Abstract
Hepatitis delta virus (HDV) is a small RNA virus that contains one 1.7-kb single-stranded circular RNA of negative polarity. The HDV particle also contains two isoforms of hepatitis delta antigen (HDAg), small (SHDAg) and large HDAg. SHDAg is required for the replication of HDV, which is presumably carried out by host RNA-dependent RNA polymerases. The localization and the HDAg and host RNA polymerase responsible for HDV replication remain important issues to be addressed. In this study, using recombinant SHDAg fused with a heterologous nucleolar localization sequence (NoLS) to confine its subcellular localization in nucleoli, we aimed to study the effect of SHDAg subcellular localization on HDV RNA replication. The initiation of genomic RNA synthesis from antigenomic template was hardly detectable when SHDAg was fused with the NoLS motif and localized mainly in nucleoli. In contrast, the initiation of antigenomic RNA synthesis was not affected. Drug treatment to release a SHDAg-NoLS mutant from nucleoli could partially restore the replication of HDV genomic RNA from antigenomic RNA. This also recovered the cointeraction between SHDAg and RNA polymerase II. These data strongly suggest that nuclear polymerase (RNA polymerase II) is involved in the synthesis of genomic RNA and that the synthesis of antigenomic RNA can occur in nucleoli. Our results support the idea that the replication of HDV genomic RNA or antigenomic RNA is likely to be carried out by different machineries in different subcellular localizations.
Hepatitis delta virus (HDV) is a small negative-strand RNA virus containing a 1.7-kb single-stranded circular RNA. Its RNA genome folds into an unbranched rod-like structure due to a high degree of intramolecular base pairing (for reviews, see references 18 and 32). This virus requires hepatitis B virus as a helper to supply the envelope proteins for viral particle assembly and infection (29). Apart from its genome and hepatitis B virus envelope, the HDV particle also contains hepatitis delta antigen (HDAg), the only known protein encoded by HDV. HDAg includes two protein species of 24 kDa (195 amino acid residues) and 27 kDa (214 amino acid residues), known as small and large HDAg, respectively (SHDAg and LHDAg, respectively). These two isoforms of HDAg share similar amino acid sequences, except for the additional 19-amino-acid extension in the C terminus of LHDAg, but exhibit different functions in the HDV life cycle. SHDAg is essential for HDV RNA replication (17). LHDAg is synthesized in the late stage of the viral replication cycle due to an RNA editing event that alters the termination codon of the open reading frame (ORF) for SHDAg (21). LHDAg is necessary for virion assembly (4).
The exact molecular mechanism for HDV RNA replication is still obscure. A double rolling-circle mechanism for HDV replication has been proposed (for reviews, see references 18 and 32). In this model, HDV genomic RNA undergoes RNA-dependent RNA synthesis to produce a multimeric intermediate, and it is then processed to the circular, unit-length complement of the genomic RNA, termed the antigenome. The antigenomic RNA serves as a template for the synthesis of genomic RNA via the same rolling-circle mechanism. Since HDV does not encode its RNA-dependent RNA polymerase (Pol) and HDAg does not possess any RNA Pol activity, the host RNA Pol is likely to be redirected for HDV replication. Some recent studies suggested that replication of HDV RNA is carried out by two different cellular RNA Pols, Pol II and Pol I (20, 22, 24). The two RNA Pols reside in different subcellular locations, and HDV replication has to move accordingly. In fact, during the replication cycle, HDV ribonucleoprotein (RNP) indeed shuttles between the nucleolus, nucleus, nucleoplasm, and cytoplasm (31). The observation that HDV RNA accumulated only in the cytoplasm and was translocated into the nucleus in the presence of HDAg suggested that viral RNA movement is mediated by HDAg (8). To study the effect of intranuclear locations of HDAg on viral RNA replication, we introduced a heterogeneous nucleolar localization signal (NoLS) from the human immunodeficiency virus (HIV) Rev protein (3, 15) to SHDAg (Fig. 1) to restrict its localization and then analyzed its effect on HDV replication. When SHDAg was fused with the NoLS motif and mainly localized in nucleoli, we found that the initiation of genomic RNA synthesis, but not that of antigenomic RNA synthesis, was abolished. Drug treatment by actinomycin D to release the SHDAg-NoLS mutant from nucleoli could partially restore the replication of genomic RNA. These data suggest that subcellular localization of SHDAg affects its ability to support the initiation of HDV RNA replication and that the replication of the two polarities of HDV viral RNA occurs in different subcellular sites.
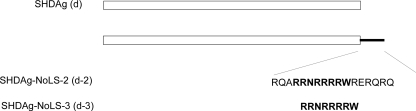
FIG. 1.
Schematic diagram of SHDAg and its mutants. Clones d-2 and d-3 were derived from SHDAg with the C terminus fused with the NoLS of the HIV Rev protein.
MATERIALS AND METHODS
Cell lines and plasmid construction.
Huh7, a human hepatocellular carcinoma cell line, was cultured as described elsewhere (16). NIH 3T3 cells, a high-anchorage-dependent cell line established from NIH Swiss mouse embryo cultures, was cultured in similar culture conditions as Huh7 cells. Plasmid pCD2G contains a tandem dimer of wild-type HDV cDNA (1.7-kb XbaI fragment, genomic sense) under the control of the cytomegalovirus immediate-early promoter (26). This construct transcribed dimeric HDV genomic RNA. pCD2AG, like pCD2G, also contains an HDV cDNA dimer but in a different orientation. Plasmids pCDm2G and pCDm2AG are similar to wild-type pCD2G and pCD2AG except for a two-base deletion in the HDAg ORF. pCDAg-S contained the HDAg ORF and expressed SHDAg while pCDAg-S(−) contained the SHDAg ORF in opposite orientation and was used as a template for in vitro transcription of HDAg mRNA under control of the T7 promoter (26).
Site-directed mutagenesis of SHDAg.
Site-directed mutagenesis was performed by a QuickChange site-directed mutagenesis lit (Stratagene). Plasmid pCDAg-S was used as mutagenesis template for SHDAg-NoLS mutants. The mutagenic primers used for the addition of Rev NoLS to the 3′ terminus of SHDAg CDNA (Fig. 1, clone d-2) were 5′-CGAAGGAATAGAAGAAGAAGGTGGAGAGAGAGACAGAGATAGGATATACTCTTCCCAGCC-3′ and 5′-CTCTCTCCACCTTCTTCTTCTATTCCTTCGAGCCTGTCTTGGAAATCCCTGGTTTCCCCTGATG-3′. After DNA synthesis by Pfu Ultra DNA Pol, the reaction mixture was treated with restriction enzyme DpnI to digest template plasmids and then transformed to E. coli competent cells. These pCDAg-S-d-2 mutant clones were confirmed by sequence analysis. Plasmid pCDAg-S-d-3 was derived from pCDAg-S-d-2 with the primer set 5′-CGAAGGAATAGAAGAAGAAGGTGGTAGGATATACTCTTCCCAGCC-3′and 5′-CCACCTTCTTCTTCTATTCCTTCGTGGAAATCCCTGGTTTCCCCTGATG-3′. pCDAg-S-d-2(−) and pCDAg-S-d-3(−) contained the d-2 ORF and d-3 ORF in opposite orientations, respectively, and were used for in vitro transcription of d-2 mRNA and d-3 mRNA.
DNA transfection and RNA transfection.
DNA transfection was performed with using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions as described previously (27). For cotransfection, the ratio of cDNA of SHDAg or its NoLS mutants to HDV RNA templates pCDm2G or pCDm2AG was 1:3. For the RNA transient transfection experiment, in vitro transcribed RNAs were transfected into Huh7 cells by DMRIE-C transfection reagent (Invitrogen), according to the manufacturer's instructions, as described elsewhere (23). An experiment for the initiation of HDV RNA replication was performed according to the method of a previous study (22) with some modifications. Briefly, 10 μg of in vitro transcribed HDV genomic or antigenomic RNAs together with 0.25 μg of capped mRNA of SHDAg or 0.5 μg of capped mRNA of SHDAg-NoLS mutants was used to transfected 2 × 106 Huh7 cells in a 60-mm dish. Transfected cells were incubated overnight and passaged equally to two dishes. Cells were then either left untreated or were treated with 0.1 μg/ml actinomycin D for 8 h. After drug treatment, cells were washed twice with phosphate-buffered saline (PBS), which was replaced with fresh medium without actinomycin D. Cells were harvested for analysis at day 6 posttransfection.
Coimmunoprecipitation.
Huh7 cells transfected with SHDAg or its mutants were harvested and lysed with radioimmunoprecipitation assay buffer (25 mM Tris-HCl pH 7.6, 150 mM NaCl, 1% NP-40, 1% sodium deoxycholate, 0.1% sodium dodecyl sulfate [SDS]) at 48 h posttransfection. For actinomycin D treatment, 0.1 μg/ml drug was added at 42 h posttransfection. Lysates were precipitated with rabbit anti-HDAg antibody-conjugated protein G-Sepharose (packed volume, 20 μl; Pierce). The immunoprecipitates were washed with PBS buffer containing 0.3% NP-40 and processed for SDS-polyacrylamide gel electrophoresis and immunoblotting analysis.
Western blot analysis.
Lysates of transfected cells or immunoprecipitated proteins were subjected to electrophoresis in SDS-polyacrylamide gels, followed by a Western blotting procedure, as described previously (27). HDAg was detected with rabbit polyclonal antibody or mouse monoclonal antibody against HDAg. RNA Pol II was detected with a monoclonal antibody against the C-terminal domain of RNA Pol II (8WG16; Abcam). RNA Pol I was detected with a polyclonal antibody against N-terminal amino acids 1 to 300 of RPA194 (Santa Cruz). Cellular actin was detected with a mouse monoclonal antibody against C-terminal actin (Santa Cruz). After incubation with the secondary antibody conjugated to horseradish peroxidase, the blots were developed by a Western Lightning ECL blot detection system (Perkin Elmer).
Indirect immunofluorescence.
Huh7 cells that transiently expressed wild-type or SHDAg-NoLS mutants were grown on coverslips and then fixed with 3.7% paraformaldehyde at 48 h posttransfection. For actinomycin D treatment, 0.1 μg/ml drug was added at 42 h posttransfection. After extraction with CSK buffer (50 mM NaCl, 300 mM sucrose, 10 mM PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid] pH 6.8, 3 mM MgCl2, 0.5% Triton X-100) for 5 min, cells were blocked in PBS containing 1% bovine serum albumin for 1 h at room temperature. Cells were then immunostained with rabbit anti-HDAg antiserum together with monoclonal anti-nucleolin or anti-heterogeneous nuclear RNP (hnRNP) C1/C2 antibodies for 1 h at room temperature, followed by fluorescein isothiocyanate-conjugated rabbit anti-mouse immunoglobulin G or rhodamine-conjugated rabbit anti-rabbit immunoglobulin G for another 1 h. After PBS washing, the coverslips were mounted on glass slides with Vectashield (Vector Laboratory, Inc., Burlingame, CA) and observed with an immunofluorescence microscope (Leica).
In vitro transcription.
For RNA transfection of HDV RNA, genomic RNAs and antigenomic RNAs were transcribed in vitro from HindIII-linearized plasmids pCD2G and pCD2AG, respectively, with a T7 MEGAscript transcription kit (Ambion) (26). Capped mRNAs for SHDAg and its mutants were prepared by in vitro transcription using the T7 mMESSAGE mMACHINE (Ambion) transcription kit. For detection of HDV genomic and antigenomic RNA, digoxigenin (DIG)-labeled strand-specific probes for HDV genomic and antigenomic RNAs were prepared by in vitro transcriptions of HindIII-linearized pCD2G and pCD2AG using T7 Pol with a DIG Northern starter kit (Roche).
RNA preparation and Northern blotting.
Total RNAs from Huh7 cells were extracted by using TRIzol reagent (Invitrogen) according to the instructions of the supplier. For Northern blot analysis, total cellular RNA samples were subjected to electrophoresis on a 6% formaldehyde-1% agarose RNA gel and then transferred into a nylon filter and cross-linked. The filter was prehybridized and then hybridized with DIG-labeled HDV genomic and antigenomic RNA probes, respectively, according to the instructions of the supplier (DIG Northern starter kit; Roche).
Heterokaryon assay.
Expression plasmids for the d-2 mutant were used to transfect Huh7 cells seeded on coverslips. Transfected cells were treated with 40 μg/ml emetine at 40 h posttransfection and then mixed with mouse NIH 3T3 cells. After coculturing for 3 h, cell fusion was induced by rinsing cells in PBS and then adding a drop of polyethylene glycerol (PEG 1500; Roche); cells were incubated for 2 min and washed twice in PBS. Cells were then further incubated in fresh medium in the presence of 40 μg/ml emetine. Five hours after heterokaryon formation, cells were fixed with 3.7% paraformaldehyde for indirect immunofluorescence staining and analyzed with immunofluorescence microscopy.
RESULTS
SHDAg-NoLS chimera is localized in the nucleoli.
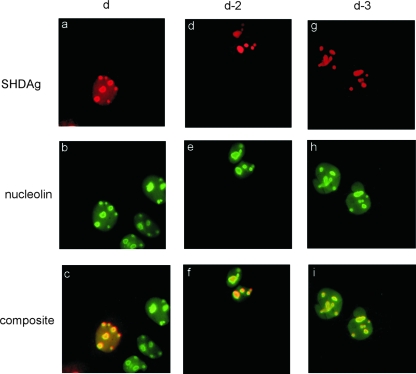
It had been demonstrated that SHDAg was distributed in both the nucleus and nucleoli (2, 7, 16, 19, 31) (Fig. 2a to c). To discover whether protein shuttling might play a role in viral replication, we constructed a mutant HDAg, which should be confined in the nucleolus, and then studied the effect of disrupting the intranuclear shuttling of HDAg on viral replication. For this purpose, SHDAg was fused with the HIV NoLS in either the N or C terminus. To verify whether the fused NoLS could confine SHDAg to a subcellular localization, we introduced SHDAg-NoLS mutants into Huh7 cells by cDNA transfection and analyzed the distribution of these mutants. By immunofluorescence microscopy analysis, SHDAg-NoLS mutants (clones d-2 and d-3) were concentrated in nucleoli (Fig. 2d to f and g to i, respectively) while wild-type SHDAg was found in both compartments. Nucleoplasmic distribution of either mutant was not detected. Furthermore, coexpression with the HDV genomic RNA or antigenomic RNA by cDNA transfection did not alter the nucleolar distribution pattern of either mutant (data not shown). These results suggest that Rev NoLS directs the intranuclear location of SHDAg mutants to concentrate in nucleoli.
FIG. 2.
Subcellular localization of SHDAg and NoLS-fused mutants. Expression plasmids of SHDAg (clone d) or its NoLS-fused constructs (clones d-2 and d-3) were used to transfect Huh7 cells with or without the expression plasmid of the cDNA dimer of the HDV genome (pCDm2G) or antigenome (pCDm2AG) with a two-base deletion in the SHDAg ORF. Transfected cells were fixed 2 days posttransfection and analyzed with an immunofluorescence assay. Composite, merge of SHDAg and nucleolin images.
SHDAg-NoLS mutants are confined in nucleoli.
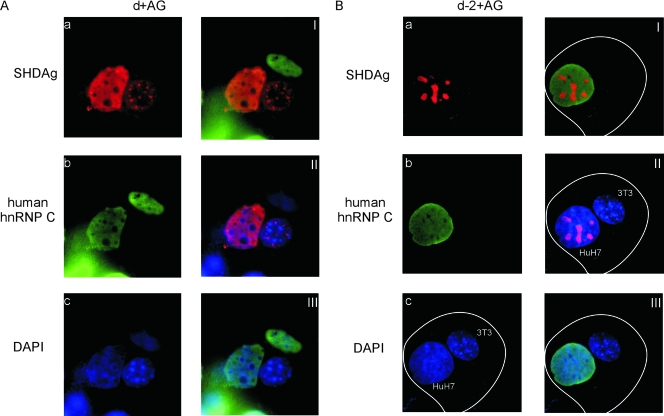
In the previous experiment, we found that SHDAg-NoLS mutants were detected in nucleoli exclusively. To analyze whether SHDAg-NoLS mutants were confined in nucleoli, Huh7 cells expressing such mutant and viral antigenomic RNAs were fused with NIH 3T3 cells, and then protein shuttling was monitored. Antibodies against human hnRNP C1/C2 were also used to differentiate human and mouse nuclei. As shown in Fig. 3A, wild-type SHDAg distributed in the nucleoplasm of the transfected Huh7 cell as well as the fused NIH 3T3 cell in the heterokaryon, indicating that HDAg was able to shuttle between nucleus/nucleolus and cytoplasm (Fig. 3A, frames I to III). Interestingly, no nucleolar distribution was detected even in the transfected Huh7 cells after heterokaryon formation (Fig. 3A, frame a), suggesting that nucleolar SHDAg has shuttled and moved out from the nucleoli. The lack of nucleolar signal may indicate that emetine blocked rRNA synthesis. However, when Huh7 cells expressing a SHDAg-NoLS mutant were fused with NIH 3T3 cells, the mutant was detected exclusively in the nucleoli of the Huh7 cell in the heterokaryon (Fig. 3B, frames I to III). These results strongly indicate that SHDAg fused to heterogeneous NoLS accumulates in the cellular nucleolus.
FIG. 3.
A heterokaryon assay demonstrated that subcellular localization of the SHDAg-NoLS mutant was confined to the nucleolus. Huh7 cells seeded on coverslips were cotransfected with 3 μg of pCDm2AG as well as 1 μg of the expression plasmid of SHDAg (d+AG) or clone d-2 (d-2+AG). The transfected cells were incubated and then fused with NIH 3T3 cells (see Materials and Methods). Five hours after heterokaryon formation, cells were fixed and analyzed by immunofluorescence microscopy. Frames I to III in panels A and B are merged images of the immunofluorescence results as follows: frames I, SHDAg and human hnRNP C (C1/C2); frames II, SHDAg and DAPI (4′,6′-diamidino-2-phenylindole) staining; and frames III, human hnRNP C (C1/C2) and DAPI staining. White lines mark cell boundaries.
A SHDAg-NoLS mutant could support the initiation of antigenomic RNA synthesis but not genomic RNA synthesis.
It was previously suggested that replication of HDV genomic RNA and antigenomic RNA is carried out in different locations of cellular nuclei (20). In that study, newly synthesized antigenomic RNA was found to localize in nucleoli, while newly synthesized genomic RNA was found in the nucleoplasm. However, the investigators could not determine whether the localizations of the synthesized RNAs were the sites where these RNAs were replicated or whether the RNAs just accumulated at these sites after being synthesized. Since SHDAg can conduct the nuclear localization of HDV RNP (8, 31) and SHDAg-NoLS mutants were confined in nucleoli, we could then investigate if confinement of SHDAg to the nucleolus would affect its ability to facilitate HDV RNA replication. However, SHDAg-NoLS mutants could not support HDV RNA replication by cotransfection with the expression plasmids containing the cDNA of HDAg-deficient HDV genomic RNA or antigenomic RNA (pCDm2G and pCDm2AG, respectively), regardless of whether the NoLS was fused to the N-terminal, internal, or the C-terminal portions of SHDAg (data not shown). A possible explanation for this result is that the SHDAg-NoLS mutant behaves as a dominant negative inhibitor.
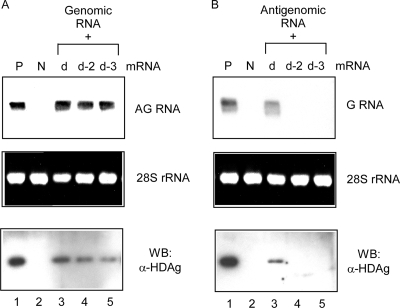
To circumvent the problem, we used an RNA transfection method to initiate replication by cointroducing in vitro transcribed SHDAg-NoLS mRNA together with dimeric HDV RNA and monitored the replication of viral RNA. Because the transfected HDV RNAs were wild type and able to express SHDAg after the RNA replication cycle, we could investigate if the SHDAg-NoLS mutant supported the initiation of HDV RNA replication. By this approach, the SHDAg-NoLS mutant was found to be able to support the initiation of antigenomic RNA synthesis and the expression of SHDAg when genomic RNA template was cotransfected (Fig. 4A, lanes 4 and 5). However, the SHDAg-NoLS mutant could not facilitate the initiation of HDV genomic RNA synthesis (Fig. 4B, lanes 4 and 5). Since the localization of the SHDAg-NoLS mutant is in nucleoli, these results suggest that the initiation of HDV antigenomic RNA, but not that of genomic RNA, occurs in nucleoli. Combined with the previous finding that newly synthesized antigenomic RNAs were localized in nucleoli (20), this result suggests that the synthesis of antigenomic RNA occurs in nucleoli.
FIG. 4.
SHDAg-NoLS mutant could support the initiation of HDV antigenomic RNA synthesis but not that of genomic RNA synthesis. (A) In vitro transcribed mRNAs of SHDAg (clone d) or SHDAg-NoLS mutants (clones d-2 and d-3) were cotransfected with HDV genomic RNA to Huh7 cells. Cellular RNAs and proteins were extracted 6 days posttransfection, and the expression of HDV antigenomic RNA and HDAg was analyzed. (B) The same as the experiment as shown in panel A except that antigenomic RNA was used for transfection. G, genomic; AG, antigenomic; P, positive control; N, untransfected cells; α, anti; WB, Western blotting.
Release of the SHDAg-NoLS mutant from nucleoli restored its ability to facilitate the initiation of genomic RNA synthesis.
Our results had indicated that an HDAg-NoLS mutant could not support the initiation of HDV genomic RNA synthesis in nucleoli. This effect might result from the inappropriate localization caused by the HDAg-NoLS mutant. However, we could not rule out the possibility that the fused fragment impaired the functions of SHDAg since some mutations or deletions in SHDAg usually affect its ability to help HDV RNA replication.
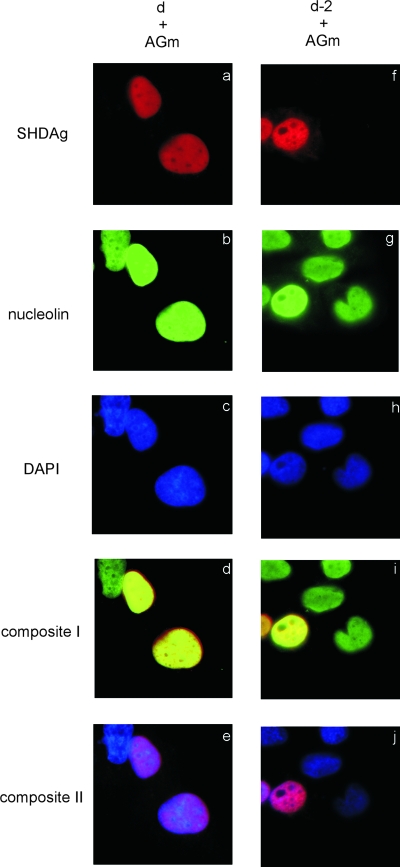
To address this question, we tried to move the SHDAg-NoLS mutant from nucleoli to nucleoplasm and then tested our hypothesis that the NoLS-fused SHDAg mutant still possessed the ability to support the initiation of HDV genomic RNA synthesis and that its failure to support HDV genomic RNA synthesis was due to its nucleolar localization and inaccessibility to the components/machinery for genomic RNA synthesis. Introducing plasmid pCDm2AG in this experiment clearly revealed the distribution patterns of SHDAg and its mutant with antigenomic RNA after drug treatment. Some reagents had been demonstrated to be able to release nucleolar proteins into the nucleoplasm (9, 10, 14, 30). Actinomycin D is one of these and does not affect HDV RNA synthesis (22). So, we first treated SHDAg-NoLS mutant-transfected cells with actinomycin D to investigate the localization of the mutant. This mutant was distributed normally in nucleoli (Fig. 2d). As expected, when transfected cells were treated with actinomycin D, the SHDAg-NoLS mutant was translocated into nucleoplasm (Fig. 5f).
FIG. 5.
Actinomycin D treatment could induce the export of SHDAg and its mutants from the nucleolus. Expression plasmids for SHDAg (clone d) or its NoLS-fused mutant (clone d-2) were used to transfect Huh7 cells with the expression plasmid of the cDNA dimer of the HDV antigenome (AGm) with a two-base deletion in the SHDAg ORF (pCDm2AG). Transfected cells were treated with 0.1 μg/ml actinomycin D at 42 h posttransfection, fixed at 48 h posttransfection, and then analyzed by immunofluorescence assay. Composite I, merged image of the results of HDAg and nucleolin staining; composite II, merged image of the results of HDAg and DAPI (4′,6′-diamidino-2-phenylindole) staining.
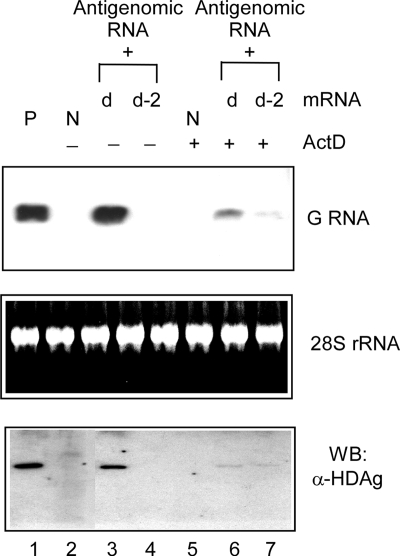
We then verified the effect of the SHDAg-NoLS mutant on the initiation of HDV genomic RNA synthesis with the treatment of actinomycin D. Cells were transfected with synthetic HDV antigenomic RNA and SHDAg mRNA or SHDAg-NoLS mutant mRNA to mimic the native RNA-direct RNA synthesis process of genomic RNA synthesis from the antigenomic RNA template. As shown in Fig. 6, the accumulation of HDV genomic RNA and SHDAg was detected from the RNA-transfected cells treated with actinomycin D (comparing lanes 3 and 4 and lanes 6 and 7), indicating that this mutant could support the initiation of HDV genomic RNA synthesis after actinomycin D treatment. This result implies that the SHDAg-NoLS mutant per se did not lose its ability to facilitate HDV genomic RNA synthesis, but NoLS-induced restriction of the mutant to nucleoli prevented from it carrying out genomic RNA replication.
FIG. 6.
Actinomycin D treatment partially rescued the ability of SHDAg-NoLS to facilitate the initiation of HDV genomic RNA (G RNA) synthesis from antigenomic RNA. In vitro transcribed mRNAs of SHDAg (clone d) or clone d-2 were cotransfected with HDV antigenomic RNAs to Huh7 cells. After overnight incubation, the transfected cells were reseeded and treated with 0.1 μg/ml actinomycin D (ActD) for 8 h. Cellular RNA and proteins were extracted 6 days posttransfection, and the expression levels of HDV antigenomic RNA and HDAg were analyzed. P, positive control; N, protein samples from untransfected cells; WB, Western blotting; α, anti.
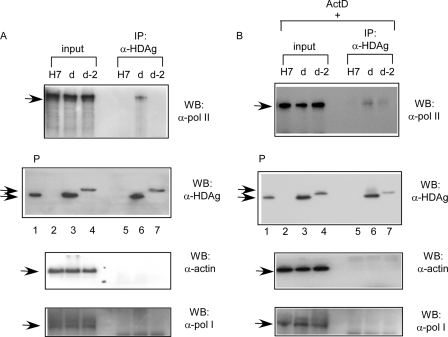
SHDAg, but not the SHDAg-NoLS mutant, could interact with cellular RNA Pol II.
Several lines of evidence indicated that cellular RNA Pol II carried out the synthesis of genomic RNA in nuclei. HDAg has also been demonstrated to physically interact with RNA Pol II (33). Since fusion with NoLS confined the SHDAg fusion protein in the nucleoli, this might also affect its interaction with RNA Pol II so as to influence its ability to facilitate the synthesis of genomic RNA. As shown in Fig. 7, cellular Pol II could be coprecipitated with wild-type SHDAg, but not the SHDAg-NoLS mutant, by anti-HDAg antibodies (Fig. 7A, lanes 6 and 7, respectively). However, when cells were treated with actinomycin D, cellular RNA Pol II could again be coprecipitated with wild-type SHDAg as well as the SHDAg-NoLS mutant by anti-HDAg antibodies (Fig. 7B, lanes 6 and 7). Cellular actin could not be coprecipitated with wild-type SHDAg or the SHDAg-NoLS mutant under the same conditions (Fig. 7). This inability of the SHDAg-NoLS mutant to support the synthesis of genomic RNA could be explained in part by the loss of interaction with RNA Pol II. Furthermore, RNA Pol I could not be coprecipitated with wild-type SHDAg or the SHDAg-NoLS mutant (Fig. 7). Although a previous study found that the Pol I-specific transcription factor SL1 interacts with SHDAg (20), our result indicates that Pol I may not interact with SHDAg.
FIG. 7.
Interaction of SHDAg-NoLS mutant and cellular RNA Pol II. (A) Huh7 cells transfected with expression plasmids for SHDAg (clone d) or clone d-2 were harvested at 48 h posttransfection. Cells were lysed with radioimmunoprecipitation assay buffer, and the lysates were immunoprecipitated with anti-HDAg antibodies. The precipitants were analyzed. (B) The same experiment as shown in panel A except that the transfected cells were treated with 0.1 μg/ml actinomycin D (ActD) at 42 h posttransfection. P, positive control; H7, untransfected Huh7 cells; WB, Western blotting; IP, immunoprecipitation; α, anti.
DISCUSSION
HDV RNA replication is mediated by cellular Pols, but the identity of such machinery is still controversial. It was previously suggested that replication of HDV genomic RNA and antigenomic RNA is carried out in different locations within cellular nuclei. Li et al. found that newly synthesized antigenomic RNA localized in nucleoli, while newly synthesized genomic RNA was found in the nucleoplasm (20). However, the investigators could not determine whether the synthesized RNAs were localized at sites where they replicated or whether they just quickly accumulated at these sites after being synthesized. In this study, we fused a nucleolar localization signal to SHDAg to study the initiation of each strand of HDV RNA in situ and thus could better characterize the location effect.
HDV RNP has been demonstrated to shuttle between the nucleus and cytoplasm (31), while HDV RNA alone (without antigen) accumulated only in the cytoplasm. However, in the presence of SHDAg, the HDV RNA was translocated into the nucleus, suggesting that nuclear import of HDV RNA is mediated by SHDAg (8). In this study, we found that SHDAg fused with NoLS did not accumulate in the nucleoplasm and could not support the synthesis of HDV genomic RNA (Fig. 4B). Since the chimeric SHDAg-NoLS may translocate HDV RNP into nucleoli, this finding suggests that such RNA synthesis does not occur in nucleoli. This is likely to be the case since the NoLS-fused mutant does not lose its ability to facilitate the initiation of genomic RNA synthesis (Fig. 6, lane 7). Furthermore, after actinomycin D treatment to induce the nucleoplasmic distribution of the mutant, the mutant is able to interact with RNA Pol II (Fig. 7B, lane 7), which, according to previous studies, carries out the synthesis of genomic RNA (5, 6, 11, 12, 24, 25, 33), after actinomycin D treatment to induce the nucleoplasmic distribution of the mutant. These data support the model that Pol II mediates the synthesis of genomic RNA in nuclei.
A SHDAg-NoLS mutant confined in the nucleoli could support the initiation of HDV antigenomic RNA synthesis (Fig. 4A, lanes 4 and 5), indicating that the machinery mediating HDV antigenomic RNA synthesis can localize in nucleoli. It is unlikely that the nucleoplasmic Pol machinery (Pol II), which is responsible for genomic RNA synthesis, can associate with the mutant to translocate into nucleoli to carry out antigenomic RNA synthesis since the coimmunoprecipitation experiment clearly demonstrated that Pol II did not interact with the SHDAg mutant when it was retained in nucleoli (Fig. 7A, lane 7). Although we cannot exclude the possibility that a small number of genomic RNA-SHDAg-NoLS RNPs were translocated to the nucleoplasm and support antigenomic RNA replication there, it is quite unlikely because the differences in the accumulated antigenomic RNAs (Fig. 4B, lanes 3 to 5) did not reflect the dramatic differences in quantity between wild-type SHDAgs and the possible contamination of SHDAg-NoLS mutants in the nucleoplasmic fractions (Fig. 2a, d, and g).
HDV is a viroid-like animal pathogen and replicates its RNA in a similar rolling-circle mechanism. It is noteworthy that the negative-strand RNA of such a viroid was localized in the nucleoplasm, whereas its positive-strand RNA was localized in the nucleolus as well as in the nucleoplasm with distinct spatial patterns (28). Several lines of evidence support the idea that replications of HDV RNAs are carried out by two different RNA Pols in different subcellular localizations. First, studies from Lai's laboratory had demonstrated that machineries that carried out the synthesis of genomic RNA and antigenomic RNA had different sensitivities to alpha-amanitin, indicating there are two different enzyme activities involved (22, 24). Second, two polarities of HDV viral RNAs showed different subcellular distribution patterns (1). Li et al. recently also demonstrated that the newly synthesized antigenomic RNA was detected mainly in nucleoli, indicating that nucleolar machinery carries out antigenomic RNA replication through genomic RNA (20). In addition, two studies also revealed that nucleolar proteins may affect HDV replication. Nucleolar proteins, such as B23 (16) and nucleolin (19), had been found to enhance HDV RNA replication. Interestingly, these two proteins can affect cellular RNA Pol I-mediated ribosome biogenesis and form complexes with HDAg. Furthermore, it had been found that RNA Pol II and the Pol I-associated transcription factor SL1 could be precipitated with HDAg, suggesting the association of HDV replication complex with the Pol I and Pol II transcription machineries (20). These data combined with ours suggest that nucleolar Pol (RNA Pol I or others) might mediate the synthesis of antigenomic RNA. Although we did not find the associations of RNA Pol I with SHDAg or its NoLS-fused mutant by a coimmunoprecipitation assay, more studies are required to judge whether Pol I is involved in HDV RNA replication. A recent report demonstrated that RNA Pol II interacts with HDV-derived RNA from both polarities of HDV RNA (13). However, the study was an interaction in vitro rather than an initiation or replication. Furthermore, the Pol II-HDV genomic RNA complex may be responsible for the synthesis of HDAg mRNA instead of the synthesis of antigenomic RNA. Moreover, it has been suggested that the transcription of the mRNA and the replication of the HDV genome are independent processes and occur concurrently (23). As synthesis of both antigenomic RNA and HDAg mRNA uses genomic RNA as a template, it is quite likely that HDV coordinates the two processes by carrying them out in different subcellular localizations with different RNA synthesis machineries.
In summary, the data are in agreement with the idea that the replication of HDV antigenomic RNA, in contrast to genomic RNA, occurs in nucleoli. RNA Pol II is suggested to be the Pol responsible for genomic RNA replication in the nucleoplasm. Further studies are required to identify the possible cellular RNA Pol responsible for HDV antigenomic RNA synthesis.
Footnotes
Published ahead of print on 7 November 2007.
REFERENCES
- 1.Bell, P., R. Brazas, D. Ganem, and G. G. Maul. 2000. Hepatitis delta virus replication generates complexes of large hepatitis delta antigen and antigenomic RNA that affiliate with and alter nuclear domain 10. J. Virol. 745329-5336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bichko, V. V., and J. M. Taylor. 1996. Redistribution of the delta antigens in cells replicating the genome of hepatitis delta virus. J. Virol. 708064-8070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bohnlein, E., J. Berger, and J. Hauber. 1991. Functional mapping of the human immunodeficiency virus type 1 Rev. RNA binding domain: new insights into domain structure of Rev and Rex. J. Virol. 657051-7055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chang, F. L., P. J. Chen, S. J. Tu, C. J. Wang, and D. S. Chen. 1991. The large form of hepatitis delta antigen is crucial for assembly of hepatitis delta virus. Proc. Natl. Acad. Sci. USA 888490-8494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang, J., and J. M. Taylor. 2002. In vivo RNA-directed transcription, with template switching, by a mammalian RNA polymerase. EMBO J. 21157-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang, J., S. O. Gudima, C. Tarn, X. Nie, and J. M. Taylor. 2006. Action of inhibitors on accumulation of processed hepatitis delta virus RNAs. J. Virol. 803205-3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang, M. F., S. C. Baker, L. H. Soe, T. Kamahora, J. G. Keck, S. Makino, S. Govindarajan, and M. M. C. Lai. 1988. Human hepatitis delta antigen is a nuclear phosphoprotein with RNA-binding activity. J. Virol. 622403-2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chou, H. C., T. Y. Hsieh, G. T. Sheu, and M. M. C. Lai. 1998. Hepatitis delta antigen mediates the nuclear import of hepatitis delta virus RNA. J. Virol. 723684-3690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.David-Pfeuty, T., Y. Nouvian-Dooghe, V. Sirri, P. Roussel, and D. Hernandez-Verdun. 2001. Common and reversible regulation of wild-type p53 function and of ribosomal biogenesis by protein kinases in human cells. Oncogene 205951-5963. [DOI] [PubMed] [Google Scholar]
- 10.Dundr, M., G. H. Leno, M. Hammarskjold, D. Rekosh, C. Helga-Maria, and M. O. Olson. 1995. The roles of nucleolar structure and function in the subcellular location of the HIV-1 Rev. protein. J. Cell Sci. 1082811-2823. [DOI] [PubMed] [Google Scholar]
- 11.Filipovska, J., and M. M. Konarska. 2000. Specific HDV RNA-templated transcription by Pol II in vitro. RNA 641-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fu, T.-B., and J. M. Taylor. 1993. The RNAs of hepatitis delta virus are copied by RNA polymerase II in nuclear homogenates. J. Virol. 676965-6972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Greco-Stewart, V. S., P. Miron, A. Abrahem, and M. Pelchat. 2007. The human RNA polymerase II interacts with the terminal stem-loop regions of the hepatitis delta virus RNA genome. Virology 35768-78. [DOI] [PubMed] [Google Scholar]
- 14.Haaf, T., and D. C. Ward. 1996. Inhibition of RNA polymerase II transcription causes chromatin decondensation, loss of nucleolar structure, and dispersion of chromosomal domains. Exp. Cell Res. 224163-173. [DOI] [PubMed] [Google Scholar]
- 15.Hope, T. J., X. Huang, D. Mcdonald, and T. G. Parslow. 1990. Steroid-receptor fusion of the human immunodeficiency virus type 1 Rev transactivator: mapping cryptic functions of the arginine-rich motif. Proc. Natl. Acad. Sci. USA 877787-7791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang, W. H., B. Y. Yung, W. J. Syu, and Y. H. W. Lee. 2001. The nucleolar phosphoprotein B23 interacts with hepatitis delta antigens and modulates the hepatitis delta virus RNA replication. J. Biol. Chem. 27625166-25175. [DOI] [PubMed] [Google Scholar]
- 17.Kuo, M. Y., M. Chao, and J. M. Taylor. 1989. Initiation of replication of the human hepatitis delta virus genome from cloned DNA: role of delta antigen. J. Virol. 631945-1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lai, M. M. C. 2005. RNA replication without RNA-dependent RNA polymerase: surprises from hepatitis delta virus. J. Virol. 797951-7958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee, C. H., S. C. Chang, C. J. Chen, and M. F. Chang. 1998. The nucleolin binding activity of hepatitis delta antigen is associated with nucleolus targeting. J. Biol. Chem. 2737650-7656. [DOI] [PubMed] [Google Scholar]
- 20.Li, Y. J., T. B. Macnaughton, L. Gao, and M. M. C. Lai. 2006. RNA-templated replication of hepatitis delta virus: genomic and antigenomic RNAs associate with different nuclear bodies. J. Virol. 806478-6486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Luo, G. X., M. Chao, S. Y. Hsieh, C. Sureau, K. Nishikura, and J. M. Taylor. 1990. A specific base transition occurs on replicating hepatitis delta virus RNA. J. Virol. 641021-1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Macnaughton, T. B., S. T. Shi, L. E. Modahl, and M. M. C. Lai. 2002. Rolling circle replication of hepatitis delta virus RNA is carried out by two different cellular RNA polymerases. J. Virol. 763920-3927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Modahl, L. E., and M. M. C. Lai. 1998. Transcription of hepatitis delta antigen mRNA continues throughout hepatitis delta virus (HDV) replication: a new model of HDV RNA transcription and regulation. J. Virol. 725449-5456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Modahl, L. E., T. B. Macnaughton, N. Zhu, D. L. Johnson, and M. M. C. Lai. 2000. RNA-dependent replication and transcription of hepatitis delta virus RNA involve distinct cellular RNA polymerases. Mol. Cell. Biol. 206030-6039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moraleda, G., and J. M. Taylor. 2001. Host RNA polymerase requirements for transcription of the human hepatitis delta virus genome. J. Virol. 7510161-10169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mu, J. J., D. S. Chen, and P. J. Chen. 2001. The conserved serine 177 in the delta antigen of hepatitis delta virus is one putative phosphorylation site and is required for efficient viral RNA replication. J. Virol. 759087-9095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mu, J. J., Y. G. Tsay, L. J. Juan, T. F. Fu, W. H. Huang, D. S. Chen, and P. J. Chen. 2004. The small delta antigen of hepatitis delta virus is an acetylated protein and acetylation of lysine 72 may influence its cellular localization and viral RNA synthesis. Virology 31960-70. [DOI] [PubMed] [Google Scholar]
- 28.Qi, Y., and D. Biao. 2003. Differential subnuclear localization of RNA strands of opposite polarity derived from an autonomously replicating viroid. Plant Cell. 152566-2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rizzetto, M., B. Hoyer, M. G. Canese, J. W. Shih, R. H. Purcell, and J. L. Gerin. 1980. delta Agent: association of delta antigen with hepatitis B surface antigen and RNA in serum of delta-infected chimpanzees. Proc. Natl. Acad. Sci. USA 776124-6128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rubbi, C. P., and J. Milner. 2003. Disruption of the nucleolus mediates stabilization of p53 in response to DNA damage and other stresses. EMBO J. 226068-6077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tavanez, J. P., C. Cunha, M. C. Silva, E. David, J. Monjardino, and M. Carmo-Fonseca. 2002. Hepatitis delta virus ribonucleoproteins shuttle between the nucleus and the cytoplasm. RNA 8637-646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Taylor, J. M. 2006. Hepatitis delta virus. Virology 34471-76. [DOI] [PubMed] [Google Scholar]
- 33.Yamaguchi, Y., J. Filipovska, K. Yano, A. Furuya, N. Inukai, T. Narita, T. Wada, S. Sugimoto, M. M. Konarska, and H. Handa. 2001. Stimulation of RNA polymerase II elongation by hepatitis delta antigen. Science 293124-127. [DOI] [PubMed] [Google Scholar]