Abstract
The relative importance of humoral and cellular immunity in the prevention or clearance of hepatitis C virus (HCV) infection is poorly understood. However, there is considerable evidence that neutralizing antibodies are involved in disease control. Here we describe the detailed analysis of human monoclonal antibodies (MAbs) directed against HCV glycoprotein E1, which may have the potential to control HCV infection. We have identified two MAbs that can strongly neutralize HCV-pseudotyped particles (HCVpp) bearing the envelope glycoproteins of genotypes 1a, 1b, 4a, 5a, and 6a and less strongly neutralize HCVpp bearing the envelope glycoproteins of genotype 2a. Genotype 3a was not neutralized. The epitopes for both MAbs were mapped to the region encompassing amino acids 313 to 327. In addition, robust neutralization was also observed against cell culture-adapted viruses of genotypes 1a and 2a. Results from this study suggest that these MAbs may have the potential to prevent HCV infection.
The World Health Organization estimates that ∼170 million people worldwide are infected with hepatitis C virus (HCV) (10), with an additional 4 million people infected every year. The majority of infected individuals progress to chronic hepatitis, which greatly increases their risk for developing cirrhosis and hepatocellular carcinoma (27). The standard treatment for HCV patients, based on alpha interferon (IFN-α) and ribavirin, is expensive and is less efficacious for infections with genotypes 1 and 4, the most common genotypes. Moreover, the treatment is associated with numerous side effects. Therefore, new therapies are urgently needed.
There is considerable evidence that neutralizing antibodies are involved in disease control. They emerge during the course of acute HCV infection in patients (25, 28, 35), and several studies have suggested that they might be involved in the control of viral loads during acute infection (17, 29). Also, polyclonal hyperimmune globulin can prevent or modify HCV infection in vivo when administered before exposure to the virus (11, 13, 20), and anti-E1 and anti-E2 polyclonal globulin have been reported to neutralize infection with HCV pseudotyped particles (HCVpp) or HCV cell culture-adapted viruses (HCVcc) in vitro (1, 7, 8, 14, 15, 24, 31, 36). However, polyclonal hyperimmune globulin preparations are subject to contamination by blood-borne viruses that can be present in human plasma pools, a problem not encountered with monoclonal antibodies (MAbs). Moreover, MAbs are more easily standardized than polyclonal hyperimmune globulin. MAbs against E2 of human or primate origin have been used successfully to neutralize HCVpp of various genotypes and subtypes (18, 34), but anti-E2 MAb-based immunotherapy may be hampered by the very high strain-to-strain variation in the immunodominant hypervariable region of E2. On the other hand, E1 displays a relatively high degree of conservation within subtypes, such as subtype 1b (26), and might show a higher degree of intergenotypic cross-neutralization than E2, as suggested in a recent publication (22).
In this study, we have demonstrated that human-derived MAbs against E1 can not only broadly neutralize infection with HCVpp of various genotypes but can also neutralize HCVcc derived from HCV strains of genotype 1a or 2a.
MATERIALS AND METHODS
Source of antibodies.
The MAbs against HCV envelope proteins were supplied by Innogenetics NV, Ghent, Belgium, and were derived from mice, chimpanzees, and healthy human volunteers who had been immunized with recombinant HCV E1 glycoprotein (23) or from patients who had been successfully treated with IFN-α for chronic HCV infection. Hybridoma technology was used to recover antibodies from peripheral blood mononuclear cells as described by Depraetere et al. (6). The mouse MAb against HCV core was purchased from Anogen (YES Biotech Laboratories Ltd., Mississauga, Ontario, Canada), and the irrelevant purified goat immunoglobulin G (IgG) (02-6202) was purchased from Zymed Laboratories, Inc., San Francisco, CA.
Sequencing of the neutralizing antibodies.
Stable subclones of IGH520 (sister clone of IGH505) and IGH526 were selected for sequencing. The heavy and light variable-chain cDNA sequences of the MAbs were determined. Amino acid sequencing was also performed on purified antibodies up to about amino acid 40. This allowed confirmation of the amino acid sequences of both antibodies as deduced from DNA sequencing up to the first complementarity-determining region (CDR) for both the light and heavy chains.
ELISA and mapping of the E1 epitope.
Screening for antibodies specific to E1 was performed by a capture enzyme-linked immunosorbent assay (ELISA). Microtiter plates were coated overnight with goat anti-human IgG (heavy plus light chains) (109-005-088; Jackson ImmunoResearch Europe Ltd., Suffolk, United Kingdom) at 0.9 μg/ml. The plates were washed once and blocked with phosphate-buffered saline (PBS)-0.1% casein. Then the plates were incubated overnight with 100 μl of supernatant from the hybridomas and 100 μl of blocking buffer supplemented with 0.4% Triton X-705. The plates were incubated with E1 at 10 ng/ml, followed by a biotinylated mouse anti-E1 MAb (IGH198). After a wash, the plates were incubated for 30 min with horseradish peroxidase (HRP)-conjugated streptavidin (100 ng/ml; Jackson), washed five times, and incubated with 3,3′,5,5′-tetramethylbenzidine in HRP substrate buffer for 30 min at room temperature. The reaction was stopped by acidification, and the optical densities were read at 450 to 595 nm.
For the epitope mapping, microtiter plates were coated overnight with streptavidin (Roche Diagnostics GmbH, Mannheim, Germany) at 1 μg/ml, washed once, and blocked with blocking buffer for 30 min. Incubation with overlapping biotinylated peptides at 100 ng/ml was then performed, followed by a wash step and a further incubation with supernatants of the hybridomas and HRP-conjugated sheep anti-human IgG (1/2,000; Amersham Biosciences). The following biotinylated peptides were used: ITGHRMAWDMMMNWSPTAAL (313-332), SIYPGHITGHRMAWDMMMNWSPTTALVVSQLLRI (307-340), SQLFTISPRRHETVQDCNCSIYPGHITGHRMAWDMMMNWS (288-327), RMAWDM (317-322), and WDMMMNW (320-326).
Affinity measurement.
The affinities of the neutralizing anti-HCV E1 envelope protein antibodies were measured using peptide ITGHRMAWDMMMNWS (313-327). The association and dissociation of this peptide with immobilized antibodies were measured using Biacore (Biacore AB, Uppsala, Sweden).
Alanine scan of the E1 epitope recognized by neutralizing antibodies IGH526 and IGH505.
An alanine scan was performed on the reference peptide ITGHRMAWDMMMNWS. Each amino acid was replaced by alanine (or glycine where alanine was present in the sequence). Each alanine (glycine) variant was synthesized with an N-terminal biotin and two additional glycine residues as a spacer between the biotin moiety and the epitope.
The binding of antibodies IGH526 and IGH505 was assessed by ELISA. In brief, biotinylated peptides were incubated on streptavidin-coated plates at 1 μg/ml. After a wash, a serial dilution of the antibody (5 to 0.0002 μg/ml) was applied. Binding of antibodies to the streptavidin-bound peptide was detected by incubation with an HRP-labeled secondary antibody specific for human immunoglobulin and subsequent incubation with 3,3′,5,5′-tetramethylbenzidine for 30 min. Curve fitting via the 4-parameter logistic fit (GraphPad Prism software, San Diego, CA) allowed calculation of log 50% effective concentrations (EC50s) for each peptide. The impact of mutating one amino acid to an alanine was expressed as the difference (Δ) in the log EC50 from the reference peptide. A positive Δlog EC50 indicated reduced binding, while a negative Δlog EC50 indicated increased binding.
Construction of natural variants of the E1 epitope.
A series of peptides representing natural variants of peptide ITGHRMAWDMMMNWS was generated. The Los Alamos HCV sequence database (http://hcv.lanl.gov/) (21) was analyzed for known variants of this region. Each sequence occurring more than once in the database was synthesized as a peptide with an N-terminal biotin and two additional glycine residues as a spacer between the biotin moiety and the epitope. The binding of the antibodies IGH526 and IGH505 was assessed using ELISA as described for the alanine mutants and was expressed as Δlog EC50s.
Generation of pseudotyped virus particles.
Pseudotyped virus particles were prepared essentially as described previously (2). Briefly, 3 million HEK-293T cells (ATCC) were seeded in 100-mm-diameter flat culture dishes and allowed to adhere overnight. Cells were transfected with Lipofectamine Plus reagents (Invitrogen, Carlsbad, CA) according to the protocol supplied. A total of 4 μg of plasmid DNA including 1.5 μg of cytomegalovirus Gag-Pol packaging construct, 1.5 μg of murine leukemia virus-green fluorescent protein (GFP) plasmid, and 1.0 μg of HCV E1E2 expression vector was diluted in 250 μl of serum-free Dulbecco's modified essential medium (DMEM). Eight microliters of Lipofectamine Plus reagent was added to the DNA solution, and the mixture was incubated at room temperature for 15 min. The DNA mixture was then added dropwise to 250 μl of serum-free DMEM containing 12 μl of Lipofectamine Plus reagent and was incubated for 15 min. Cells were washed twice and covered with 2 ml of serum-free DMEM. The DNA-Lipofectamine mixture was added dropwise to the dishes, and cells were incubated at 37°C for 3 h. The plates were washed once and cultured in 7.5 ml of complete medium for 2 days. Transfection efficiency was consistently 80 to 90%, based on visual examination of GFP expression. Culture supernatants were harvested and passed through a 0.45-μm-pore-size filter to remove cells and debris. HCVpp infections were performed on the same day. HCVpp of genotypes 2 through 6 were produced as previously described (28). The HCVpp of genotype 1b was based on BE-11, a Belgian isolate the E1E2 sequence of which was kindly provided by Innogenetics NV. The six ukn HCVpp were provided by D. Lavillette and are based on sequences deposited in GenBank by the University of Nottingham, United Kingdom. The corresponding GenBank accession numbers are AY734977 (2a ukn 1.2), AY734979 (2a ukn 2.4), AY734983 (2b ukn 2.8), AY734984 (3a ukn 1.28), AY785283 (5a ukn 14.4), and AY736194 (6a ukn 5.34).
Pseudotyped virus infectivity assays.
Huh-7 cells were seeded at 4 × 104/well in 24-well plates and cultured overnight. The next day, 100 μl of cell supernatant containing HCVpp was combined with 100 μl 2× Polybrene (Sigma, St. Louis, MO) diluted in complete medium (final concentration, 4 μg/ml). Huh-7 cell supernatants were replaced with 200 μl of HCVpp-Polybrene mixture per well and incubated for 3 h with shaking every hour. The infection medium was then replaced with 1 ml of complete medium. Three days later, cells were rinsed with 300 μl PBS (pH 7.4) and then incubated with 100 μl of trypsin solution (BioWhittaker) at 37°C for 30 min. Detached cells were transferred to a 1.5-ml Eppendorf tube, washed with 1 ml of PBS, resuspended in 300 μl of PBS, and placed on ice. Twenty thousand cells were analyzed for GFP expression using a FACScan cell sorter (BD Biosciences, Rockville, MD).
Neutralization of retrovirus pseudoparticles (pp) bearing HCV envelope glycoproteins.
All procedures were performed in the presence of 5 to 10% fetal bovine serum (FBS). Test antibody samples were incubated for 1 h at room temperature with HCVpp, added to Huh-7 cells, and incubated at 37°C. Supernatants were removed after 8 h, and the cells were incubated in DMEM-10% FBS for 72 h at 37°C. GFP-positive cells were quantified by fluorescence-activated cell sorter (FACS) analysis. The percentage of neutralization for each MAb was calculated by comparison with an irrelevant MAb. Neutralization was defined as a ≥50% reduction in the number of GFP-positive cells. Neutralization titers were determined by serial twofold dilutions of MAbs in DMEM, followed by incubation with the HCVpp. The neutralization titer was defined as the highest dilution neutralizing the HCVpp infection by 50% or more.
Cell culture and HCV production.
Virus stocks were produced as previously described (24, 40). Briefly, Huh7.5 human hepatoma cells (a generous gift from Charles Rice, The Rockefeller University) were grown in DMEM (Cellgro; Mediatech Inc., Herndon, VA) supplemented with 10% FBS. Plasmid pJFH1 contained the full-length cDNA of the JFH1 isolate (40); the chimeric plasmids pJ6/JFH1 and pH77/JFH1 contained the core through NS2 of the J6 strain or the core through NS2 of the H77S strain in the JFH1 background, respectively. Plasmid pJFH1 was a gift from T. Wakita (National Institute of Infectious Diseases, Tokyo, Japan), and pH77/JFH1 was a gift from S. Lemon (University of Texas Medical Branch at Galveston, Galveston, TX). All plasmids were linearized at the 3′ end of the HCV sequence by XbaI digestion, purified, and transcribed in vitro with the Riboprobe system T7 (Promega, Madison, WI) to generate HCV genomic RNA. Cells were transfected with the in vitro-transcribed RNA using DMRIE-C transfection reagent (Invitrogen) as recommended by the manufacturer. The cell culture supernatant was used to infect naïve Huh7.5 cells, and virus was adapted to grow in Huh7.5 cells by additional passages on naïve cells. Stocks with a titer of 1 × 105 focus-forming units (FFU)/ml were generated for JFH1, and stocks with a titer of 1 × 104 FFU/ml were generated for the J6/JFH1 and H77/JFH1 viruses.
HCVcc neutralization assays.
Huh7.5 cells were seeded on eight-well chamber slides. HCVcc was incubated with the indicated amount of MAb, purified IgG from patient H, or an irrelevant IgG in 100 μl total medium (DMEM-10% FBS) for 1 h at 37°C. The mixture was then coded and incubated with Huh7.5 cells for approximately 5 h at 37°C, when it was replaced with fresh medium and cells were further incubated for 48 h at 37°C. Each test was performed in triplicate, and the extent of neutralization by the MAbs was estimated by immunofluorescence microscopy and manual counting of foci stained for the core. The percentage of neutralization was estimated by comparison with the mean neutralization for triplicate HCVcc incubations with an irrelevant-antibody control. The EC50 (concentration [in micrograms per milliliter] of antibody required to neutralize 50% of approximately 100 FFU of virus) was determined based on a neutralization curve generated from a series of five threefold antibody dilutions tested in duplicate or triplicate as described above. The mean from six samples incubated with an irrelevant antibody was used as the control.
Indirect immunofluorescence microscopy.
Huh7.5 cells were seeded on eight-well chamber slides, and experiments were performed as described in Results. Cells were washed in PBS and fixed with 100% acetone. Both primary and secondary antibodies were incubated for 20 min at room temperature in PBS containing 5% bovine serum albumin. The Alexa Fluor 488 secondary antibody was from Invitrogen. Virus titers were determined by manually counting stained foci under code.
RESULTS
Human MAbs that neutralize HCVpp 1a infection.
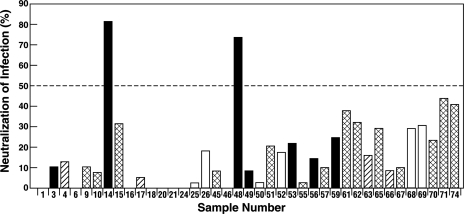
Thirty-one MAbs of human (n = 8), chimpanzee (n = 4), or mouse (n = 19) origin with specificities for E1 glycoprotein and eight control MAbs against non-HCV antigens were tested under code for their abilities to neutralize retrovirus pp bearing recombinant E1 and E2 glycoproteins of genotype 1a, HCV strain H77. A high concentration of antibody was used to ensure that all potentially neutralizing antibodies were detected. One hundred microliters of HCVpp was incubated with 10 μg of each MAb (100 μg/ml), and the percentage of neutralization was then determined as described in Materials and Methods. Only 2 of the 31 anti-HCV MAbs neutralized the pp (≥50% reduction in number of HCVpp-infected cells) (Fig. 1). The two neutralizing MAbs, MAb 14 (IGH505) and MAb 48 (IGH526), were both derived from the same human donor who had been infected previously with HCV genotype 1b but had cleared the virus after IFN-based therapy. The two MAbs neutralized the HCVpp with similar efficiencies.
FIG. 1.
Effects of anti-E1 MAbs on infectivity of HCVpp of genotype 1a. One hundred microliters of an HCVpp suspension was incubated with 10 μg of each MAb or irrelevant IgG and then inoculated into Huh-7 cells. Infected cells were detected by FACS analysis of GFP expression. The percentage of neutralization was calculated by comparison with HCVpp incubated with irrelevant IgG. Antibodies to HCV E1 were derived from humans (solid bars), chimpanzees (hatched bars), and mice (cross-hatched bars); open bars represent non-HCV antibodies. The following MAbs were studied further: MAb 14 (IGH505), MAb 15 (IGH207), MAb 45 (IGH209), MAb 46 (IGH 210), MAb 48 (IGH526), and MAb 49 (IGH450).
Epitope mapping.
Antibodies had been prescreened using ELISA for their abilities to recognize synthetic peptides corresponding to E1 sequences of the genotype 1b strain BE-11 (data not shown). Additional overlapping peptides were used in ELISA to perform finer epitope mapping. Six MAbs, including the two neutralizing anti-E1 MAbs, all reacted with a peptide encompassing amino acids 307 to 340, which contains a highly conserved region. The smallest epitope recognized by three of the four nonneutralizing MAbs could be further mapped to amino acids 317 to 322 or 320 to 326. In contrast, MAb IGH207 and the neutralizing MAbs IGH505 and IGH526 were unable to react with peptides lacking any of amino acids 313 to 327, suggesting that they all recognized noncontinuous epitopes that required this entire sequence for their formation (Table 1). Interestingly, the reactivities of murine MAb IGH207 and the two human neutralizing MAbs were almost identical in the ELISA, yet the murine MAb did not neutralize the pp (Fig. 1, sample 15).
TABLE 1.
Mapping of the E1 epitope by ELISA
| MAba | Optical densityb with peptidec:
|
||||
|---|---|---|---|---|---|
| 313-332 | 288-327 | 307-340 | 317-322 | 320-326 | |
| IGH450 | 0.086 | 0.072 | 0.508 | 0.046 | 1.194 |
| IGH505 | 1.451 | 1.548 | 1.486 | 0.054 | 0.053 |
| IGH526 | 1.442 | 1.461 | 1.441 | 0.050 | 0.043 |
| IGH207 | 1.257 | 1.410 | 1.070 | 0.053 | 0.043 |
| IGH209 | 1.895 | 1.924 | 1.792 | 1.436 | 1.672 |
| IGH210 | 0.051 | 0.046 | 0.217 | 0.05 | 0.150 |
MAb IGH450 is from a human immunized with the E1 glycoprotein; IGH505 and IGH526 are from an HCV-infected human; and IGH207, IGH209, and IGH210 are from mice immunized with the E1 glycoprotein.
Positive results (>2 × background) are boldfaced.
Amino acids from the HCV genotype 1b strain BE-11.
The anti-E1 neutralizing MAbs are similar but distinct and have high affinity.
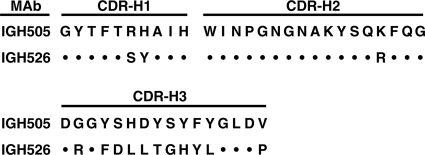
Since the smallest peptides recognized by the two neutralizing anti-E1 MAbs were almost identical, the antibody genes were sequenced to determine their relatedness. Although the CDR-H1 and -H2 regions differed by only 2 of 10 and 1 of 17 amino acids, respectively, the CDR-H3 regions differed by 11 of 16 amino acids (Fig. 2) suggesting that the two antibodies recognized overlapping, rather than identical, epitopes.
FIG. 2.
Alignment of the heavy-chain consensus amino acid sequence for anti-E1 neutralizing MAbs. Theoretically predicted CDR loops, based on consensus sequence rules, are shown. Dots indicate identity.
The affinities of each neutralizing antibody and murine MAb IGH207 were measured by Biacore using a peptide spanning amino acids 313 to 327. The two antibodies IGH505 and IGH526 exhibited similar affinities, with affinity constants (Kds) of 0.23 and 0.20 nM, respectively (Table 2), whereas the Kd of the murine MAb was almost 10-fold lower, a fact that could account for its inability to neutralize.
TABLE 2.
Affinities of MAbs for peptide 313-327a
| MAb | kon (105 m−1 S−1) | koff (10−4 S−1) | Kd (10−9 M) |
|---|---|---|---|
| IGH505 | 15.7 | 3.67 | 0.23 |
| IGH526 | 15.4 | 3.05 | 0.20 |
| IGH207 | 2.8 | 5.92 | 1.86 |
The association and dissociation of the peptide with immobilized antibodies were measured with Biacore. kon, association rate; Koff, dissociation rate; Kd, dissociation constant.
Human MAbs can broadly neutralize HCVpp of multiple genotypes.
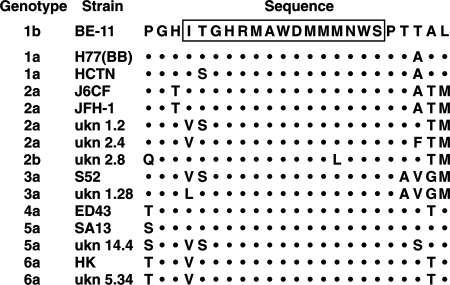
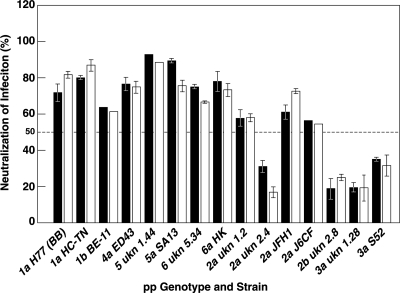
Alignment of sequences representing each of the genotypes revealed that the overlapping epitopes recognized by the two anti-E1 neutralizing MAbs were highly conserved, suggesting that these two antibodies may possess the ability to neutralize viruses of different genotypes (Fig. 3). Therefore, the two neutralizing MAbs were tested for their abilities to neutralize HCVpp representing each of the HCV genotypes compared in the alignment. A 100-μl volume of each HCVpp was incubated with 5 μg of IGH505 or IGH526, which was half the concentration used in the initial screen. At this concentration (50 μg/ml), both MAbs efficiently neutralized the infectivities of HCVpp of genotypes 1a, 1b, 4a, 5a, and 6a (Fig. 4). In contrast, neither MAb neutralized HCVpp of genotype 3a or 2b efficiently. Three of the 2a strains were neutralized, but a fourth was not. Thus, the pseudotyped particles tested segregated into two or possibly three groups according to their sensitivities to neutralization: the group that was efficiently neutralized consisted of genotypes 1, 4, 5, and 6; a second group yielded mixed results and consisted of genotype 2; and a third group was not neutralized at all and consisted of genotype 3.
FIG. 3.
Alignment of the amino acid sequences of the indicated genotypes and strains in the region of the E1 glycoprotein corresponding to the putative epitope recognized by MAbs IGH505 and IGH526. The epitope is boxed. Dots indicate identity. ukn, isolate from Nottingham, United Kingdom.
FIG. 4.
Effects of MAbs IGH505 and IGH526 on the infectivities of HCVpp of diverse genotypes. Plasmids encoding the appropriate HCV envelope proteins were used to generate HCVpp of the indicated genotypes and subtypes. One hundred microliters of an HCVpp suspension was incubated for 1 h at 37°C with 5 μg of MAb IGH505 or IGH526. Infected cells were detected by FACS analysis of GFP expression. The percentage of neutralization was calculated by comparison with an HCVpp of the corresponding genotype and subtype preincubated with 5 μg of irrelevant IgG. Each pp type was tested in at least two independent experiments, with very similar results. Error bars are included for those pp tested in three or more independent experiments. Open bars, IGH505; solid bars, IGH526. ukn, isolate from Nottingham, United Kingdom.
Neutralization titers.
The neutralization titers of the two MAbs were determined against the various HCVpp. Both MAbs reacted strongly against pseudotyped particles of genotypes 1a, 1b, 4a, 5a, and 6a, with EC50 titers ranging from 25 μg/ml to <2 μg/ml (Table 3). Although both anti-E1 MAbs recognized the same small region of E1, they differed somewhat in their reactivities: MAb IGH505 was significantly more potent for neutralization of genotypes 4a and 5a, while MAb IGH526 was the more potent for neutralization of genotype 1a. Again, neutralization of genotype 3a could not be demonstrated. Genotype 2a was somewhat more diverse: strain J6CF was neutralized only at the highest concentration of antibody tested, whereas strain JFH1 was neutralized more efficiently (EC50 titers, 25 and 12.5 μg/ml).
TABLE 3.
Neutralization titers of anti-E1 MAbs for HCVpp
| HCVppa | EC50b (μg/ml) of MAb:
|
|
|---|---|---|
| IGH505 | IGH526 | |
| H77 (1a) | 6 | <2 |
| BE-11 (1b) | 12.5 | 25 |
| ED43 (4a) | <2 | 12.5 |
| SA13 (5a) | <2 | 3 |
| HK (6a) | 12.5 | 6 |
| JFH1 (2a) | 25 | 12.5 |
| J6CF (2a) | 100 | 100 |
| S52 (3a) | >100 | >100 |
Given as strain (genotype).
EC50, lowest concentration providing 50% or greater neutralization. Serial twofold dilutions of MAbs were tested.
It must be cautioned that these EC50 titers are approximate. The HCVpp lose infectivity when frozen, so new HCVpp are made for each experiment and tested immediately. Any variation in the yield of infectious HCVpp would obviously have some effect on the EC50. However, since the error bars in Fig. 4 demonstrate that the results are quite reproducible, the effect is not expected to be greater than twofold, which was the dilution factor tested.
MAbs that neutralize HCVpp genotypes 1a and 2a also neutralize the infectivity of HCVcc.
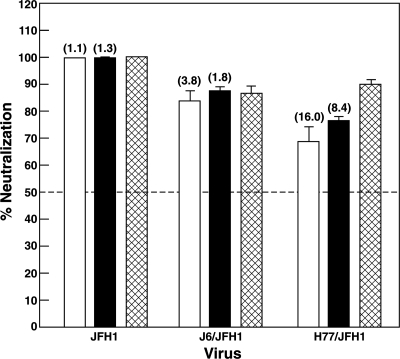
If the neutralization of HCVpp infectivity demonstrated for the MAbs was biologically relevant, these MAbs should similarly neutralize authentic HCV virions. Two cell-cultured strains of genotype 2a, JFH1 and chimeric J6/JFH1, and one of genotype 1a, chimeric H77/JFH1, were available for testing. Therefore, neutralization tests were repeated with HCVcc of these strains. Prior to incubation with Huh7.5 cells, a cell culture supernatant containing HCVcc particles (∼500 FFU of JFH1; ∼100 FFU of J6/JFH1 or H77/JFH1) was mixed with 5 μg of an irrelevant goat-purified IgG or of one of the MAbs. Five micrograms of IgG purified from the chronic-phase serum of patient H (infected with genotype 1a) was also tested: late-phase serum from this patient neutralizes HCVpp of various genotypes (28). Quantification of foci suggested that both MAbs strongly neutralized JFH1 infectivity (99 to 100%) at a level comparable to that of IgG from patient H (Fig. 5). However, the chimeric J6/JFH1 virus appeared to be less efficiently neutralized (84 to 88% neutralization) than JFH1 by either of the MAbs or by IgG from patient H (Fig. 5), even though the putative epitope has the identical sequence in both. Note that the envelope proteins of the genotype 2a HCVcc are the same as those in the HCVpp; thus, the relative resistance of J6/JFH1 to neutralization by IGH505 and IGH526 and also by patient H polyclonal antibodies was consistent between the two systems (Fig. 4 and 5; Table 3) (21). In contrast, whereas the H77pp were neutralized to the same extent as the JFH1pp, the H77/JFH1cc were more resistant to neutralization by the MAbs (70 to 77% neutralization compared to 99% neutralization for JFH1) as well as by the IgG from patient H (90% compared to 100%). This result was especially surprising because the H77 strain was actually isolated from patient H.
FIG. 5.
Effects of MAbs IGH505 and IGH526 on the infectivities of HCVcc strain JFH1 and chimeras J6/JFH1 and H77/JFH1. HCVcc from infected Huh7.5 cell supernatants were incubated with 5 μg of MAb IGH505, MAb IGH526, purified IgG (from patient H), or irrelevant IgG, and coded triplicate samples were tested for the ability to infect naïve Huh7.5 cells. Infected cells were detected at day 2 postinfection by immunofluorescence microscopy after staining of cells with an anti-HCV core antibody. Focus-forming units were counted manually, and the percentage of neutralization was estimated by comparison with HCVcc preincubated with irrelevant IgG. Numbers of foci with irrelevant IgG were 466 (JFH1),112 (J6/JFH1), and 90 (H77/JFH1). Open bars, IGH505; solid bars, IGH526; cross-hatched bars, patient H IgG. The EC50 (in micrograms per milliliter), as determined in a titration experiment with all three viruses tested in parallel at a concentration of approximately 100 FFU/well, is given in parentheses above each bar.
Effects of E1 amino acid changes on antibody recognition.
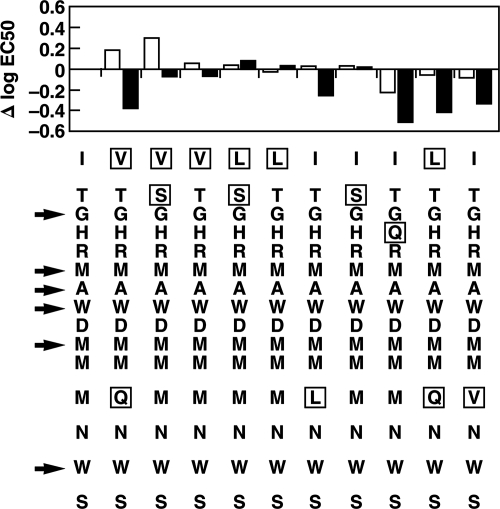
It was intriguing that HCVpp and HCVcc of different genotype 2a strains were neutralized with different efficiencies even when the sequences forming the putative epitope were identical. An alignment of amino acids 313 to 327 was performed on the 143 sequences present in the Los Alamos HCV sequence database on 5 January 2006 in order to determine the extent of natural variation in this region (data not shown). Six amino acids were universally conserved, and six others were changed in only one case. The first one or two amino acids (IT) were mutated in a majority of strains, as predicted by Fig. 3, and an L appeared in place of the M at position 12 in all 13 genotype 2b strains in the database (data not shown). Each sequence occurring more than once in the database was then synthesized and tested by ELISA for recognition by the two neutralizing anti-E1 MAbs. The EC50 (antibody concentration at which half-maximal binding was observed) was determined, and the changes from the EC50 for the sequence of strain BE-11 were plotted (Fig. 6). The profiles for the two anti-E1 MAbs were generally similar, but whereas naturally occurring changes at the amino terminus of the peptide increased the binding of IGH526, they decreased the binding of IGH505, a result supporting the conclusion that the two antibodies recognized epitopes that were overlapping but not identical.
FIG. 6.
Effects of natural sequence variation on the binding of neutralizing anti-E1 MAbs to the epitope. The difference in log EC50 from strain BE-11 (sequence in first column) for each of the natural sequence variants is shown. A positive Δlog EC50 indicates reduced binding, and a negative Δlog EC50 indicates increased binding. Open bars, IGH505; solid bars, IGH526. Mutated amino acids are boxed. Arrows point to universally conserved amino acids.
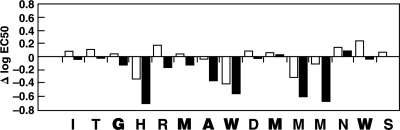
Alanine scanning, in which each amino acid was changed one at a time to alanine (or to glycine for the one alanine residue already present), was performed, and the peptides containing these unnatural substitutions were tested as described above. Once again, the patterns for the two anti-E1 antibodies were similar but not identical (Fig. 7). A number of changes actually increased antibody binding somewhat, but in no case did the amino acid substitution abolish binding.
FIG. 7.
Effects of Ala (or Gly) substitutions on the binding of neutralizing anti-HCV MAbs IGH505 and IGH526 to the epitope. Universally conserved amino acids (based on information in the 2006 database) are boldfaced. Open bars, IGH505; solid bars, IGH526. See the legend to Fig. 6.
DISCUSSION
The relative importance of humoral and cellular immunity in the prevention and/or clearance of HCV infection is controversial and poorly understood. This is due, in part, to limitations in studying HCV infections imposed by a single animal model, the chimpanzee, and the complete absence, until recently, of a cell culture system for replicating the virus. Many studies have pointed to a predominant influence of cellular immunity on clearance, and possibly also on prevention or modification of HCV infections following the initial exposure or vaccination. However, almost as many studies have pointed to a major role for antibodies in the prevention and even resolution of such infections.
One aspect of this debate is what role quasispecies of the virus play in resistance to clearance, whether through the humoral or the cellular immune arm of the host. There is considerable evidence that antibodies can neutralize HCV, but escape from neutralization has also been well documented (3, 11-13, 30, 38). Antibodies directed against the E2 glycoprotein of HCV have been more widely studied, and both polyclonal and monoclonal antibodies have been shown to neutralize HCV in the limited systems available for analysis (1, 9, 12, 13, 18, 25, 34, 36). Convincing data for prevention of HCV infection by in vitro neutralization of the virus, followed by the inoculation of chimpanzees, has been obtained (11, 12). Vaccination of chimpanzees with expressed recombinant E2 (as a heterodimer with E1) has also resulted in protection of chimpanzees that correlated with titers of antibody against E2 (5). Surrogate tests of neutralization, based on the neutralization of pseudotyped virus particles bearing the envelope glycoproteins of HCV, have confirmed and/or identified neutralization epitopes in the HVR1 region of E2, as well as other epitopes elsewhere in E2 (1, 15, 25, 28, 34, 42). However, such antibodies have not been broadly reactive, in part because of the genetic heterogeneity of the E2 glycoprotein, which is among the most heterogeneous of HCV gene products.
Less is known about the immune response to the E1 glycoprotein of HCV (19, 22, 31). Mouse polyclonal serum raised against modified genotype 1a HCVpp harboring only E1 glycoprotein neutralized HCVcc of genotype 2a by a mechanism distinct from that of polyclonal sera from chronically infected patients (7); however, since neither the generation nor the characterization of the mouse antibodies has been published, the data are difficult to interpret at this time. Until recently it was thought that E1 did not contain neutralization epitopes, and indeed, neutralization of HCV by polyclonal or monoclonal antibodies directed against this glycoprotein has not been confirmed in the chimpanzee model. However, at least two regions of E1 believed to contain putative neutralization epitopes have been identified. One of these is located at the N terminus of E1 (amino acids 192 to 202) and was identified by mapping the specificity of a MAb recovered from a patient chronically infected with genotype 1b HCV (19). The antibody, designated H-111, reacted in ELISA with expressed E1 proteins representing genotypes 1a, 1b, 2b, and 3a but not with genotype 2a or 4a (genotypes 5 and 6 were not tested). MAb H-111 was also shown to inhibit the entry of baculovirus-derived “HCV-like particles” into MOLT-4 cells and to block incompletely the infection of Raji cells by an HCV genotype 2b strain (SB) derived from a non-Hodgkin's B-cell lymphoma patient. However, the MAb was unable to significantly (>50%) neutralize HCVpp at 5 μg/ml (11).
A second E1 region of interest was first identified by Bukh et al. (4) as a universally conserved region of E1 spanning amino acids 315 through 328 (except for one mostly conserved amino acid at position 324). Siemoneit et al. (37) constructed a synthetic peptide representing this sequence and tested human sera from chronic HCV patients for reactivity to it by ELISA. Thirty of 92 sera (32%) reacted with the peptide. The authors also recovered IgM and IgG MAbs that reacted with the peptide. The two IgG MAbs identified overlapping but different epitopes that were not recognized by the neutralizing MAbs IGH505 and IGH526. Thirty-four percent of antibodies to approximately the same epitope-containing region were found in human sera by Ishida et al. (16) and in 92% of sera by Ray et al. (32). In contrast, Sällberg et al. (33) found such antibodies in only 9% of 34 sera. A possible reason for this discrepancy is discussed below.
Although others have reported the blocking of binding of HCV to HepG2 cells by polyclonal rabbit sera raised against a synthetic peptide representing part of the conserved E1 region (8), the development of pseudotyped-particle technology for HCV has permitted more-extensive testing of antibody specificity and neutralizing activity, the results of which correlate with neutralization results obtained with chimpanzees. However, the correlation is not perfect. Interestingly, only 2 (6.5%) of our 31 MAbs directed against the HCV E1 glycoprotein neutralized a genotype 1a HCVpp, thus confirming the paucity of such neutralizing antibodies. The epitope reacting with these MAbs was mapped to a 15-amino-acid region (amino acids 313 to 327) near the C terminus of E1, a region first identified as a conserved sequence and as an epitope-containing region in 1993 (4). In fact, the region apparently consists of multiple overlapping epitopes. The two MAbs described here, IGH526 and IGH505, react with this epitope of E1 with similar affinities, even though they have significantly different CDR sequences.
Modifying the sequence of the epitope to match the epitope sequences of other strains in the Los Alamos HCV sequence database (21) altered the reactivity of the epitope with the two MAbs in patterns that were unique to each antibody, but it did not abolish reactivity altogether. In fact, in some cases, reactivity was enhanced. Similarly, alanine scanning of the epitope resulted in enhanced binding of IGH526 at eight amino acid positions and of IGH505 at four amino acid positions. Virtually the opposite of this pattern was observed for diminished binding: the reactivity of IGH526 was diminished at only one position, but binding of IGH505 was diminished at eight positions. The significance of this is not known, but the results confirm the lack of functional identity of these two antibodies.
The broad heterotypic neutralization by these two antibodies is similar to that observed with late-convalescent-phase serum from patient H, the well-known patient chronically infected with a genotype 1a HCV strain, H77. Like the serum from patient H, these MAbs reacted strongly with HCVpp representing genotypes 1a, 1b, 4a, 5a, and 6a but weakly with HCVpp bearing envelope proteins of genotype 2a or 2b and virtually not at all with HCVpp of genotype 3a (28). These data suggest a serologic classification of HCV genotypes in which genotypes 1, 4, 5, and 6 comprise one serotype and genotypes 2 and 3 comprise one or two additional serotypes. However, neutralization data for sera from patient H obtained by others are not in full agreement with this (25). The distribution of antibodies to this conserved E1 epitope in various populations also suggests (but does not prove) a role for serologic differences in the immune response to HCV: HCV carriers with high prevalences of such antibodies were found among blood donor populations in Japan (39) and the United States (32), whose most prevalent HCV genotype is 1, whereas chronically infected patients with low prevalences of such antibodies were principally Swedish drug addicts more likely to have been infected with the genetically quite distant genotype 3 (41).
The difference in the neutralization of different genotypes by MAbs IGH526 and IGH505 is difficult to explain on the basis of the identified extended epitope, since the epitope sequences of genotypes that are neutralized and those that are not neutralized either are identical or contain amino acid changes that are not discriminatory (Fig. 3). However, certain amino acids flanking the epitope, especially the methionine at position 332 (typical for genotype 2 or 3 sequences), precisely define poorly neutralizing genotypes, suggesting that there are conformational components to this linear epitope. Furthermore, four other MAbs, one of which could not be mapped to a smaller region, were not able to neutralize HCVpp at all. This further underlines the conformational nature of this small region, which apparently contains many antigenic determinants.
Neutralization of HCVcc qualitatively confirmed the results obtained with HCVpp. In fact, the genotype 2a HCVcc strains available for testing were neutralized somewhat more robustly than HCVpp bearing the identical glycoprotein sequences. In contrast, neutralization of genotype 1a HCVcc was less efficient than that of genotype 1a HCVpp, an observation the reason for which remains unclear. In addition, all of the neutralizations reported here were performed with HCVpp and HCVcc that were derived from clones of the viruses and were, therefore, essentially monoclonal. The effects of quasispecies on such neutralization remain to be determined.
In summary, this study has shown that MAbs to a highly conserved region of the E1 glycoprotein can strongly neutralize HCVpp bearing the envelope glycoproteins of genotypes 1a, 1b, 4a, 5a, and 6a and less strongly neutralize particles bearing envelope glycoproteins of some strains of genotype 2a and 2b but cannot neutralize HCVpp of genotype 3a. Neutralization was also observed against HCVcc of genotype 1a or 2a. These MAbs may overcome the disadvantages of immunoprophylaxis and immunotherapy for hepatitis C resulting from the marked heterogeneity of HCV strains and the limited and generally strain-specific neutralizing capabilities of most antibodies studied to date. They may also provide insights into the development of vaccines that maintain the conformation necessary to display the neutralization epitope in an immunodominant configuration.
Acknowledgments
We thank Takaji Wakita (National Institute of Infectious Diseases, Tokyo, Japan) for kindly providing the infectious molecular clone of JFH1 and Dimitri Lavillette (INSERM, Lyon, France) for kindly providing HCVpp constructs of genotypes 1 to 6.
This work was supported in part by the Intramural Program of the National Institute of Allergy and Infectious Diseases, National Institutes of Health, and in part by grant IWT000405 from the Instituut voor de Aanmoediging van Innovatie door Wetenschap en Technologie in Vlaanderen (IWT).
The authors have no conflicting financial interests.
Footnotes
Published ahead of print on 31 October 2007.
REFERENCES
- 1.Bartosch, B., J. Bukh, J. C. Meunier, C. Granier, R. E. Engle, W. C. Blackwelder, S. U. Emerson, F. L. Cosset, and R. H. Purcell. 2003. In vitro assay for neutralizing antibody to hepatitis C virus: evidence for broadly conserved neutralization epitopes. Proc. Natl. Acad. Sci. USA 10014199-14204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartosch, B., J. Dubuisson, and F. L. Cosset. 2003. Infectious hepatitis C virus pseudo-particles containing functional E1-E2 envelope protein complexes. J. Exp. Med. 197633-642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bowen, D. G., and C. M. Walker. 2005. Adaptive immune responses in acute and chronic hepatitis C virus infection. Nature 436946-952. [DOI] [PubMed] [Google Scholar]
- 4.Bukh, J., R. H. Purcell, and R. H. Miller. 1993. At least 12 genotypes of hepatitis C virus predicted by sequence analysis of the putative E1 gene of isolates collected worldwide. Proc. Natl. Acad. Sci. USA 908234-8238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Choo, Q. L., G. Kuo, R. Ralston, A. Weiner, D. Chien, G. Van Nest, J. Han, K. Berger, K. Thudium, C. Kuo, et al. 1994. Vaccination of chimpanzees against infection by the hepatitis C virus. Proc. Natl. Acad. Sci. USA 911294-1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Depraetere, S., L. Verhoye, G. Leclercq, and G. Leroux-Roels. 2001. Human B cell growth and differentiation in the spleen of immunodeficient mice. J. Immunol. 1662929-2936. [DOI] [PubMed] [Google Scholar]
- 7.Dreux, M., T. Pietschmann, C. Granier, C. Voisset, S. Ricard-Blum, P. E. Mangeot, Z. Keck, S. Foung, N. Vu-Dac, J. Dubuisson, R. Bartenschlager, D. Lavillette, and F. L. Cosset. 2006. High density lipoprotein inhibits hepatitis C virus-neutralizing antibodies by stimulating cell entry via activation of the scavenger receptor BI. J. Biol. Chem. 28118285-18295. [DOI] [PubMed] [Google Scholar]
- 8.El-Awady, M. K., A. A. Tabll, K. Atef, S. S. Yousef, M. H. Omran, Y. El-Abd, N. G. Bader-Eldin, A. M. Salem, S. F. Zohny, and W. T. El-Garf. 2006. Antibody to E1 peptide of hepatitis C virus genotype 4 inhibits virus binding and entry to HepG2 cells in vitro. World J. Gastroenterol. 122530-2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eren, R., D. Landstein, D. Terkieltaub, O. Nussbaum, A. Zauberman, J. Ben-Porath, J. Gopher, R. Buchnick, R. Kovjazin, Z. Rosenthal-Galili, S. Aviel, E. Ilan, Y. Shoshany, L. Neville, T. Waisman, O. Ben-Moshe, A. Kischitsky, S. K. Foung, Z. Y. Keck, O. Pappo, A. Eid, O. Jurim, G. Zamir, E. Galun, and S. Dagan. 2006. Preclinical evaluation of two neutralizing human monoclonal antibodies against hepatitis C virus (HCV): a potential treatment to prevent HCV reinfection in liver transplant patients. J. Virol. 802654-2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.European Association for the Study of the Liver. 1999. EASL International Consensus Conference on hepatitis C. Paris, 26-27 February 1999. Consensus statement. J. Hepatol. 31(Suppl. 1)3-8. [PubMed] [Google Scholar]
- 11.Farci, P., H. J. Alter, D. C. Wong, R. H. Miller, S. Govindarajan, R. Engle, M. Shapiro, and R. H. Purcell. 1994. Prevention of hepatitis C virus infection in chimpanzees after antibody-mediated in vitro neutralization. Proc. Natl. Acad. Sci. USA 917792-7796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Farci, P., A. Shimoda, D. Wong, T. Cabezon, D. De Gioannis, A. Strazzera, Y. Shimizu, M. Shapiro, H. J. Alter, and R. H. Purcell. 1996. Prevention of hepatitis C virus infection in chimpanzees by hyperimmune serum against the hypervariable region 1 of the envelope 2 protein. Proc. Natl. Acad. Sci. USA 9315394-15399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Féray, C., M. Gigou, D. Samuel, B. Ducot, P. Maisonneuve, M. Reynes, A. Bismuth, and H. Bismuth. 1998. Incidence of hepatitis C in patients receiving different preparations of hepatitis B immunoglobulins after liver transplantation. Ann. Intern. Med. 128810-816. [DOI] [PubMed] [Google Scholar]
- 14.Grollo, L., J. Torresi, H. Drummer, W. Zeng, N. Williamson, and D. C. Jackson. 2006. Exploiting information inherent in binding sites of virus-specific antibodies: design of an HCV vaccine candidate cross-reactive with multiple genotypes. Antivir. Ther. 111005-1014. [PubMed] [Google Scholar]
- 15.Hsu, M., J. Zhang, M. Flint, C. Logvinoff, C. Cheng-Mayer, C. M. Rice, and J. A. McKeating. 2003. Hepatitis C virus glycoproteins mediate pH-dependent cell entry of pseudotyped retroviral particles. Proc. Natl. Acad. Sci. USA 1007271-7276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ishida, C., K. Matsumoto, K. Fukada, K. Matsushita, H. Shiraki, and Y. Maeda. 1993. Detection of antibodies to hepatitis C virus (HCV) structural proteins in anti-HCV-positive sera by an enzyme-linked immunosorbent assay using synthetic peptides as antigens. J. Clin. Microbiol. 31936-940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ishii, K., D. Rosa, Y. Watanabe, T. Katayama, H. Harada, C. Wyatt, K. Kiyosawa, H. Aizaki, Y. Matsuura, M. Houghton, S. Abrignani, and T. Miyamura. 1998. High titers of antibodies inhibiting the binding of envelope to human cells correlate with natural resolution of chronic hepatitis C. Hepatology 281117-1120. [DOI] [PubMed] [Google Scholar]
- 18.Keck, Z. Y., T. K. Li, J. Xia, B. Bartosch, F. L. Cosset, J. Dubuisson, and S. K. Foung. 2005. Analysis of a highly flexible conformational immunogenic domain a in hepatitis C virus E2. J. Virol. 7913199-13208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Keck, Z. Y., V. M. Sung, S. Perkins, J. Rowe, S. Paul, T. J. Liang, M. M. Lai, and S. K. Foung. 2004. Human monoclonal antibody to hepatitis C virus E1 glycoprotein that blocks virus attachment and viral infectivity. J. Virol. 787257-7263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Knodell, R. G., M. E. Conrad, A. L. Ginsberg, and C. J. Bell. 1976. Efficacy of prophylactic gamma-globulin in preventing non-A, non-B post-transfusion hepatitis. Lancet i557-561. [DOI] [PubMed] [Google Scholar]
- 21.Kuiken, C., K. Yusim, L. Boykin, and R. Richardson. 2005. The Los Alamos hepatitis C sequence database. Bioinformatics 21379-384. [DOI] [PubMed] [Google Scholar]
- 22.Leroux-Roels, G., A. H. Batens, I. Desombere, B. Van Den Steen, C. Vander Stichele, G. Maertens, and F. Hulstaert. 2005. Immunogenicity and tolerability of intradermal administration of an HCV E1-based vaccine candidate in healthy volunteers and patients with resolved or ongoing chronic HCV infection. Hum. Vaccin. 161-65. [DOI] [PubMed] [Google Scholar]
- 23.Leroux-Roels, G., E. Depla, F. Hulstaert, L. Tobback, S. Dincq, J. Desmet, I. Desombere, and G. Maertens. 2004. A candidate vaccine based on the hepatitis C E1 protein: tolerability and immunogenicity in healthy volunteers. Vaccine 223080-3086. [DOI] [PubMed] [Google Scholar]
- 24.Lindenbach, B. D., M. J. Evans, A. J. Syder, B. Wolk, T. L. Tellinghuisen, C. C. Liu, T. Maruyama, R. O. Hynes, D. R. Burton, J. A. McKeating, and C. M. Rice. 2005. Complete replication of hepatitis C virus in cell culture. Science 309623-626. [DOI] [PubMed] [Google Scholar]
- 25.Logvinoff, C., M. E. Major, D. Oldach, S. Heyward, A. Talal, P. Balfe, S. M. Feinstone, H. Alter, C. M. Rice, and J. A. McKeating. 2004. Neutralizing antibody response during acute and chronic hepatitis C virus infection. Proc. Natl. Acad. Sci. USA 10110149-10154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maertens, G., and L. Stuyver. 1997. Genotypes and genetic variation of hepatitis C virus, p. 182-233. In A. Zuckerman and T. Harrison (ed.), Molecular medicine of hepatitis. Molecular medical science series. Wiley, Chichester, United Kingdom.
- 27.Mast, E. E., M. J. Alter, and H. S. Margolis. 1999. Strategies to prevent and control hepatitis B and C virus infections: a global perspective. Vaccine 171730-1733. [DOI] [PubMed] [Google Scholar]
- 28.Meunier, J. C., R. E. Engle, K. Faulk, M. Zhao, B. Bartosch, H. Alter, S. U. Emerson, F. L. Cosset, R. H. Purcell, and J. Bukh. 2005. Evidence for cross-genotype neutralization of hepatitis C virus pseudo-particles and enhancement of infectivity by apolipoprotein C1. Proc. Natl. Acad. Sci. USA 1024560-4565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Orland, J. R., T. L. Wright, and S. Cooper. 2001. Acute hepatitis C. Hepatology 33321-327. [DOI] [PubMed] [Google Scholar]
- 30.Pestka, J. M., M. B. Zeisel, E. Blaser, P. Schurmann, B. Bartosch, F. L. Cosset, A. H. Patel, H. Meisel, J. Baumert, S. Viazov, K. Rispeter, H. E. Blum, M. Roggendorf, and T. F. Baumert. 2007. Rapid induction of virus-neutralizing antibodies and viral clearance in a single-source outbreak of hepatitis C. Proc. Natl. Acad. Sci. USA 1046025-6030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pietschmann, T., A. Kaul, G. Koutsoudakis, A. Shavinskaya, S. Kallis, E. Steinmann, K. Abid, F. Negro, M. Dreux, F. L. Cosset, and R. Bartenschlager. 2006. Construction and characterization of infectious intragenotypic and intergenotypic hepatitis C virus chimeras. Proc. Natl. Acad. Sci. USA 1037408-7413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ray, R., A. Khanna, L. M. Lagging, K. Meyer, Q. L. Choo, R. Ralston, M. Houghton, and P. R. Becherer. 1994. Peptide immunogen mimicry of putative E1 glycoprotein-specific epitopes in hepatitis C virus. J. Virol. 684420-4426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sällberg, M., U. Ruden, B. Wahren, and L. O. Magnius. 1993. Antigenic regions within the hepatitis C virus envelope 1 and non-structural proteins: identification of an IgG3-restricted recognition site with the envelope 1 protein. Clin. Exp. Immunol. 91489-494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schofield, D. J., B. Bartosch, Y. K. Shimizu, T. Allander, H. J. Alter, S. U. Emerson, F. L. Cosset, and R. H. Purcell. 2005. Human monoclonal antibodies that react with the E2 glycoprotein of hepatitis C virus and possess neutralizing activity. Hepatology 421055-1062. [DOI] [PubMed] [Google Scholar]
- 35.Shimizu, Y. K., M. Hijikata, A. Iwamoto, H. J. Alter, R. H. Purcell, and H. Yoshikura. 1994. Neutralizing antibodies against hepatitis C virus and the emergence of neutralization escape mutant viruses. J. Virol. 681494-1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shimizu, Y. K., H. Igarashi, T. Kiyohara, T. Cabezon, P. Farci, R. H. Purcell, and H. Yoshikura. 1996. A hyperimmune serum against a synthetic peptide corresponding to the hypervariable region 1 of hepatitis C virus can prevent viral infection in cell cultures. Virology 223409-412. [DOI] [PubMed] [Google Scholar]
- 37.Siemoneit, K., S. Cardoso Mda, K. Koerner, A. Wolpl, and B. Kubanek. 1995. Human monoclonal antibodies for the immunological characterization of a highly conserved protein domain of the hepatitis C virus glycoprotein E1. Clin. Exp. Immunol. 101278-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Takaki, A., M. Wiese, G. Maertens, E. Depla, U. Seifert, A. Liebetrau, J. L. Miller, M. P. Manns, and B. Rehermann. 2000. Cellular immune responses persist and humoral responses decrease two decades after recovery from a single-source outbreak of hepatitis C. Nat. Med. 6578-582. [DOI] [PubMed] [Google Scholar]
- 39.Takeuchi, K., Y. Kubo, S. Boonmar, Y. Watanabe, T. Katayama, Q. L. Choo, G. Kuo, M. Houghton, I. Saito, and T. Miyamura. 1990. Nucleotide sequence of core and envelope genes of the hepatitis C virus genome derived directly from human healthy carriers. Nucleic Acids Res. 184626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wakita, T., T. Pietschmann, T. Kato, T. Date, M. Miyamoto, Z. Zhao, K. Murthy, A. Habermann, H. G. Krausslich, M. Mizokami, R. Bartenschlager, and T. J. Liang. 2005. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat. Med. 11791-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Westin, J., M. Lindh, L. M. Lagging, G. Norkrans, and R. Wejstal. 1999. Chronic hepatitis C in Sweden: genotype distribution over time in different epidemiological settings. Scand. J. Infect. Dis. 31355-358. [DOI] [PubMed] [Google Scholar]
- 42.Zhang, P., C. G. Wu, K. Mihalik, M. L. Virata-Theimer, M. Y. Yu, H. J. Alter, and S. M. Feinstone. 2007. Hepatitis C virus epitope-specific neutralizing antibodies in Igs prepared from human plasma. Proc. Natl. Acad. Sci. USA 1048449-8454. [DOI] [PMC free article] [PubMed] [Google Scholar]