi. 1994. Variation of DNA sequence in immediate-early gene of human herpesvirus 6 and variant identification by PCR. J. Clin. Microbiol. 32473-476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yeo, W. M., and V. T. Chow. 2007. The VP1 structural protein of enterovirus 71 interacts with human ornithine decarboxylase and gene trap ankyrin repeat. Microb. Pathog. 42129-137. [DOI] [PubMed] [Google Scholar]