Abstract
In retroviruses, the first nucleotide added to the tRNA primer defines the end of the U5 region in the right long terminal repeat, and the subsequent removal of this tRNA primer by RNase H exactly defines the U5 end of the linear double-stranded DNA. In most retroviruses, the entire tRNA is removed by RNase H cleavage at the RNA/DNA junction. However, the RNase H domain of human immunodeficiency virus type 1 (HIV-1) reverse transcriptase cleaves the tRNA 1 nucleotide from the RNA/DNA junction at the U5/primer binding site (PBS) junction, which leaves an rA residue at the U5 terminus. We made sequence changes at the end of the U5 region adjacent to the PBS in HIV-1 to determine whether such changes affect the specificity of tRNA primer cleavage by RNase H. In some of the mutants, RNase H usually removed the entire tRNA, showing that the cleavage specificity was shifted by 1 nucleotide. This result suggests that the tRNA cleavage specificity of the HIV-1 RNase domain H depends on sequences in U5.
Reverse transcription is initiated from a cellular tRNA primer that is base paired at the primer binding site (PBS) near the 5′ end of the viral genome. The tRNA used to prime reverse transcription in human immunodeficiency virus type 1 (HIV-1) is tRNALys (8, 25, 28, 31, 26, 29). After the 5′ end of genomic RNA is copied, the freshly copied RNA is degraded by the RNase H activity of reverse transcriptase (RT) and minus-strand DNA is transferred to the 3′ end of a viral genome, allowing minus-strand DNA synthesis to continue. The synthesis of the plus-strand DNA is initiated from the polypurine tract (PPT) primer that is generated by specific RNase H cleavages of the RNA genome adjacent to U3. Plus-strand synthesis proceeds until the first 18 nucleotides of the tRNA are reverse transcribed. DNA synthesis stops at a 1-methyl-adenosine residue in the tRNA that cannot serve as a template for reverse transcription (29, 36). This creates an RNA/DNA duplex, and the tRNA primer is removed from the minus-strand DNA by RNase H, which defines the right (U5) long terminal repeat (LTR) terminus of the linear double-stranded DNA. In most retroviruses, RNase H removes the entire tRNA. Most retroviruses have two nucleotides between the tRNA PBS and the CA dinucleotide found at the end of the integrated provirus, and in general, the last two nucleotides are removed from each of the 3′ ends of the linear DNA intermediate during integration. However, in the case of HIV-1, there is only one nucleotide (G) between the tRNA PBS and the CA dinucleotide. The RNase H domain of HIV-1 RT cleaves the tRNALys primer between the terminal rA and the adjacent rC, 1 nucleotide from the RNA-DNA junction, which leaves the rA residue at the U5 terminus of the linear DNA (8, 20, 25, 28, 33). HIV-1 integrase (IN) then removes the last two nucleotides from the 3′ end of the plus strand at the U5 terminus, one of which, the terminal nucleotide dT, is base paired with the rA nucleotide on the minus strand.
What determines the specificity of the HIV-1 RNase H domain, causing it to cleave 1 nucleotide from the end of the tRNALys primer? The RNase H domain of HIV-1 RT can cleave at an RNA/DNA junction; the RNase H domain cleaves the PPT primer at the RNA/DNA junction which defines the left (U3) end of the linear viral DNA (1, 4, 15). Furthermore, at least in vitro, the RNase H domain of HIV-1 RT can cleave a model substrate in which the 3′-terminal AMP of the tRNA primer was changed from rA to dA at the RNA/DNA junction (immediately after the dA) (28).
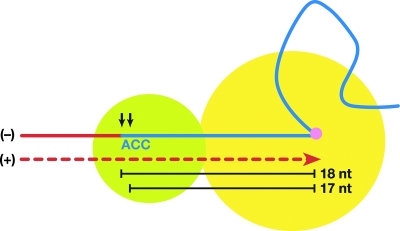
There is a simple model which could explain the specificity of tRNA cleavage by retroviral RTs (Fig. 1). The crystal structure and biochemical studies of HIV-1 RT show that the polymerase (Pol) and RNase H active sites are 17 or 18 bp apart (9, 10, 11, 12, 13, 14, 16, 26, 27, 34). The distance between the 1-methyladenosine on the tRNA primer and the U5/PBS junction is 18 base pairs (29, 36). In the model, the cleavage of the tRNA occurs when the Pol is stalled at the 1-methyladenosine and the position of the cleavage is determined by the tRNA/PBS sequence and the physical properties of RT. If this model is correct, the U5 sequences are not expected to have a significant impact on the site of cleavage. This idea is supported by the structure of HIV-1 RT in a complex with an RNA/DNA substrate (26): the vast majority of the contacts between the nucleic acid and the enzyme are 5′ (on the template strand) of the RNase H cleavage site. We previously showed that tRNA cleavages by the RNase H domain of Rous sarcoma virus (RSV) RT conformed to this model: as expected, changing the sequences in U5 had no effect on where RNase H cleaved the tRNA primer. We replaced the sequence (TT) that is normally present between the PBS and the CA dinucleotide of the RSV-derived vector RSVP(A)Z so that it would match the HIV-1 sequence (G) and the HIV-2 sequence (GGT). In all of the mutants, the RNase H domain of the RSV RT consistently removed the entire tRNA, which suggests that mutations in the U5 terminus did not affect tRNA primer cleavage by RSV RNase H (23).
FIG. 1.
A simple model for the specificity of the tRNA cleavage by RNase H. Pol is shown in a yellow circle, and RNase H is shown in a green circle. The tRNA primer is indicated by a blue line, and last three nucleotides (nt), CCA, are shown. A 1-methyladenosine residue in the tRNA is indicated by a purple circle. DNA strands are shown as red lines, and minus (−)/plus (+) strands are indicated. The two alternative RNase H cleavages, at the RNA/DNA junction and in the tRNA 1 nucleotide from the junction, are shown as black arrows. The distance between the 1-methyladenosine on the tRNA primer and the RNase H active sites are shown as black lines.
We show here that, when the single nucleotide (G) that is between the PBS and the CA dinucleotide of HIV-1 was changed, the specificity of the RNase H cleavage was affected. In some of the HIV-1 U5 mutants, RNase H removed the entire tRNA in the majority of the cleavage events. This suggests, contrary to what was proposed in the simple model, that the specificity of the cleavage of the tRNA primer by the RNase H domain of HIV-1 RT depends on sequences on both sides of the DNA/RNA junction at the U5/PBS junction. Sequence analysis of circular viral DNAs derived from autointegration events shows that HIV-1 IN can remove either a single nucleotide or three nucleotides beyond the CA dinucleotide from the U5 terminus of the linear viral DNA. In addition, IN can remove two nucleotides if the canonical CA is mutated to AT. In all of the mutants, the processed DNAs were properly inserted into the viral DNA, creating a 5-bp duplication at the target site in concerted autointegration events, showing that the conserved CA is not essential for IN-mediated viral integration.
MATERIALS AND METHODS
Plasmid construction.
pHIV1-SH was derived from pNLNgoMIVR-E-.HSA (17). A cassette containing the zeocin resistance gene, a lac operator sequence, and a ColE1 replication origin was PCR amplified from the RSVP plasmid (24) by using two primers, Not-Cla (5′-GCATGCGGCCGCATCGATTTGTTGACAATT) and Xho-Mlu (5′-GATCCTCGAGACGCGTCCCGTAG). The resulting PCR fragment was digested with NotI and XhoI and used to replace the NotI/XhoI fragment containing the HSA coding sequence of pNLNgoMIVR-E-.HSA, which generated pNLNgoMIVR-E-(Cass). The murine RNA Pol II promoter was cloned into the NotI-to-ClaI site of pNLNgoMIVR-E-(Cass), which generated pNLNgoMIVR-E-(Pol-Cass). To remove a nonviral region from the parental plasmid, pNLNgoMIVR-E-(Pol-Cass) was digested with PstI and BspMI and a 6.7-kb BspMI fragment containing the pol-env-Pol II-cassette-LTR region of pNLNgoMIVR-E-(Pol-Cass) was ligated with two complementary oligonucleotide sequences (FSP1, 5′-GGCCGAATTCCTGCAGAGTACG, and FSP2, 5′-TGTTCGTACGCTGCAGGAATTC) that included FspI sites, indicated by the underlines, which generated HIV1-SH(part1). The FspI site was used in a subsequent cloning step. To remove most of the env gene, pNLNgoMIVR-E-.HSA was digested with SalI and NotI and ligated with two sets of PCR-amplified products. The first PCR product was amplified with the For1 primer (5′-TTACGGGGATACTTGGGCAGGAGTGG), which anneals at position 5701 (upstream of SalI), and the Rev1 primer (5′-GCTTAAGCTAGCTCTATTAGTGGTGGTTGCTTCCTTCCACACAGG), which anneals at position 6374 (150 bp downstream of the env start codon). The second PCR product was amplified with the For2 primer (5′-GTCAATGCTAGCGGAGGCTTGGTAGGTTTAAGAATAG), which anneals to position 8281, and the Rev2 primer (5′-TCTTGTCTTCTTTGGGAGTGAATTAGCC), which anneals to position 9111 in U3. The Rev1 and For2 primers contained NheI sites (underlined), which were used in a subsequent three-piece ligation step. PCR product set 1 was digested with SalI and NheI, and PCR product set 2 was digested with NheI and NotI. The PCR products were then ligated into the parental vector pNLNgoMIVR-E-.HSA, which was previously digested with SalI and NotI, to generate pNLNgoMIVR-E-.HSA(Δenv). The RRE sequence was inserted as a NheI-to-NotI fragment derived from pNLNgoMIVR-E-.HSA, which replaced the NheI-to-NotI fragment of pNLNgoMIVR-E-.HSA(Δenv), to generate HIV1-SH(part2). Finally, HIV-SH(part1) was digested with NotI and FspI and ligated to the FspI-to-NotI fragment of HIV1-SH(part2) to generate pHIV1-SH.
Construction of the U5 end mutants.
KS(AatII) was derived from the pBluescript II KS phagemid by replacing a SacI-to-SpeI segment with two complementary oligonucleotide sequences that included AatII sites. To introduce mutations at the end of the U5 sequence, a 3-kb AatII-to-SpeI fragment containing a nonviral sequence, the 5′ LTR, the PBS, and the gag region of pNLNgoMIVR-E-.HSA was inserted into the AatII-to-SpeI site of KS(AatII), which generated KS/HIV1. In vitro site-directed mutagenesis was carried out, with KS/HIV1 as a template and the appropriate set of primers, by using a QuikChange site-directed mutagenesis kit (Stratagene), following the manufacturer's recommendations. Sequence analysis of the resulting plasmids (KS/HIV1-mu) was performed to ensure that only desired mutations were introduced. The 1.6-kb FspI-to-SpeI fragment of KS/HIV1-mu was used to replace the corresponding FspI-to-SpeI fragment in pHIV1-SH, which generated the viral vectors containing the appropriate mutations.
Cells, transfection, and infection.
The human embryonal kidney cell line 293 and the human osteosarcoma cell line HOS were maintained in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 5% fetal bovine serum, 5% newborn calf serum, 100 U of penicillin per ml, and 100 μg of streptomycin per ml (Quality Biological). 293 cells were plated in 100-mm-diameter dishes at a density of 1.8 × 106 cells per plate on the day prior to transfection. 293 cells were transfected with 5 to 10 μg of the pHIV-SH vectors and 2 μg of pHCMV-g (obtained from Jane Burns, University of California at San Diego) by using a calcium phosphate transfection kit (Invitrogen), following the manufacturer's recommendations. The precipitate was added to 293 cells drop-wise. The medium was changed 24 h after transfection. The 48-h supernatants were harvested, subjected to low-speed centrifugation, and filtered through a Millex-GS 0.22-μm filter (Millipore) to remove cellular debris. A portion of the filtered infectious virions was used to infect HOS cells.
Determination of relative virus titer.
Immediately after the virus-containing supernatant was harvested, the 293 cells were lysed in 5 ml Glo-lysis buffer (Promega) and the luciferase activity in the 293 cells was determined using Steady-Glo (Promega). The luciferase activity in the transfected cells was used to normalize the amount of virus. HOS cells were plated in six-well cluster plates the day before infection at a density of 0.8 × 105 cells per well. The virions were allowed to adsorb to the HOS cells for 2 h, and then fresh medium was added. Forty-eight hours after infection, the cells were washed once with 2 ml phosphate-buffered saline and then lysed in 0.5 ml of Glo-Lysis buffer (Promega). The luciferase activity was measured using the Steady-Glo reagent system (Promega). The relative virus titers were determined from the linear range of dilution.
Unintegrated viral DNA analysis.
The circularized unintegrated viral DNA was isolated from the HOS cells ca. 24 h after infection by using a DNeasy tissue kit (Qiagen). A portion of the recovered DNA was used to transform ElectroMax DH10B (Invitrogen) by electroporation. Electroporation was performed as described previously (24). The transformed bacteria were plated onto Luria-Bertani (LB) plates containing 50 μg of zeocin per ml. Recovered plasmids were directly sequenced using the following primers: PBS-UP (5′-TGGCGTACTCACCAGTCGCCG) for the U3 junction and LTR-5 (5′-CATGGAGCAATCACAAGTAGC) for the U5 junction. The PBS-UP primer was also used to confirm that the U5 end mutations that we introduced were present in the recovered plasmids.
Sequencing the integration sites.
Recovered plasmids were directly sequenced using the following primers: the PBS primer for the U3 integration sites (5′-ACTATCACGTCGGGGTCACCA) and the Ori-2 primer for the U5 integration sites (5′-AGGAGTCCCCTTAGGATATAG). The Ori-2 primer was also used to confirm that the PPT mutations that we introduced were present in the recovered plasmids. In those cases in which the viral DNA was flanked by human genomic sequences, they were analyzed by BLAT searches (http://genome.ucsc.edu/cgi-bin/hgBlat).
RESULTS AND DISCUSSION
Construction of pHIV1-SH.
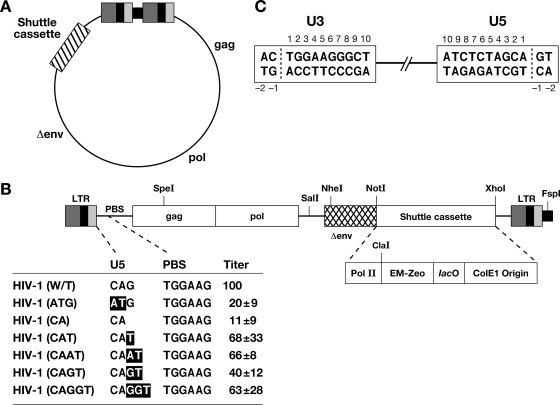
We constructed pHIV1-SH by inserting a cassette (RNA Pol II promoter-zeocin resistance gene-lac operator-ColE1) into the pNLNgoMIVR-E-.HSA vector, which was derived from the pNL4-3 isolate of HIV-1 (17) (Fig. 2). Most of the nonviral sequences present in the parental plasmid, pNLNgoMIVR-E-.HSA, including the ampicillin resistance gene and a bacterial origin of replication, were removed. pHIV1-SH contains a selectable drug resistance gene (zeocin) and a bacterial origin of replication (ColE1) inserted into the viral genome (Fig. 2). In mammalian cells, the zeocin resistance gene is expressed from a Pol II promoter; when the viral DNA replicates as a plasmid in bacteria, the same gene is expressed from the synthetic EM-7 bacterial promoter. We deleted a portion of the env gene, limiting the vector to a single cycle of replication; the deleted env gene cannot revert if pHIV1-SH is complemented with vesicular stomatitis virus G glycoprotein (VSV-G).
FIG. 2.
Schematic drawing showing the structure of the pHIV1-SH vector. (A) pHIV1-SH. The viral genes gag and pol and partially deleted env are shown (not to scale). The cassette is indicated by a box with slashes. (B) Schematic representation of the cassette and the mutants. Mutations are indicated in bold. The virus titers, normalized to the luciferase activity, are shown relative to that of the WT virus. These results are the averages from three independent experiments. Pol II, murine RNA Pol II promoter; EM-Zeo, EM-7 promoter-zeocin resistance gene; lacO, lac operator. A nonviral sequence derived from the parental vector is indicated by a small black box at the right end of the drawing. (C) Schematic numbering of the nucleotide positions in the LTR termini of the circles derived from autointegration events.
The U5 end mutations and virus titer.
We changed the sequence of pHIV1-SH (G) between the PBS and the CA dinucleotide normally found at the U5 end of the provirus to determine whether the U5 end mutations affect the cleavage specificity of the HIV-1 RNase H domain. We also changed the CA dinucleotide to AT to determine how this mutation affects RNase H cleavage and autointegration. To determine the effects of the U5 end mutations on relative virus titer, we inserted the luciferase gene into the nef reading frame of the pHIV1-SH-based vectors. Viruses were generated by cotransfecting 293 cells with the HIV-1 vectors and pHMCV-g, a plasmid that expresses the VSV glycoprotein (3, 35). This generated VSV-G-pseudotyped virions that were harvested and used to infect human osteosarcoma (HOS) cells as described in Materials and Methods. The infected HOS cells were lysed, and the luciferase activity in the cell lysates was measured to determine the virus infectivity. The relative titers were normalized using the level of luciferase expression in the transfected 293 cells. The U5 mutations have different effects on virus titers. The HIV-1(ATG) and HIV-1(CA) mutations decreased virus titers to 20% and 11% of the wild-type (WT) level, respectively. The result with the HIV(ATG) mutant agrees with published data showing that mutating the conserved CA in U5 had a relatively modest effect on viral titer (5, 22). The HIV-1(CAT), HIV-1(CAAT), HIV-1(CAGT), and HIV-1(CAGGT) mutations had smaller effects on the relative titers; the titers of these mutants were from 40% to 68% of the WT level (Fig. 2).
Effects of the mutations on the initiation of viral DNA synthesis.
Because the mutations were in the region of the viral genome where minus-strand DNA synthesis is initiated, it is possible that the mutations had an impact on initiation. Cells were infected with the version of the mutant vectors that carried the luciferase gene so that the titer and the initiation of viral DNA synthesis could be measured on the same samples by using real-time PCR to measure the level of RU5 DNA in the infected cells at various times after infection (19). The limit of accuracy for this assay is approximately twofold, so that for the mutations with relative titers within twofold of the WT level [HIV-1(CAT), HIV-1(CAAT), and HIV-1(CAGT)], there were no obvious differences in the level of RU5 DNA made in cells infected with the mutant relative to that in cells infected with the WT vector. The HIV-1(CAGGT) mutant had a titer of 40% of the WT vector level. This is near the limit of accuracy for the assay; however, in two separate assays, the level of RU5 DNA was slightly lower than the WT level (two- to threefold) at 2, 4, 6, and 24 h after infection. This suggests that the HIV-1(CAGGT) mutation has a modest effect on the initiation of viral DNA synthesis and that this may account for the reduced titer seen with the vector carrying this mutation. The level of RU5 DNA for the HIV-1(CA) mutant was, in two separate assays, approximately fivefold lower than the WT level at 2, 4, 6, and 24 h after infection, which suggests that this mutation has an effect on the initiation of viral DNA and contributes to the reduced titer of the mutant virus. The level of the RU5 DNA seen in cells infected with the HIV-1(ATG) mutant was not reproducibly lower than that in cells infected with the WT vector (data not shown).
Recovery and analysis of 2-LTR circle junctions.
To recover 2-LTR circle junctions, 293 cells were cotransfected with the pHIV1-SH vectors containing the U5 end mutations and with pHCMV-g. The virions were harvested and used to infect HOS cells. Total DNA was prepared from the infected cells 24 h after infection and used to transform Escherichia coli; zeocin-resistant cells were selected. The 2-LTR circles that we isolated were different from the starting plasmid that was used for transfection. The starting vector contained a nonviral sequence derived from the parental plasmid, pNLNgoMIVR-E-.HSA, between the two LTRs. This nonviral sequence was absent when we recovered the 2-LTR circles from infected cells, showing that the 2-LTR circles that we recovered came from successful replication of pHIV-SH RNA and were not carryovers of the pHIV-SH plasmid used in the initial transfection.
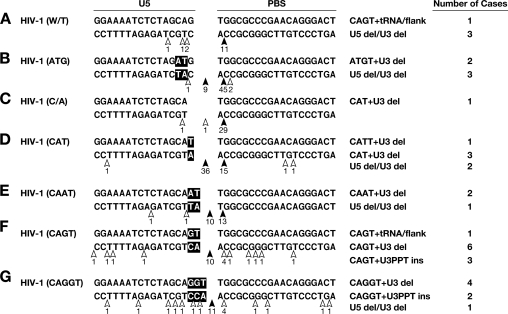
Analysis of the 2-LTR circle junctions obtained from infections with the WT and the mutant vectors is summarized in Fig. 3. In the case of the WT, as expected, HIV-1 RNase H usually cleaved the tRNA between the terminal rA and the adjacent rC, 1 nucleotide from the RNA-DNA junction (Fig. 3A). This cleavage specificity did not change when the single nucleotide (G) that lies between the CA dinucleotide and the PBS was deleted (Fig. 3C). The RNase H cleavage specificity was modestly altered when the CA dinucleotide was changed to AT (Fig. 3B) (most of the cleavages occurred 1 nucleotide from the end of the tRNA primer); however, in 9 of 62 2-LTR circles derived from infection with the HIV-1(ATG) mutant, it appeared that cleavage occurred at the RNA/DNA junction and thus that RNase H removed the entire tRNA. The RNase H cleavage that removed the entire tRNA was seen more frequently with some of the other mutants. In the case of HIV-1(CAT), in which the sequence (G) between the CA and the PBS was changed to T, RNase H removed the entire tRNA in 39 of 60 of the 2-LTR circle junctions (Fig. 3D). This was twice the number of circle junctions that retained the A from the tRNA (16 of 60). There were, in addition, 2-LTR circle junctions that contained insertions and/or deletions. However, when the G was replaced by AT in HIV-1(CAAT), changing not only the spacing but also the sequence (Fig. 3E), RNase H removed the entire tRNA in 12 of the 28 circle junctions and cleaved the tRNA 1 nucleotide from the RNA-DNA junction in 13 of 28 circle junctions. When the G was changed to GT in the mutant HIV-1(CAGT), RNase H removed the entire tRNA in 19 of 33 circle junctions and cleaved the tRNA 1 nucleotide from the RNA-DNA junction in only 4 of 33 circle junctions (Fig. 3F). Similar results were obtained with HIV-1(CAGGT), in which the G was changed to GGT, increasing the spacing between the CA and the PBS (Fig. 3G). RNase H removed the entire tRNA in 17 of 33 circle junctions and cleaved the tRNA 1 nucleotide from the RNA-DNA junction in 4 of 33 circle junctions.
FIG. 3.
Cleavage of the tRNA primer in the WT and mutant vectors by HIV-1 RT. Mutations are indicated in bold. The arrows depict the positions of the cleavage. The filled arrows denote more-frequent cleavages. Each of the mutant viruses also gave rise to some aberrant 2-LTR circle junctions. These aberrant junctions were classified according to the nature of the aberration (del is deletion, ins is insertion); these data are shown on the right side of the figure for each of the mutants.
As has already been mentioned in the introduction, there is a simple model that has been proposed to explain the incomplete removal of the tRNALys primer by HIV-1 RNase H. In this model, the RNase H domain cleaves the tRNALys primer a fixed distance from the Pol active site after RT has copied the first 18 nucleotides of the tRNA primer (Fig. 1) (2, 13). Crystallographic data show that the Pol and RNase H active sites are 18 bp apart for an HIV-1 RT-RNA DNA complex and 17 bp apart for an HIV-1 RT-DNA DNA complex; this is in good agreement with where RNase H cleaves model substrates in vitro (9, 10, 11, 12, 13, 14, 15, 26, 27, 34). The overall structures of the nucleic acid template primers in the RT-DNA-DNA and RT-RNA-DNA complexes (PPT versus PBS) are remarkably similar. Both have a bend in the nucleic acid that occurs in a segment 5 to 9 bp from the Pol active site. The majority of the protein-nucleic acid contacts occur near the RT Pol active site in the Pol domain and in the thumb; there are additional contacts in the connection domain and in the RNase H primer grip in the RNase H domain (6, 7, 14, 16, 26). The RNase H primer grip serves to position the DNA primer strand near the RNase H active site and makes several contacts with the nucleic acid near the scissile phosphate (26). However, there is little contact on either strand of the nucleic acid beyond (3′ on the RNA strand) the RNase H active site. As described in the introduction, this suggests a model in which the tRNA cleavage specificity of retroviral RNase H should depend on the distance between the two active sites and on the portion of the template primer that lies between the Pol and RNase H active sites where there are direct contacts between the nucleic acid and the protein (Fig. 1). In this model, RNase H cleavage specificity is not expected to depend on the sequences in U5, which lie beyond the cleavage site. The model is supported by data obtained from RSV mutants (23). In the case of RSV, mutations in U5 did not affect the specificity of the cleavage that removes the tRNA. However, in the case of HIV-1, when the U5 sequence was changed, the primary site of tRNA cleavage was shifted by 1 nucleotide, suggesting that the tRNA cleavage specificity of the HIV-1 RNase H domain does depend on sequences in U5 beyond the RNase H cleavage site.
Based on the crystallographic data, HIV-1 RT residues that contact the RNA template strand near the scissile phosphate include the side chains of R448, N474, Q475, and H539 of p66 (26). Mutations in amino acids R448 and N474 had minimal effects on titer; H539 is part of the active site. Q475 was the only other amino acid that contacts the nucleic acid near the active site where a mutation had a significant effect on viral titer (18). However, Q475 contacts the RNA template 5′ of the scissile phosphate. Given the lack of significant contacts 3′ of the scissile phosphate (on the RNA strand), it is not obvious why/how the mutations in U5 affect the specificity of the tRNA cleavage. One possibility is that altering the U5 sequence affects the structure of the adjacent RNA/DNA duplex, which in turn affects the specificity of the cleavage. However, making similar mutations in the U5 sequence of an RSV had no measurable effect of the specificity of RNase H cleavage. There are two obvious differences in these two situations: the tRNA primers are different (and hence the two nucleic acids have different sequences), and the RTs, while related, are also different. In terms of the sequences, all tRNAs have the same three nucleotides, CCA, at their 3′ ends. This suggests, but does not prove, that the critical difference lies either in the two RTs or, more likely, in differences in the ways in which the RTs interact with their respective tRNA/plus-strand cleavage substrates.
Recovery of unusual circular viral DNAs.
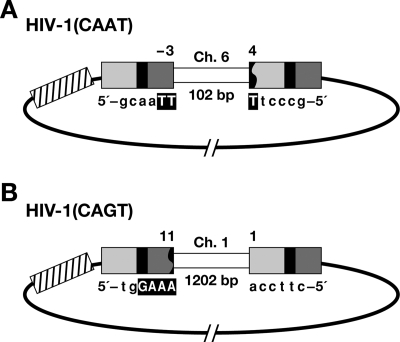
We recovered two circular viral DNAs that contained host DNA sequences (Fig. 4). In one of the viral DNAs recovered from an infection with HIV-1(CAAT), three nucleotides of the U5 LTR terminus were retained beyond the CA sequence and were joined to a segment from human chromosome 6. However, we cannot define the exact junction, because the last two nucleotides at the U5 LTR terminus were identical to the first two nucleotides of the host DNA. The U3 terminus was deleted, and the truncated U3 was joined to the segment from human chromosome 6. The segment of the host DNA between the ends of the viral DNA was 102 bp long (Fig. 4A). We also recovered a circular DNA from an infection with HIV-1(CAGT) in which the U3 terminus was properly processed and joined to a segment from human chromosome 1. The U5 terminus was deleted and joined to the segment from human chromosome 1. The host DNA segment captured between the ends of the viral DNA was 1,202 bp long (Fig. 4B). Both of these captured segments probably arose from abortive integration events in which one end of the viral DNA was properly integrated and the other end was then joined to the host DNA in a way that captured the host DNA between the two LTRs (23).
FIG. 4.
Structure of unusual circular viral DNAs. (A, B) U3 is indicated by a light gray box; R is indicated by a black box; U5 is shown by a dark gray box. The cassette is indicated by a box with slashes. Deletions in the LTR are indicated by a black jagged end. Six nucleotides at the viral/host DNA junctions are shown, and a microhomology with the host DNA is indicated by a bold capital letter. The captured host DNA segments are indicated by white boxes.
Recovery and analysis of autointegrations.
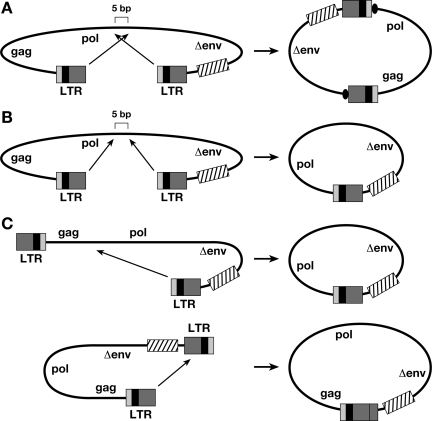
When we recovered the 2-LTR viral DNA circles, in addition to 1-LTR circles, we also obtained circular viral DNAs derived from autointegration events (Table 1). The circles appear to arise from two types of autointegration events (Fig. 5). The first is a concerted autointegration event in which both ends are processed correctly and inserted into the viral DNA in a reaction that generates a 5- or 6-bp duplication at the target site (Fig. 5A and Table 2). This type of event requires that the LTRs be inserted into the viral DNA in an orientation opposite to their own polarity. The second type is a one-ended autointegration event. This event can arise from a reaction in which one end is processed correctly and is inserted into the viral DNA, generating a smaller circle that contains only one LTR (Fig. 5C), or from a concerted integration event in which both LTRs are inserted into the viral DNA in a concerted reaction with the same polarity as the LTRs (Fig. 5B). Although the reactions catalyzed by IN do not give rise to the final circular forms shown in Fig. 5, the IN-mediated joining reactions take place in the nucleus. The nucleus contains host enzymes that can complete the circularization reactions in the same way that host enzymes complete the IN-mediated joining reactions that give rise to the mature provirus. As expected, we recovered more single-end insertions that involved U5 end joinings, because the cassette containing the drug resistance gene and the bacterial origin of replication is nearer the U5 LTR terminus of the vector.
TABLE 1.
Recovered circles
| Virus | No. (%) of circles
|
|||||
|---|---|---|---|---|---|---|
| Total | 1 LTR | 2 LTR | Other | Sequenceda | Autointegrationb | |
| HIV-1 WT | 208 | 91 (44) | 19 (9) | 98 (47) | 36 | 29 |
| HIV-1(ATG) | 212 | 113 (53) | 62 (29) | 37 (18) | 13 | 9 |
| HIV-1(CA) | 126 | 63 (50) | 32 (25) | 31 (25) | 18 | 4 |
| HIV-1(CAT) | 217 | 110 (50) | 60 (28) | 47 (22) | 20 | 13 |
| HIV-1(CAAT) | 295 | 128 (43) | 28 (10) | 139 (47) | 46 | 29 |
| HIV-1(CAGT) | 337 | 174 (52) | 33 (10) | 130 (38) | 59 | 44 |
| HIV-1(CAGGT) | 252 | 142 (56) | 29 (12) | 81 (32) | 46 | 31 |
For circles designated “Other,” a portion of the circles was sequenced.
Of those sequenced, the number of circles resulting from autointegration is indicated; this number is different from the total number of sequenced circles because there were sequence failures.
FIG. 5.
Two types of autointegrations. (A, B) Concerted autointegrations. A 5-bp duplication of viral sequence is indicated by a black oval. (C) One-end autointegration. U3 is indicated by a light-gray line; R is indicated by a black box; U5 is shown by a dark gray box. The cassette is indicated by a box with slashes.
TABLE 2.
Concerted autointegrations
| Virus | Position of the integrationa
|
Length of duplication (bp) | No. of casesb | |
|---|---|---|---|---|
| U3 | U5 | |||
| HIV-1 WT | 1 | 1 | 6 | 1 (pol-env) |
| 1 | 1 | 5 | 18 (7 pol, 4 pol-env, 6 env, 1 Pol II) | |
| HIV-1(ATG) | 1 | 1 | 5 | 4 (3 pol, 1 pol-env) |
| HIV-1(CA) | 1 | 1 | 5 | 2 (1 pol, 1 env) |
| HIV-1(CAT) | 1 | 1 | 5 | 8 (3 pol, 2 pol-env, 2 env, 1 Pol II) |
| HIV-1(CAAT) | 1 | 1 | 5 | 18 (10 pol, 3 pol-env, 3 env, 2 Pol II) |
| HIV-1(CAGT) | 1 | 1 | 5 | 22 (13 pol, 1 pol-env, 7 env, 1 Pol II) |
| HIV-1(CAGGT) | 1 | 1 | 5 | 17 (10 pol, 4 pol-env, 3 Pol II) |
| 1 | 1 | 9 | 1 (pol) | |
The last nucleotide at each LTR end of the circles is indicated by a number, using the numbering system in Fig. 1C.
The positions of autointegrations are indicated. pol-env, between pol and env; Pol II, RNA Pol II promoter.
Circles recovered from the HIV-1(CAT) and HIV-1(CA) mutants showed that HIV-1 IN can remove a single nucleotide beyond the CA and properly integrate the processed end (Table 1 and Table 3). This result is in agreement with previous in vitro observations showing that HIV-1 IN can cleave 1 nucleotide 3′ of the CA dinucleotide (30). The HIV-1(CA) mutation reduced the virus titer to 10% of the WT level (Fig. 2). The reduction in the initiation of viral DNA synthesis is sufficient to account for at least some of the reduction in titer; however, as shown in Table 1, for this mutant the 2-LTR circles make up a larger proportion of the total circular DNA forms (25%, compared to 9% for the WT), suggesting that the linear DNA produced by the HIV-1(CA) mutant, which, based on the analysis of the 2-LTR circle junctions, has a single nucleotide beyond the CA, is not an efficient substrate for integration. It is likely that the higher titer of the HIV-1(CAT) mutant [than of the HIV-1(CA) mutant] is due to the fact that the HIV-1(CAT) mutant produces, in addition to linear DNAs with one nucleotide beyond the CA, a significant number of linear DNAs with two nucleotides beyond the CA. The HIV-1(CAT) mutant also showed an increase in the proportion of 2-LTR circles relative to the total number of circles (28%), which is presumably due to the fact that the linear DNAs with one nucleotide beyond the CA are integrated poorly. The only other mutant that showed an elevated proportion of 2-LTR circles is the HIV-1(ATG) mutant (29%), and because its linear DNA lacks the canonical CA at the U5 end, the linear DNA of this mutant is not expected to be a good substrate for integration. When we changed the two nucleotides (TT) of the RSVP(A)Z vector to match the HIV-1 sequence (G), RSV IN removed a single nucleotide and properly integrated the processed ends in about 70% of the proviruses that we isolated. This mutant decreased virus titer to about 46% of the WT level (23).
TABLE 3.
One-end autointegrations
| Virus | Position of the integrationa
|
No. of casesb | |
|---|---|---|---|
| U3 | U5 | ||
| HIV-1 WT | Variable | 1 | 9 (4 gag, 3 pol, 2 pol-env) |
| gag | −2 | 1 | |
| HIV-1(ATG) | 1 | U3 | 1 |
| Variable | 1 | 4 (1 U3, 1 gag, 2 pol) | |
| HIV-1(CA) | Variable | 1 | 2 (2 pol) |
| HIV-1(CAT) | 1 | U5 | 1 |
| Variable | 1 | 4 (1 gag, 2 pol, 1 pol-env) | |
| HIV-1(CAAT) | 1 | U3 | 1 |
| Variable | 1 | 10 (1 U3, 5 gag, 3 pol, 1 pol-env) | |
| HIV-1(CAGT) | 1 | Variable | 2 (1 R, 1 U5) |
| Variable | 1 | 20 (7 gag, 12 pol, 1 pol-env) | |
| HIV-1(CAGGT) | Variable | 1 | 13 (1 U3, 4 gag, 6 pol, 2 pol-env) |
The last nucleotide at each LTR end of the circles is indicated by a number, using the numbering system in Fig. 1C.
The positions of autointegrations are indicated. pol-env, between pol and env.
For one of the mutant viruses, HIV-1(CAGGT), we recovered circles from both concerted autointegration events and one-end autointegration events (Table 2 and Table 3), suggesting that HIV IN is able to remove three nucleotides beyond the CA sequence from the U5 LTR terminus and insert the processed DNA correctly, creating a 5-bp duplication at the target site. The titer of this mutant is relatively high, 63% of the WT level, which means that HIV-1 IN can efficiently remove three nucleotides from the U5 end of the viral DNA. This result agrees with the in vitro data showing that HIV-1 IN can remove three nucleotides 3′ of the canonical CA dinucleotide (21, 30). It is possible that HIV-1 IN is relatively efficient in removing three nucleotides from the U5 end because the IN of the related virus HIV-2 normally removes three nucleotides from the U5 end of its linear DNA (32). In one concerted autointegration event from an infection with HIV-1(CAGGT), both ends were processed correctly and inserted into the viral sequence; however, there was a 9-bp duplication at the target site instead of a normal 5-bp duplication (Table 2). Because we have seen only one such event, we do not know whether it was caused by the mutation or was the result of a rare error by IN. We previously reported the effects of changing the sequence (TT) between the PBS and the CA dinucleotide of the RSV-derived vector RSVP(A)Z to match the HIV-2 sequence (GGT) (23). Proviruses recovered from this mutant showed that RSV IN was able to remove three nucleotides beyond the CA and properly insert the processed end. However, the titer of this mutant virus was reduced to only 3.5% of the WT level, and although it is possible that the mutation has some effect on the initiation of viral DNA synthesis, the substantial reduction in titer suggests that, although RSV IN can remove three nucleotides, it does so relatively inefficiently.
We also obtained DNA circles from infections with the HIV-1(ATG) mutant, in which the canonical CA sequence was mutated to AT (Table 2). Although, based on published reports, we were not surprised that this mutant had a reasonably high titer (5, 22), we expected, based on the results of the experiments with the RSV-based vector, that IN would insert the normal (U3) end correctly and that, because there was no CA in U5, this end would be inserted by host factors (23). Sequence analysis of the autointegrants produced by infection with the HIV-1(ATG) mutant suggests that HIV-1 IN can remove two nucleotides from the U5 LTR terminus beyond the (mutated) AT and can properly insert the processed DNA, creating a 5-bp duplication at the target site. In contrast to our in vivo result, it has been previously reported, in an in vitro experiment with HIV-1 IN, that the replacement of the CA dinucleotide with TG significantly impaired both 3′ processing and strand transfer, suggesting that, at least in vitro, the CA dinucleotide is required for integration (21). However, Zhou et al. previously showed that sequences other than the canonical CA were successfully integrated in an in vitro assay using RSV IN (37). In these experiments, substrate oligonucleotides were inserted into a circular DNA target. A large pool of oligonucleotides was used, and the terminal five nucleotides of the oligonucleotides were randomized. This mixture was subjected to a competitive integration reaction catalyzed by RSV IN. Integrated substrates were then PCR amplified to determine the substrate/target DNA junction. After one round of selection, sequence analysis of selected clones showed that a diverse group of terminal sequences was found, suggesting that, at least in an in vitro reaction, RSV IN does not require the canonical CA for integration. Our results show that, in vivo, HIV-1 can integrate successfully using sequences other than CA. This suggests that, despite the evolutionary conservation of the CA dinucleotide, this dinucleotide is not required for appropriate integration of the U5 end of HIV-1 linear DNA; however, as has already been discussed, the increase in the proportion of 2-LTR circles suggests that the linear DNA from the HIV-1 mutant that lacks the CA at the U5 end, HIV-1(ATG), is not an efficient substrate for integration, and because this mutation does not have a clear effect on the initiation of viral DNA synthesis, the decrease in titer (to 20% of the WT level) is primarily due to the effect of the mutation on integration.
Acknowledgments
We are grateful to Hilda Marusiodis and Terri Burdette for help in preparing the manuscript.
This research was supported by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
Footnotes
Published ahead of print on 7 November 2007.
REFERENCES
- 1.Arnold, E., J. Ding, S. H. Hughes, and Z. Hostomsky. 1995. Structures of DNA and RNA polymerases and their interactions with nucleic acid substrates. Curr. Opin. Struct. Biol. 527-38. [DOI] [PubMed] [Google Scholar]
- 2.Artzi, H. B., J. Shemesh, E. Zeelon, B. Amit, L. Keiman, M. Gorecki, and A. Panet. 1996. Ribonuclease H activity during initiation of reverse transcription using tRNAlys/RNA primer/template of human immunodeficiency virus. Arch. Biochem. Biophys. 325201-216. [DOI] [PubMed] [Google Scholar]
- 3.Bartz, S. R., and M. A. Vodicka. 1997. Production of high-titer human immunodeficiency virus type 1 pseudotyped with vesicular stomatitis virus glycoprotein. Methods 12337-342. [DOI] [PubMed] [Google Scholar]
- 4.Boyer, P. L., A. L. Ferris, P. Clark, J. Whitmer, P. Frank, C. Tantillo, E. Arnold, and S. H. Hughes. 1994. Mutational analysis of the fingers and palm subdomains of human immunodeficiency virus type 1 (HIV-1) reverse transcriptase. J. Mol. Biol. 243472-483. [DOI] [PubMed] [Google Scholar]
- 5.Brown, H. E., H. Chen, and A. Engelman. 1999. Structure-based mutagenesis of the human immunodeficiency virus type 1 DNA attachment site: effects on integration and cDNA synthesis. J. Virol. 739011-9020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ding, J., K. Das, Y. Hsiou, S. G. Sarafianos, A. D. Clark, A. Jacobo-Molina, C. Tantillo, S. H. Hughes, and E. Arnold. 1998. Structure and functional implications of the polymerase active site region in a complex of HIV-1 RT with double-stranded DNA and an antibody Fab fragment at 2.8 A resolution. J. Mol. Biol. 2841095-1111. [DOI] [PubMed] [Google Scholar]
- 7.Federoff, O. Y., M. Salazar, and B. R. Reid. 1996. Structural variation among retroviral primer-DNA junctions: solution of the HIV-1 (-)-strand Okazaki fragment r(gca)d(GCAGTGGC). Biochemistry 3511070-11080. [DOI] [PubMed] [Google Scholar]
- 8.Furfine, E. S., and J. E. Reardon. 1991. Human immunodeficiency virus reverse transcriptase ribonuclease H: specificity of tRNALys3-primer excision. Biochemistry 307041-7046. [DOI] [PubMed] [Google Scholar]
- 9.Gao, H. Q., P. L. Boyer, E. Arnold, and S. H. Hughes. 1998. Effects of mutations in the polymerase domain on the polymerase, RNase H and strand transfer activities of HIV-RT. J. Mol. Biol. 277559-572. [DOI] [PubMed] [Google Scholar]
- 10.Gao, H. Q., S. G. Sarafianos, E. Arnold, and S. H. Hughes. 1999. Similarities and differences in the RNase H activities of HIV-RT and Moloney murine leukemia virus RT. J. Mol. Biol. 2941097-1113. [DOI] [PubMed] [Google Scholar]
- 11.Ghosh, M., K. J. Howard, C. E. Cameron, S. J. Benkovic, S. H. Hughes, and S. F. Le Grice. 1995. Truncating α-helix E′ pf p66 HIV RT modulates RNase H function and impairs DNA strand transfer. J. Biol. Chem. 2707068-7076. [DOI] [PubMed] [Google Scholar]
- 12.Gopalakrishnan, V., J. A. Peliska, and S. J. Benkovic. 1992. HIV-1 RT: Spatial and temporal relationship between the polymerase and RNase H activities. Proc. Natl. Acad. Sci. USA 8910763-10767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gotte, M., S. Fackler, T. Hermann, E. Perola, L. Cellai, H. J. Gross, S. F. Le Grice, and H. Heumann. 1995. HIV-1 RT-associated RNase H cleaves RNA/RNA in arrested complexes: implications for mechanism by which RNase H discriminates between RNA/RNA and RNA/DNA. EMBO J. 14833-841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang, H., R. Chopra, G. L. Verdine, and S. C. Harrison. 1998. Structure of a covalently trapped catalytic complex of HIV-1 RT: implications for drug resistance. Science 2821669-1675. [DOI] [PubMed] [Google Scholar]
- 15.Huber, H. E., and C. C. Richardson. 1990. Processing of the primer for plus strand DNA synthesis by human immunodeficiency virus 1 reverse transcriptase. J. Biol. Chem. 26510565-10573. [PubMed] [Google Scholar]
- 16.Jacobo-Molina, A., J. Ding, R. G. Nanny, A. D. Clark, X. Lu, C. Tantillo, R. L. Williams, G. Kamer, A. L. Ferris, P. Clark, A. Hizi, S. H. Hughes, and E. Arnold. 1993. Crystal structure of HIV-1 RT complexed with double-stranded DNA at 3.0 Å resolution shows bent DNA. Proc. Natl. Acad. Sci. USA 906320-6324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Julias, J. G., A. L. Ferris, P. L. Boyer, and S. H. Hughes. 2001. Replication of phenotypically mixed human immunodeficiency virus type 1 virions containing catalytically active and catalytically inactive reverse transcriptase. J. Virol. 756537-6546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Julias, J. G., M. J. McWilliams, S. G. Sarafianos, W. G. Alvord, E. Arnold, and S. H. Hughes. 2004. Effects of mutations in the G tract of the human immunodeficiency virus type 1 polypurine tract on virus replication and RNase H cleavage. J. Virol. 7813315-13324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Julias, J. G., M. J. McWilliams, S. G. Sarafianos, E. Arnold, and S. H. Hughes. 2002. Mutations in the RNase H domain of HIV-1 reverse transcriptase affect the initiation of DNA synthesis and the specificity of RNase H cleavage in vivo. Proc. Natl. Acad. Sci. USA 999515-9520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kulkosky, J. R., A. Katz, and A. M. Skalka. 1990. Terminal nucleotides of the preintegrative linear form of HIV-1 DNA deduced from the sequence of circular DNA junctions. J. Acquir. Immune Defic. Syndr. 3852. [PubMed] [Google Scholar]
- 21.Leavitt, A. D., R. B. Rose, and H. E. Varmus. 1992. Both substrate and target oligonucleotide sequences affect in vitro integration mediated by human immunodeficiency virus type 1 integrase protein produced in Saccharomyces cerevisiae. J. Virol. 662359-2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Masuda, T., M. J. Kuroda, and S. Harada. 1998. Specific and independent recognition of U3 and U5 att sites by human immunodeficiency virus type 1 integrase in vivo. J. Virol. 728396-8402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oh, J., K. W. Chang, and S. H. Hughes. 2006. Mutations in the U5 sequences adjacent to the primer binding site do not affect tRNA cleavage by Rous sarcoma virus RNase H but do cause aberrant integration in vivo. J. Virol. 80451-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oh, J., J. G. Julias, A. L. Ferris, and S. H. Hughes. 2002. Construction and characterization of a replication-competent retroviral shuttle vector plasmid. J. Virol. 761762-1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pullen, K. A., L. K. Ishimoto, and J. J. Champoux. 1992. Incomplete removal of the RNA primer for minus-strand DNA synthesis by human immunodeficiency virus type 1 reverse transcriptase. J. Virol. 66367-373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sarafianos, S. G., K. Das, C. Tantillo, A. D. Clark, J. Ding, J. M. Whitcomb, P. L. Boyer, S. H. Hughes, and E. Arnold. 2001. Crystal structure of HIV-1 reverse transcriptase in complex with a polypurine tract RNA:DNA. EMBO J. 201449-1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schatz, O., J. Mous, and S. F. Le Grice. 1990. HIV-1 RT-associated ribonuclease H displays both endonuclease and 3′→5′ exonuclease activity. EMBO J. 91171-1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smith, J. S., and M. Roth. 1992. Specificity of human immunodeficiency virus-1 reverse transcriptase-associated ribonuclease H in removal of the minus-strand primer, tRNA(Lys3). J. Biol. Chem. 26715071-15079. [PubMed] [Google Scholar]
- 29.Swanstrom, R., H. E. Varmus, and J. M. Bishop. 1981. The terminal redundancy of the retroviral genome facilitates chain elongation by reverse transcriptase. J. Biol. Chem. 1561115-1121. [PubMed] [Google Scholar]
- 30.Vink, C., D. C. van Gent, Y. Elgersma, and R. A. Plasterk. 1991. Human immunodeficiency virus integrase protein requires a subterminal position of its viral DNA recognition sequence for efficient cleavage. J. Virol. 654636-4644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wain-Hobson, S., P. Sonigo, O. Danos, S. Cole, and M. Alizon. 1985. Nucleotide sequence of the AIDS virus, LAV. Cell 409-17. [DOI] [PubMed] [Google Scholar]
- 32.Whitcomb, J. M., and S. H. Hughes. 1991. The sequence of human immunodeficiency virus type 2 circle junction suggests that integration protein cleaves the ends of linear DNA asymmetrically. J. Virol. 653906-3910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Whitcomb, J. M., R. Kumar, and S. H. Hughes. 1990. Sequence of the circle junction of human immunodeficiency virus type 1: implications for reverse transcription and integration. J. Virol. 644903-4906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wohrl, B. M., and K. Moelling. 1990. Interaction of HIV-1 RNase H with polypurine tract containing RNA-DNA hybrids. Biochemistry 2910141-10147. [DOI] [PubMed] [Google Scholar]
- 35.Yee, J. K., T. Friedmann, and J. C. Burns. 1994. Generation of high-titer psedotyped retroviral vectors with very broad host range. Methods Cell Biol. 4399-112. [DOI] [PubMed] [Google Scholar]
- 36.Youvan, D. C., and J. E. Hearst. 1979. Reverse transcriptase pauses at N2-methylguanine during in vitro transcription of Escherichia coli 16s ribosomal RNA. Proc. Natl. Acad. Sci. USA 763751-3754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou, H., J. Rainey, S. K. Wong, and J. M. Coffin. 2001. Substrate sequence selection by retroviral integrase. J. Virol. 751359-1370. [DOI] [PMC free article] [PubMed] [Google Scholar]