Abstract
Human astroviruses (HAstVs) belong to a family of nonenveloped, icosahedral RNA viruses that cause noninflammatory gastroenteritis, predominantly in infants. Eight HAstV serotypes have been identified, with a worldwide distribution. While the HAstVs represent a significant public health concern, very little is known about the pathogenesis of and host immune response to these viruses. Here we demonstrate that HAstV type 1 (HAstV-1) virions, specifically the viral coat protein (CP), suppress the complement system, a fundamental component of the innate immune response in vertebrates. HAstV-1 virions and purified CP both suppress hemolytic complement activity. Hemolytic assays utilizing sera depleted of individual complement factors as well as adding back purified factors demonstrated that HAstV CP suppresses classical pathway activation at the first component, C1. HAstV-1 CP bound the A chain of C1q and inhibited serum complement activation, resulting in decreased C4b, iC3b, and terminal C5b-9 formation. Inhibition of complement activation was also demonstrated for HAstV serotypes 2 to 4, suggesting that this phenomenon is a general feature of these human pathogens. Since complement is a major contributor to the initiation and amplification of inflammation, the observed CP-mediated inhibition of complement activity may contribute to the lack of inflammation associated with astrovirus-induced gastroenteritis. Although diverse mechanisms of inhibition of complement activation have been described for many enveloped animal viruses, this is the first report of a nonenveloped icosahedral virus CP inhibiting classical pathway activation at C1.
Human astroviruses (HAstVs) are a significant cause of acute gastroenteritis in young children. While second only to rotavirus in the incidence of virally induced gastroenteritis in children (7), HAstVs are recognized as the leading cause of viral diarrhea in infants (27). Eight distinct HAstVs have been identified to date, with serotype 1 being the most prevalent worldwide (18). In addition to humans, members of this virus family infect a variety of other young mammals and birds, causing diseases ranging from diarrhea to nephritis (18). Astroviruses are small, nonenveloped, icosahedral particles with a single-stranded mRNA genome (∼7 kb) that is organized into three open reading frames (ORFs); ORFs 1a and 1b encode nonstructural proteins (11, 16, 34), whereas ORF2 encodes the coat protein (CP) precursor (16, 32). CP precursors assemble into particles, encapsidating the viral genome, and are subsequently cleaved extracellularly by trypsin, which renders the virion infectious (1, 20).
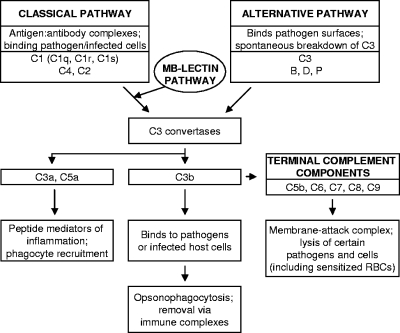
The pathogenesis of and immunity to HAstVs are poorly understood. Virions have been identified in intestinal epithelial cells of children with diarrhea, correlating with fecal shedding of the virus (reviewed in reference 21). However, HAstV-induced diarrhea appears not to result in significant cell death or inflammation in humans (26), and astrovirus animal models have suggested that the innate immune system may contribute to pathogenesis (reviewed in reference 13). In an attempt to determine if HAstVs disrupt the innate immune system in humans, we tested whether HAstV virions had an effect on the complement system. Complement is a fundamental component of the innate immune response in vertebrates and represents a frontline defense against invading pathogens. It is comprised of an ancient family of soluble and cell surface proteins that detect bacteria and viruses through one of three pathways, the classical (primarily activated by antigen-antibody interactions), mannose-binding lectin (binds specific polysaccharides on pathogen surfaces), and alternative (neither antibody nor lectin dependent) pathways (28, 31) (Fig. 1). Upon activation of complement, infectious agents are subsequently destroyed through multiple mechanisms, including the lysis of infected cells and certain pathogens, opsonization and phagocytosis, removal via immune complexes, enhanced priming of T and B cells, and mediation of a robust inflammatory response via leukocyte chemotaxis to the site of infection (Fig. 1) (31). The limited inflammatory response or cell death observed in both human infection (26) and an avian astrovirus small-animal model (14) led us to speculate that astroviruses may act upon the complement system during infection.
FIG. 1.
Overview of the pathways of serum complement activation. The main protein factors and their effector actions are indicated.
Here we demonstrate that the CP of HAstVs specifically suppresses activation of the human complement system through an interaction with C1, the first component of the classical pathway (Fig. 1). The mechanism of suppression is inhibitory in nature, as assessed by the lack of C4d, iC3b, and terminal C5b-9 membrane attack complex formation. Since it lacks any sequence homology to known complement regulators or other host genes that modulate immune function, HAstV CP appears to possess a novel viral immune evasion activity that directly inhibits complement activation and downstream effector functions.
MATERIALS AND METHODS
Cell lines and viruses.
CaCo-2 cells were cultivated in minimum essential medium with 10 to 20% heat-inactivated fetal bovine serum according to the instructions of the ATCC. A cell-adapted strain of HAstV type 1 (HAstV-1; Oxford) (kindly provided by D. K. Mitchell, Eastern Virginia Medical School, Norfolk, VA) was propagated in CaCo-2 cells (33). Briefly, CaCo-2 cell monolayers in minimum essential medium lacking fetal bovine serum were infected with a viral inoculum containing 10 μg/ml of trypsin type IX (Sigma), and virus was allowed to adhere for 1 h at 37°C. The inoculum was removed, and medium containing 2 μg/ml of trypsin was added; cells were then incubated for approximately 48 h at 37°C. Following incubation, viral suspensions were released from the cells by three freeze-thaw cycles. Cell debris was then removed in a low-speed spin, and the cell lysate containing virus was aliquoted and stored at −80°C. Cell-adapted strains of HAstV types 2, 3, and 4, previously propagated as described above, were provided by D. K. Mitchell.
Spodoptera frugiperda cells (line IPLB-Sf21) were propagated in TC100 medium supplemented with 10% heat-inactivated fetal bovine serum, and viral stocks of a recombinant baculovirus carrying the wild-type HAstV-1 CP gene were prepared using standard methodology (24). To produce recombinant CP, 2 × 108 IPLB-Sf21 cells in a 50-ml conical vial were infected with the recombinant baculovirus at a multiplicity of infection of 5 per cell. After incubation for 1 h at room temperature with rocking, the infected cells were transferred to a spinner flask containing cell growth medium and antibiotics. The spinner flasks were then allowed to stir at 27°C. At 5 days postinfection, cells were pelleted in a low-speed spin and the medium was discarded. Cell pellets were then frozen at −20°C until needed.
Recombinant CP isolation.
The following protocol was developed to purify soluble CP from infected cells. Unless otherwise indicated, all of the following steps were carried out at 4°C with prechilled buffers and protease inhibitors (BD Pharmingen). Frozen pellets were resuspended in 2 volumes of TNE (50 mM Tris [pH 7.0], 0.1 M NaCl, 10 mM EDTA) buffer and lysed by three freeze-thaw cycles. The lysate was centrifuged for 10 min at 13,300 × g, and the supernatant was discarded. The pellet, which contained aggregates of CP, was resuspended in 1 ml TNE buffer containing 2% NP-40 and incubated on ice for 30 min. The resulting suspension was centrifuged at 13,300 × g for 5 min, and the supernatant was discarded. The pellet was resuspended in 1 ml TNM (50 mM Tris [pH 7.0], 0.1 M NaCl, 20 mM MgSO4) buffer plus 2 μl of 10-mg/ml DNase I (Sigma-Aldrich), incubated for 30 min at room temperature, and centrifuged for 5 min at 13,000 × g, after which the supernatant was discarded. The pellet was resuspended in 1 ml TNE buffer and pelleted through a 1-ml 30% (wt/vol) sucrose cushion in TNE buffer containing protease inhibitors (BD Pharmingen) at 234,000 × g in an SW50.1Ti rotor (Beckman) for 1 h at 4°C. The supernatant was discarded, and the pellet was then resuspended with a syringe and needle in 500 μl of dissociation buffer (100 mM Tris [pH 9.0], 0.5 M NaCl, 100 mM urea, 10 mM EDTA, 5 mM dithiothreitol, protease inhibitors) and frozen overnight at −20°C. This dissociation buffer step dissolved the CP aggregates and was developed previously to dissociate flock house virus (FHV) provirions (25). The solubilized protein was then centrifuged at 13,300 × g for 10 min, and 500 μl of the supernatant was loaded onto a 5 to 25% (wt/vol) sucrose gradient made in dissociation buffer lacking protease inhibitors and spun at 274,000 × g in an SW41Ti rotor (Beckman) for 10 h at 4°C. The gradient was then fractionated with a gradient fractionator (ISCO) at 0.75 ml/min and 0.5 min/fraction. Fractions containing CP (typically fractions 6 to 10), as determined by 7.5% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Coomassie blue staining, were pooled, aliquoted, and stored at −70°C. CP was quantified by SDS-PAGE followed by Coomassie blue staining, along with known quantities of albumin (Amersham Biosciences), to generate a standard curve. Densitometric analysis to quantify and determine CP purity was performed on a Versadoc instrument (Bio-Rad) using QuantityOne software.
NHS, buffers, and cells for complement assays.
Normal human serum (NHS) for red blood cell (RBC) lysis experiments was isolated from the blood of healthy human volunteers in accordance with an Institutional Review Board-approved protocol (IRB 02-06-EX-0216; Eastern Virginia Medical School) and was pooled, aliquoted, and frozen at −80°C as a stock for all experiments, as previously described (6). Sheep RBCs (MP Biomedicals) were sensitized with 0.72 mg/ml anti-sheep RBC antibody (Rockland) and standardized to 5 × 108 cells/ml with GVBS++ (Veronal-buffered saline, 0.1% gelatin, 0.15 mM CaCl2, and 1.0 mM MgCl2). Complement activation experiments were stopped by the addition of ice-cold EDTA-GVBS−− (GVBS, 0.01 M EDTA).
Complement assays with HAstVs, CVF, and recombinant CP.
Twenty microliters of NHS was incubated alone, with 1 μg cobra venom factor (CVF; CompTech), with titrating amounts of cell lysate containing HAstV-1 virions at the indicated genome copy number, or with uninfected cell lysate in GVBS++ for 1 h at 37°C. The total reaction volume was then brought up to 0.75 ml with GVBS++ including 100 μl of sensitized sheep RBCs and again incubated for 1 h at 37°C. The experiment was stopped by the addition of 4.0 ml GVBS−−, and the samples were centrifuged for 5 min at 1,620 × g. The absorbance of the supernatants was read in a spectrophotometer at 412 nm, and the percent lysis of each sample was standardized to that of the NHS-only control. Cell lysates containing HAstV serotypes 2 to 4 were also tested in the complement assay at 2.92 × 108 genome copies each, as determined by real-time reverse transcription-PCR (RT-PCR). For HAstV-1 CP, titrating amounts of purified CP, 1 μg of CVF, or 15 μg of bovine serum albumin (BSA) was incubated with 20 μl NHS in a total volume of 50 μl with GVBS++ for 1 h at 37°C. The reaction mix was brought up to a volume of 800 μl, of which 250 μl was then combined with 100 μl sensitized sheep RBCs and 450 μl GVBS++ for 1 h at 37°C. The reaction was quenched with 4.0 ml of GVBS−−, and the samples were then centrifuged and read at 412 nm as described above. To test specifically for classical pathway activity, factor B-depleted serum (fBD; CompTech) and NHS were incubated with either the indicated amounts of CP or 85 μl of cell lysate containing HAstV-1 virions (2.92 × 108 genome copies), followed by incubation with sensitized sheep RBCs as described above.
To assay for alternative pathway activity, rabbit RBCs (CompTech Inc., Tyler, TX) were standardized to 1.5 × 108 cells/ml in Mg-EGTA-GVBS (Veronal-buffered saline, 0.1% gelatin, 8 mM EGTA, and 5 mM MgCl2). Thirty microliters of NHS or C2-depleted serum (C2D; CompTech) was incubated with 6.3 μg CP or 30 μl of cell lysate containing HAstV-1 virions (1.03 × 108 genome copies) and adjusted to an equal volume with Mg-EGTA-GVBS for 1 h at 37°C. The total volume was then brought up to 0.75 ml with Mg-EGTA-GVBS including 375 μl rabbit RBCs and again incubated for 1 h at 37°C. The reaction was stopped with the addition of 0.75 ml of ice-cold GVBS−−, and the samples were centrifuged. The absorbance of the supernatants was read in a spectrophotometer at 412 nm as described above.
C3 immunoblotting and enzyme-linked immunosorbent assays (ELISAs) to detect iC3b, SC5b-9, and C4d formation.
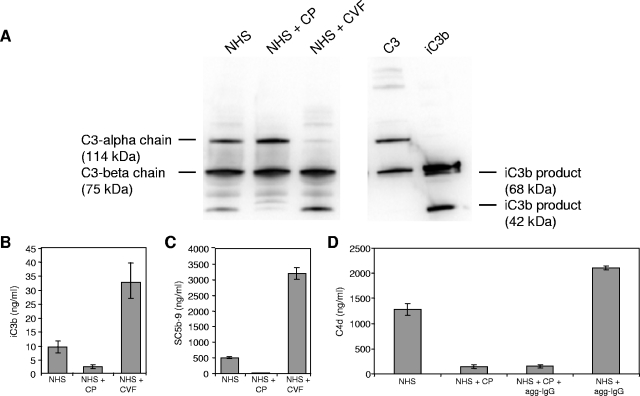
Twenty microliters of NHS was preincubated alone, with 5.4 μg CP, or with 1 μg CVF at 37°C for 3 h. The total volume was then brought up to 800 μl in GVBS++, and aliquots of each reaction mix were boiled, reduced, run in an SDS-PAGE gel, and transferred to a polyvinyl difluoride membrane. The blot was probed with goat anti-human C3 antibody (CompTech) at a 1:10,000 dilution, followed by horseradish peroxidase (HRP)-conjugated rabbit anti-goat antibody (Sigma-Aldrich) at a 1:4,000 dilution. Signal detection by enhanced chemiluminescence (ECL) was performed on a Versadoc instrument (Bio-Rad). Positive controls of purified C3 (alpha [114 kDa] and beta [75 kDa] chains) along with purified iC3b (α′1 [68 kDa] and α′2 [42 kDa] chains) were included in the gel.
Duplicate aliquots of the reaction mixtures described above were used to quantify iC3b and SC5b-9 levels by ELISA. For the iC3b ELISA, a monoclonal antibody recognizing a neoantigen of iC3b (Quidel) was utilized. The absorbance of the supernatants was read in a spectrophotometer at 450 nm. A standard curve was generated using purified iC3b. Similar methodology was used to determine SC5b-9 and C4d levels, using commercial kits (Quidel). For the C4d ELISA, samples were prepared as described above, except that the incubation time was 1 h. Heat-aggregated human immunoglobulin G (IgG) (50 μg/ml; Miles, Inc.), a classical pathway inhibitor, was prepared by dialysis in phosphate-buffered saline (PBS) overnight, followed by heating at 63°C for 20 min. Heat-aggregated IgG (25 μl of a 1:250 dilution), CP, or both were added to NHS simultaneously.
CP overlay blots.
Duplicate aliquots (1 μg) of the purified complement factors C1, C2, C3, and C4 (CompTech) were mixed with 1× PBS and an equal volume of 2× SDS sample buffer lacking reducing agents and loaded onto a 7.5% SDS-PAGE gel without being boiled. Proteins were transferred to nitrocellulose and blocked in 5% nonfat dried milk-PBS-0.01% Tween 20 for 1 h at room temperature. Purified CP (∼2 μg) was then added to one blot, whereas the second blot did not receive the probe; the blots were allowed to incubate for 1 h, after which they were washed with PBS-0.01% Tween 20. The rest of the procedure was carried out as for a standard immunoblot, using a rabbit polyclonal antibody to HAstV-1 particles (2) at a 1:1,000 dilution and an HRP-conjugated goat anti-rabbit IgG secondary antibody (Pierce) at a 1:2,000 dilution. For BSA, C1, C1q, C1r, and C1s, 3 μg of each sample was mixed with 1× PBS as described above, except for the 2× SDS sample buffer containing reducing agents, and the samples were boiled and loaded onto a 12% SDS-PAGE gel. Overlay blots were then prepared as described above. Both blots were stripped with Restore Western blot stripping buffer (Pierce) according to the manufacturer's guidelines and probed with polyclonal antibodies to C1q, C1r, and C1s (Santa Cruz) followed by the appropriate HRP-conjugated secondary antibodies. Signal detection by ECL was performed on a Versadoc instrument (Bio-Rad).
C1 reconstitution and zymosan assays.
NHS or heat-inactivated NHS was incubated alone, with 6.3 μg CP, or with 1 μg CVF for 1 h at 37°C as described above. This amount of CP was used to achieve roughly 50% RBC lysis. After the incubation, 2 μg of C1 or 10 μg of BSA was added to the indicated samples. RBCs were then added to all samples, and hemolysis was determined as described above.
To measure the number of C3 fragments deposited on zymosan, 20 μl NHS was incubated alone, with 9 μg CP, or with 1 μg CVF, and all samples were brought up to 70 μl with GVBS++ buffer for 1 h at 37°C. After incubation, the volume was adjusted to 1 ml with GVBS++ buffer, and then 450 μl was aliquoted, 3 μg C1 was added to the indicated samples, and 25 μl of activated zymosan (Sigma-Aldrich) was added to all samples. After a 10-min incubation at 37°C, the samples were washed twice with GVBS−− buffer and incubated with 25 mM methylamine for 1 h at 37°C to release bound C3 fragments, and the supernatant was collected. A C3 ELISA was performed on the samples, using a polyclonal antibody to C3, and analyzed as described above.
Real-time RT-PCR.
To quantify HAstV stocks, a real-time PCR method was developed. To isolate total RNA, 40-μl CaCo-2 cell lysates infected with HAstV-1, -2, -3, and -4 were diluted 1:5 in 1× minimum essential medium. RNAs were then extracted using Trizol (Invitrogen) per the manufacturer's instructions. Following isolation, RNAs were treated with DNase I (Promega) for 30 min at 37°C, and the enzyme was then inactivated at 65°C for 10 min. RNAs were stored at −80°C.
One-step real-time RT-PCR was performed using an iCycler IQ system (Bio-Rad). The real-time RT-PCR mixture was assembled using an iScrip one-step RT-PCR kit with SYBR green (Bio-Rad). Briefly, a reaction mixture was made, containing 12.5 μl 2× SYBR green RT-PCR mix, 0.5 μl each of 10 μM forward primer ORF1a-F1 and reverse primer ORF1a-R1 (targeting a conserved portion of the serine protease gene of the HAstVs; final concentration, 200 nM) (primer sequences will be provided upon request), 0.5 μl iScript reverse transcriptase for one-step RT-PCR, 6.0 μl sterile water, and 5.0 μl of the total RNA (isolated as described above). cDNA synthesis was achieved by incubating the reaction mixture for 10 min at 50°C, followed by inactivation of iScript reverse transcriptase at 95°C for 5 min. PCR cycling and detection included 45 cycles of incubation at 95°C for 10 s, 55°C for 30 s, and 72°C for 30 s. For the melt curve, samples were incubated at 95°C for 1 min and 57°C for 1 min, followed by 80 cycles of incubation for 10 s each, starting at 57°C and increasing at 0.5°C increments with each successive cycle. To generate a standard curve, an RNA standard was prepared by T7-mediated in vitro transcription (Ambion) of a genome-length cDNA clone (pAVIC) of HAstV-1 (10). The RNA standard was serially diluted from 1010 to 100, and the standard curve was established by plotting the cycle threshold versus log starting copy number for each dilution. The log starting copy number of the viral RNA contained in the CaCo-2 cell lysate was then extrapolated from the equation of the standard curve line, i.e., y = mx + b, where y is the cycle threshold, m is the slope of the standard curve line, x is the log starting copy number, and b is the y intercept or threshold fluorescence value.
Construction of recombinant baculoviruses.
A recombinant baculovirus containing the full-length HAstV-1 (Newcastle) CP gene was generated with a BacPAK baculovirus expression system kit (Clontech). Sequencing of this CP gene cDNA (kindly provided by M. J. Carter, University of Surrey, England) (32) verified 14 amino acid differences in HAstV-1 CP between the Oxford and Newcastle strains. The DNA fragment carrying the cDNA of the full-length wild-type protein was amplified by PCR with Pfu polymerase (Stratagene) and primers harboring BamHI and XbaI restriction sites at the 5′ and 3′ ends of the PCR product, respectively. The PCR product was then purified by agarose gel electrophoresis and a Gene Clean II kit (Qbiogene), digested with BamHI and XbaI, and ligated into BamHI/XbaI-digested transfer vector pBacPAK9. Following transformation of JM109 competent cells (Promega), plasmid DNA was isolated from several clones, and the presence of the inserted DNA was determined by diagnostic restriction endonuclease mapping. Positive clones harboring the CP gene were then completely sequenced across the inserted DNA, using a Big Dye Terminator v. 3.1 sequencing kit in an automated sequencing instrument (Applied Biosystems).
Generation of the recombinant baculovirus was carried out according to the manufacturer's protocols (Clontech). Briefly, the transfer vector pBacPAK9 containing the HAstV-1 CP gene was separately mixed with Bsu36I-linearized BacPAK6 baculovirus DNA and transfected into IPLB-Sf21 cells. At 3 days posttransfection, cell supernatants were harvested and putative recombinant viruses were isolated by growth of plaques once on IPLB-Sf21 cell monolayers. Individual plaque isolates were amplified and titrated according to standard procedures following confirmation of the presence and expression of the inserted gene by immunoblot analysis of the infected cell lysates, using a rabbit polyclonal antibody to HAstV-1 particles at a 1:1,000 dilution and an HRP-conjugated anti-rabbit IgG secondary antibody (Pierce) at a 1:2,000 dilution. Signal detection by ECL was performed on a Versadoc instrument (Bio-Rad).
Statistical analysis.
Results of replicate experiments were averaged, and means ± standard errors were calculated (Microsoft Excel 98). Comparisons were made by Student's t test (parametric) or the Mann-Whitney test (nonparametric), as appropriate for the specific data set (GraphPad In Stat, version 3.0), with P values of <0.05 being considered statistically significant.
RESULTS
HAstV-1 virions suppress serum hemolytic complement activity and display similar kinetics to CVF.
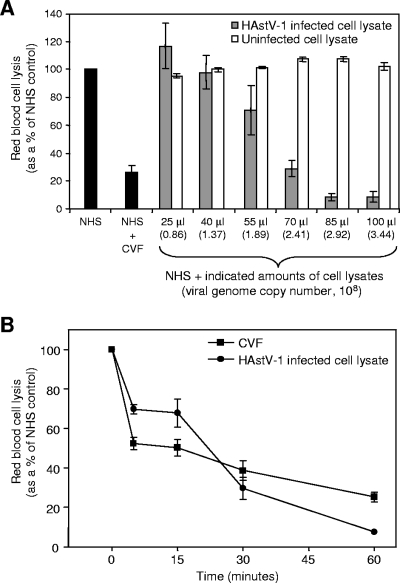
To investigate whether HAstV-1 virions affect serum complement activity, we utilized a standard hemolytic complement assay. In this procedure, sheep RBCs are sensitized with antibody and incubated with 2% NHS, and hemolytic complement activity is measured (Fig. 2A, NHS, black bar). As a positive control for the inhibition of hemolysis, we utilized CVF. CVF is a structural and functional homolog to C3b that acts as a C3/C5 convertase to rapidly deplete C3 and the terminal complement cascade components, thus depleting functional serum complement activity (Fig. 2A, NHS + CVF) (30). As increasing amounts of infected cell lysate containing HAstV-1 virions were added to NHS, a dose-dependent response was seen, with 70 μl of the virus-containing lysate (corresponding to 2.41 × 108 genome copies) inhibiting lysis to approximately the same extent as 1 μg (4.27 × 1012 molecules) of CVF (Fig. 2A, shaded bars). At 85 μl of virus-containing lysate (2.92 × 108 genome copies), inhibition of hemolysis surpassed that of CVF (P = 0.0143). To demonstrate that the inhibition of hemolytic complement activity was due to HAstV-1 virions, NHS was incubated with equivalent amounts of cell lysate from uninfected cells to show that this activity was virus specific (Fig. 2A, white bars). In addition, 7.64 × 1010 particles of sucrose gradient-purified FHV, a heterologous icosahedral RNA virus that infects insects (15), were found not to inhibit hemolysis (RBC lysis, 107% ± 6.57%; n = 3).
FIG. 2.
HAstV-1 virions suppress complement activity in a hemolytic complement assay, with similar kinetics to those of CVF. (A) Antibody-sensitized sheep RBCs were incubated with 20 μl NHS alone or in the presence of 1 μg CVF (black bars), along with increasing amounts of infected CaCo-2 cell lysates containing HAstV-1 virions (shaded bars) or uninfected CaCo-2 cell lysates (white bars). Hemolysis was standardized to 100% for NHS alone. One microliter of infected cell culture lysate corresponds to 3.44 × 106 genome copies, as determined by real-time RT-PCR. (B) CaCo-2 cell lysates containing HAstV-1 virions (85 μl of cell lysate, corresponding to 2.92 × 108 genome copies) and 1 μg CVF were incubated for 0 to 60 min in the presence of 20 μl NHS. At 0, 5, 15, 30, and 60 min, aliquots were removed and incubated with sensitized sheep RBCs to test hemolytic complement activity. Data are the means from five independent experiments. Error bars denote standard errors of the means (SEM).
To characterize the kinetics of activity of the virions on NHS in the hemolytic complement assay, a time course comparing HAstV-1 virions and CVF was conducted. Infected cell lysates containing HAstV-1 particles (2.92 × 108 genome copies) and 1 μg CVF were incubated for 0 to 60 min in the presence of 2% NHS. At increasing time intervals, aliquots were removed and exposed to sensitized RBCs. Both CVF and the virus showed similar kinetics in the inhibition of hemolysis (Fig. 2B). While CVF displayed a more pronounced suppression of hemolytic activity at earlier time points, at the 60-min time point the virus demonstrated a stronger inhibitory effect, of 93%, than the 75% suppression for CVF (P = 0.0001).
Other HAstV serotypes suppress hemolytic complement activity.
To determine whether HAstV serotypes other than type 1 exhibit the same effects on complement activity, equivalent amounts of cell lysates infected with serotypes 1 to 4 were analyzed in the hemolytic complement assay, with all four serotypes demonstrating nearly identical levels of hemolysis inhibition (Table 1). These findings suggest that the complement-suppressing effect reported here is a conserved property of the HAstVs.
TABLE 1.
Hemolytic assay of HAstV serotypes 1 to 4
A total of 2.92 × 108 genome copies from infected cell lysates was utilized for each serotype tested.
n = 3.
Soluble HAstV-1 CP suppresses hemolytic complement activity.
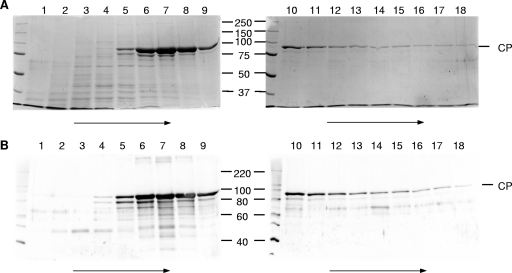
While HAstV-infected cell lysates demonstrated suppression of hemolytic complement activity, one cannot rule out the possibility that a newly induced/overproduced cellular protein or viral nonstructural protein was responsible for the observed complement suppression activity. To address this contingency, we isolated soluble HAstV-1 CP produced from a recombinant baculovirus. While small numbers of intact virus-like particles were visualized in infected IPLB-Sf21 cells by thin-section electron microscopy, most of the CP was in the form of aggregates (data not shown). To produce soluble CP, we utilized a four-step purification procedure in which CP aggregates were initially isolated in crude form by extraction of cells with 2% NP-40 followed by 0.5 mg of DNase I per ml (22). After high-speed pelleting, the CP-containing pellet was resuspended in dissociation buffer with freezing overnight to solubilize the aggregated CP (25) and then purified further by sucrose gradient ultracentrifugation in the same buffer. During centrifugation, the majority of the CP was found to peak in fractions 6 to 8, along with some CP degradation products, as analyzed by SDS-PAGE followed by Coomassie blue staining (Fig. 3A) and immunoblotting using antibody to HAstV-1 particles (2) (Fig. 3B).
FIG. 3.
SDS-PAGE and immunoblot analysis of fractions obtained during the sucrose gradient ultracentrifugation step of HAstV-1 CP purification. The first 18 fractions of the gradient were run in SDS-PAGE gels and stained with Coomassie blue (A) or subjected to immunoblotting with antibody to HAstV-1 virions (B). Numbers refer to gradient fractions, with arrows representing the direction of sedimentation. Molecular mass markers (in kDa) are indicated in the middle, while the position of CP is indicated to the right.
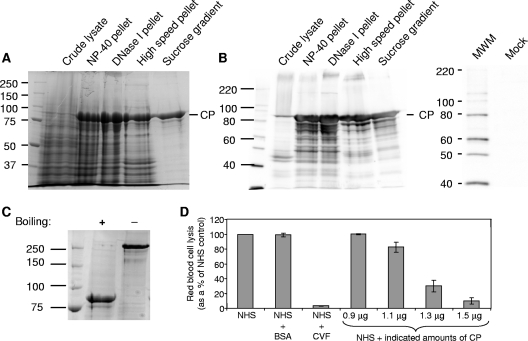
SDS-PAGE analysis of the CP-containing fractions at each step of the purification procedure was monitored by Coomassie blue staining (Fig. 4A) and immunoblotting (Fig. 4B). Densitometric scanning of the Coomassie blue-stained gel demonstrated that CP constituted >94% of the protein present in the peak fraction (in multiple preparations). A small portion of this signal was due to a CP-specific degradation product that migrated slightly below the full-length 87-kDa band. When gradient-purified CP was boiled in the presence of β-mercaptoethanol and run in a 7.5% SDS-PAGE gel, CP was detected at 87 kDa, as expected (Fig. 4C). However, CP that was not boiled gave rise to a band that migrated just above the 250-kDa molecular size standard (Fig. 4C). The size of this band is consistent with a trimer of CP molecules, which would be predicted to run at approximately 261 kDa (87 kDa × 3), and possibly represents an assembly intermediate. However, fully assembled virus-like particles were not detected under these dissociation conditions by negative-stain electron microscopy (data not shown). Consistent with our findings, HAstV serotype 8 CP was recently reported to form trimers in translation experiments performed in vitro (19). We also subjected the purified CP to digestion with trypsin, which is normally required to cleave the viral capsid into proteins of 34, 29, and 26 kDa, as resolved by SDS-PAGE (1, 20). Trypsin cleavage of the CP resulted in digestion products of <25 kDa, further demonstrating that the soluble CP was structurally different from intact virus-like particles (data not shown).
FIG. 4.
Analysis of HAstV-1 CP purification, oligomerization state, and activity in the hemolytic complement assay. Coomassie blue staining (A) and immunoblot analysis (B) of the CP-containing fraction were performed at each stage of the purification procedure. An IPLB-Sf21 mock-infected cellular lysate was also analyzed by immunoblotting to demonstrate that there was no cross-reactivity to the HAstV-1 antibody. Molecular size markers (MWM; in kDa) are indicated to the left, with the position of CP denoted on the right. (C) Aliquots of sucrose-purified CP were either boiled or not boiled in the presence of 2-mercaptoethanol, resolved by SDS-PAGE, and then stained with Coomassie blue. The boiled protein migrated at 87 kDa, the expected mass of the uncleaved CP precursor (monomer), whereas the unboiled sample migrated above 250 kDa, possibly representing a trimer. Molecular size markers (in kDa) are indicated to the left. (D) Antibody-sensitized sheep RBCs were incubated with 2% NHS alone or in the presence of 15 μg BSA, 1 μg CVF, or the indicated amounts of purified CP. Hemolysis was standardized to 100% for NHS alone. Data are the means for four independent experiments. Error bars denote SEM.
To ascertain whether soluble CP displays complement-suppressing activity, as demonstrated for authentic virus, we again utilized the hemolytic complement assay (Fig. 4D). As expected, NHS alone and NHS plus purified BSA protein demonstrated equivalent amounts of RBC lysis, whereas NHS plus CVF inhibited lysis significantly. As increasing amounts of CP were added to NHS, a dose-dependent response in hemolysis demonstrated decreased complement activity, as seen with HAstV-1-infected cell lysates. A total of 1.5 μg CP (3.51 × 1012 molecules of trimer) suppressed hemolysis to a similar degree as did 1 μg (4.27 × 1012 molecules) of CVF (see Fig. 7D), suggesting that CP possesses potent complement-suppressing activity. Equivalent amounts of sucrose gradient buffer alone or sucrose gradient buffer from a recombinant baculovirus expressing a soluble, heterologous viral protein (the RNA polymerase of FHV) (15) that was subjected to an identical purification procedure to that used for HAstV-1 CP demonstrated no effect on hemolytic complement activity (data not shown).
FIG. 7.
HAstV-1 CP inhibits iC3b formation, terminal complement cascade activation, and C4d formation. Reaction mixtures containing NHS were preincubated alone, with 5.4 μg CP, or with 1 μg CVF at 37°C for 3 h and then aliquoted. CVF is a positive control for complement activation. (A) Reaction products were resolved by SDS-PAGE and subsequently analyzed by immunoblotting using antisera to C3. CVF is a positive control for iC3b formation and corresponding C3 alpha chain depletion. Purified standards for C3 (alpha [114 kDa] and beta [75 kDa] chains) and iC3b (α′1 [68 kDa] and α′2 [42 kDa]) products were included in the gel, as indicated to the left and right of the gel, respectively. Under these gel conditions, the 68-kDa iC3b band comigrated with the C3 beta chain product. Aliquots of the above reaction mixtures were analyzed by ELISA (ng/ml) for iC3b (B) and SC5b-9 (C) formation. (D) NHS was incubated for 1 h in the presence of heat-aggregated IgG (agg-IgG; a classical pathway activator), CP, or both and measured for C4 cleavage by C4d ELISA. Standard curves were generated using purified iC3b, SC5b-9, and C4d. Data are the means for four independent experiments for each ELISA. Error bars denote SEM.
HAstV-1 CP specifically suppresses the classical pathway.
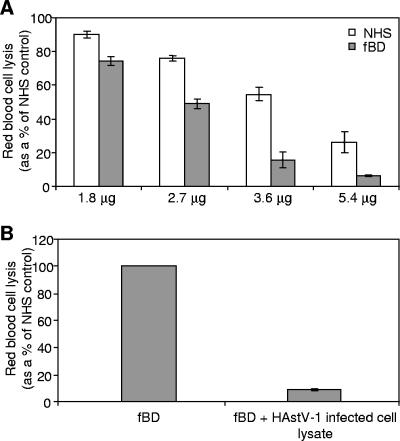
To determine if CP suppresses complement activation of the classical or alternative pathway, sera depleted of complement factors specific for each pathway were utilized in the hemolytic complement assay. To evaluate suppression of classical pathway activation by CP, NHS and fBD serum were compared. Factor B is essential for alternative pathway activation, and utilizing fBD serum allows the specific testing of classical pathway activation. Both NHS and fBD serum demonstrated a dose-dependent suppression of hemolysis by CP (Fig. 5A). However, CP was considerably more effective at suppressing fBD serum than NHS. For 3.6 μg of CP, hemolysis was suppressed 16% for fBD serum, compared with 55% for NHS (P = 0.0159). Increased inhibition when the classical pathway was isolated (fBD serum) compared with that when activation could occur by any or all pathways (NHS) suggested that CP may more strongly suppress activation of the classical pathway than the alternative pathway. As with CP, infected cell lysates containing HAstV-1 particles (2.92 × 108 genome copies) also inhibited hemolysis in fBD serum (Fig. 5B). It should be noted that fBD serum retains the same capacity for hemolysis as NHS because C3 and C5 to C9 are intact. It is possible that the lectin pathway may also be activated under these conditions, but it is currently unknown whether astrovirus capsids are glycosylated.
FIG. 5.
HAstV-1 CP and virions strongly suppress classical pathway activity. (A) Antibody-sensitized sheep RBCs were incubated with NHS (white bars) or fBD serum (shaded bars) in the presence of the indicated amounts of CP. (B) Antibody-sensitized sheep RBCs were incubated with fBD serum alone or in the presence of HAstV-1 virions (85 μl of cell lysate, corresponding to 2.92 × 108 genome copies). Hemolysis was standardized to 100% for each serum in the absence of CP or virus. Data are the means for four (A) or three (B) independent experiments. Error bars denote SEM.
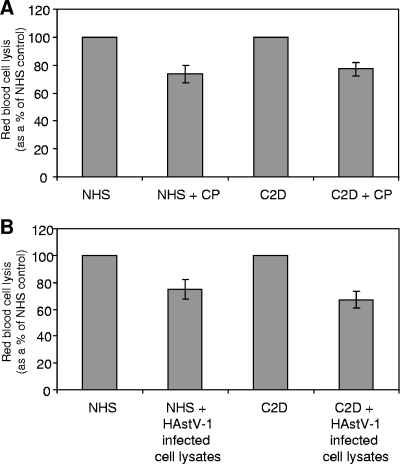
We tested the effects of CP and virus on the alternative pathway as well. As demonstrated in Fig. 6A, in an assay to test for alternative pathway activation (i.e., the rabbit RBCs were not sensitized with antibody and only the alternative pathway may activate in Mg-EGTA-GVBS buffer), NHS lysed rabbit RBCs as expected. When 6.3 μg of CP was added to NHS, there was only a modest effect on the suppression of hemolysis (Fig. 6A, NHS + CP). To confirm that activation was limited to the alternative pathway, C2D serum was utilized in place of NHS. By depleting NHS of C2 in this assay, the classical and lectin pathways are blocked and any RBC lysis is due exclusively to alternative pathway activity. As with NHS, C2D serum in the presence of CP suppressed hemolysis approximately 25% compared to that with C2D serum alone (Fig. 6A, C2D and C2D + CP). The same results were obtained when HAstV-1-infected cell lysates (2.92 × 108 genome copies) were tested for alternative pathway activity (Fig. 6B). In both of these alternative pathway activation assays, CP appeared to only partially block activation, suggesting that HAstV-1 CP suppresses hemolytic complement activity via the classical pathway more than via the alternative pathway.
FIG. 6.
HAstV-1 CP and virions have a modest effect on alternative pathway activity. NHS or C2D serum was incubated with rabbit RBCs in Mg-EGTA-GVBS buffer alone or in the presence of 6.3 μg CP (A) or HAstV-1 virions (30 μl of cell lysate, corresponding to 1.03 × 108 genome copies) (B). Hemolysis was standardized to 100% for each serum in the absence of CP or virus. Data are the means for three (A) or six (B) independent experiments. Error bars denote SEM.
HAstV-1 CP inhibits iC3b, C5b-9 membrane attack complex, and C4d formation.
While CP was found to suppress complement in hemolytic assays, it was unclear whether this effect was due to activation and depletion of serum complement components, as occurs with CVF, or as the result of inhibition of activation. To test these competing hypotheses, aliquots of NHS alone, NHS plus CP, or NHS plus CVF were incubated for 3 h at 37°C, and the amount of iC3b produced was measured by immunoblotting and an iC3b-specific ELISA. Upon activation of the three pathways of the complement system, C3 is cleaved to C3a and C3b. Some C3b is further cleaved to the inactive form iC3b by soluble complement regulators in NHS (factor I plus factor H). Utilizing an immunoblot with an antibody specific for C3 fragments, two bands were present for the uncleaved C3 control, namely, C3-alpha (114 kDa) and C3-beta (75 kDa) (Fig. 7A, lane C3). Cleavage of C3b to iC3b results in two cleavage products from the alpha chain, of 68 kDa (α′1) and 42 kDa (α′2), while the C3-beta chain remains invariant (Fig. 7A, lane iC3b). NHS alone produced only a small amount of the 42-kDa iC3b product after a 3-h incubation, consistent with low-level, spontaneous activation of C3 (Fig. 7A, lane NHS). In the CP-containing sample, very little of the 42-kDa iC3b product was observed (Fig. 7A, lane NHS + CP lane). In contrast, CVF depleted the C3-alpha chain, generating large amounts of the 42-kDa iC3b band (Fig. 7A, lane NHS + CVF). To more precisely quantify the amount of iC3b generated, duplicate aliquots of these samples were measured by iC3b-specific ELISA. Consistent with the immunoblot data, CP suppressed iC3b formation to a greater extent than did NHS alone (P = 0.0314), whereas NHS plus CVF generated significant amounts of iC3b, consistent with the known actions of CVF to activate and deplete the complement system (Fig. 7B). Thus, even though HAstV-1 virions displayed similar kinetics to those of CVF (Fig. 2B), these data show that the CP of this virus does not cause complement activation resulting in cleavage of C3, suggesting that suppression of serum complement activation occurs through an inhibitory mechanism.
To measure the effect of CP inhibition of serum complement on the terminal complement cascade, aliquots of the samples used above were assayed for formation of the membrane attack complex, utilizing an ELISA specific for SC5b-9 complex formation. As shown in Fig. 7C, for NHS alone, a modest amount of SC5b-9 complex was detected, whereas virtually no complex was present in NHS treated with CP (P = 0.0286), suggesting no terminal complement cascade activation. In CVF-treated serum, large amounts of SC5b-9 complex were generated, as expected for relentless activation and depletion.
The ability of CP to inhibit iC3b and SC5b-9 formation, coupled with its strong suppression of the classical pathway (Fig. 5), led us to investigate if CP suppressed the complement system at the level of C4, the second component of the classical pathway, after C1. Upon activation of the C1 complex, C4 is cleaved and then C2 is cleaved to form the classical pathway C3 convertase (31). An ELISA that detects a specific by-product of C4 activation (C4d) demonstrated that in the presence of CP, serum generated very low levels of C4d, in contrast to that in the presence of NHS alone (P = 0.0286) (Fig. 7D), suggesting an inhibition of spontaneous classical pathway activation. The classical pathway activator heat-aggregated IgG greatly increased C4d generation, yet when CP was added simultaneously with heat-aggregated IgG to NHS, C4d formation was greatly inhibited (P = 0.0286) (Fig. 7D). These results show that HAstV-1 CP can inhibit classical pathway activation by a potent classical activator, heat-aggregated IgG, before C4 cleavage can occur, suggesting that CP is a strong classical pathway inhibitor and that inhibition likely occurs at C1.
HAstV-1 CP binds to the A chain of C1q.
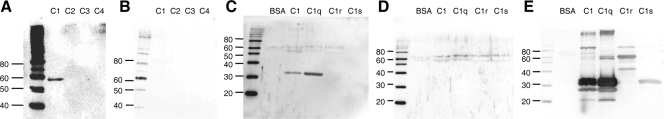
CP inhibition of the classical pathway led us to speculate that CP interacts with one of the classical pathway factors (C1 complex, C4, or C2). To ascertain whether CP binds to specific complement factors, we utilized a modified virus overlay protein binding assay approach (3). To this end, C1 complex and highly purified C2, C3, and C4 were mixed with SDS sample buffer lacking reducing agents and loaded onto a 7.5% SDS-PAGE gel without being boiled. Following electrophoresis, proteins were transferred to nitrocellulose, blocked, and then probed with or without purified CP in blocking solution for 1 h, after which the blots were washed. CP binding was then detected with antiserum to HAstV-1 (2) followed by a labeled secondary antibody. As shown in Fig. 8A, a specific band of approximately 59 kDa, consistent with a dimer of C1 chains, was present in the C1 lane, while no binding was detected for C2 to C4. In the absence of the CP probe, no signal was detected for C1 (Fig. 8B). The fact that C1 is a multimolecular complex of C1q, C1r, and C1s along with the fact that the gels were run under nonreducing conditions without boiling the samples made it difficult to determine exactly which component of C1 interacts with the viral CP. To address this issue, all three of the highly purified constituents of the C1 complex (C1q, C1r, and C1s) were boiled, reduced, loaded onto 12% SDS-PAGE gels, transferred to nitrocellulose, and probed with or without CP as described above. The blot probed with CP showed a band of ∼34 kDa in the C1 and C1q lanes, whereas there was no signal for BSA, C1r, or C1s (Fig. 8C). As expected, no signal was detected in the duplicate blot that did not receive the CP probe (Fig. 8D). The blots were stripped and reprobed with antisera to C1q, C1r, and C1s to reveal the individual protein constituents of C1 (Fig. 8E). C1q is composed of six subunits, each of which contains three polypeptide chains, i.e., A, B, and C. As shown in the C1 and C1q lanes, all three C1q chains react with C1q antisera, and under reducing conditions, chain C runs at 27.5 kDa, chain B runs at 31.6 kDa, and chain A runs at 34.8 kDa (23). CP was found to overlay precisely with the 34-kDa band corresponding to the A chain of C1q in both the C1 and C1q lanes. We believe that the band detected by CP at approximately 59 kDa in the nonreduced C1 lanes (Fig. 8A) corresponds to disulfide-bonded A-B C1q dimers.
FIG. 8.
HAstV-1 CP binds to the A chain of C1q. (A) The indicated complement factors were loaded onto a 7.5% SDS-PAGE gel in the absence of boiling or reduction. Proteins were then transferred to nitrocellulose, blocked, and probed with CP for 1 h. (B) An identical blot that did not receive the CP probe. Blots were subsequently washed and probed with antibody to HAstV-1 particles. (C and D) The indicated proteins were boiled, reduced, resolved in 12% SDS-PAGE gels, and transferred to nitrocellulose. Overlay blotting was then carried out as described above, with one blot receiving the probe (C), while the other did not (D). (E) Both overlay blots were stripped and reprobed with antibodies to C1q, C1r, and C1s. Only one reprobed blot is shown, as both blots yielded identical results. Molecular size markers (in kDa) are indicated to the left.
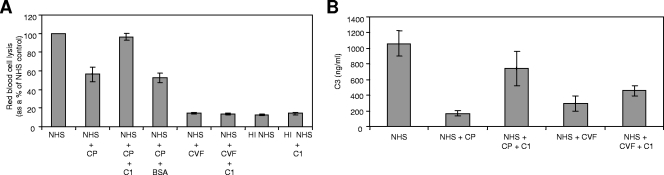
Exogenous C1 can reconstitute hemolytic activity and C3 activation of CP-treated NHS.
Based on the overlay blot data, CP appears to interact with the A chain of C1q. If C1 interacts with CP, then additional exogenous C1 should reconstitute hemolytic complement activity. Thus, enough CP was added to NHS for 1 h to inhibit lysis approximately 50% (Fig. 9A). Exogenous C1 (10 μg) was then added to CP-treated NHS, and hemolytic activity was fully restored, from 56% to 97% (P = 0.0286). Reconstitution of hemolytic activity did not occur when BSA was substituted for C1 or when additional C1 was added to CVF-treated NHS, suggesting that CP reversibly inhibits complement activation via C1.
FIG. 9.
Exogenous C1 reconstitutes hemolytic activity and C3 activation for CP-treated NHS. (A) NHS was incubated alone, with 6.3 μg CP, or with 1 μg CVF for 1 h at 37°C. Heat-inactivated NHS (HI NHS) was used as an additional control. After the incubation, 2 μg of C1 or 10 μg BSA was added to the indicated samples. Sensitized RBCs were then added to all samples, and hemolysis was determined. (B) NHS was incubated alone, with 9 μg CP, or with 1 μg CVF for 1 h at 37°C. After the incubation, 3 μg C1 was added to the indicated samples, followed by the addition of 25 μl of zymosan to all samples for 10 min at 37°C. Samples were washed and incubated with 25 mM methylamine for 1 h at 37°C to remove bound C3 fragments, and the supernatant was collected. A C3 ELISA was performed on the samples, using a polyclonal antibody to C3, and a standard curve was utilized to determine the value of C3. For both experiments, data are the means for four independent experiments. Error bars denote SEM.
To confirm the reversal of CP inhibition of complement activity by C1, the deposition of C3 on zymosan was measured (Fig. 9B). Zymosan particles activate complement and serve as a target for C3 fragment binding. Duplicate aliquots of NHS were incubated with CP or CVF for 1 h at 37°C. After the incubation, C1 was added to the indicated samples, followed by the addition of zymosan to all samples. After 10 min at 37°C, the samples were treated with methylamine to cleave off bound C3 fragments, which were measured in a C3-specific ELISA. NHS alone bound substantial amounts of C3 fragments on zymosan, as expected, whereas CP-treated NHS bound much lower levels of C3 fragments (Fig. 9B). The addition of exogenous C1 to CP-treated NHS increased C3 fragment binding, again showing the ability of excess C1 to reconstitute complement activity. As expected, the addition of excess C1 to CVF-treated NHS did not significantly increase C3 deposition. Taken together, these experiments suggest that CP inhibits complement activation at C1 and that the inhibition of the complement activation cascade is reversible.
DISCUSSION
The complement system comprises a group of related plasma proteins that, when activated, generate an extremely potent immunological cascade. In humans, complement is the first line of defense against viral and bacterial pathogens, and to avoid complement-mediated destruction, many pathogens have devised mechanisms to thwart this innate immune response. Viruses have been shown to utilize one of the following three basic strategies for evasion of the complement response: (i) avoidance of complement binding to antibody-antigen complexes (classical pathway) by removing these antibody-antigen complexes from the surface of the infected cell or by expression of Fc receptors (Herpesviridae and Coronoviridae family members), (ii) encoding viral proteins that mimic the activities of complement regulatory proteins (Poxviridae and Herpesviridae family members), and (iii) incorporation of host complement regulatory proteins into the viral envelope or upregulation of these regulators on the infected cell surface (Herpesviridae, Poxviridae, Retroviridae, and Togaviridae family members) (reviewed in reference 9). In addition to these evasion strategies, it was recently demonstrated that West Nile virus (a member of the Flaviviridae) nonstructural protein 1 binds and recruits complement regulatory protein factor H to the surface of the infected cell, resulting in decreased complement activation against the infected cell (4). Each example of complement evasion listed above involves enveloped virus families, and to our knowledge, there are no reports of complement evasion by nonenveloped, icosahedral viruses. The findings reported here demonstrate that members of the nonenveloped Astroviridae family specifically suppress serum complement activity via the viral CP.
Soluble CP and virus both inhibit complement activity.
Like infectious HAstV-1 virions, soluble CP was also effective in suppressing hemolytic complement activity. While the viral capsid consists of multiple copies of trypsin-cleaved CP molecules, soluble CP was purified as uncleaved (precursor) protein in the form of trimers. Oligomers of CP were also observed recently for HAstV-8, suggesting that trimer formation may represent an assembly intermediate in the formation of the viral particle (19). While it is not clear from these experiments if the oligomerization state of the protein is critical for its observed activity on complement, it is apparent that CP retains complement inhibition activity regardless of its state as a soluble protein or in the context of an infectious, trypsin-cleaved virus particle.
CP interaction with C1 is a novel method of complement inhibition by a virus.
C1 initiates activation of the classical pathway and links the adaptive and innate immune systems by recognizing antibody-bound pathogens (Fig. 1). It consists of a multimolecular complex formed by the pattern recognition protein C1q associated with a calcium-dependent tetramer of the serine proteases C1r and C1s (reviewed in reference 5). The six subunits of C1q each consist of the three protein chains, A, B, and C, which compose an N-terminal, triple-helical, collagen-like (stalk) domain followed by a globular (head) domain. The head domain is the moiety responsible for binding of the conventional activating molecules, clustered IgG and IgM. However, other, nonimmunoglobulin molecules, such as C-reactive protein, serum amyloid P, lipopolysaccharide, and DNA, may also bind C1q (29). Binding of the globular head of C1q by the appropriate activating molecule is thought to induce a conformational change in C1q that activates C1r, which in turn cleaves C1s to initiate classical pathway activation (5). In serum, activation of C1 is tightly regulated by C1 inhibitor, a protein of the serpin family which inactivates C1 by forming an extremely stable complex with activated C1r and C1s (5). Our data suggest that CP inhibits complement activity at C1 and directly binds the A chain of C1q, but the precise mechanism by which this inhibition occurs is unknown. Unlike C1 inhibitor, CP does not have homology to serpin proteins and appears to bind exclusively with C1q. Thus, CP appears to inhibit C1 by a novel mechanism that is under investigation.
Insights into astrovirus pathogenesis.
Many enteric pathogens, such as rotavirus (12), initiate diarrhea by destroying enterocytes in the epithelium and inducing an increased inflammatory response. However, astrovirus-induced diarrhea does not appear to result in significant cell death or inflammation in humans or in an avian astrovirus animal model (reviewed in reference 21). One of the important roles of complement is to trigger robust inflammatory (leukocyte) cell migration to infected tissues via the peptide mediators C3a and C5a, which are generated by cleavage of C3 and C5, respectively (8). Inhibition of iC3b and SC5b-9 formation by CP (Fig. 7) reveals that CP would also likely inhibit formation of these chemoattractants and perhaps contribute to limited leukocyte migration to the site of infection. Interestingly, in vivo, C1 components are synthesized mainly in columnar intestinal epithelial cells and are secreted into the lumen of the gastrointestinal tract (reviewed in reference 17). The nearly identical levels of complement suppression demonstrated by equivalent amounts of four HAstV serotypes (Table 1) suggest that this novel activity may be a conserved mechanism utilized by the Astroviridae to modify the host immune response. This new mechanism of HAstV-host defense interaction may eventually lead to antiastrovirus therapies; currently, there are none for preventing or modifying disease in humans as well as other mammals and birds.
Acknowledgments
We thank M. Carter (University of Surrey, United Kingdom) for the HAstV-1 CP gene, D. Bass (Stanford University, CA) for antibody to HAstV-1 virions, D. K. Mitchell (Eastern Virginia Medical School, Norfolk, VA) for providing HAstV 1-4 viral stocks, A. Schneemann (The Scripps Research Institute, La Jolla, CA) for IPLB-Sf21 cells and authentic FHV, K. Bok for amplification of HAstV-1 stocks, and E. Mendez (Universidad Nacional Autónoma de México) for providing the pAVIC plasmid.
This work was supported in part by National Institutes of Health grant R21 AI060874 to N.K.K. and by a grant from The Children's Hospital of The King's Daughters Research Endowment to K.M.C.
Footnotes
Published ahead of print on 24 October 2007.
REFERENCES
- 1.Bass, D. M., and S. Qiu. 2000. Proteolytic processing of the astrovirus capsid. J. Virol. 741810-1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bass, D. M., and U. Upadhyayula. 1997. Characterization of human serotype 1 astrovirus-neutralizing epitopes. J. Virol. 718666-8671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Borrow, P., and M. B. A. Oldstone. 1992. Characterization of lymphocytic choriomeningitis virus-binding protein(s): a candidate cellular receptor for the virus. J. Virol. 667270-7281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chung, K. M., M. K. Liszewski, G. Nybakken, A. E. Davis, R. R. Townsend, D. H. Fremont, J. P. Atkinson, and M. S. Diamond. 2006. West Nile virus nonstructural protein NS1 inhibits complement activation by binding the regulatory protein factor H. Proc. Natl. Acad. Sci. USA 10319111-19116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cooper, N. R. 1985. The classical complement pathway: activation and regulation of the first complement component. Adv. Immunol. 37151-216. [DOI] [PubMed] [Google Scholar]
- 6.Cunnion, K. M., P. S. Hair, and E. S. Buescher. 2004. Cleavage of complement C3b to iC3b on the surface of Staphylococcus aureus is mediated by serum complement factor I. Infect. Immun. 722858-2863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dennehy, P. H., S. M. Nelson, S. Spangenberger, J. S. Noel, S. S. Monroe, and R. I. Glass. 2001. A prospective case-control study of the role of astrovirus in acute diarrhea among hospitalized young children. J. Infect. Dis. 18410-15. [DOI] [PubMed] [Google Scholar]
- 8.Ember, J. A., M. A. Jagels, and T. E. Hugli. 1998. Characterization of complement anaphylatoxins and their biological responses, p. 241-284. In J. E. Volanakis and M. M. Frank (ed.), The human complement system in health and disease, 1st ed. Marcel Dekker, Inc., New York, NY.
- 9.Favoreel, H. W., G. R. Van de Walle, H. J. Nauwynck, and M. B. Pensaert. 2003. Virus complement evasion strategies. J. Gen. Virol. 841-15. [DOI] [PubMed] [Google Scholar]
- 10.Geigenmüller, U., N. H. Ginzton, and S. M. Matsui. 1997. Construction of a genome-length cDNA cone for human astrovirus serotype 1 and synthesis of infectious RNA transcripts. J. Virol. 721713-1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jiang, B., S. S. Monroe, E. V. Koonin, S. E. Steine, and R. I. Glass. 1993. RNA sequence of astrovirus: distinctive genomic organization and a putative retrovirus-like ribosomal frameshifting signal that directs the viral replicase synthesis. Proc. Natl. Acad. Sci. USA 9010539-10543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kapikian, A. Z., Y. Hoshino, and R. M. Chanock. 2001. Rotaviruses, p. 1787-1833. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed., vol. 2. Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 13.Koci, M. D. 2005. Immunity and resistance to astrovirus infection. Viral Immunol. 1811-16. [DOI] [PubMed] [Google Scholar]
- 14.Koci, M. D., L. A. Moser, L. A. Kelley, D. Larson, C. C. Brown, and S. Schultz-Cherry. 2003. Astrovirus induces diarrhea in the absence of inflammation and cell death. J. Virol. 7711798-11808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krishna, N. K., D. Marshall, and A. Schneemann. 2003. Analysis of RNA packaging in wild-type and mosaic protein capsids of flock house virus using recombinant baculovirus vectors. Virology 30510-24. [DOI] [PubMed] [Google Scholar]
- 16.Lewis, T. L., H. B. Greenberg, J. E. Hermann, L. S. Smith, and S. M. Matsui. 1994. Analysis of astrovirus serotype 1 RNA, identification of the viral RNA-dependent RNA polymerase motif, and expression of a viral structural protein. J. Virol. 6877-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Loos, M. 1983. Biosynthesis of the collagen-like C1q molecule and its receptor functions for Fc and polyanionic molecules on macrophages. Curr. Top. Microbiol. Immunol. 1021-56. [DOI] [PubMed] [Google Scholar]
- 18.Matsui, S. M., and H. B. Greenberg. 2001. Astroviruses, p. 875-893. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed., vol. 1. Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 19.Méndez, E., G. Aguirre-Crespo, G. Zavala, and C. F. Arias. 2007. Association of the astrovirus structural protein VP90 with membranes plays a role in virus morphogenesis. J. Virol. 8110649-10658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Méndez, E., T. Fernández-Luna, S. López, M. Méndez-Toss, and C. F. Arias. 2002. Proteolytic processing of a serotype 8 human astrovirus ORF2 polyprotein. J. Virol. 767996-8002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moser, L. A., and S. Schultz-Cherry. 2005. Pathogenesis of astrovirus infection. Viral Immunol. 184-10. [DOI] [PubMed] [Google Scholar]
- 22.Newcomb, W. W., R. M. Juhas, D. R. Thomsen, F. L. Homa, A. D. Burch, S. K. Weller, and J. C. Brown. 2001. The UL6 gene product forms the portal for entry of DNA into the herpes simplex virus capsid. J. Virol. 7510923-10932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reid, K. B. M., D. M. Lowe, and R. R. Porter. 1972. Isolation and characterization of C1q, a subcomponent of the first component of complement, from human and rabbit sera. Biochem. J. 130749-763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schneemann, A., R. Dasgupta, J. E. Johnson, and R. R. Rueckert. 1993. Use of recombinant baculoviruses in synthesis of morphologically distinct virus-like particles of flock house virus, a nodavirus. J. Virol. 672756-2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schneemann, A., T. M. Gallagher, and R. R. Rueckert. 1994. Reconstitution of flock house provirions: a model system for studying structure and assembly. J. Virol. 684547-4556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sebire, N. J., M. Malone, N. Shah, G. Anderson, H. B. Gaspar, and W. D. Cubitt. 2004. Pathology of astrovirus associated diarrhoea in a paediatric bone marrow transplant recipient. J. Clin. Pathol. 571001-1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shastri, S., A. M. Doane, J. Gonzales, U. Upadhyayula, and D. M. Bass. 1998. Prevalence of astroviruses in a children's hospital. J. Clin. Microbiol. 362571-2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thiel, S., T. Vorup-Jensen, C. M. Stover, W. Schwaeble, S. B. Laursen, K. Poulsen, A. C. Willis, P. Eggleton, S. Hansen, U. Holmskov, K. B. M. Reid, and J. C. Jensenius. 1997. A second serine protease associated with mannan-binding lectin that activates complement. Nature 386506-510. [DOI] [PubMed] [Google Scholar]
- 29.Trinder, P. K. E., M. J. Maeurer, M. Kaul, F. Petry, and M. Loos. 1993. Functional domains of the human C1q A-chain. Behring Inst. Mitt. 93180-188. [PubMed] [Google Scholar]
- 30.Vogel, C.-W., C. A. Smith, and H. J. Müller-Eberhard. 1984. Cobra venom factor: structural homology with the third component of complement. J. Immunol. 1333235-3241. [PubMed] [Google Scholar]
- 31.Volanakis, J. E. 1998. Overview of the complement system, p. 9-32. In J. E. Volanakis and M. M. Frank (ed.), The human complement system in health and disease, 1st ed. Marcel Dekker, Inc., New York, NY.
- 32.Willcocks, M. M., and M. J. Carter. 1993. Identification and sequence determination of the capsid protein gene of human astrovirus serotype 1. FEMS Microbiol. Lett. 1141-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Willcocks, M. M., M. J. Carter, F. R. Laidler, and C. R. Madeley. 1990. Growth and characterization of human faecal astrovirus in a continuous cell line. Arch. Virol. 11373-81. [DOI] [PubMed] [Google Scholar]
- 34.Willcocks, M. M., T. D. K. Brown, C. R. Madeley, and M. J. Carter. 1994. The complete sequence of a human astrovirus. J. Gen. Virol. 751785-1788. [DOI] [PubMed] [Google Scholar]