Abstract
Arenaviruses are rodent-borne viruses, with five members of the family capable of causing severe hemorrhagic fevers if transmitted to humans. To date, two distinct cellular receptors have been identified that are used by different pathogenic viruses, α-dystroglycan by Lassa fever virus and transferrin receptor 1 (TfR1) by certain New World clade B viruses. Our previous studies have suggested that other, as-yet-unknown receptors are involved in arenavirus entry. In the present study, we examined the use of TfR1 by the glycoproteins (GPs) from a panel of New World clade B arenaviruses comprising three pathogenic and two nonpathogenic strains. Interestingly, we found that TfR1 was only used by the GPs from the pathogenic viruses, with entry of the nonpathogenic strains being TfR1 independent. The pathogenic GPs could also direct entry into cells by TfR1-independent pathways, albeit less efficiently. A comparison of the abilities of TfR1 orthologs from different species to support arenavirus entry found that the human and feline receptors were able to enhance entry of the pathogenic strains, but that neither the murine or canine forms were functional. Since the ability to use TfR1 is a characteristic feature of the human pathogens, this interaction may represent an important target in the treatment of New World hemorrhagic fevers. In addition, the ability to use TfR1 may be a useful tool to predict the likelihood that any existing or newly discovered viruses in this family could infect humans.
Arenaviruses are enveloped RNA viruses that enter host cells through the action of a fusion glycoprotein (GP) displayed on the surface of the viral particle (3). While the natural hosts of the viruses are rodents, several members of the family can also be transmitted to humans, causing severe hemorrhagic fevers. The combination of high morbidity/mortality rates, airborne transmission capability, and person-to-person spread has resulted in their classification by the Centers for Disease Control and Prevention as category A bioterrorism agents.
Phylogenetic analyses have divided the arenaviruses into Old World and New World strains. Lymphocytic choriomeningitis virus (LCMV) and Lassa fever virus (LASV) comprise the human pathogens in the Old World group, but only LASV causes a hemorrhagic fever. The New World arenaviruses are further subdivided into clades A, B, and C (2), with all of the human pathogens being found in clade B. These include Junin virus (JUNV), Machupo virus (MACV), and Guanarito virus (GTOV), which are carried by New World rodents and cause similar pathologies and mortality rates in humans exposed via contact with contaminated rodent waste (12, 15-18, 33). Clade B also includes Sabia virus (SABV), which has been reported to have caused only a single human infection outside of laboratory-acquired infections (24). It also contains three viruses that are not associated with any known natural human infections, Cupixi virus (CPXV), Tacaribe virus (TCRV), and Amapari virus (AMAV), although TCRV has been reported to have caused a single case of laboratory-acquired febrile illness with mild central nervous system symptoms (24).
The arenavirus GP is a typical class I fusion protein, comprising two noncovalently associated subunits, GP1 and GP2, which are cleaved from a precursor protein, GPC. When present on the surface of retroviral vector particles, arenavirus GPs are able to direct entry into a broad range of cell types in vitro (21, 26-28). The GP1 subunit contains the receptor binding site (22) and GP1-Fc immunoadhesin fusion proteins bind to cells in a manner that recapitulates the tropism of GP-pseudotyped retroviral vectors (21, 25, 28).
The arenaviruses display a complex pattern of receptor use. To date, two different cellular receptors have been identified, and it is likely that others exist (21, 26-28). LASV, certain strains of LCMV (5), and New World clade C viruses (32) use α-dystroglycan (α-DG) to enter cells, although some strains of LCMV can infect cells independently of α-DG (14, 27). More recently, transferrin receptor 1 (TfR1; also called TFRC) was reported as a receptor for the New World clade B arenaviruses that cause hemorrhagic fevers in humans (25).
We are interested in understanding the determinants of pathogenicity in the New World clade B arenaviruses and, in particular, the differences between the pathogenic and nonpathogenic members of this group. Previously, we reported that pseudotyped retroviral vectors displaying GPs from the pathogenic clade B1 viruses, JUNV and MACV, exhibited markedly different properties from vectors carrying the GP from the related, but nonpathogenic, virus TCRV (21). These findings prompted us to consider whether the ability to use alternate host receptors and/or entry pathways could underlie the differences in the ability of the clade B viruses to cause human disease.
In the present study, we have expanded the analysis of pathogenic and nonpathogenic clade B arenaviruses to include members of the B2 lineage, GTOV and AMAV. We examined dependence on TfR1 for entry into both human and rodent cells and observed a correlation between the ability to use human TfR1 (hTfR1) and known human pathogenicity. These observations also held true for the feline ortholog of TfR1, but the murine and canine receptors were unable to enhance entry by any GP. Finally, we discovered that the nonpathogenic clade B members could infect cells efficiently in a completely TfR1-independent manner, suggesting that the clade B viruses can use additional, unknown receptors.
MATERIALS AND METHODS
Cell lines.
293A, 293T, NIH 3T3, CHO-K1, and T1B27 cells were maintained in Dulbecco's modified Eagle's medium (DMEM) (Mediatech, Herndon, VA) supplemented with 10% fetal bovine serum (FBS) (HyClone, Logan, UT) and 2 mM glutamine (Gemini Bio-Products, West Sacramento, CA). CEM and BHK21 cells were maintained in RPMI (Mediatech, Herndon, VA) supplemented with 10% FBS and 2 mM glutamine. α-DG knockout (−/−) and heterozygous control (+/−) R1 murine embryonic stem (mES) cells (35) were generously provided by Kevin Campbell (University of Iowa) and cultured in DMEM supplemented with 20% FBS, 2 mM glutamine, 1 mM nonessential amino acids (Chemicon, Temecula, CA), 0.001% β-mercaptoethanol (Sigma, St. Louis, MO) and 103 U/ml of murine leukemia inhibiting factor (Millipore, Billerica, MA). All cells were maintained in 5% CO2 atmosphere except T1B27 and BHK21 cells, which were maintained at 10% CO2.
GP-pseudotyped retroviral vectors.
Expression plasmids for the GPs from JUNV (Parodi strain), MACV (Carvallo strain), TCRV (TRVL 11598 strain), LCMV (Armstrong 53b strain), LASV (Josiah strain), and vesicular stomatitis virus (VSV) (Indiana strain) have been described previously (21, 27). An expression plasmid for the GTOV GP (INH-95551 strain) was kindly provided by Stefan Kunz (The Scripps Research Institute) (28). A human codon-optimized form of AMAV GP (BeAn 70563 strain) was chemically synthesized and cloned into the pCAGGS expression vector (20). Pseudotyped retroviral vectors displaying GPs were generated by cotransfection of 293T cells with expression plasmids for the specific GP, together with plasmids expressing murine leukemia virus Gag-Pol and an enhanced green fluorescent protein (EGFP)-expressing retroviral vector genome, pMND-eGFP, as described previously (21, 27).
The efficiency of entry of the GP-pseudotyped retroviral vectors into cells was measured as previously described (21, 27). Briefly, vectors were incubated with target cells for 4 h and then replaced with fresh medium. After 48 h, the cells were trypsinized, washed in phosphate-buffered saline, and analyzed for EGFP expression using a FACScan flow cytometer (BD, Franklin Lakes, NJ). The efficiency of entry (titer) was determined by multiplying the percentage of EGFP-positive cells by the number of cells initially incubated with the vectors. Titers were expressed as transducing units (TU) per ml of vector-containing supernatant. In experiments with control and experimental arms (e.g., small interfering RNA [siRNA] treatment and antibody blocking), the titers were made relative to the control (mock treatment) cells and were expressed as a percentage of the control titer.
Antibody pretreatment assay.
293A cells were seeded in 12-well plates 1 day prior to transduction with GP-pseudotyped vectors. The cells were pretreated with 5 nM or 50 nM of mouse anti-human TfR1 antibody (clone M-A712; BD Biosciences, San Jose, CA) or medium alone for 30 min at 37°C, followed by transduction with GP-pseudotyped vectors for 4 h in the presence of the anti-hTfR1 antibody. The antibody-vector mixtures were then replaced with fresh media, and the cells were allowed to recover for 48 to 72 h before fluorescence-activated cell sorter (FACS) analysis for EGFP expression. Titers were determined by measurement of the percentage of EGFP-positive cells, as described above.
TfR1 knockdown by siRNA.
hTfR1 and murine TfR1 (mTfR1)-specific siRNAs were purchased as ON-TARGET plus SMART pools (Dharmacon, Lafayette, CO; catalog no. L-003941-00 and L-055550-01). A nontargeted siRNA (5′-AACACAGCAACCUCUACUUGG-3′) was used as a control. 293A or NIH 3T3 cells were seeded at 3 × 106 cells per 10-cm plate for 12 to 14 h before transfection. Forty microliters of Lipofectamine 2000 (Invitrogen) was mixed with 110 μl of Opti-MEM I (Invitrogen), incubated for 10 min at room temperature, and then added to 825 μl of Opti-MEM I plus 25 μl of 20 μM siRNA, followed by incubation for 20 min at room temperature. Opti-MEM I was added to the siRNA mixture to give a final volume of 5 ml and then added to cells prerinsed with Opti-MEM I. The cells were incubated for 4 h, when the siRNA mixture was replaced with fresh DMEM plus 20% FBS, for overnight incubation. The following day, the cells were trypsinized and seeded into six-well plates at a density of 1 × 105 cells per well. Twenty-four hours later, aliquots of cells from both control- and TfR1-siRNA-treated plates were harvested using sodium dodecyl sulfate-lysis buffer, in order to assay the efficiency of knockdown of TfR1 expression by Western blotting. The remaining cells were transduced with GP-pseudotyped vectors, and entry efficiency was determined by FACS analysis for EGFP expression, as described previously.
TfR1 expression was detected by Western blotting of cell lysates using a 1:500 dilution of mouse anti-TfR1 antibody, clone H68.4 (Invitrogen). This antibody recognizes a conserved epitope in chicken, mouse, rat, Chinese hamster, and human TfR1. Specific bands were detected using horseradish peroxidase-conjugated goat anti-mouse immunoglobulin G (IgG) (Pierce, Rockford, IL), diluted 1:10,000, followed by incubation with the ECL enhanced chemiluminescence detection reagent (Amersham Biosciences, Piscataway, NJ) and exposure to Kodak BIOMAX XAR film (Sigma). To ensure equal protein loading, membranes were also probed with mouse anti-actin monoclonal antibody (Sigma), diluted 1:10,000.
Transient transfection of CHO-K1 cells with TfR1.
Expression plasmids for murine, human, canine, and feline TfR1 orthologs, as well as a series of chimeric human/murine receptors (34), were generously provided by Susan Ross (University of Pennsylvania). CHO-K1 cells were transiently transfected with the expression plasmids using Lipofectamine 2000 (Invitrogen). Briefly, CHO-K1 cells were plated to be 95 to 100% confluent in 10-cm plates at the time of transfection and were transfected with 24 μg of TfR1 expression plasmids. Cells were incubated in transfection mixture for 4 h and then cultured overnight in DMEM plus 10% FBS. The following day, cells were trypsinized, plated into six-well plates at 40% confluence and incubated overnight. Cells were then transduced with GP-pseudotyped vectors for 4 h and the titers determined 48 h later by FACS analysis for EGFP expression, as described above. Cell samples were also collected at the time of transduction and analyzed by Western blotting to determine levels of TfR1 expression, as described above.
Immunoadhesin production and binding assay.
An immunoadhesin construct comprising the MACV GP1 sequence fused to a rabbit IgG heavy-chain Fc sequence has previously been described (21). Immunoadhesin-containing culture supernatants were generated by transfection of 293T cells, followed by concentration of culture supernatants using centrifugal filter units (21). The ability of the immunoadhesin to bind to CHO-K1 cells (control) or cells previously transfected with hTfR1 or mTFR1 expression plasmids was assessed by incubating 30 μl of the MACV immunoadhesin stock with 5 × 105 cells on ice for 30 min, followed by washing and incubation with 50 μl of a 1:50 dilution of fluorescein isothiocyanate (FITC)-conjugated anti-rabbit IgG (BD Pharmingen, San Jose, CA) for 20 min on ice. Cells were analyzed for binding on a FACScan flow cytometer, as described previously (21). As controls, aliquots of all cell populations were also stained with only the secondary antibody.
RESULTS
Consistent tropism differences are observed between pathogenic and nonpathogenic clade B GPs.
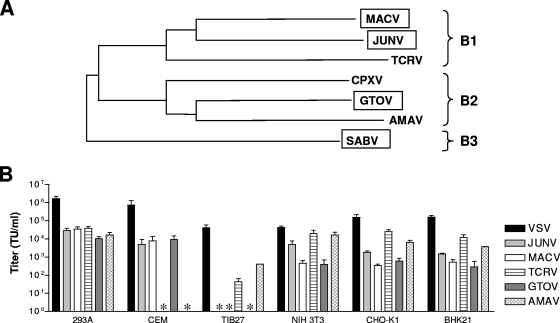
The New World clade B arenaviruses have been divided into subgroups B1, B2, and B3, based on phylogenetic relationships (Fig. 1A) (8). Previously, we reported differences in the characteristics of the entry pathways used by GPs from the pathogenic clade B1 viruses, JUNV and MACV, versus the related but nonpathogenic B1 virus, TCRV (21). Specifically, JUNV and MACV GP-pseudotyped retroviral vectors could transduce human (CEM) but not murine (TIB27) T-lymphocyte cell lines, while TCRV GP vectors demonstrated the opposite pattern. In addition, we also found that JUNV and MACV GP vectors were consistently less efficient at transducing cell lines derived from both mice (NIH 3T3) and Chinese hamsters (CHO-K1) compared to TCRV vectors. These observations led us to speculate that different cellular receptors and entry pathways were being used by the pathogenic versus nonpathogenic clade B1 viruses and that this characteristic might reflect an essential determinant of human pathogenicity.
FIG. 1.
Tropism of clade B GPs for different cell lines. (A) Phylogenetic relationships among the New World clade B arenaviruses based on sequence analysis of GP. Viruses that exhibit pathogenicity in humans (boxed) are distributed throughout all three sublineages (adapted from reference 8). (B) Titers of GP-pseudotyped retroviral vectors on various cell lines. The values shown are means ± standard errors from two to eight independent experiments. All vector-cell combinations that gave no titer (*) were confirmed using 10× concentrated stocks of vectors.
To examine whether these patterns were more generally true across the clade B viruses, we expanded the panel to include GPs from pathogenic (GTOV) and nonpathogenic (AMAV) clade B2 viruses. GP-pseudotyped vectors were generated and standardized on human kidney epithelial 293A cells, where we observed that all five of the clade B vectors produced similar titers (approximately 104 TU/ml of unconcentrated culture supernatant) (Fig. 1B). As a control, we included vectors pseudotyped with the VSV glycoprotein (VSV-G). This glycoprotein has very broad tropism, and the resulting vectors efficiently transduce diverse cell lines, thereby providing a control for the level of transduction directed by the retroviral vector machinery in each cell line. The vectors were then examined on the panel of indicator cell lines (CEM, TIB27, NIH 3T3, and CHO-K1 cells) that have previously revealed differences between the pathogenic and nonpathogenic B1 viruses (21). In addition, we also included a Syrian hamster cell line, BHK21.
The results of these studies revealed that GTOV GP had a tropism pattern that was most similar to those of the pathogenic B1 viruses, JUNV and MACV, while the AMAV GP vectors were more similar to the nonpathogenic TCRV vectors (Fig. 1B). Specifically, we observed that GTOV but not AMAV GP vectors could transduce CEM cells, while the opposite pattern was true on T1B27 cells. Furthermore, we found that the JUNV, MACV, and GTOV GP vectors transduced the three rodent cell lines (NIH 3T3, CHO-K1, and BHK21 cells) with significantly less efficiency than either TCRV or AMAV GP vectors. In agreement with our previous studies (21), we found that JUNV vectors gave higher titers on these three rodent cell lines than the MACV vectors and we also noted that the GTOV vectors behaved more like the MACV vectors than the JUNV vectors. Taken together, these data indicate that the characteristics of the entry pathways directed by the clade B GPs are more closely correlated with human pathogenicity than with close phylogenetic relationships.
hTfR1 use by clade B viruses.
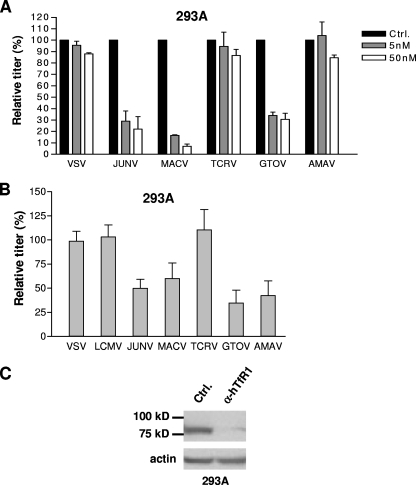
Recently, Radoshitzky et al. (25) demonstrated that hTfR1 plays a role in the entry of JUNV, MACV, GTOV, and SABV into human cell lines, since pretreatment of 293T cells with anti-TfR1 antibody was able to reduce infectivity by all four viruses and pretreatment of 293T cells with either soluble hTfR1 or anti-hTFR1 antibody reduced the titer of MACV and JUNV GP-pseudotyped retroviral vectors. We performed similar antibody blocking experiments using 293A cells and the expanded panel of GP vectors, with VSV-G-pseudotyped vectors serving as a control. We observed that while incubation with 50 nM of anti-hTfR1 antibody caused significant reductions in titers for the JUNV, MACV, and GTOV GP vectors (78, 93, and 70% reductions, respectively), there was no significant inhibition of entry for either TCRV or AMAV GP vectors beyond the level that we observed with the control VSV-G-pseudotyped vectors (Fig. 2A). These data therefore suggest that although the pathogenic clade B viruses require hTfR1 for entry into 293A cells, the nonpathogenic viruses TCRV and AMAV do not.
FIG. 2.
Role of human TfR1 in clade B viral entry. (A) Human 293A cells were pretreated with anti-hTfR1 antibody at two different concentrations and then challenged with the indicated GP-pseudotyped vectors. The relative titers were calculated as a percentage of the control (Ctrl.; no antibody pretreatment). Data are reported as means ± standard errors of two independent experiments. (B) 293A cells were treated with either a control or anti-hTfR1 siRNA and challenged with pseudotyped vectors 48 h later. The titers obtained in the presence of the anti-hTfR1 siRNA were made relative to the titers obtained with the control siRNA and are expressed as a percentage of the control. The values shown are means ± standard error from two to six independent experiments. (C) hTfR1 knockdown by anti-hTfR1 (α-hTfR1) siRNA was confirmed by Western blotting of cell lysates. Ctrl., cells treated with the control siRNA. Lysates were also probed for actin to ensure equal loading.
To confirm these results, we treated 293A cells with hTfR1-targeted siRNA prior to incubation with pseudotyped vectors. As controls, we used both VSV-G and LCMV GP-pseudotyped vectors. Under conditions in which the endogenous hTfR1 expression was greatly reduced by the siRNA treatment, we found that the titers of the JUNV, MACV, and GTOV GP vectors were reduced by 35 to 65% compared to treatment with control siRNA (Fig. 2B and C). This was a less complete inhibition than we observed with the antibody pretreatment studies and may reflect the fact that even very low levels of surface TfR1 are sufficient to support significant virus entry. In contrast, the TCRV GP vector titers were unaffected by hTfR1 knockdown. Surprisingly, the AMAV GP vectors also appeared to be sensitive to the loss of hTfR1, despite the fact that we had not observed such an effect when in the presence of the anti-hTfR1 antibody. This observation could reflect some usage of hTFR1 by the AMAV GP or could be the result of differences between the two methods of blocking TfR1 usage, where the more long-term consequences of siRNA knockdown of TfR1 could have an indirect effect on AMAV entry through effects on other cellular activities.
Utilization of α-DG by AMAV and TCRV GPs.
Since we had observed that the entry of TCRV and AMAV GP vectors into 293A cells was either fully independent of (TCRV) or possibly only partially dependent on (AMAV) the presence of hTFR1, we next considered whether these nonpathogenic GPs could instead be utilizing α-DG for entry into cells. Our previous studies with a DG-knockout mES cell line have shown that TCRV vectors can efficiently transduce cells in the absence of α-DG (21). Similarly, Spiropoulou et al. (32) reported that AMAV can readily infect DG−/− mES cells. To confirm these findings, we examined the titers of TCRV and AMAV GP vectors on DG+/− versus DG−/− mES cells and confirmed that TCRV vectors gave equivalent titers on the two cell lines when normalized to control VSV-G vectors. In contrast, the AMAV vectors appeared to be somewhat sensitive to the loss of α-DG in this system since the relative titers on the knockout cells were only 31% ± 6% of those of the control cells (data not shown). While it is possible to interpret this finding as meaning that AMAV entry is enhanced by the presence of α-DG, we hesitate to draw this conclusion since the mES cells are clonal in origin. An alternative explanation is that variations in the levels of another unknown factor(s) that AMAV requires for entry underlie these differences. However, when combined with the data from the hTfR1 siRNA studies, these observations do suggest that some differences may exist between the entry pathways used by the two nonpathogenic viruses, TCRV and AMAV.
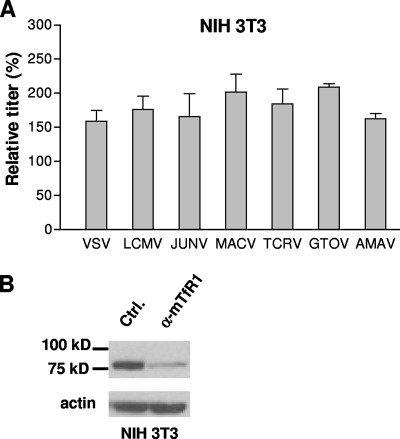
All clade B viruses enter murine cells independently of TfR1.
Our observations so far support a role for hTfR1 in the entry of the pathogenic clade B arenaviruses into human cells. However, these viruses are found predominantly in rodent hosts in nature. We therefore asked whether TfR1 was also being used for entry into rodent cells, by looking at viral entry into murine NIH 3T3 cells. As shown in Fig. 3, siRNA knockdown of mTfR1 in NIH 3T3 cells resulted in no reduction in titer relative to the control siRNA treatment, suggesting that mTfR1 plays no role in entry into these cells, even by the pathogenic viruses. A caveat of these findings is that Old World mice are not natural hosts for the New World arenaviruses, which are carried by New World rodents in the Cricetidae family.
FIG. 3.
Vector titer is unaffected by mTfR1 knockdown. (A) NIH 3T3 cells were treated with either a control or anti-mTfR1 siRNA and challenged with pseudotyped vectors 48 h later. The titers obtained in the presence of the anti-mTfR1 siRNA were made relative to the titers obtained with the control siRNA and are expressed as a percentage of the control. The values shown are means ± standard errors for two to seven independent experiments. (B) siRNA knockdown of mTfR1 was confirmed by Western blotting of cell lysates. α-mTfR1, anti-mTfR1; Ctrl., cells treated with the control siRNA. Lysates were also probed for actin to ensure equal loading.
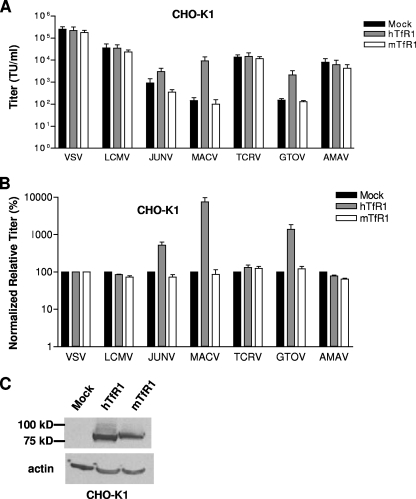
Addition of hTfR1, but not mTfR1, enhances entry into CHO-K1 cells.
Radoshitzky et al. (25) demonstrated that the susceptibility of CHO-K1 cells to transduction by JUNV and MACV GP vectors could be increased four- to eightfold by the transient expression of hTfR1. This suggested that entry into CHO-K1 cells via the endogenous receptor(s) was limiting. We therefore asked whether entry could be increased into CHO-K1 cells by the transient expression of either hTfR1 or mTfR1 prior to challenge with pseudotyped vectors. As shown in Fig. 4, we found that expression of hTfR1 in CHO-K1 cells had no effect on the titers of VSV-G- or LCMV GP-pseudotyped vectors but caused significant increases in the titer of vectors pseudotyped with the GPs from JUNV (1/2 log; P = 0.01), MACV (2 logs; P < 0.01), and GTOV (1 log; P < 0.01). In contrast, the titers of TCRV and AMAV vectors were unaffected by the expression of hTfR1. In agreement with the findings from the siRNA knockdown experiments in NIH 3T3 cells, we found that addition of the mTfR1 had no effect on the titers of any of the pseudotyped vectors tested. Taken together, these findings suggest that although the entry of the pathogenic viruses is enhanced by TfR1, this is only true for the human receptor, and not for the murine ortholog.
FIG. 4.
hTfR1, but not mTfR1, increases the titer of pathogenic GP vectors on CHO-K1 cells. (A) Titers (TU/ml) of pseudotyped retroviral vectors were obtained on mock-transfected CHO-K1 cells or cells transiently transfected with expression plasmids for hTfR1 or mTfR1. The values shown are means ± standard errors from three independent experiments. (B) Vector titers are displayed as normalized relative titers (%), where each experimental value was first made relative to the value obtained on the mock-transfected cells and then normalized to the ratios obtained with the VSV-G vectors. This normalizes the data for any toxic effects of the siRNAs. (C) Western blotting confirmed expression of both hTfR1 and mTfR1.
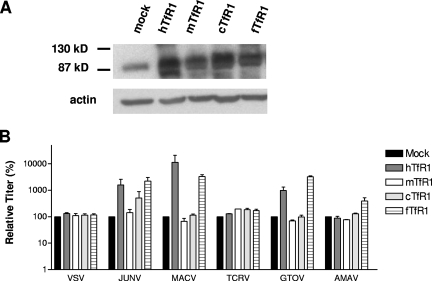
Species specificity in the ability of TfR1 to function as a clade B receptor.
We expanded our studies to examine the use of the canine and feline TfR1 orthologs (23) by expressing these receptors in CHO-K1 cells. These analyses revealed further specificities in the GP-TfR1 interaction, since the feline receptor was able to enhance entry, while the canine form could not. Similar to our findings with hTfR1, significant enhancement by the feline receptor was restricted to GPs from the three pathogenic strains (Fig. 5). The same results were also observed when NIH 3T3 cells were transfected (data not shown).
FIG. 5.
Ability of TfR1 orthologs from different species to enhance entry into CHO-K1 cells. (A) CHO-K1 cells were transfected with the TfR1 orthologs from humans, mice, dogs (cTfR1), and cats (fTFR1), and expression was confirmed by Western blotting. (B) Mock- and TfR1-transfected CHO-K1 cells were challenged with the indicated pseudotyped vectors. Vector titers were made relative to the value obtained for the mock-transfected cells and are expressed as a percentage of that control. The values shown are means ± standard errors from two to four independent experiments.
We also noted differences in the effectiveness of the receptors from different species that were GP specific. For example, MACV GP-directed entry, which was the most strongly enhanced by hTfR1 expression in both CHO-K1 and NIH 3T3 cells, showed a preference for the human receptor over the feline receptor, while JUNV and GTOV, which exhibited lower overall levels of enhancement by hTfR1, displayed a slight preference for the feline receptor. JUNV GP vectors alone displayed a small enhancement of entry in the presence of the canine TfR1. Finally, we observed that AMAV GP vector entry was slightly enhanced by the feline TfR1, again suggesting that AMAV behaves differently from TCRV and may have an intermediate phenotype in regards to TfR1 usage.
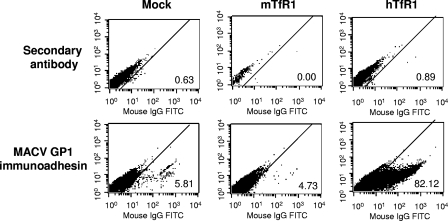
hTfR1 enhances entry at the level of binding.
While our data show that expression of hTfR1 can enhance entry into CHO-K1 cells for pathogenic GP vectors, it is unclear at what stage this enhancement is occurring. The arenavirus GP is a class I viral fusion protein, comprising noncovalently linked subunits designated GP1 and GP2. GP1 is responsible for host receptor binding (22), while GP2 is responsible for fusion of viral and cell membranes (4). Immunoadhesins, which are fusions between the receptor binding subunit of a viral fusion protein and an immunoglobulin Fc domain, are useful tools to measure the binding of a viral fusion protein to a cell. Previously we have shown that immunoadhesins containing the GP1 sequences of MACV, TCRV, or LASV bind only to those cells that are able to support entry (21). We therefore compared MACV GP1 immunoadhesin binding to CHO-K1, CHO-K1(mTfR1), and CHO-K1(hTfR1) cells. These data (Fig. 6) revealed that the expression of hTFR1 in CHO-K1 cells resulted in a significant increase in the ability of the MACV immunoadhesin to bind to these cells, while the expression of mTfR1 did not. Since it is likely that the increase in binding we observed can fully account for the increase in titers when hTfR1 is present, our data are most consistent with the hTfR1 molecule functioning as a primary attachment molecule for MACV.
FIG. 6.
Binding of MACV GP1 immunoadhesin to CHO-K1 cells transfected with hTFR1 or mTFR1. CHO-K1 cells were mock transfected or transfected with expression plasmids for hTfR1 or mTfR1. The binding of MACV GP1 immunoadhesin to each cell line was detected by incubation with the immunoadhesin, followed by an FITC-labeled goat anti-rabbit secondary antibody and FACS analysis (bottom panel). As a control, cells were also treated with just the secondary antibody (top panel). The percentage of cells staining positive in each sample is indicated.
hTfR1/mTfR1 chimeras reveal complex interaction with GPs.
It has previously been reported that mouse mammary tumor virus (MMTV) uses TfR1 for entry and, in an opposite pattern from what we observe for the arenaviruses, is able to use the murine but not the human version of this molecule (34). Through the use of human/murine chimeras, the residues responsible for this species difference in MMTV utilization have been mapped to two regions that are spatially close on the surface of TfR1, at the outer edge of the TfR1 homodimer, in a region that is distinct from the binding sites for either of its natural ligands, transferrin or the hereditary hematochromatosis protein, HFE (34). Since it has also been demonstrated that soluble transferrin does not compete with MACV GP vectors for entry into human cells (25), we asked if the clade B GPs could be binding to a region of hTFR1 similar to the one MMTV uses on mTfR1.
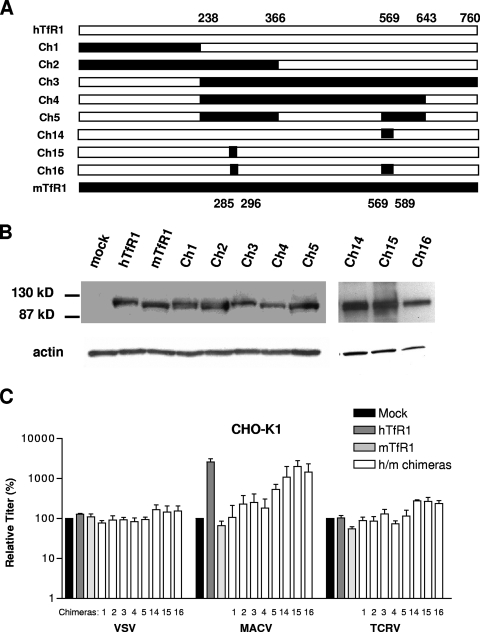
To address this question, we used MACV GP-pseudotyped vectors, since these vectors displayed the greatest increase (fold) in titer (2 orders of magnitude) when hTfR1 was transfected into CHO-K1 cells (Fig. 4). We analyzed the ability of the vectors to enter CHO-K1 cells transfected with a panel of reciprocal chimeras between hTfR1 and mTfR1 (Fig. 7A), with chimeras 3 to 5 and 16 containing substitutions in hTfR1 of the murine sequences necessary to confer full MMTV entry (34). As controls, we used VSV-G and TCRV GP vectors, neither of which is affected by the expression of either hTfR1 or mTfR1 in CHO-K1 cells (Fig. 4).
FIG. 7.
Entry directed by hTfR1/mTfR1 chimeras in CHO-K1 cells. (A) Schematic showing the extent of human (open) and mouse (filled) sequences in the TfR1 chimeras (Ch1 to -5 and Ch14 to -16). Numbers delineate the regions exchanged, based on the amino acid sequence. (B) Western blotting confirmed expression of all TfR1 chimeras. (C) The ability of VSV-G, MACV GP and TCRV GP-pseudotyped vectors to transduce cells transfected with the indicated TfR1 constructs is shown as relative titers (%), where each experimental value was normalized to the value obtained on mock-transfected cells. The values shown are means ± standard errors from three independent experiments.
We first demonstrated that all of the chimeric proteins used were expressed at similar levels to the wild-type hTfR1 and mTfR1 molecules (Fig. 7B). We then confirmed that expression of the hTfR1 and mTfR1 parental molecules, as well as the chimeras, had no significant effect on the titers of either VSV-G or TCRV GP vectors. In contrast, the MACV GP vectors exhibited a range of responses to the chimeric proteins, with a general increase in vector titer being observed as the extent of human sequence in the molecules was increased (Fig. 7C). However, the pattern obtained did not highlight any particular region as being essential for hTfR1 receptor function. For example, since neither of the reciprocal chimeras 1 and 3 displayed significant MACV receptor activity, it is likely that the MACV GP binding site on hTfR1 is not a simple linear epitope. Interestingly, chimera 16, which is an hTFR1 receptor containing murine sequence substitutions that are sufficient to support full MMTV entry, produced the second-highest titer of all of the chimeras tested. It is therefore probable that MACV GP does not use the same binding site on hTfR1 as MMTV uses on mTfR1 or, if it does, the species specificity that we observed is determined by residues outside of this region.
DISCUSSION
Arenaviruses are found predominantly in rodent reservoirs, with humans acting as incidental hosts. The host range of the viruses is determined largely by the habitat of the rodent vector. For the New World clade B arenaviruses, among which are four known human pathogens, the rodent hosts are New World rats and mice from the Sigmodontinae subfamily. A possible exception is TCRV, which was originally isolated from Artibeus bats in Trinidad (10), although the true natural host remains in question (3).
An understanding of the elements that allow arenaviruses to transition between chronic, nonlethal infections in rodent hosts to acute, pathogenic infections in humans would be of significant interest. Studies with LCMV have highlighted the important role played by the viral GP in determining the in vivo distribution and pathogenic outcome of arenavirus infections (29, 31). The clade B New World arenaviruses offer an important opportunity to study pathogenic determinants, as this clade is characterized by evolutionarily related members that may be subdivided according to pathogenicity.
Receptor usage in the arenaviruses appears complex. At least two molecules have so far been identified as cellular receptors: α-DG is used by LASV, certain strains of LCMV, and two clade C viruses (8, 13, 27, 32), while TfR1 is used by members of the clade B New World lineage (25). In our previous studies of the clade B1 viruses, we noted striking differences in entry pathway characteristics that separated the pathogenic and nonpathogenic members of the group. In the present study, we have extended those observations to include the B2 lineage. Together, our data reveal that the ability of a GP to use hTfR1 correlates with being from a strain designated as pathogenic for humans.
Despite the evidence supporting the use of hTfR1 as an arenavirus receptor, it is also clear that the clade B viruses can use TfR1-independent entry pathways. Together with our previous findings (21, 27), we ruled out α-DG as being involved in the entry of these viruses, suggesting that other, as-yet-unidentified receptors exist. We found three distinct situations in which TfR1-independent pathways were being used. First, the entry of the nonpathogenic viruses, TCRV and AMAV, into both human and rodent cell lines was both TfR1 and α-DG independent. Second, we observed that the entry of the pathogenic viruses, JUNV, MACV, and GTOV, into murine NIH 3T3 cells did not depend on TfR1, since siRNA knockdown of mTfR1 had no effect on vector titer, while the addition of mTfR1 to CHO-K1 cells was unable to increase the entry of MACV GP-pseudotyped vectors. In addition, the fact that MACV entry into CHO-K1 cells could be stimulated by almost 2 orders of magnitude when hTfR1 was present implies that entry into CHO-K1 cells is suboptimal for MACV, which in turn suggests that the endogenous Chinese hamster TfR1 cannot facilitate clade B entry. The third situation we observed was the entry of pathogenic viruses into human cells that were depleted for hTfR1, either by siRNA knockdown or antibody blockage, although it is difficult to rule out that entry was occurring due to incomplete removal of hTfR1. Overall, we do not yet understand the relationship between these different TfR-1-independent pathways and, specifically, whether the non-TfR1 receptor that is used by TCRV and AMAV is the same molecule that is used by the pathogenic viruses under conditions in which hTfR1 is unavailable.
It is now appreciated that several different roles can be played by the cellular factors that are recruited by viral fusion proteins to mediate the entry of enveloped viruses into cells. In the simplest case, the fusion protein binds to a single receptor molecule and this interaction triggers conformational changes in the fusion protein that expose the fusion peptide and initiate virus-cell fusion (9). Alternatively, as is the case with the influenza virus hemagglutinin protein, binding to the receptor can allow the virus to be endocytosed into a low-pH environment that provides the trigger for the necessary conformational change. Other viruses, such as avian leukosis virus, appear to use a combination of these features (19). Viruses also vary in the number of receptor molecules with which they can interact. Some viruses have evolved the ability to bind to more than one cellular receptor (alternate primary receptors), such as the 10A1 variant of the murine leukemia virus Env protein (11) that shows expanded tropism. Another variation is the use of both a primary and secondary (or co-) receptor. The prototype example of this is human immunodeficiency virus type 1 (HIV-1) entry, where binding to CD4 triggers initial conformational changes in the viral Env protein that expose the coreceptor (e.g., CCR5) binding site (36), with engagement of the coreceptor necessary for virus-cell fusion. Finally, some viruses clearly make use of less-specific interactions with attachment factors to facilitate binding to the cell surface before interaction with the true receptor. An example of this type of interaction is the binding of HIV-1 to syndecans, which enhances virus entry in macrophages (30).
At present, it is not clear which of these roles is being played by hTfR1 in arenavirus entry. It could be acting as a classic viral receptor that is itself sufficient to orchestrate the entry of MACV, JUNV, and GTOV into cells. Alternatively, the fact that entry can occur in certain situations in the absence of hTfR1 leads us to consider that an interaction with hTfR1 could reflect an initial virus-cell interaction that occurs before handing the GP onto a second receptor molecule that actually directs virus-cell fusion. Certainly, our immunoadhesin binding studies demonstrated a strong interaction between MACV GP1 and hTfR1. Such a scenario could therefore resemble either the receptor/coreceptor interactions of the HIV Env with CD4 and CCR5 or the less-specific interaction of HIV-1 Env with syndecans, but with hTfR1 binding representing an additional step, superimposed at the beginning of an existing entry pathway.
TfR1 normally transitions between the cell surface and endosomes. At extracellular pH 7.4, TfR1 binds iron-laden transferrin and the resulting complex is endocytosed via clathrin-coated pits. As the endosome acidifies to pH 5.5, the iron is released, and the apo-transferrin/TfR1 complex is recycled to the cell surface, where the transferrin is released (1). Our previous studies with NH4Cl, an inhibitor of endosomal acidification, have revealed that clade B entry requires endosomal acidification, although we identified at least one cell line, K562, in which JUNV and MACV entry did not (21). Interestingly, it has been reported that the pH required to trigger cell-cell fusion directed by the JUNV GP is between 5.0 and 5.5 (6, 37). Although TfR1 typically recycles through early endosomes that only acidify to pH 6.5, it is possible that engagement of the receptor with the arenavirus GP could divert TfR1 from the recycling pathway to deeper in the endosomal pathway, where it would encounter a lower-pH environment.
Our analysis of the functionality of TfR1 from different species revealed that both the human and feline receptors could be effectively used, while the murine and canine forms were not. mTfR1 and hTfR1 are 76.8% identical, and the human and canine receptors are 78.6% identical. However, despite this homology, no clade B arenaviruses could use mTfR1, and only JUNV GP appeared able to use the canine TfR1, albeit with significantly less efficiency than either the human or feline forms. Similar species-specific restrictions are seen with other viruses that use TfR1 for entry; MMTV can use murine but not human, cat, dog, or hamster TfR1 (34), while canine parvovirus can use human, canine, and feline TfR1, but not quail or hamster (23). Studies of the interaction of MMTV with mTfR1 have shown that this virus binds to the outer edge of the TfR1 dimer, distant from the binding site for either transferrin or HFE. Our preliminary studies with human/mouse chimeric receptors did not identify any particular region of hTfR1 as being sufficient to allow high-level entry of MACV vectors, but we noted that substitution for the hTfR1 with the murine segments that confer susceptibility to MMTV infection did not abolish MACV entry.
A consistent observation that we made with the pseudotyped vectors carrying the hTfR1-using GPs (MACV, JUNV, and GTOV) was that they resulted in lower titers on all of the nonlymphocytic rodent cell lines that we examined (NIH 3T3, CHO-K1, and BHK21 cells) compared to the titers obtained with the TCRV and AMAV vectors. Such differences were not seen on nonlymphocytic cell lines of human origin (21). This suggests that, similar to the situation in murine cells, neither the Chinese hamster nor Syrian hamster TfR1 receptor can promote clade B entry. This is an intriguing result as hamsters are more closely related to the New World rodents that are the natural hosts for these viruses. However, a definitive analysis of whether TfR1 is ever used in a rodent host will require studies using the specific species’ TfR1.
We also noted that while the addition of hTfR1 to CHO-K1 cells could increase the titer of MACV vectors, it merely served to increase titers to the same levels that AMAV and TCRV vectors achieved in the absence of this molecule (Fig. 4A). This suggests that, in the absence of a functional TfR1, entry of pathogenic viruses into rodent cells is limited, while this is not the case for the non-hTfR1-using viruses. These findings are consistent with a model whereby acquisition of the ability to use TfR1 comes at a price of reduced ability to access any TfR1-independent pathways (at least in the rodent cell lines we have examined). We further speculate that the ancestral clade B virus may have used a receptor other than TfR1, but that certain viruses in this lineage subsequently evolved the ability to use TfR1. These viruses are now also able to jump species and infect humans, but whether the acquisition of human TfR1 binding activity is the root cause of the ability to jump species, or an accident of the similarity with a receptor species used in the natural host, awaits further studies.
In the opposite scenario, it is also possible that the ancestral clade B virus used TfR1 as a receptor but that certain members of the lineage evolved to use an alternate receptor(s) and thereby lost the ability to infect humans as accidental hosts. However, any such model must account for the fact that both hTfR1-dependent and -independent viruses are found throughout the different clade B sublineages (Fig. 1A), suggesting that any such changes could have occurred more than once. Indeed, it will be interesting to determine if any other arenaviruses that can infect humans are also using TfR1. Our preliminary investigations with the putative human pathogen Whitewater Arroyo virus (7) have shown that this New World clade A virus does not use hTfR1 (26).
In summary, the finding that TfR1 use by clade B arenaviruses is correlated with the ability to cause severe hemorrhagic fevers in humans has clear implications for therapies aimed at blocking this interaction, as discussed by Radoshitzky et al. (25). However, it also has wider implications for characterizing these viruses, as it suggests that a good predictor of human pathogenicity would be the ability to use hTfR1. If this hypothesis holds true, hTfR1 receptor use could serve as a diagnostic test applied to both existing and newly identified members of this family.
Acknowledgments
We thank En xiu Wang, Susan Ross, and Stefan Kunz for reagents and helpful discussions.
This work was supported by PHS grant 1U54 AI065359 to the Pacific Southwest Regional Center of Excellence for Biodefense and Emerging Infectious Diseases (P.M.C.) and a Saban Research Institute Career Development Award (T.R.).
Footnotes
Published ahead of print on 14 November 2007.
REFERENCES
- 1.Aisen, P. 2007. Transferrin receptor 1. Int. J. Biochem. Cell Biol. 362137-2143. [DOI] [PubMed] [Google Scholar]
- 2.Bowen, M. D., C. J. Peters, and S. T. Nichol. 1996. The phylogeny of New World (Tacaribe complex) arenaviruses. Virology 219285-290. [DOI] [PubMed] [Google Scholar]
- 3.Buchmeier, M. J., J.-C. de la Torre, and C. J. Peters. 2006. Arenaviridae: the viruses and their replication, p. 1-37. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 5th ed. Lippincott Williams & Wilkins, Philadelphia, PA.
- 4.Burns, J. W., and M. J. Buchmeier. 1991. Protein-protein interactions in lymphocytic choriomeningitis virus. Virology 183620-629. [DOI] [PubMed] [Google Scholar]
- 5.Cao, W., M. D. Henry, P. Borrow, H. Yamada, J. H. Elder, E. V. Ravkov, S. T. Nichol, R. W. Compans, K. P. Campbell, and M. B. A. Oldstone. 1998. Identification of alpha-dystroglycan as a receptor for lymphocytic choriomeningitis virus and Lassa fever virus. Science 2822079-2081. [DOI] [PubMed] [Google Scholar]
- 6.Castilla, V., and S. E. Mersich. 1996. Low-pH-induced fusion of Vero cells infected with Junin virus. Arch. Virol. 1411307-1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Centers for Disease Control and Prevention. 2000. Fatal illnesses associated with a New World arenavirus—California, 1999-2000. Morb. Mort. Wkly. Rep. 49709-711. [PubMed] [Google Scholar]
- 8.Charrel, R. N., H. Feldmann, C. F. Fulhorst, R. Khelifa, R. de Chesse, and X. K. de Lamballerie. 2002. Phylogeny of New World arenaviruses based on the complete coding sequences of the small genomic segment identified an evolutionary lineage produced by intrasegmental recombination. Biochem. Biophys. Res. Commun. 2961118-1124. [DOI] [PubMed] [Google Scholar]
- 9.Colman, P. M., and M. C. Lawrence. 2003. The structural biology of type I viral membrane fusion. Nat. Rev. Mol. Cell Biol. 4309-319. [DOI] [PubMed] [Google Scholar]
- 10.Downs, W. G., C. R. Anderson, L. Spence, T. H. G. Aitken, and A. M. Greenhall. 1963. Tacaribe virus, a new agent isolated from Artibeus bats and mosquitoes in Trinidad, West Indies. Am. J. Trop. Med. Hyg. 12640-646. [DOI] [PubMed] [Google Scholar]
- 11.Han, J.-Y., P. M. Cannon, K.-M. Lai, Y. Zhao, M. V. Eiden, and W. F. Anderson. 1997. Identification of envelope protein residues required for the expanded host range of 10A1 murine leukemia virus. J. Virol. 718103-8108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kunz, S., and J. C. de la Torre. 2005. Novel antiviral strategies to combat human arenavirus infections. Curr. Mol. Med. 5735-751. [DOI] [PubMed] [Google Scholar]
- 13.Kunz, S., J. M. Rojek, M. Kanagawa, C. F. Spiropoulou, R. Barresi, K. P. Campbell, and M. B. A. Oldstone. 2005. Posttranslational modification of α-dystroglycan, the cellular receptor for arenaviruses, by the glycosyltransferase LARGE is critical for virus binding. J. Virol. 7914282-14296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kunz, S., N. Sevilla, J. M. Rojek, and M. B. A. Oldstone. 2004. Use of alternative receptors different than α-dystroglycan by selected isolates of lymphocytic choriomeningitis virus. Virology 325432-445. [DOI] [PubMed] [Google Scholar]
- 15.Mackenzie, R. B. 1965. Epidemiology of Machupo virus infection. I. Pattern of human infection, San Joaquin, Bolivia, 1962-1964. Am. J. Trop. Med. Hyg. 14808-813. [DOI] [PubMed] [Google Scholar]
- 16.Maiztegui, J., M. Feuillade, and A. Briggiler. 1986. Progressive extension of the endemic area and changing incidence of Argentine hemorrhagic fever. Med. Microbiol. Immunol. 175149-152. [DOI] [PubMed] [Google Scholar]
- 17.Mills, J. N., B. A. Ellis, J. E. Childs, K. T. McKee, Jr., J. I. Maiztegui, C. J. Peters, T. G. Ksiazek, and P. B. Jahrling. 1994. Prevalence of infection with Junin virus in rodent populations in the epidemic area of Argentine hemorrhagic fever. Am. J. Trop. Med. Hyg. 51554-562. [PubMed] [Google Scholar]
- 18.Mills, J. N., B. A. Ellis, K. T. McKee, Jr., G. E. Calderon, J. I. Maiztegui, G. O. Nelson, T. G. Ksiazek, C. J. Peters, and J. E. Childs. 1992. A longitudinal study of Junin virus activity in the rodent reservoir of Argentine hemorrhagic fever. Am. J. Trop. Med. Hyg. 47749-763. [DOI] [PubMed] [Google Scholar]
- 19.Mothes, W., A. L. Boerger, S. Narayan, J. M. Cunningham, and J. A. Young. 2000. Retroviral entry mediated by receptor priming and low pH triggering of an envelope glycoprotein. Cell 103679-689. [DOI] [PubMed] [Google Scholar]
- 20.Niwa, H., K. Yamamura, and J. Miyazaki. 1991. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 108193-199. [DOI] [PubMed] [Google Scholar]
- 21.Oldenburg, J., T. Reignier, M. L. Flanagan, G. A. Hamilton, and P. M. Cannon. 2007. Differences in tropism and pH dependence for glycoproteins from the clade B1 arenaviruses: implications for receptor usage and pathogenicity. Virology 364132-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parekh, B. S., and M. J. Buchmeier. 1986. Proteins of lymphocytic choriomeningitis virus: antigenic topography of the viral glycoproteins. Virology 153168-178. [DOI] [PubMed] [Google Scholar]
- 23.Parker, J. S., W. J. Murphy, D. Wang, S. J. O'Brien, and C. R. Parrish. 2001. Canine and feline parvoviruses can use human or feline transferrin receptors to bind, enter, and infect cells. J. Virol. 753896-3902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peters, C. J. 1995. Arenavirus diseases, p. 227-246. In J. Portfield (ed.), Exotic viral infections. Chapman & Hall Medical, London, United Kingdom.
- 25.Radoshitzky, S. R., J. Abraham, C. F. Spiropoulou, J. H. Kuhn, D. Nguyen, W. Li, J. Nagel, P. J. Schmidt, J. H. Nunberg, N. C. Andrews, M. Farzan, and H. Choe. 2007. Transferrin receptor 1 is a cellular receptor for New World haemorrhagic fever arenaviruses. Nature 44692-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reignier, T., J. Oldenburg, M. Flanagan, G. A. Hamilton, V. K. Martin, and P. M. Cannon. 9 November 2007. Receptor use by the Whitewater Arroyo virus glycoprotein. Virology. [Epub ahead of print.] [DOI] [PMC free article] [PubMed]
- 27.Reignier, T., J. Oldenburg, B. Noble, E. Lamb, V. Romanowski, M. J. Buchmeier, and P. M. Cannon. 2006. Receptor use by pathogenic arenaviruses. Virology 353111-120. [DOI] [PubMed] [Google Scholar]
- 28.Rojek, J. M., C. F. Spiropoulou, and S. Kunz. 2006. Characterization of the cellular receptors for the South American hemorrhagic fever viruses Junin, Guanarito, and Machupo. Virology 349476-491. [DOI] [PubMed] [Google Scholar]
- 29.Salvato, M., P. Borrow, E. Shimomaye, and M. B. A. Oldstone. 1991. Molecular basis of viral persistence: a single amino acid change in the glycoprotein of lymphocytic choriomeningitis virus is associated with suppression of the antiviral cytotoxic T-lymphocyte response and establishment of persistence. J. Virol. 651863-1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saphire, A. C. S., M. D. Bobardt, Z. Zhang, G. David, and P. A. Gallay. 2001. Syndecans serve as attachment receptors for human immunodeficiency virus type 1 on macrophages. J. Virol. 759187-9200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smelt, S. C., P. Borrow, S. Kunz, W. Cao, A. Tishon, H. Lewicki, K. P. Campbell, and M. B. A. Oldstone. 2001. Differences in affinity of binding of lymphocytic choriomeningitis virus strains to the cellular receptor α-dystroglycan correlate with viral tropism and disease kinetics. J. Virol. 75448-457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Spiropoulou, C. F., S. Kunz, P. E. Rollin, K. P. Campbell, and M. B. A. Oldstone. 2002. New World arenavirus clade C, but not clade A and B viruses, utilizes α-dystroglycan as its major receptor. J. Virol. 765140-5146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vainrub, B., and R. Salas. 1994. Latin American hemorrhagic fever. Infect. Dis. Clin. N. Am. 847-59. [PubMed] [Google Scholar]
- 34.Wang, E., L. Albritton, and S. R. Ross. 2006. Identification of the segments of the mouse transferrin receptor 1 required for mouse mammary tumor virus infection. J. Biol. Chem. 28110243-10249. [DOI] [PubMed] [Google Scholar]
- 35.Williamson, R. A., M. D. Henry, K. J. Daniels, R. F. Hrstka, J. C. Lee, Y. Sunada, O. Ibraghimov-Beskrovnaya, and K. P. Campbell. 1997. Dystroglycan is essential for early embryonic development: disruption of Reichert's membrane in Dag1-null mice. Hum. Mol. Genet. 6831-841. [DOI] [PubMed] [Google Scholar]
- 36.Wu, L., N. P. Gerard, R. Wyatt, H. Choe, C. Parolin, N. Ruffing, A. Borsetti, A. A. Cardoso, E. Desjardin, W. Newman, C. Gerard, and J. Sodroski. 1996. CD4-induced interaction of primary HIV-1 gp120 glycoproteins with the chemokine receptor CCR-5. Nature 384179-183. [DOI] [PubMed] [Google Scholar]
- 37.York, J., and J. H. Nunberg. 2006. Role of the stable signal peptide of Junin arenavirus envelope glycoprotein in pH-dependent membrane fusion. J. Virol. 807775-7780. [DOI] [PMC free article] [PubMed] [Google Scholar]