Abstract
VifIIIB, which has been a standard model for the viral infectivity factor of human immunodeficiency virus type 1 (HIV-1), binds the cytidine deaminase APOBEC3G (A3G) and induces its degradation, thereby precluding its lethal incorporation into assembling virions. Additionally, VifIIIB less efficiently degrades A3F, another potent anti-HIV-1 cytidine deaminase. Although the APOBEC3 paralogs A3A, A3B, and A3C have weaker anti-HIV-1 activities and are only partially degraded by VifIIIB, we found that VifIIIB induces their emigration from the nucleus to the cytosol and thereby causes net increases in the cytosolic concentrations and anti-HIV-1 activities of A3A and A3B. In contrast, some other Vifs, exemplified by VifHXB2 and VifELI-1, much more efficiently degrade and thereby neutralize all APOBEC3s. Studies focused mainly on A3F imply that it occurs associated with mRNA-PABP1 in translationally active polysomes and to a lesser extent in mRNA processing bodies (P-bodies). A3F appears to stabilize the P-bodies with which it is associated. A correspondingly small proportion of VifIIIB also localizes in P-bodies in an A3F-dependent manner. Stress causes A3A, A3B, A3C, and A3F to colocalize efficiently with VifIIIB and mRNA-PABP1 complexes in stress granules in a manner that is prevented by cycloheximide, an inhibitor of translational elongation. Coimmunoprecipitation studies suggest that Vifs from different HIV-1 isolates associate with all tested APOBEC3s. Thus, Vifs interact closely with structurally diverse APOBEC3s, with effects on their subcellular localization, degradation rates, and antiviral activities. Cytosolic APOBEC3-Vif complexes are predominantly bound to mRNAs that dynamically move between translationally active and storage or processing pools.
The viral infectivity factor (Vif) encoded by human immunodeficiency virus type 1 (HIV-1) neutralizes a potent antiretroviral defense that occurs in lymphocytes and macrophages (29, 39). A major component of this defense system is APOBEC3G (A3G) (40), a deoxycytidine deaminase that is incorporated into assembling virion cores, where it lethally hypermutates nascent HIV-1 reverse transcripts (22, 23, 26, 55). VifIIIB, which has been used as a standard model (the vifIIIB gene also occurs in the widely used recombinant HIV-1 strain NL4-3), binds to A3G and causes its rapid polyubiquitination and proteasomal degradation (27, 28, 41, 45, 53). Vif has an amino-terminal domain that binds A3G and a carboxyl-terminal region with adjacent cullin-5 and BC-box motifs that recruit a multisubunit ubiquitin ligase complex (27, 29, 39, 45). This complex contains elongins B and C, cullin 5, and the ubiquitin-protein isopeptide ligase Rbx-1 plus associated factors (18, 53, 54). By ridding infected cells of A3G, Vif precludes its incorporation into progeny HIV-1.
A3G is encoded on chromosome 22 in tandem linkage to six related but structurally diverse cytidine deaminases: A3A, A3B, A3C, A3DE, A3F, and A3H. Early reports suggested that the A3D gene was not expressed and that the A3E locus is a pseudogene, but recent evidence showed that A3D and A3E form the N- and C-terminal regions of one protein, A3DE (7, 17, 48). All members of this family have one (A3A, A3C, and A3H) or two (A3B, A3DE, A3F, and A3G) cytidine deaminase motifs containing a conserved consensus sequence that coordinates Zn2+, a glutamic acid residue involved in proton shuttling, and critical aromatic residues involved in RNA binding (17). Although A3G has the most potent anti-HIV-1 activity, several other APOBEC3s inhibit HIV-1 to lesser extents. Specifically, A3F and A3DE weakly inactivate vif-deleted (Δvif) HIV-1 and are partially degraded by VifIIIB (9, 56), whereas A3B inhibits both wild-type (wt) and Δvif HIV-1 and is not neutralized by VifIIIB (1, 10, 52). Although one group reported that VifIIIB binds to A3C and downmodulates its intracellular concentration (21), another reported that VifIIIB neither binds nor degrades A3B or A3C (10).
Generally, the anti-HIV-1 activities of APOBEC3s have been analyzed by using 293T cells that have been transiently transfected with wt or Δvif HIV-1 vectors in the presence or absence of APOBEC3s and subsequently measuring the relative infectivities of the released virions (1, 37, 50, 52). While this system has been very useful, it has serious limitations because the APOBEC3s being compared are expressed in different amounts in these assays and because the levels of protein expression in 293T cells are often greater than amounts in normal cells. In this context, it is notable that A3B, A3F, and A3G mRNAs are widely but differentially expressed in HIV-1-susceptible cells and that relatively small but significant amounts of A3C mRNA also occur in monocytes and macrophages (52). With the exception of A3G, which has been detected in cell extracts by using a specific antiserum (45), the amounts of other APOBEC3 proteins in HIV-1-susceptible cells are unknown. Moreover, A3B and A3G mRNA levels are greatly enhanced in T-cell lines by mitogenic factors that activate extracellular signal-regulated kinase (37, 38). In macrophages but not in T cells, A3A and A3G mRNA levels are induced by type I interferons (2, 3, 34, 46). As shown here, the intracellular localizations of APOBEC3s are also regulated. These findings imply that APOBEC3s in addition to A3F and A3G might become induced or activated sufficiently to inhibit HIV-1 replication in some tissues or in damaged and inflamed microenvironments.
Recent evidence has demonstrated that A3G enzymatic activity is inhibited intracellularly by binding to RNA (5). During reverse transcription within the virion cores, the template RNA is degraded, and A3G then becomes activated and able to deaminate cytidines in the negative single-strand DNA substrate (44). Immunofluorescence microscopy and proteomic analyses have indicated that A3G is principally associated with mRNAs that shuttle between translationally active polyribosomes, mRNA processing bodies (P-bodies), and dormant stress granules (SGs) (6, 13, 19, 49). Here we extend this analysis by investigating the associations of the APOBEC3 family members A3A, A3B, A3C, and A3F with these mRNA metabolic sites and with VifIIIB. Like A3G, A3F associates with active polysomes and P-bodies in a Vif-independent manner. A3A and A3C, which occur in both the nucleus and the cytoplasm, and A3B, which has a predominantly nuclear localization, become exclusively cytoplasmic in the presence of VifIIIB. All these APOBEC3s accumulate within SGs along with Vif and mRNA-PABP1 in response to stress. Consistent with these results, Vifs from different HIV-1 isolates bind promiscuously to all tested APOBEC3s, which is surprising in view of the substantive diversities in sizes and sequences of these cytidine deaminases (17). Although all tested Vifs degrade A3G, they have surprisingly distinct abilities to degrade A3F and other APOBEC3s. In particular, it has been shown that Vifs from patient isolates of HIV-1 differ substantially in their relative abilities to degrade A3F versus A3G (43). We show that VifIIIB and VifJR-CSF are weaker and relatively specific for A3G, whereas other variants, such as VifHXB2 and VifELI-1, degrade them all more efficiently.
MATERIALS AND METHODS
Plasmids and viruses.
Previously described vectors were used to express the following proteins: A3G-Myc and LacZ-Myc (27); A3B-Myc (37); A3A-HA, A3C-HA, and A3F-HA, generously donated by B. Cullen (50); A3B-HA from the NIH AIDS Research and Reference Reagent Program, donated by B. Cullen (10, 11); and a codon-optimized derivative of VifIIIB (HVifIIIB), a kind gift from S. Bour (30). The pHIV-gpt(wt) and pHIV-gpt(Δvif) (24, 25, 32) vectors were used to express VifHXB2. Plasmid pIIIB was used to express full-length HIV-1 HXB3 (16), and a variant with a stop codon in vif that eliminates its expression was made by mutagenesis and generously donated by S. Kuhmann (Weill Medical College of Cornell University). Plasmids for expression of full-length HIV-1 strains 89.6, JR-CSF, and YU-2 were obtained from the NIH AIDS Research and Reference Reagent Program (donated, respectively, by R. Collman; I. Chen and Y. Koyanagi; and B. Hahn and G. Shaw). ELI-1 was a gift from K. Peden (Center for Biologics Evaluation and Research, FDA). GenBank accession numbers for Vifs are as follows: M19921 (IIIB), K03455 (HXB2), K03454 (ELI-1), M38429 (JR-CSF), and AY569174 (YU-2).
Protein analyses.
293T cells (ATCC) were maintained according to supplier specifications. They were cotransfected using PolyFect reagent (Qiagen) with equimolar ratios of plasmids and were lysed in radioimmunoprecipitation assay buffer (50 mM Tris-Cl [pH 7.4], 1% Nonidet P-40, 0.1% sodium deoxycholate, and 150 mM NaCl) with Complete protease inhibitors (Roche) 36 h posttransfection as previously described (27, 37). The extracts were centrifuged at 1,500 × g for 5 min at 4°C to sediment the nuclei. Postnuclear extracts adjusted to equivalent protein concentrations using Bradford reagent (Bio-Rad) were used for immunoprecipitations and/or Western immunoblotting (27, 37). Some extracts were treated with 20 μg/ml DNase-free RNase A (Roche) at 25°C for 20 min prior to analysis. The monoclonal antibodies used were anti-Myc clone 9E10 and anti-hemagglutinin (HA) clone HA-7 (Sigma). A rabbit antiserum to Vif, 2221, was from the NIH AIDS Research and Reference Reagent Program (donated by D. Gabuzda). Protein loading was assessed with a mouse antibody against α-tubulin (Sigma).
Immunofluorescence microscopy.
293T cells were cultured in Permanox chamber slides (Nalge Nunc International) treated with 0.1 mg/ml polylysine for 20 min at 37°C. They were transfected using FuGene 6 transfection reagent (Roche). At 30 h posttransfection, the cells were fixed in 5% formaldehyde and 2% sucrose in phosphate-buffered saline (Invitrogen) at room temperature for 20 min and then permeabilized with 0.25% Triton X-100 in phosphate-buffered saline for 10 min. Where indicated, cultures were preincubated with a 50 μM concentration of the proteasome inhibitor ALLN for 6 h (Calbiochem). For stress response experiments, prior to fixation, the cells were treated with 0.5 mM sodium meta-arsenite (J. T. Baker) for 1 h at 37°C. Some cultures were treated with 0.1 mg/ml cycloheximide (Sigma) for 20 min at 37°C prior to and during the 1-h arsenite treatment. For analysis of P-bodies, cells were treated with 0.1 mg/ml cycloheximide for 1 h at 37°C prior to fixation. Untreated cells were used as a negative control. Primary antibodies were anti-Vif antiserum 2221 (1:200), the Myc-specific monoclonal antibody 9E10 (1:500), the rabbit polyclonal anti-Myc antibody C3956 (1:500), the anti-HA monoclonal antibody HA-7 (1:1,000), the anti-HA polyclonal antibody H6908 (1:1,000) (Sigma), the mouse anti-PABP1 antibody 10E10 (1:300) (Abcam Inc.), rabbit anti-DDX6 (Rck/p54) (1:100) (Bethyl Laboratories), and chicken anti-LSM1 (1:200) (GenWay Biotech Inc.). Staining with anti-PABP1 was performed overnight at 4°C. Incubations with all other primary antibodies were performed for 2 h at room temperature. Secondary fluorescent antibodies were Alexa Fluor 488 goat anti-rabbit immunoglobulin G (heavy plus light chains) [IgG(H+L)], Alexa Fluor 594 goat anti-mouse IgG(H+L), Alexa Fluor 350 goat anti-rabbit IgG(H+L) (1:500) (Molecular Probes Inc.), and fluorescein isothiocyanate-conjugated goat anti-chicken IgY (1:160) (GenWay Biotech Inc.), which reacted only with their species-specific primary antibodies. Secondary antibodies were used at a dilution of 1:1,000 unless otherwise indicated, and incubations were performed for 1 h at room temperature. Slides were mounted in FluoroGuard antifade reagent (Bio-Rad Laboratories) and observed with a Zeiss Axiovert 200M deconvolution microscope. Images were acquired using a 40× objective unless otherwise indicated.
Infectivity assays.
Viruses HXB2 and IIIB were harvested from cotransfected 293T cultures 36 h posttransfection and were titered by the focal infectivity method (24) on HeLa-CD4-CCR5 cells. For HIV-gpt viruses (HXB2), the transfections included pSVIIIenv (14) for pseudotyping, and the virus-infected colonies of HeLa-CD4 cells were counted after mycophenolic acid selection (32, 33, 35).
RESULTS
HIV-1 Vif differentially affects the concentrations and subcellular localizations of APOBEC3 paralogs.
Previously, we used a combination of biochemical methods and immunofluorescence microscopic analyses of fixed cells to demonstrate that A3G is efficiently downmodulated by HIV-1 Vif (27, 39). To further evaluate our immunofluorescence methods, we did a comparative fluorescence microscopy analysis of fixed versus live cells. For fixed cells, 293T cells transiently cotransfected with expression vectors encoding either wt or vif deletion HIV-1 mutants and A3G-Myc were used. Consistent with our previous report (27), the number of A3G-Myc-positive cells in the cultures was dramatically reduced by coexpression of wt Vif, and cells that coexpressed both proteins were rare (Fig. 1A, upper panel). In contrast, A3G was frequently coexpressed in single cells with Vif(Δ10), an inactive BC-box mutant that binds A3G but is incapable of inducing its degradation (Fig. 1A, lower panel) (27). Indeed, in this field all cells with Vif(Δ10) also contained A3G. Similar results were obtained with other Vif BC-box mutants and with all tested Vif mutants that lacked biological activity (data not shown). Negative controls established that the antibody staining was specific. Moreover, treatment with proteasome inhibitors of cultures cotransfected with wt HIV-1 and A3G-Myc caused A3G to accumulate specifically in the cells that contained Vif (27). Thus, the elimination of A3G tightly correlates with Vif biological activity, in agreement with other evidence (12). A dramatic decrease in the number of A3G-Myc-positive 293T cells was also observed in cultures transiently cotransfected with codon-optimized HVifIIIB (30) in the absence of other HIV-1 proteins relative to the number in control cultures cotransfected with pcDNA3.1 (Fig. 1B). In this case, only a few Vif-positive cells had visible traces of A3G. Live cell analyses used 293T cells cotransfected with expression vectors for either wt or Δvif HIV-1 and yellow fluorescent protein (YFP)-A3G-Myc. At 24 h posttransfection, some cultures were treated with the proteasome inhibitor ALLN for an additional 6 h, and the viable cultures were then examined for fluorescence to detect YFP-A3G-Myc-positive cells and by phase-contrast microscopy to visualize all cells (Fig. 1C, left). The presence of Vif decreased the percentage of YFP-A3G-Myc-positive cells fivefold compared to that for Δvif HIV-1 cultures. The proteasome inhibitor ALLN reversed the Vif-induced downmodulation of A3G (Fig. 1C, right). Similar results were obtained using HeLa cells stably expressing YFP-A3G-Myc and HVifIIIB (data not shown). These controlled analyses confirm the mechanism of Vif function and establish that our methods for immunofluorescence microscopy reliably monitor the effects of Vif on APOBEC3s.
FIG. 1.
A3G is downmodulated by HIV-1 Vifs. (A and B) 293T cells transiently coexpressing A3G-Myc and either wt HIV-1, Vif(Δ10) HIV-1, HVifIIIB, or the negative-control plasmid pcDNA3.1 were fixed and stained for A3G-Myc and Vif. (A) In cultures cotransfected with wt HIV-1 and A3G-Myc, fewer A3G-positive cells are present than in Vif(Δ10) samples. (B) A significant decrease in A3G-positive cells is observed in cultures cotransfected with A3G-Myc and HVifIIIB. (C) 293T cells were cotransfected with YFP-A3G-Myc and either HIV-gpt(wt) or HIV-gpt(Δvif). Some cultures coexpressing YFP-A3G-Myc and HIV-gpt(wt) were treated with the proteasome inhibitor ALLN. (Left) Micrographs showing results of analysis of live cells for YFP fluorescence in order to detect A3G-positive cells (left panels) and phase-contrast microscopy to detect the total number of cells in each field (right panels). wt Vif caused a significant decrease in the number of YFP-A3G-Myc-positive cells (top left), and the effect of Vif was blocked when cells were treated with ALLN (bottom left). The ΔVif mutant did not decrease the number of A3G-positive cells (center left). (Right) Bar graph showing the percentages of YFP-A3G-Myc-positive cells in the presence of wt Vif, the ΔVif mutant, and wt Vif plus ALLN. For panels A and C, the images were acquired using a 20× objective, and for panel B, a 40× objective was used.
To determine whether A3A, A3B, A3C, and A3F are affected by Vif, immunofluorescence microscopy studies were done using 293T cells cotransfected with epitope-tagged APOBEC3s and HVifIIIB expression vectors or the negative-control plasmid pcDNA3.1. Statistical analysis of three large fields (each consisting of approximately 100 cells) from two independent experiments revealed that the presence of HVifIIIB did not decrease the number of cells positive for the APOBEC3s tested, suggesting that unlike A3G, these proteins were not efficiently eliminated from cells by HVifIIIB (data not shown). However, HVifIIIB altered the subcellular localizations of A3A-HA, A3B-Myc, and A3C-HA. In the absence of HVifIIIB, A3A and A3C were detected in both the nuclei and cytosol of 293T cells whereas A3B was detected predominantly in the nuclei. In the presence of HVifIIIB, all three proteins became exclusively cytosolic, suggesting that they interact with HVifIIIB (Fig. 2A). Western blot analyses of cytosolic extracts strongly supported these results. Thus, VifIIIB efficiently eliminated A3G but caused a net increase in the cytosolic concentration of A3B (Fig. 2D). Under the conditions of our assays, the VifIIIB and HVifIIIB vectors were expressed at similar levels.
FIG. 2.
HVifIIIB affects the intracellular distributions of A3A, A3B, and A3C and colocalizes in P-bodies with A3F. (A and B) 293T cells were transiently transfected with either A3A-HA, A3B-Myc, A3C-HA, or A3F-HA and an HVifIIIB expression vector or the negative-control plasmid pcDNA3.1. Cells were fixed and probed with mouse anti-HA or mouse anti-Myc and rabbit anti-Vif antibodies. (A) In the absence of HVifIIIB, A3A and A3C are present in both the nucleus and the cytoplasm, whereas A3B is predominantly intranuclear (control panels). In the presence of HVifIIIB, A3A, A3B, and A3C migrate from the nuclei to the cytoplasm (HVifIIIB panels). (B) Control panel shows A3F staining throughout the cytoplasm as well as in discrete cytoplasmic granules. HVifIIIB colocalizes in the cytoplasmic granules with A3F (HVifIIIB panel). (C) 293T cells transiently expressing A3F-HA were stained for A3F and Rck/p54. Cycloheximide (CHX) treatment did not disperse the A3F-Rck/p54-containing P-bodies. In contrast, CHX substantially dispersed the Rck/p54-containing P-bodies in cells that lacked A3F. (D) Western blot analyses of cytosolic extracts from 293T cells transiently cotransfected with A3B-Myc (left) or A3G-Myc (right) and either wt HIV-1IIIB, Δvif HIV-1IIIB, or codon-optimized VifIIIB (HVifIIIB). α-Tubulin (α-tub), loading control.
A3F-HA was predominantly present in a diffuse cytosolic distribution in the absence or presence of HVifIIIB (Fig. 2B), although approximately 65% of the A3F-positive cells also contained brightly fluorescing clusters that were found to be P-bodies (see below). Importantly, HVifIIIB did not occur in similar clusters when expressed independently (data not shown) or with A3B or A3C (Fig. 2A), but it colocalized with A3F-HA in these clusters in cells that coexpressed both proteins, suggesting that A3F recruits Vif to these sites (Fig. 2B). A proportion of the cytosolic A3A also was clustered, although these clusters were more diffuse and less sharply delimited than the A3F granules (Fig. 2A). VifIIIB also tended to colocalize in these A3A clusters. Considered together, these results strongly suggest that VifIIIB interacts intracellularly with all these APOBEC3s.
Recently, we reported that A3F and A3G associate with PABP1 and other mRNA binding proteins and that A3G moves with mRNA-PABP1 complexes from actively translating polyribosomes into SGs under conditions of stress (19). Additionally, other groups found that small proportions of A3G and A3F in normal cells occur in P-bodies, which are sites of mRNA processing and degradation (13, 49). To determine whether the A3F granules seen in Fig. 2B might be P-bodies, 293T cells transiently expressing A3F-HA were stained for A3F and endogenous Rck/p54 (a protein that occurs in P-bodies as well as in dispersed cytosolic regions) (Fig. 2C, control panels). The majority of A3F clusters contained Rck/p54 (Fig. 2C) and lacked PABP1 (data not shown), indicating that they were P-bodies. Similar results were obtained using LSM1, another P-body protein (data not shown). Treatment with cycloheximide, an inhibitor of translational elongation, is known to disperse P-bodies either by causing their mRNAs to shift into polysomes or by retarding the entry of mRNAs into P-bodies and allowing the resident mRNAs to be degraded (20, 42). Consistent with this expectation, cycloheximide caused a dramatic decrease in the number of P-bodies, especially in cells lacking A3F. Surprisingly, however, this treatment had no significant effect on the number of A3F clusters that contained Rck/p54 (Fig. 2C, CHX panel). This implies that A3F stabilizes P-bodies, thereby preventing their dispersal in the presence of cycloheximide.
Natural HIV-1 Vif variants have significant but distinct effects on the antiviral activities and quantities of diverse APOBEC3s.
The results reported above imply that VifIIIB interacts intracellularly with all tested APOBEC3s. To address the potential antiviral consequences of these interactions and possible differences between Vif isolates, we cotransfected the APOBEC3 expression vectors with vectors encoding either wt or Δvif HIV-1 into 293T cells, and we subsequently harvested the released virions and analyzed their infectivities in HeLa-CD4-CCR5 cells (clone JC-53) (36). The infectivity of HIV-1HXB2(Δvif) was inhibited strongly by A3G and to lesser but significant extents by A3A, A3B, A3C, and A3F (Fig. 3A, right). Interestingly, these inhibitory effects were all substantially or completely eliminated by VifHXB2, as seen by the much greater infectivities of the corresponding wt virus. Consistent with evidence described below, this implied that VifHXB2 might neutralize all these APOBEC3s rather than only A3F and A3G. Similarly, A3C, A3F, and A3G inhibited the infectivity of HIV-1IIIB(Δvif) and were substantially neutralized by VifIIIB (Fig. 3A, left). However, A3A and A3B were reproducibly much more inhibitory to HIV-1IIIB(wt) than to HIV-1IIIB(Δvif). Indeed, A3B was almost as inhibitory to HIV-1IIIB(wt) as A3G was to HIV-1IIIB(Δvif) (Fig. 3A). Statistical analyses using the paired-comparison t test confirmed that VifHXB2 significantly inhibits the anti-HIV-1 activities of A3B (P ≤ 0.05; n = 5) and A3A (P ≤ 0.03; n = 5) whereas VifIIIB significantly enhances the anti-HIV-1 activities of the same enzymes (P ≤ 0.03; n = 7 and n = 4, respectively). Although the enhancements of A3A and A3B anti-HIV-1 activities by VifIIIB initially seemed counterintuitive, we note that data shown but not emphasized by Yu and coworkers also suggested that VifIIIB enhances the anti-HIV-1 activity of A3B (52). Moreover, as shown above, VifIIIB induces the emigration of A3A, A3B, and A3C from nuclei to the cytosol (Fig. 2A). Importantly, these infectivity data support our immunofluorescence microscopic evidence that Vifs functionally interact to a significant degree with all APOBEC3s.
FIG. 3.
HIV-1 Vif variants have distinct effects on the cytosolic levels and anti-HIV-1 activities of APOBEC3s. 293T cells were cotransfected with expression vectors encoding APOBEC3s and wt or Δvif HIV-1. (A) Single-cycle infectivity assay. The infectivities of wt and Δvif derivatives of HIV-1IIIB (left) or HIV-1HXB2 (right) in the presence of APOBEC3s were measured relative to the same viruses in the absence of APOBEC3. Error bars, standard deviations. (B) Western blot analyses of APOBEC3s in cytosolic extracts of 293T cells that also expressed wt or Δvif HIV-1. VifHXB2 downmodulated all APOBEC3s. VifIIIB strongly downmodulated A3G and downmodulated A3F and A3C to lesser degrees, but it increased the expression of A3A and A3B. (C) Effects of wt HIV-1 JR-CSF, ELI-1, YU-2, and 89.6 isolates on cellular quantities of APOBEC3s in comparison to the effect of HIV-1HXB2(Δvif). α-Tubulin (α-tub) blots in panels B and C show equivalent protein loading.
Based on these results, we analyzed the influences of these Vifs on the quantities of APOBEC3s in the cytosolic extracts from cotransfected 293T cultures. Consistent with previous evidence (27, 28, 41, 45, 53) and with Fig. 3A, A3G was downmodulated (decreased in quantity) by both Vifs in comparison to the Δvif negative controls (Fig. 3B). Similarly, A3F and A3C were also downmodulated by both Vifs, although these downmodulations were weaker with VifIIIB than with VifHXB2. In contrast, A3A and A3B were downmodulated by VifHXB2, but their concentrations in the cytosolic extracts were strongly increased by VifIIIB. Similar increases in the cytosolic amounts of A3A and A3B occurred when we expressed codon-optimized HVifIIIB in the absence of other HIV-1 proteins (see Fig. S1 in the supplemental material at http://www.ohsu.edu/biochem/faculty/MarinetalSupplement.pdf). Thus, the opposite effects of VifIIIB and VifHXB2 on the antiviral activities of A3A and A3B (Fig. 3A) were paralleled by their correspondingly opposite effects on the cytosolic quantities of these proteins. The data of Bishop and coworkers also implied an inductive effect of VifIIIB on the quantity of A3B in cytosolic extracts (1). These results are consistent with our finding that Vifs induce emigration of A3A, A3B, and A3C from nuclei into the cytosol (Fig. 2), thus increasing their concentrations at sites of viral production. Additionally, VifHXB2 degrades all APOBEC3s more efficiently, resulting in net decreases in their cytosolic concentrations, whereas VifIIIB degrades them less efficiently, resulting in net increases in the cytosolic concentrations of A3A and A3B. In contrast to these results, Vifs had no significant effect on the expression of other proteins encoded by the same vectors. Dilutions of our vectors also indicated that the distinct effects of VifHXB2 and VifIIIB were not caused by differences in their expression levels in the 293T cell system (data not shown).
We expanded this analysis using a broader panel of wt HIV-1 strains including JR-CSF, ELI-1, YU-2, and 89.6 in addition to HXB2 (Fig. 3C). In comparison to the HIV-1HXB2(Δvif) negative control, the Vif-expressing viruses differed dramatically in their abilities to alter the cytosolic concentrations of APOBEC3s. For example, ELI-1 strongly downmodulated all these cytidine deaminases to a greater extent than HXB2, whereas JR-CSF downmodulated A3G but significantly increased the cytosolic amounts of A3A, A3B, and A3C. Thus, different HIV-1 Vifs at similar expression levels broadly but differentially alter the cytosolic concentrations of all the APOBEC3s tested. In all cases, the antiviral activities of the APOBEC3s were correlated with their net cytosolic concentrations in the virus-producing cells and not with their total cellular or intranuclear concentrations.
Natural HIV-1 Vif variants bind promiscuously to APOBEC3s.
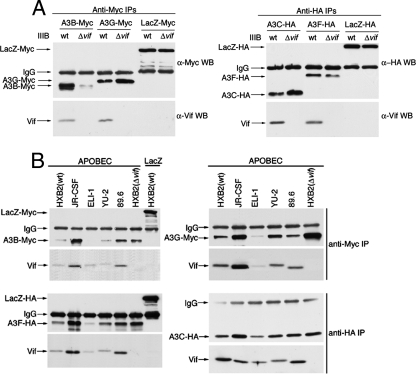
Our finding that HIV-1 Vifs strongly influence the intracellular localizations, concentrations, and activities of all APOBEC3s suggested that these proteins must directly or indirectly interact. To initially test this, we cotransfected 293T cells with epitope-tagged APOBEC3 expression vectors or negative-control plasmids encoding identically tagged LacZ along with vectors encoding HIV-1IIIB(wt) or HIV-1IIIB(Δvif). Proteins immunoprecipitated from the cytosolic extracts with anti-tag antibodies were subsequently analyzed by Western immunoblotting for the presence of the tagged proteins and for VifIIIB (Fig. 4A). VifIIIB coimmunoprecipitated with A3B-Myc, A3C-HA, A3F-HA, and A3G-Myc but not with LacZ-Myc or LacZ-HA by using the same anti-tag antibodies. Identical results were obtained in assays using expression vectors for HVifIIIB in the absence of other HIV-1 proteins (results not shown) (see Fig. S6A in the supplemental material at http://www.ohsu.edu/biochem/faculty/MarinetalSupplement.pdf). Similar studies using the wt virus JR-CSF, ELI-1, YU-2, 89.6, or HXB2 or the negative control HXB2(Δvif) indicated that all of these Vifs also coimmunoprecipitated with A3B-Myc, A3C-HA, A3F-HA, and A3G-Myc but not with LacZ-Myc or LacZ-HA (Fig. 4B). Additional studies suggested that the Vifs also associate significantly but less strongly with A3A-HA (results not shown). We conclude that HIV-1 Vifs bind to all of the tested human APOBEC3s rather than only to A3F and A3G.
FIG. 4.
HIV-1 Vifs bind promiscuously to all APOBEC3 paralogs. Extracts of cotransfected 293T cells that expressed HIV-1 molecular clones plus A3B-Myc, A3G-Myc, A3F-HA, or A3C-HA or the LacZ-Myc or LacZ-HA negative control were immunoprecipitated with anti-Myc or anti-HA antibodies, and the immunoprecipitates were analyzed by Western immunoblotting with either anti-Myc, anti-HA, or anti-Vif antibodies, as indicated. Vifs coimmunoprecipitated with all APOBEC3s.
A concern with the results reported above derives from our use of the 293T cell system, which could potentially result in high levels of protein expression and in adventitious protein interactions that would not occur under more physiological conditions. We addressed this by using small amounts of APOBEC3 expression vectors (0.3 μg per 35-mm-diameter culture) in all of our 293T cell transfections and by establishing that the resulting A3G level was similar to the amount in H9 T cells, even when the latter were cultured in the absence of extracellular signal-regulated kinase activators, which induce A3G and A3B (see Fig. S2 in the supplemental material at http://www.ohsu.edu/biochem/faculty/MarinetalSupplement.pdf). Moreover, our Vif vectors lack a simian virus 40 ori, and the Vif levels in 293T cells were also similar to the amounts in infected T cells.
In any case, the binding results shown in Fig. 4 did not depend on protein overexpression, because they persisted in the dilute cell extracts, and because the associations formed rapidly when highly diluted cell extracts that separately contain either Vifs or APOBEC3s were mixed at 4°C (see Fig. S6B in the supplemental material at http://www.ohsu.edu/biochem/faculty/MarinetalSupplement.pdf).
After we had completed the studies for which results are shown in Fig. 3 and 4, Langlois et al. (21) reported that VifIIIB binds to A3C and induces its downmodulation, whereas Doehle et al. (10) reported that neither A3B nor A3C binds to VifIIIB. To obtain additional evidence concerning the association of A3B with VifIIIB, we compared results using our A3B-Myc vector with the A3B-HA vector used by Doehle et al. (10). This was important because our A3B sequence has two amino acid substitutions compared to that used by the other group and because they used a different buffer to lyse the cells. We found that VifIIIB increased the concentration of their A3B-HA protein in cytosolic extracts of 293T cells and also associated with it, as seen by coimmunoprecipitation (see Fig. S3 in the supplemental material at http://www.ohsu.edu/biochem/faculty/MarinetalSupplement.pdf).
APOBEC3s and HVifIIIB accumulate within cytoplasmic SGs.
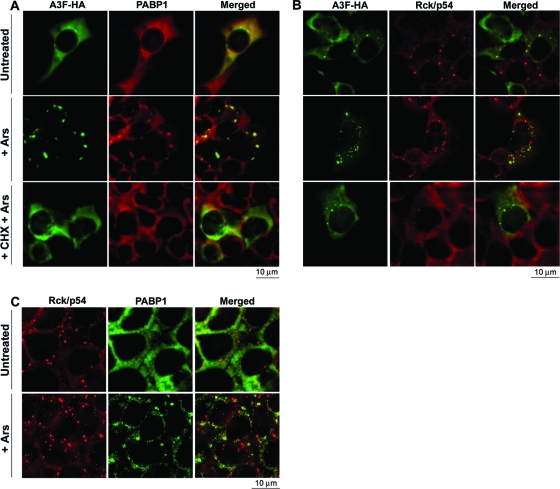
In response to cellular stressors such as sodium arsenite, A3G-mRNA-PABP1 complexes translocate from translationally active polysomes to SGs, a category of cytoplasmic microdomain that stores 5′ cap-dependent mRNAs whose translational initiation is temporarily suppressed (13, 19). We previously found that A3F binds to PABP1 and YB-1, defined markers for active polysomes and SGs, which are absent in P-bodies (19). In agreement with these considerations, sodium arsenite caused nearly all of the cellular A3F to accumulate in SGs that contained PABP1, and this accumulation was blocked when translational elongation was inhibited by cycloheximide (Fig. 5A). Although a significant amount of A3F occurred in P-bodies in unstressed cells (Fig. 2C and 5B), stress caused A3F to move slowly from the Rck/p54-containing P-bodies and into SGs, resulting in a substantial separation between these proteins (Fig. 5B, center row). Addition of cycloheximide to the stressed cells caused dispersal of SGs (Fig. 5A) and of Rck/p54-containing P-bodies that lacked A3F (Fig. 5B, bottom row). Notably, however, the few P-bodies that contained residual A3F were relatively resistant to dispersal by cycloheximide, in agreement with our other evidence that A3F stabilizes P-bodies (Fig. 2C).
FIG. 5.
Stress causes A3F and PABP1 to accumulate in SGs. (A and B) 293T cells transiently transfected with the expression vector for A3F-HA were treated with sodium arsenite (Ars) in the presence or absence of cycloheximide (CHX) prior to immunostaining for A3F-HA and PABP1 (A) or for A3F-HA and Rck/p54 (B). (C) 293T cells transiently transfected with the negative-control vector pcDNA3.1 were treated with sodium arsenite prior to staining for Rck/p54 and PABP1. Untreated cultures were used as negative controls. (A) Arsenite causes A3F to accumulate in SGs along with PABP1 (center). CHX blocks this effect of arsenite (bottom). (B) A3F moves from P-bodies to SGs in arsenite-treated cells (center). (C) Rck/p54 P-bodies coexist with and are distinct from PABP1-containing SGs.
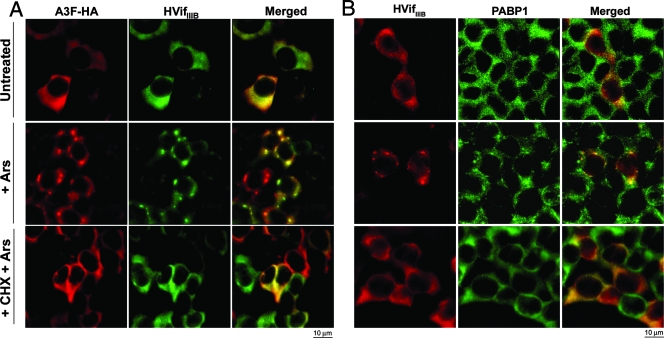
As shown above, HVifIIIB partially colocalizes with A3F in P-bodies (Fig. 2B). To determine whether HVifIIIB has an effect on the accumulation of A3F in SGs, 293T cells transiently expressing A3F-HA and HVifIIIB were stained for A3F and HVifIIIB or PABP1. Arsenite induced the accumulation of A3F in SGs in a manner similar to that observed in the absence of HVifIIIB (Fig. 6A). Furthermore, HVifIIIB colocalized with A3F in these SGs (Fig. 6A). Importantly, HVifIIIB also localized in SGs in arsenite-treated cells lacking A3F (Fig. 6B). However, analysis of large-field images showed that A3F substantially enhances the recruitment of HVifIIIB into SGs (see Fig. S4 in the supplemental material at http://www.ohsu.edu/biochem/faculty/MarinetalSupplement.pdf).
FIG. 6.
HVifIIIB colocalizes with A3F and PABP1 in SGs. 293T cells transiently transfected with expression vectors for A3F-HA and HVifIIIB (A) or HVifIIIB alone (B) were treated with sodium arsenite (Ars) in the presence or absence of cycloheximide (CHX) prior to immunostaining for A3F-HA and HVifIIIB (A) or HVifIIIB and PABP1 (B). Untreated cultures were used as negative controls. (A) A3F and HVifIIIB colocalize in SGs in arsenite-treated cells (center). (B) HVifIIIB also localizes in SGs when expressed alone (center).
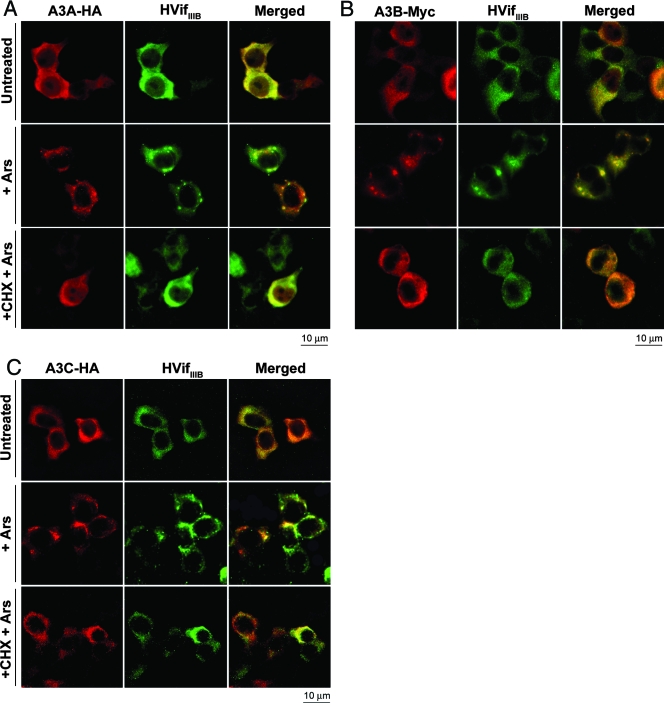
We analyzed the effects of stress on the localizations of A3A, A3B, and A3C in the presence and absence of HVifIIIB. Consistent with the results shown in Fig. 2, in the presence of HVifIIIB, a major portion of these APOBEC3 proteins were in the cytosol. Arsenite caused substantial proportions of these cytosolic APOBEC3s to accumulate with HVifIIIB in SGs, and in all cases these accumulations were prevented by cycloheximide (Fig. 7). Control studies established that the arsenite-induced cytosolic APOBEC3 accumulations contained PABP1, indicating that they were SGs (data not shown). In cells expressing the APOBEC3s in the absence of HVifIIIB, arsenite treatment caused most of the cytoplasmic APOBEC3 proteins to accumulate within SGs, whereas the nuclear levels of the APOBEC3 proteins remained uniformly high, suggesting that only the cytoplasmic APOBEC3 proteins were translocated to the SGs (see Fig. S5 in the supplemental material at http://www.ohsu.edu/biochem/faculty/MarinetalSupplement.pdf).
FIG. 7.
A3A, A3B, and A3C colocalize with HVifIIIB in SGs. 293T cells transiently coexpressing HVifIIIB and A3A-HA (A), A3B-Myc (B), or A3C-HA (C) were fixed and stained for Vif and the corresponding APOBEC3s. Some cultures were treated with sodium arsenite (Ars) in the presence or absence of cycloheximide (CHX) prior to fixation. Untreated cultures were used as negative controls. A3A, A3B, and A3C move to the cytoplasm in the presence of HVifIIIB. In arsenite-treated cells, all three APOBEC3s accumulate within SGs along with HVifIIIB. CHX prevents the formation of SGs in all cases.
RNA differentially modulates APOBEC3-Vif interactions.
Although the results reported above strongly suggest that Vifs interact closely and specifically with APOBEC3s, it remains uncertain whether these interactions are direct or mediated by mRNA or other factors. Previously, we found that efficient degradation of RNA with RNase A caused an increase in A3G-Vif coimmunoprecipitation, suggesting that RNA binding may sterically interfere with their association (19). However, we could not exclude the possibility that fragments of RNA might be protected from degradation by association with the proteins and that these protected RNAs might mediate the Vif-A3G binding. To address these issues, we coexpressed different epitope-tagged derivatives of A3B, A3C, and A3F or negative-control plasmids encoding identically tagged LacZ along with an HVifIIIB expression vector in 293T cells and performed coimmunoprecipitation assays from untreated or RNase A-treated cell extracts (see Fig. S6A in the supplemental material at http://www.ohsu.edu/biochem/faculty/MarinetalSupplement.pdf). In addition, we prepared the APOBEC3 and HVifIIIB proteins in separate cell extracts that were treated with RNase A before the samples were mixed and the complexes were allowed to form at 4°C (see Fig. S6B in the supplemental material at http://www.ohsu.edu/biochem/faculty/MarinetalSupplement.pdf). The results confirmed the evidence in Fig. 4 that all APOBEC3s associate with HVifIIIB in undigested samples. The effects of RNase A were complex, with some APOBEC3s binding more extensively after digestion (i.e., A3B and A3G) and some binding to a lesser extent (i.e., A3C and A3F). However, the most striking result was that the efficient and documented elimination of RNA by RNase A caused only modest two- to threefold changes in the extents of APOBEC3s associations with HVifIIIB. While these results might have different interpretations as described above, they are incompatible with the idea that the proteins bind at different positions on large RNAs and coimmunoprecipitate as a consequence.
DISCUSSION
APOBEC3 proteins are components of a powerful cellular defense network that inhibits the replication and amplification of a broad number of exogenous viruses and endogenous retroelements (4, 8, 15). Most previous studies characterized the effects of HIV-1 VifIIIB on the APOBEC3 paralogs A3G and A3F. Here we show that HIV-1 Vifs have a broader impact on APOBEC3s than previously reported and that different Vif-isolates have distinct abilities to degrade and neutralize the anti-HIV-1 activities of the APOBEC3s tested (Fig. 3). Moreover, Vifs induce several APOBEC3s to emigrate from nuclei into the cytosol (Fig. 2A). All the cytosolic APOBEC3s appear to associate with mRNA-PABP1 complexes that shuttle with Vif between different sites of mRNA metabolism (Fig. 2, 5, 6, and 7).
HIV-1 Vifs functionally interact with diverse APOBEC3s.
Our immunocytochemical, biochemical, and infectivity analyses all strongly suggest that HIV-1 Vifs from different HIV-1 isolates interact closely with all of the tested APOBEC3 paralogs and not just with A3F and A3G. These associations occur intracellularly, as indicated by the effects of Vifs on APOBEC3 subcellular localizations (Fig. 1 and 2), anti-HIV-1 activities (Fig. 3A), and concentrations (Fig. 3B and C). Surprisingly, the biological outcomes of these interactions depend on the HIV-1 Vif isolate. VifHXB2 and VifELI-1 strongly downmodulate all tested APOBEC3s, consequently diminishing their anti-HIV-1 activities. Similarly, VifIIIB degrades and neutralizes A3G, A3F, and A3C to different degrees. However, VifIIIB does not significantly degrade A3A or A3B, although it induces their movement into the cytosol (Fig. 2) and thereby causes net increases in their anti-HIV-1 activities (Fig. 3A). These results are in agreement with other studies, which were consistent with the idea that VifIIIB enhances the cytosolic concentration and anti-HIV-1 activity of A3B (1, 52). The different activities of Vifs are especially intriguing. Although Vifs contain many conserved sequences, the overall identity of the Vifs we examined was only 76%. Moreover, many of the amino acid differences occur at positions that are highly polymorphic, with high proportions of nonsynonymous substitutions indicative of positive selection (31, 51). We are currently attempting to identify the Vif polymorphisms that influence its activities against different APOBEC3s.
The broad range of Vif-APOBEC3 interactions is surprising because the APOBEC3s are structurally diverse, having only regions of segmental identity interspersed by divergent sequences (17). Moreover, the segments of identity differ for the APOBEC3s being compared. Indeed, the enormous diversities of APOBEC3s imply that Vif might have more than one binding site for their association or that their associations with Vif might be mediated by one or more bridging factors. Recently, we found that RNase A treatment caused an increase in A3G-Vif coimmunoprecipitation, suggesting that RNA can interfere with their interaction (19). In this context, it should be understood that Vif and APOBEC3s might potentially associate directly as well as indirectly and that RNA could interfere with as well as mediate some of these interactions. The mild sensitivities of the Vif-APOBEC3 associations to RNase A treatment suggest that these interactions are not completely RNA dependent (see Fig. S6 in the supplemental material at http://www.ohsu.edu/biochem/faculty/MarinetalSupplement.pdf). A parsimonious interpretation is that Vifs associate with APOBEC3s in a manner that is sufficiently close and stable to enable their coimmunoprecipitations from RNase A-digested extracts as well as to permit the Vif-dependent subcellular redistribution and degradation of APOBEC3s. Presumably, if Vifs have more than one site for binding APOBEC3s, groups of these cytidine deaminases might bind to these different sites by direct and/or indirect mechanisms. In agreement with this idea, recent studies suggest that different APOBEC3s might bind to distinct sites on Vif (47; also data not shown). Further investigations are required to elucidate this issue.
APOBEC3s and Vif associate with mRNA metabolic sites.
An important outcome of our immunofluorescence studies is the detection of A3F in cytoplasmic clusters under steady-state conditions in cells (Fig. 2 and 5). The majority of these clusters were identified as P-bodies due to their colocalization with Rck/p54 (Fig. 2C). Interestingly, HVifIIIB colocalizes in all the A3F-containing clusters (Fig. 2B). The finding that HVifIIIB localizes in P-bodies only in the presence of A3G (49) and A3F strongly suggests that Vif is recruited to P-bodies due to its interactions with these APOBEC3s. Wichroski et al., recently reported the localization of A3F and A3G in cytoplasmic clusters that were entirely classified as P-bodies (49). In contrast, Gallois-Montbrun et al. found a heterogeneous population of A3G granules, with the majority being P-bodies and the remainder considered to be either a different class of P-bodies or other cytoplasmic microdomains (13). Our data concerning the A3F clusters are in close agreement with the latter report, although approximately 95% of the A3F clusters that we detected contained the P-body marker Rck/p54.
Cycloheximide, an inhibitor of translation elongation known to decrease the number and size of P-bodies (42), caused substantial decreases in the numbers and sizes of P-bodies lacking A3F but had no effect on any of the A3F clusters (Fig. 2C). This result implies that A3F alters the dynamics of P-body function, possibly by preventing or delaying the processing or export of mRNAs. Furthermore, the association of A3F with components of polysomes and SGs (6, 19) (as discussed below), coupled with its shuttling between these sites and P-bodies in response to protein synthesis inhibitors and/or stress, is compatible with the hypothesis that A3F plays an active role in the function of these messenger ribonucleoprotein complexes.
An important finding is the localization of A3A, A3B, A3C, A3F, and HVifIIIB along with mRNA-PABP1 complexes within SGs in a manner preventable by cycloheximide (Fig. 6 and 7; also Fig. S5 in the supplemental material at http://www.ohsu.edu/biochem/faculty/MarinetalSupplement.pdf). Although this movement of HVifIIIB into SGs occurs to a degree in cells lacking APOBEC3s, the presence of A3F substantially increased the accumulation of HVifIIIB in SGs (Fig. 6; also Fig. S4 in the supplemental material at http://www.ohsu.edu/biochem/faculty/MarinetalSupplement.pdf). In contrast, the other three APOBEC3s do not noticeably enhance the accumulation of HVifIIIB in SGs, implying differences in the avidities and/or functions of these HVifIIIB-APOBEC3 complexes. Interestingly, arsenite treatment does not induce the movement of A3A, A3B, and A3C from nuclei into the cytosol, suggesting that their translocation to the cytosol is a specific effect of Vif rather than a generalized stress response (see Fig. S5 in the supplemental material at http://www.ohsu.edu/biochem/faculty/MarinetalSupplement.pdf). Our results demonstrate that the tested APOBEC3s and HVifIIIB colocalize at multiple cytosolic sites of mRNA metabolism including P-bodies, SGs, and polysomes. Thus, the battle between Vifs and APOBEC3s occurs predominantly in the context of mRNA, including HIV-1 genomic RNA, although APOBEC3s also associate with other RNAs (6, 19). Moreover, APOBEC3s appear to function in part as a mobile and inducible defense network rather than simply as individuals.
Acknowledgments
This research was supported by grant AI49729 from the U.S. National Institutes of Health.
We thank Bryan Cullen for several APOBEC3 expression vectors, Michael Malim for Vif mutants, and the NIH AIDS Research and Reference Reagent Program for anti-Vif and anti-A3G antibodies.
Footnotes
Published ahead of print on 31 October 2007.
REFERENCES
- 1.Bishop, K. N., R. K. Holmes, A. M. Sheehy, N. O. Davidson, S. J. Cho, and M. H. Malim. 2004. Cytidine deamination of retroviral DNA by diverse APOBEC proteins. Curr. Biol. 141392-1396. [DOI] [PubMed] [Google Scholar]
- 2.Chen, H., C. E. Lilley, Q. Yu, D. V. Lee, J. Chou, I. Narvaiza, N. R. Landau, and M. D. Weitzman. 2006. APOBEC3A is a potent inhibitor of adeno-associated virus and retrotransposons. Curr. Biol. 16480-485. [DOI] [PubMed] [Google Scholar]
- 3.Chen, K., J. Huang, C. Zhang, S. Huang, G. Nunnari, F. X. Wang, X. Tong, L. Gao, K. Nikisher, and H. Zhang. 2006. Alpha interferon potently enhances the anti-human immunodeficiency virus type 1 activity of APOBEC3G in resting primary CD4 T cells. J. Virol. 807645-7657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chiu, Y. L., and W. C. Greene. 2006. Multifaceted antiviral actions of APOBEC3 cytidine deaminases. Trends Immunol. 27291-297. [DOI] [PubMed] [Google Scholar]
- 5.Chiu, Y. L., V. B. Soros, J. F. Kreisberg, K. Stopak, W. Yonemoto, and W. C. Greene. 2005. Cellular APOBEC3G restricts HIV-1 infection in resting CD4+ T cells. Nature 435108-114. [DOI] [PubMed] [Google Scholar]
- 6.Chiu, Y. L., H. E. Witkowska, S. C. Hall, M. Santiago, V. B. Soros, C. Esnault, T. Heidmann, and W. C. Greene. 2006. High-molecular-mass APOBEC3G complexes restrict Alu retrotransposition. Proc. Natl. Acad. Sci. USA 10315588-15893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Conticello, S. G., C. J. Thomas, S. K. Petersen-Mahrt, and M. S. Neuberger. 2005. Evolution of the AID/APOBEC family of polynucleotide (deoxy)cytidine deaminases. Mol. Biol. Evol. 22367-377. [DOI] [PubMed] [Google Scholar]
- 8.Cullen, B. R. 2006. Role and mechanism of action of the APOBEC3 family of antiretroviral resistance factors. J. Virol. 801067-1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dang, Y., X. Wang, W. J. Esselman, and Y. H. Zheng. 2006. Identification of APOBEC3DE as another antiretroviral factor from the human APOBEC family. J. Virol. 8010522-10533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Doehle, B. P., A. Schafer, and B. R. Cullen. 2005. Human APOBEC3B is a potent inhibitor of HIV-1 infectivity and is resistant to HIV-1 Vif. Virology 339281-288. [DOI] [PubMed] [Google Scholar]
- 11.Doehle, B. P., A. Schafer, H. L. Wiegand, H. P. Bogerd, and B. R. Cullen. 2005. Differential sensitivity of murine leukemia virus to APOBEC3-mediated inhibition is governed by virion exclusion. J. Virol. 798201-8207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fang, L., and N. R. Landau. 2007. Analysis of Vif-induced APOBEC3G degradation using an alpha-complementation assay. Virology 359162-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gallois-Montbrun, S., B. Kramer, C. M. Swanson, H. Byers, S. Lynham, M. Ward, and M. H. Malim. 2007. Antiviral protein APOBEC3G localizes to ribonucleoprotein complexes found in P bodies and stress granules. J. Virol. 812165-2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Helseth, E., M. Kowalski, D. Gabuzda, U. Olshevsky, W. Haseltine, and J. Sodroski. 1990. Rapid complementation assays measuring replicative potential of human immunodeficiency virus type 1 envelope glycoprotein mutants. J. Virol. 642416-2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Holmes, R. K., M. H. Malim, and K. N. Bishop. 2007. APOBEC-mediated viral restriction: not simply editing? Trends Biochem. Sci. 32118-128. [DOI] [PubMed] [Google Scholar]
- 16.Hwang, S. S., T. J. Boyle, H. K. Lyerly, and B. R. Cullen. 1991. Identification of the envelope V3 loop as the primary determinant of cell tropism in HIV-1. Science 25371-74. [DOI] [PubMed] [Google Scholar]
- 17.Jarmuz, A., A. Chester, J. Bayliss, J. Gisbourne, I. Dunham, J. Scott, and N. Navaratnam. 2002. An anthropoid-specific locus of orphan C to U RNA-editing enzymes on chromosome 22. Genomics 79285-296. [DOI] [PubMed] [Google Scholar]
- 18.Kobayashi, M., A. Takaori-Kondo, Y. Miyauchi, K. Iwai, and T. Uchiyama. 2005. Ubiquitination of APOBEC3G by an HIV-1 Vif-Cullin5-Elongin B-Elongin C complex is essential for Vif function. J. Biol. Chem. 28018573-18578. [DOI] [PubMed] [Google Scholar]
- 19.Kozak, S. L., M. Marin, K. M. Rose, C. Bystrom, and D. Kabat. 2006. The anti-HIV-1 editing enzyme APOBEC3G binds HIV-1 RNA and messenger RNAs that shuttle between polysomes and stress granules. J. Biol. Chem. 28129105-29119. [DOI] [PubMed] [Google Scholar]
- 20.Kshirsagar, M., and R. Parker. 2004. Identification of Edc3p as an enhancer of mRNA decapping in Saccharomyces cerevisiae. Genetics 166729-739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Langlois, M. A., R. C. Beale, S. G. Conticello, and M. S. Neuberger. 2005. Mutational comparison of the single-domained APOBEC3C and double-domained APOBEC3F/G anti-retroviral cytidine deaminases provides insight into their DNA target site specificities. Nucleic Acids Res. 331913-1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lecossier, D., F. Bouchonnet, F. Clavel, and A. J. Hance. 2003. Hypermutation of HIV-1 DNA in the absence of the Vif protein. Science 3001112. [DOI] [PubMed] [Google Scholar]
- 23.Liu, B., X. Yu, K. Luo, Y. Yu, and X. F. Yu. 2004. Influence of primate lentiviral Vif and proteasome inhibitors on human immunodeficiency virus type 1 virion packaging of APOBEC3G. J. Virol. 782072-2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Madani, N., and D. Kabat. 2000. Cellular and viral specificities of human immunodeficiency virus type 1 Vif protein. J. Virol. 745982-5987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Madani, N., and D. Kabat. 1998. An endogenous inhibitor of human immunodeficiency virus in human lymphocytes is overcome by the viral Vif protein. J. Virol. 7210251-10255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mangeat, B., P. Turelli, G. Caron, M. Friedli, L. Perrin, and D. Trono. 2003. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature 42499-103. [DOI] [PubMed] [Google Scholar]
- 27.Marin, M., K. M. Rose, S. L. Kozak, and D. Kabat. 2003. HIV-1 Vif protein binds the editing enzyme APOBEC3G and induces its degradation. Nat. Med. 91398-1403. [DOI] [PubMed] [Google Scholar]
- 28.Mehle, A., B. Strack, P. Ancuta, C. Zhang, M. McPike, and D. Gabuzda. 2004. Vif overcomes the innate antiviral activity of APOBEC3G by promoting its degradation in the ubiquitin-proteasome pathway. J. Biol. Chem. 2797792-7798. [DOI] [PubMed] [Google Scholar]
- 29.Navarro, F., and N. R. Landau. 2004. Recent insights into HIV-1 Vif. Curr. Opin. Immunol. 16477-482. [DOI] [PubMed] [Google Scholar]
- 30.Nguyen, K. L., M. Llano, H. Akari, E. Miyagi, E. M. Poeschla, K. Strebel, and S. Bour. 2004. Codon optimization of the HIV-1 vpu and vif genes stabilizes their mRNA and allows for highly efficient Rev-independent expression. Virology 319163-175. [DOI] [PubMed] [Google Scholar]
- 31.Oberste, M. S., and M. A. Gonda. 1992. Conservation of amino-acid sequence motifs in lentivirus Vif proteins. Virus Genes 695-102. [DOI] [PubMed] [Google Scholar]
- 32.Page, K. A., N. R. Landau, and D. R. Littman. 1990. Construction and use of a human immunodeficiency virus vector for analysis of virus infectivity. J. Virol. 645270-5276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Page, K. A., S. M. Stearns, and D. R. Littman. 1992. Analysis of mutations in the V3 domain of gp160 that affect fusion and infectivity. J. Virol. 66524-533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peng, G., K. J. Lei, W. Jin, T. Greenwell-Wild, and S. M. Wahl. 2006. Induction of APOBEC3 family proteins, a defensive maneuver underlying interferon-induced anti-HIV-1 activity. J. Exp. Med. 20341-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Platt, E. J., S. E. Kuhmann, P. P. Rose, and D. Kabat. 2001. Adaptive mutations in the V3 loop of gp120 enhance fusogenicity of human immunodeficiency virus type 1 and enable use of a CCR5 coreceptor that lacks the amino-terminal sulfated region. J. Virol. 7512266-12278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Platt, E. J., K. Wehrly, S. E. Kuhmann, B. Chesebro, and D. Kabat. 1998. Effects of CCR5 and CD4 cell surface concentrations on infections by macrophagetropic isolates of human immunodeficiency virus type 1. J. Virol. 722855-2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rose, K. M., M. Marin, S. L. Kozak, and D. Kabat. 2005. Regulated production and anti-HIV type 1 activities of cytidine deaminases APOBEC3B, 3F, and 3G. AIDS Res. Hum. Retrovir. 21611-619. [DOI] [PubMed] [Google Scholar]
- 38.Rose, K. M., M. Marin, S. L. Kozak, and D. Kabat. 2004. Transcriptional regulation of APOBEC3G, a cytidine deaminase that hypermutates human immunodeficiency virus. J. Biol. Chem. 27941744-41749. [DOI] [PubMed] [Google Scholar]
- 39.Rose, K. M., M. Marin, S. L. Kozak, and D. Kabat. 2004. The viral infectivity factor (Vif) of HIV-1 unveiled. Trends Mol. Med. 10291-297. [DOI] [PubMed] [Google Scholar]
- 40.Sheehy, A. M., N. C. Gaddis, J. D. Choi, and M. H. Malim. 2002. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature 418646-650. [DOI] [PubMed] [Google Scholar]
- 41.Sheehy, A. M., N. C. Gaddis, and M. H. Malim. 2003. The antiretroviral enzyme APOBEC3G is degraded by the proteasome in response to HIV-1 Vif. Nat. Med. 91404-1407. [DOI] [PubMed] [Google Scholar]
- 42.Sheth, U., and R. Parker. 2003. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science 300805-808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Simon, V., V. Zennou, D. Murray, Y. Huang, D. D. Ho, and P. D. Bieniasz. 2005. Natural variation in Vif: differential impact on APOBEC3G/3F and a potential role in HIV-1 diversification. PLoS Pathog. 1e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Soros, V. B., W. Yonemoto, and W. C. Greene. 2007. Newly synthesized APOBEC3G is incorporated into HIV virions, inhibited by HIV RNA, and subsequently activated by RNase H. PLoS Pathog. 3e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stopak, K., C. de Noronha, W. Yonemoto, and W. C. Greene. 2003. HIV-1 Vif blocks the antiviral activity of APOBEC3G by impairing both its translation and intracellular stability. Mol. Cell 12591-601. [DOI] [PubMed] [Google Scholar]
- 46.Stopak, K. S., Y. L. Chiu, J. Kropp, R. M. Grant, and W. C. Greene. 2007. Distinct patterns of cytokine regulation of APOBEC3G expression and activity in primary lymphocytes, macrophages, and dendritic cells. J. Biol. Chem. 2823539-3546. [DOI] [PubMed] [Google Scholar]
- 47.Tian, C., X. Yu, W. Zhang, T. Wang, R. Xu, and X. F. Yu. 2006. Differential requirement for conserved tryptophans in human immunodeficiency virus type 1 Vif for the selective suppression of APOBEC3G and APOBEC3F. J. Virol. 803112-3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wedekind, J. E., G. S. Dance, M. P. Sowden, and H. C. Smith. 2003. Messenger RNA editing in mammals: new members of the APOBEC family seeking roles in the family business. Trends Genet. 19207-216. [DOI] [PubMed] [Google Scholar]
- 49.Wichroski, M. J., G. B. Robb, and T. M. Rana. 2006. Human retroviral host restriction factors APOBEC3G and APOBEC3F localize to mRNA processing bodies. PLoS Pathog. 2e41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wiegand, H. L., B. P. Doehle, H. P. Bogerd, and B. R. Cullen. 2004. A second human antiretroviral factor, APOBEC3F, is suppressed by the HIV-1 and HIV-2 Vif proteins. EMBO J. 232451-2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang, W., J. P. Bielawski, and Z. Yang. 2003. Widespread adaptive evolution in the human immunodeficiency virus type 1 genome. J. Mol. Evol. 57212-221. [DOI] [PubMed] [Google Scholar]
- 52.Yu, Q., D. Chen, R. Konig, R. Mariani, D. Unutmaz, and N. R. Landau. 2004. APOBEC3B and APOBEC3C are potent inhibitors of simian immunodeficiency virus replication. J. Biol. Chem. 27953379-53386. [DOI] [PubMed] [Google Scholar]
- 53.Yu, X., Y. Yu, B. Liu, K. Luo, W. Kong, P. Mao, and X. F. Yu. 2003. Induction of APOBEC3G ubiquitination and degradation by an HIV-1 Vif-Cul5-SCF complex. Science 3021056-1060. [DOI] [PubMed] [Google Scholar]
- 54.Yu, Y., Z. Xiao, E. S. Ehrlich, X. Yu, and X. F. Yu. 2004. Selective assembly of HIV-1 Vif-Cul5-ElonginB-ElonginC E3 ubiquitin ligase complex through a novel SOCS box and upstream cysteines. Genes Dev. 182867-2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang, H., B. Yang, R. J. Pomerantz, C. Zhang, S. C. Arunachalam, and L. Gao. 2003. The cytidine deaminase CEM15 induces hypermutation in newly synthesized HIV-1 DNA. Nature 42494-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zheng, Y. H., D. Irwin, T. Kurosu, K. Tokunaga, T. Sata, and B. M. Peterlin. 2004. Human APOBEC3F is another host factor that blocks human immunodeficiency virus type 1 replication. J. Virol. 786073-6076. [DOI] [PMC free article] [PubMed] [Google Scholar]