Abstract
The objective of this study was to characterize the mutations selected by darunavir (DRV) use in protease inhibitor (PI)-experienced patients and the associated factors. We analyzed treatment failure in 54 PI-experienced human immunodeficiency virus (HIV)-infected patients on a DRV- and ritonavir-containing regimen. Viral genotyping was carried out at the baseline, at between 1 and 3 months of treatment, and at between 3 and 6 months of treatment to search for the selection of mutations conferring resistance to PIs. The median baseline HIV RNA level was 4.9 log10 copies/ml, and the median CD4 count was 87 cells/mm3. At the baseline, the median numbers of resistance mutations were as follows: 3 DRV resistance mutations, 4 major PI resistance mutations, and 10 minor PI resistance mutations. The most common mutations that emerged at rebound included V32I (44%), I54M/L (24%), L33F (25%), I84V (21%), and L89V (12%). Multivariate analysis showed that higher baseline HIV RNA levels and smaller numbers of nucleoside reverse transcriptase inhibitor simultaneously used with DRV were associated with a higher risk of DRV resistance mutation selection. By contrast, L76V, a known DRV resistance mutation, was found to decrease the risk of selection of another DRV resistance mutation. The occurrence of virological failure while a patient was on DRV was associated with the selection of mutations that increased the level of DRV resistance without affecting susceptibility to tipranavir (TPV). In these PI-treated patients who displayed treatment failure while they were on a DRV-containing regimen, we confirmed the set of emerging mutations associated with DRV failure and identified the factors associated with the selection of these mutations. TPV susceptibility does not seem to be affected by the selection of a DRV resistance mutation.
Virological rebound during antiretroviral treatment is often associated with the emergence of drug resistance, compromising both current and future therapy. The resistance of human immunodeficiency virus (HIV) to protease inhibitors (PIs) generally involves the accumulation of primary and secondary mutations within and outside of the active site of the retroviral protease, sometimes accompanied by mutations in one or more gag cleavage sites (3, 11, 19, 25). The patterns of mutation selected by most of the PIs currently available have been characterized (3, 12, 19, 25, 29). In general, individual viral mutations generate only modest changes in phenotypic susceptibility to PIs. However, more than 20% of the 99 amino acids comprising the HIV protease homodimer have been shown to mutate under the selection pressure exerted by drugs (10, 24). Patients with PI-resistant HIV therefore often have viruses with highly complex genotypic patterns. Different PIs may select different primary mutations, but the secondary mutations observed tend to be common to the entire PI class, potentially limiting the success of subsequent PI treatment following the failure of any PI-containing regimen (3, 12, 19, 25, 29).
The HIV type 1 (HIV-1) protease is essential for the correct processing of viral precursor proteins and the maturation of infectious virus and is therefore an important target for antiretroviral therapy (17). The inclusion of drugs that inhibit this protease in highly active antiretroviral therapy for patients with HIV infection has been shown to result in sustained virological suppression and to decrease the morbidity and mortality associated with HIV disease considerably (8, 27, 32). More effective antiretroviral therapies are urgently required for patients with HIV-1 infection who have already received other treatments, as the extensive drug resistance observed in these patients tends to restrict their treatment options, making it extremely difficult to manage the infection. New compounds need to be highly selective and potent to limit both the toxicity and the number of pills taken by patients, thereby encouraging patient compliance.
Darunavir (TMC114), a new PI with a strong binding affinity for the HIV-1 protease, is highly potent in vitro against wild-type and multidrug-resistant HIV-1 strains (6). It has been suggested that HIV-1 is unlikely to develop significant resistance in patients treated with DRV, particularly if these patients have not previously been treated with other PIs (Y. Koh, T. Towata, A. K. Ghosh, and H. Mitsuya, presented at the 14th Conference on Retroviruses and Opportunistic Infections, poster 606). The POWER (Performance of TMC114/r When Evaluated in Treatment-Experienced Patients with PI Resistance) 1 and 2 studies (TMC114-C213 and TMC114-C202, respectively) are randomized, multinational, phase IIB trials comparing the efficacy and safety of DRV administered in combination with low-dose ritonavir (RTV) (DRV-RTV) with those of currently available PIs in treatment-experienced HIV-1-infected patients. In both studies, the twice-daily administration of 600/100 mg of DRV-RTV, respectively, was the most effective, with 53% of the patients in the POWER 1 study and 39% of those in the POWER 2 study showing decreases in viral loads to less than 50 copies per ml, whereas only 18% and 7% of the patients receiving control PIs in the two studies, respectively, showed such decreases in viral loads (9, 14). This dose has recently been approved for use in the United States (Tibotec, October 2006) and several other countries for the treatment of HIV infection in antiretroviral treatment-experienced adult patients, including those with HIV-1 strains resistant to more than one PI. We investigated the basis of the diminished virological and immunological responses to darunavir by characterizing the mutations selected by this drug and the factors associated with the selection of these mutations.
(This study was presented at the 11th European AIDS Conference, Madrid, Spain, 24 to 27 October 2007.)
MATERIALS AND METHODS
Patients and antiretroviral regimens.
We retrospectively selected 54 PI-experienced patients displaying treatment failure while they were on a DRV-containing regimen. All were treated with nucleoside reverse transcriptase inhibitors (NRTIs) and RTV (100 mg twice a day) plus DRV (600 mg twice a day). The main characteristics of the study population are reported in Tables 1 to 4.
TABLE 1.
Characteristics of patientsa
| Characteristic | Value |
|---|---|
| Sex (no. [%] male/female) | 48 (89)/6 (11) |
| No. (%) of patients infected | 47 (87) |
| with subtype B | |
| Median (range) no. of plasma HIV-1 RNA log10 copies/ml | |
| D0b | 4.9 (2.8-6) |
| M1-M3c | 4.2 (1.5-5.9) |
| M3-M6 | 4.4 (1.6-5.9) |
| Median (range) CD4 cell count/mm3 | |
| D0 | 87 (0-812) |
| M1-M3 | 161 (2-943) |
| M3-M6 | 125 (1-845) |
Data are for a total of 54 patients.
D0, day of initiation of a DRV-RTV-containing regimen.
M1, M3, and M6, months of a DRV-RTV-containing regimen.
TABLE 4.
Number of patients using one to four NRTIs concurrently with DRV
| No. of NRTIs used concurrently with DRV | No. (%) of patients |
|---|---|
| 1 | 1 (2) |
| 2 | 9 (16) |
| 3 | 28 (52) |
| 4 | 16 (30) |
HIV-1 RNA quantification.
Plasma HIV-1 RNA levels were determined at the baseline, at between 1 and 3 months of treatment (M1-M3), and at between 3 and 6 months of treatment (M3-M6) by using the Amplicor Monitor assay (Cobas 1.5; Roche Diagnostics, Basel, Switzerland), which has a detection limit of 200 copies/ml.
Genotypic resistance testing.
Plasma samples for viral genotyping were collected at the baseline, M1-M3, and M3-M6. The reverse transcriptase and protease gene sequences were determined by population sequencing, according to the Agence Nationale de Recherches sur le SIDA (ANRS; the French National Agency for AIDS Research) consensus method, with an ABI 3100 Genetic Analyzer (PE Applied Biosystems). The sequences were analyzed with Sequence Navigator software (PE Applied Biosystems), and the differences in the amino acid sequences with respect to the sequence of wild-type virus strain HXB2 were noted. Resistance was defined according to the current ANRS algorithm (http://www.hivfrenchresistance.org/tab2006.html).
Determination of plasma DRV concentration.
Blood samples were collected and placed into heparin-coated tubes at steady state, which was at M3-M6. Plasma samples (500 μl) were prepared and treated by a solid-phase extraction method (C18 cartridge; 3 ml; J. T. Baker). The DRV concentration was then determined by reverse-phase high-performance liquid chromatography (Ultrasphere octyldecyl silane column, 4.6 by 250 mm, Beckman Coulter) coupled with spectrofluorimetric detection (λ excitation, 245 nm; λ emission; 340 nm). This method was validated within a concentration range of from 5 to 1,000 ng/ml and has a quantification limit of 5 ng/ml. The between-assay bias was less than 7.7% for all quality controls (25, 200, 400, and 800 ng/ml).
Statistical methods.
Descriptive statistics were used to summarize the characteristics of the patients. McNemar's test was used to compare the changes in the resistance profiles between the baseline and the time of viral rebound. Logistic regression analysis was used to search for factors predictive of the occurrence of one or more DRV resistance mutations.
RESULTS
We studied 54 patients displaying either the incomplete suppression of plasma RNA or a rebound from maximal suppression to characterize the mutations selected by DRV (Tables 1 to 4). The changes in the median numbers of DRV resistance mutations and the major and minor PI resistance mutations between the baseline and later periods (M1-M3 and M3-M6) were investigated as recommended by the International AIDS Society—USA guidelines (13). The median number of DRV resistance mutations (V11I, V32I, L33F, I47V, I50V, I54L/M, G73S, L76V, I84V, L89V) was found to have increased by one from the number at the baseline (from three to four). The median number of major PI resistance mutations was found to have increased by one from the number at the baseline (from four to five). The median number of minor PI resistance mutations increased by 0.5 at M3-M6 (from 10 to 10.5).
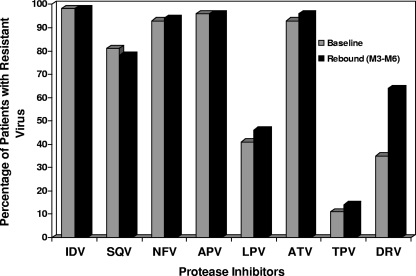
The change in the PI resistance profile between the baseline and the last observed period of rebound is displayed in Fig. 1. According to the ANRS algorithm, our population presented resistance (more than 80%) to indinavir (IDV), nelfinavir (NFV), amprenavir (APV), atazanavir (ATV), and saquinavir (SQV) without any significant possible increase. Lower levels of resistance to lopinavir (LPV) and tipranavir (TPV) were observed at the baseline, at 41% for LPV and 11% for TPV, with a slight increase observed at rebound. Finally, DRV resistance levels were about 35% at the baseline and increased to 64% at M3-M6 (P = 0.0002, McNemar's test).
FIG. 1.
Changes in PI resistance profiles between the baseline and rebound in patients treated with a regimen containing DRV. IDV, indinavir; SQV, saquinavir; NFV, nelfinavir; APV, amprenavir; LPV, lopinavir; ATV, atazanavir; TPV, tipranavir.
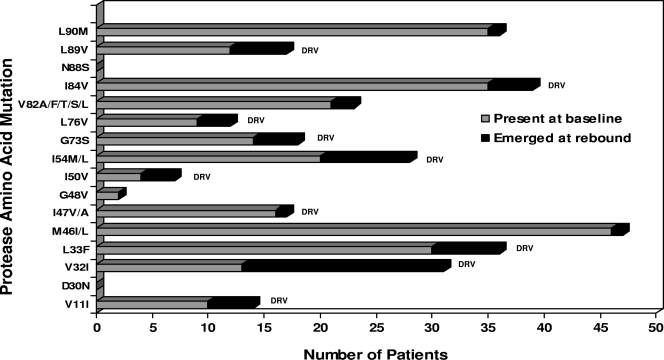
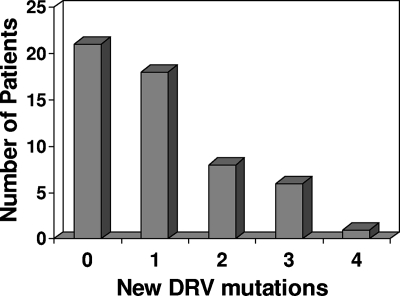
We carried out a genotypic analysis of the HIV strains selected by DRV-RTV in vivo and in PI-experienced patients. The genotypic changes between the baseline and the latest available rebound isolates from the subjects demonstrating changes in PI and DRV resistance are shown in Fig. 2. At the baseline the most common major mutations associated with PI resistance were at positions M46I/L (46 subjects), I84V (35 subjects), L90M (35 subjects), L33F (30 subjects), I54M/L (20 subjects), and V82A/F/T/S/L (27 subjects). The L33F and I84V mutations are also associated with DRV resistance, and the I54M/L mutation is a major mutation associated exclusively with DRV resistance. The median number of major PI mutations after rebound was five (range, four to six). The most common mutations to emerge at rebound included V32I (in 18 of 41 subjects, corresponding to 44% of the patients without V32I at the baseline), L33F (6 of 24 subjects, corresponding to 25% at the baseline), I54M/L (8 of 34 subjects, corresponding to 24% at the baseline), I84V (4 of 19 subjects, corresponding to 21% at the baseline), and L89V (5 of 42 subjects, corresponding to 12% at the baseline). However, it should be borne in mind that the high prevalence of some of these mutations at the baseline would have made their emergence during treatment less likely. For example, V32I was present in 13 (24%) patients at the baseline, whereas I84V was already present in 35 (65%) patients. A number of other PI or DRV resistance mutations, including V11I, M46I/L, I47V/A, I50V, G73S, L76V, V82A/F/T/S/L, and L90M, also emerged in one to four subjects at rebound. From the baseline to the latest period of viral rebound assessed, 21 patients displayed no new DRV mutations, whereas 18, 8, 6, and 1 patients experienced the occurrence of one, two, three, and four new DRV resistance mutations, respectively (Fig. 3).
FIG. 2.
Mutations present at the baseline or rebound isolates from 54 patients in which resistance to DRV was selected. Gray bars, mutations at the given amino acid positions are present in the baseline samples; black bars, mutations emerging during DRV-RTV therapy; DRV, DRV resistance mutations.
FIG. 3.
Number of patients in whom new DRV resistance mutations were detected during the last period of viral rebound assessed.
A series of univariate logistic regression models was generated for the identification of factors associated with the selection of at least one DRV resistance mutation (Table 5). Higher baseline HIV RNA levels, the presence of the I84V mutation, and the prior use of atazanavir were significantly associated with a higher risk of occurrence of at least one DRV resistance mutation. Conversely, a larger number of NRTI simultaneously used with DRV and the mutation at position I54M/L or L76V protected against the selection of at least one DRV resistance mutation. On the basis of multivariate analysis (Table 5), the following independent risk factors for the occurrence of at least one DRV mutation were retained: higher baseline HIV RNA levels, a small number of NRTI simultaneously used with DRV and the presence of the wild-type codon at position 76.
TABLE 5.
Logistic models
| Type of analysis and variablea | P value | Odds ratio |
|---|---|---|
| Univariate analysis | ||
| Baseline HIV RNA level | 0.034 | 2.9 |
| Baseline CD4 count | 0.55 | 1.12 |
| Sex (female vs male) | 0.26 | 3.57 |
| Subtype B vs non-subtype B | 0.82 | 1.21 |
| Minimum DRV concnb | 0.2 | 0.986 |
| No. of major resistance mutations at baselineb | 0.21 | 0.797 |
| No. of DRV resistance mutations at baselineb | 0.63 | 0.914 |
| No. of associated NRTIsb | 0.06 | 0.441 |
| TMC125 | 0.13 | 5.39 |
| T-20 | 0.82 | 1.15 |
| Presence of mutations at baseline | ||
| Mutation V11Ib | 0.94 | 0.944 |
| Mutation V32I | 0.49 | 1.59 |
| Mutation L33F | 0.71 | 1.23 |
| Mutation I47V/Ab | 0.28 | 0.52 |
| Mutation I50Vb | 0.16 | 0.188 |
| Mutation I54M/Lb | 0.017 | 0.24 |
| Mutation G73S | 0.13 | 3 |
| Mutation L76Vb | 0.007 | 0.05 |
| Mutation I84V | 0.039 | 3.44 |
| Mutation L89V | 0.27 | 2.25 |
| Mutation M46I/L | 0.49 | 1.71 |
| Mutation V82A/F/T/S/L | 0.53 | 1.42 |
| Mutation L90Mb | 0.42 | 0.61 |
| Prior use of PIs | ||
| LPVb | 0.18 | 0.412 |
| SQV | 0.43 | 1.75 |
| IDV | 0.71 | 1.32 |
| APVb | 0.1 | 0.28 |
| NFVb | 0.5 | 0.679 |
| ATV | 0.08 | 5.39 |
| TPV | 0.65 | 1.3 |
| No. of PIs used previouslyb | 0.75 | 0.947 |
| Multivariate analysis | ||
| Baseline HIV RNA level | 0.008 | 5.59 |
| Mutation L76Vb | 0.005 | 0.024 |
| No. of associated NRTIsb | 0.03 | 0.26 |
TMC125, etravirine; T20, enfuvirtide; LPV, lopinavir; SQV, saquinavir; IDV, indinavir; APV, amprenavir; NFV, nelfinavir; ATV, atazanavir; TPV, tipranavir.
Factors associated with a decrease in the risk of DRV resistance mutation selection.
DISCUSSION
DRV is a new PI effective against viruses carrying amino acid substitutions which confer resistance to the earlier PIs. In vitro data have shown DRV to be extremely active against both wild-type HIV strains (50% effective concentration, 1 to 5 nM) and variant forms of HIV highly cross-resistant to the PIs in current use. Moreover, in vitro selection experiments, starting from wild-type HIV-1, have shown that there is a strong genetic barrier to the development of DRV resistance (6, 7). However, HIV-1 can develop high levels of resistance to DRV in the presence of superinfections with HIV-1 variants resistant to multiple PIs, and homologous recombination may subsequently occur (Y. Koh, T. Towata, A. K. Ghosh, and H. Mitsuya, presented at the 14th Conference on Retroviruses and Opportunistic Infections, poster 606). This potent activity against PI-resistant HIV indicates that this new antiretroviral molecule may be particularly useful for the treatment of patients who experience treatment failure with other PI-containing regimens. In the POWER randomized clinical studies, DRV-RTV (600/100 mg twice a day) plus an optimized background regimen was more effective than regimens containing control PIs at 24 weeks (9, 14). Furthermore, these responses were sustained, lasting at least until week 48 (2), with satisfactory safety and tolerability in treatment-experienced patients. However, in cases of treatment failure in PI-experienced patients, this drug was shown to induce PI resistance mutations. The specific baseline PI resistance mutations associated with a poor response to DRV-RTV (600/100 mg) were V11I, V32I, L33F, I47V, I50V, I54L/M, G73S, L76V, I84V, and L89V. The V32I, L33F, I47V, I54L/M, and L89V mutations have been shown to be associated with lower levels of susceptibility to DRV and are selected in 10% of patients displaying treatment failure with this drug (7). These mutations selected by DRV were also found to have an impact on the virological response when they were present at the baseline. Our study confirmed these results for all mutations except I47V, which was present at the baseline but which was not significantly more prevalent at rebound. These results are based on genotyping and would therefore be confirmed in phenotypic tests.
We found that the presence of a valine residue at position 76 was associated with the emergence of a smaller number of DRV resistance mutations. The L76V mutation is selected by IDV and LPV, together with other major mutations, mainly, M46I, I54V, V82F, I84A, and L90M with IDV (1, 15, 21, 23, 28, 30, 31) and M46I, I54V, V82F, I84A, L90M, I50V, and N88G with LPV (1, 15, 21-23, 30). However, the L76V mutation itself seems to have an important impact on resistance, at least for LPV. Patients with treatment failure on an LPV-based regimen displayed the selection of only the L76V mutation since the baseline (4, 26). The association between L76V at the baseline and a lower risk of accumulating DRV resistance mutations may be accounted for by the strong impact of this mutation on DRV resistance, implying that the selection of other mutations is not required for the failure of DRV-containing regimens.
Some of the mutations selected by DRV, including V32I, I47V, I50V, I54L/M, G73S, and I84V, are also selected by APV. This finding is consistent with the similarities of the chemical structures of the two drugs (16). Another study suggested that APV-specific resistance profiles, such as those with I50V alone or those with V32IR and I47V, might affect the efficacy of DRV, at least in patients with a suboptimal minimum concentration of DRV (5). In the POWER studies, the previous use of APV had no effect on the virological response to DRV-RTV (18). It is thus difficult to determine whether prior APV treatment could lead to DRV treatment failure. Only a very small number of patients have been treated with APV alone, and these patients are probably at risk of developing DRV and APV resistance mutations. These findings cannot be extrapolated to patients receiving APV after several lines of PI treatment. Previous studies have shown that the use of APV in such patients leads to the selection of other PI resistance mutation patterns that are different from those associated with cross-resistance to DRV (20).
One of the major findings of this study is the lack of an effect of the selection of PI resistance mutations due to DRV failure on susceptibility to TPV. This suggests that TPV may remain active after DRV use, in most cases. This finding has potentially important implications for the strategic use of PIs. Furthermore, the prior use of TPV seems to have no effect on in vitro and in vivo susceptibilities to DRV (18).
TABLE 2.
Prior treatment
| PIa | No. (%) of patients with prior use |
|---|---|
| ATV | 8 (15) |
| IDV | 45 (83) |
| SQV | 44 (81) |
| NFV | 33 (61) |
| LPV | 38 (70) |
| APV | 43 (80) |
| TPV | 20 (37) |
ATV, atazanavir; IDV, indinavir; SQV, saquinavir; NFV, nelfinavir; LPV, lopinavir; APV, amprenavir; TPV, tipranavir.
TABLE 3.
Concurrent treatment
| Antiretroviral drug class and druga | No. (%) of patients with concurrent use with DRV |
|---|---|
| NRTIs | |
| AZT | 26 (48) |
| 3TC | 50 (93) |
| ddI | 11 (20) |
| d4T | 2 (4) |
| TDF | 35 (65) |
| ABC | 39 (72) |
| FTC | 4 (7) |
| NNRTIs | |
| NVP | 0 (0) |
| EFV | 0 (0) |
| TMC125 | 8 (15) |
| T20 (FI) | 37 (69) |
AZT, zidovudine; 3TC, lamivudine; ddI, didanosine; d4T, stavudine; TDF, tenofovir; ABC, abacavir; FTC, emtricitabine; NVP, nevirapine; EFV, efavirenz; TMC125, etravirine; T20, enfuvirtide; FI, fusion inhibitor.
Acknowledgments
This work was supported by ANRS.
Footnotes
Published ahead of print on 26 November 2007.
REFERENCES
- 1.Baxter, J. D., J. M. Schapiro, C. A. Boucher, V. M. Kohlbrenner, D. B. Hall, J. R. Scherer, and D. L. Mayers. 2006. Genotypic changes in human immunodeficiency virus type 1 protease associated with reduced susceptibility and virologic response to the protease inhibitor tipranavir. J. Virol. 80:10794-10801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Clotet, B., N. Bellos, J. M. Molina, D. Cooper, J. C. Goffard, A. Lazzarin, A. Wohrmann, C. Katlama, T. Wilkin, R. Haubrich, C. Cohen, C. Farthing, D. Jayaweera, M. Markowitz, P. Ruane, S. Spinosa-Guzman, and E. Lefebvre. 2007. Efficacy and safety of darunavir-ritonavir at week 48 in treatment-experienced patients with HIV-1 infection in POWER 1 and 2: a pooled subgroup analysis of data from two randomised trials. Lancet 369:1169-1178. [DOI] [PubMed] [Google Scholar]
- 3.Condra, J. H., W. A. Schleif, O. M. Blahy, L. J. Gabryelski, D. J. Graham, J. C. Quintero, A. Rhodes, H. L. Robbins, E. Roth, M. Shivaprakash, et al. 1995. In vivo emergence of HIV-1 variants resistant to multiple protease inhibitors. Nature 374:569-571. [DOI] [PubMed] [Google Scholar]
- 4.Delaugerre, C., P. Flandre, M. L. Chaix, P. Dellamonica, F. Raffi, H. Jäger, D. Schürmann, Ngo M. Van Norton, I. Cohen Codar, J. F. Delfraissy, and C. Rouzioux. 2007. XVI Int. HIV Drug Resistance Workshop, abstr. 75.
- 5.Delaugerre, C., D. Mathez, G. Peytavin, H. Berthe, K. Long, T. Galperine, and P. de Truchis. 2007. Key amprenavir resistance mutations counteract dramatic efficacy of darunavir in highly experienced patients. AIDS 21:1210-1213. [DOI] [PubMed] [Google Scholar]
- 6.De Meyer, S., H. Azijn, D. Surleraux, D. Jochmans, A. Tahri, R. Pauwels, P. Wigerinck, and M. P. de Bethune. 2005. TMC114, a novel human immunodeficiency virus type 1 protease inhibitor active against protease inhibitor-resistant viruses, including a broad range of clinical isolates. Antimicrob. Agents Chemother. 49:2314-2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.De Meyer, S., A. Hill, I. De Baere, L. Rimsky, H. Azijn, B. VanBaelen, et al. 2006. 13th Conf. Retrovir. Opportunistic Infect., abstr. TUPE 0083.
- 8.Gulick, R. M., J. W. Mellors, D. Havlir, J. J. Eron, A. Meibohm, J. H. Condra, F. T. Valentine, D. McMahon, C. Gonzalez, L. Jonas, E. A. Emini, J. A. Chodakewitz, R. Isaacs, and D. D. Richman. 2000. 3-year suppression of HIV viremia with indinavir, zidovudine, and lamivudine. Ann. Intern. Med. 133:35-39. [DOI] [PubMed] [Google Scholar]
- 9.Haubrich, R., D. Berger, P. Chiliade, A. Colson, M. Conant, J. Gallant, T. Wilkin, J. Nadler, G. Pierone, M. Saag, B. van Baelen, and E. Lefebvre. 2007. Week 24 efficacy and safety of TMC114/ritonavir in treatment-experienced HIV patients. AIDS 21:F11-F18. [DOI] [PubMed] [Google Scholar]
- 10.Hirsch, M. S., F. Brun-Vezinet, R. T. D'Aquila, S. M. Hammer, V. A. Johnson, D. R. Kuritzkes, C. Loveday, J. W. Mellors, B. Clotet, B. Conway, L. M. Demeter, S. Vella, D. M. Jacobsen, and D. D. Richman. 2000. Antiretroviral drug resistance testing in adult HIV-1 infection: recommendations of an International AIDS Society—USA Panel. JAMA 283:2417-2426. [DOI] [PubMed] [Google Scholar]
- 11.Ives, K. J., H. Jacobsen, S. A. Galpin, M. M. Garaev, L. Dorrell, J. Mous, K. Bragman, and J. N. Weber. 1997. Emergence of resistant variants of HIV in vivo during monotherapy with the proteinase inhibitor saquinavir. J. Antimicrob. Chemother. 39:771-779. [DOI] [PubMed] [Google Scholar]
- 12.Jacobsen, H., M. Hanggi, M. Ott, I. B. Duncan, S. Owen, M. Andreoni, S. Vella, and J. Mous. 1996. In vivo resistance to a human immunodeficiency virus type 1 proteinase inhibitor: mutations, kinetics, and frequencies. J. Infect. Dis. 173:1379-1387. [DOI] [PubMed] [Google Scholar]
- 13.Johnson, V. A., F. Brun-Vezinet, B. Clotet, D. R. Kuritzkes, D. Pillay, J. M. Schapiro, and D. D. Richman. 2006. Update of the drug resistance mutations in HIV-1: fall 2006. Top. HIV Med. 14:125-130. [PubMed] [Google Scholar]
- 14.Katlama, C., R. Esposito, J. M. Gatell, J. C. Goffard, B. Grinsztejn, A. Pozniak, J. Rockstroh, A. Stoehr, N. Vetter, P. Yeni, W. Parys, and T. Vangeneugden. 2007. Efficacy and safety of TMC114/ritonavir in treatment-experienced HIV patients: 24-week results of POWER 1. AIDS 21:395-402. [DOI] [PubMed] [Google Scholar]
- 15.Kempf, D. J., J. D. Isaacson, M. S. King, S. C. Brun, Y. Xu, K. Real, B. M. Bernstein, A. J. Japour, E. Sun, and R. A. Rode. 2001. Identification of genotypic changes in human immunodeficiency virus protease that correlate with reduced susceptibility to the protease inhibitor lopinavir among viral isolates from protease inhibitor-experienced patients. J. Virol. 75:7462-7469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.King, N. M., M. Prabu-Jeyabalan, E. A. Nalivaika, P. Wigerinck, M. P. de Bethune, and C. A. Schiffer. 2004. Structural and thermodynamic basis for the binding of TMC114, a next-generation human immunodeficiency virus type 1 protease inhibitor. J. Virol. 78:12012-12021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kohl, N. E., E. A. Emini, W. A. Schleif, L. J. Davis, J. C. Heimbach, R. A. Dixon, E. M. Scolnick, and I. S. Sigal. 1988. Active human immunodeficiency virus protease is required for viral infectivity. Proc. Natl. Acad. Sci. USA 85:4686-4690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lefebvre, E., M. P. de Bethune, S. De Meyer, T. Vangeneugden, M. De Pauw, M. Cefalone, et al. 2006. Abstr. 46th Intersci. Conf. Antimicrob. Agents Chemother. abstr. H-1387.
- 19.Maguire, M., D. Shortino, A. Klein, W. Harris, V. Manohitharajah, M. Tisdale, R. Elston, J. Yeo, S. Randall, F. Xu, H. Parker, J. May, and W. Snowden. 2002. Emergence of resistance to protease inhibitor amprenavir in human immunodeficiency virus type 1-infected patients: selection of four alternative viral protease genotypes and influence of viral susceptibility to coadministered reverse transcriptase nucleoside inhibitors. Antimicrob. Agents Chemother. 46:731-738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marcelin, A. G., D. Affolabi, C. Lamotte, H. A. Mohand, C. Delaugerre, M. Wirden, D. Voujon, P. Bossi, N. Ktorza, F. Bricaire, D. Costagliola, C. Katlama, G. Peytavin, and V. Calvez. 2004. Resistance profiles observed in virological failures after 24 weeks of amprenavir/ritonavir containing regimen in protease inhibitor experienced patients. J. Med. Virol. 74:16-20. [DOI] [PubMed] [Google Scholar]
- 21.Margot, N. A., E. Isaacson, I. McGowan, A. K. Cheng, R. T. Schooley, and M. D. Miller. 2002. Genotypic and phenotypic analyses of HIV-1 in antiretroviral-experienced patients treated with tenofovir DF. AIDS 16:1227-1235. [DOI] [PubMed] [Google Scholar]
- 22.Mo, H., M. S. King, K. King, A. Molla, S. Brun, and D. J. Kempf. 2005. Selection of resistance in protease inhibitor-experienced, human immunodeficiency virus type 1-infected subjects failing lopinavir- and ritonavir-based therapy: mutation patterns and baseline correlates. J. Virol. 79:3329-3338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mo, H., N. Parkin, K. D. Stewart, L. Lu, T. Dekhtyar, D. J. Kempf, and A. Molla. 2007. Identification and structural characterization of I84C and I84A mutations that are associated with high-level resistance to human immunodeficiency virus protease inhibitors and impair viral replication. Antimicrob. Agents Chemother. 51:732-735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Molla, A., G. R. Granneman, E. Sun, and D. J. Kempf. 1998. Recent developments in HIV protease inhibitor therapy. Antivir. Res. 39:1-23. [DOI] [PubMed] [Google Scholar]
- 25.Molla, A., M. Korneyeva, Q. Gao, S. Vasavanonda, P. J. Schipper, H. M. Mo, M. Markowitz, T. Chernyavskiy, P. Niu, N. Lyons, A. Hsu, G. R. Granneman, D. D. Ho, C. A. Boucher, J. M. Leonard, D. W. Norbeck, and D. J. Kempf. 1996. Ordered accumulation of mutations in HIV protease confers resistance to ritonavir. Nat. Med. 2:760-766. [DOI] [PubMed] [Google Scholar]
- 26.Nijhuis, M., A. M. Wensing, W. Bierman, D. de Jong, W. J. M. van Rooyen, R. Kagan, C. Jaspers, K. Schurink, L. Lu, T. Pilot-Mathias, A. Molla, M. A. van Agtmael, and C. A. B. Boucher. 2007. XVI Int. HIV Drug Resist. Workshop, abstr. 127.
- 27.Palella, F. J., Jr., K. M. Delaney, A. C. Moorman, M. O. Loveless, J. Fuhrer, G. A. Satten, D. J. Aschman, S. D. Holmberg, et al. 1998. Declining morbidity and mortality among patients with advanced human immunodeficiency virus infection. N. Engl. J. Med. 338:853-860. [DOI] [PubMed] [Google Scholar]
- 28.Parkin, N. T., S. G. Deeks, M. T. Wrin, J. Yap, R. M. Grant, K. H. Lee, D. Heeren, N. S. Hellmanna, and C. J. Petropoulos. 2000. Loss of antiretroviral drug susceptibility at low viral load during early virological failure in treatment-experienced patients. AIDS 14:2877-2887. [DOI] [PubMed] [Google Scholar]
- 29.Patick, A. K., M. Duran, Y. Cao, D. Shugarts, M. R. Keller, E. Mazabel, M. Knowles, S. Chapman, D. R. Kuritzkes, and M. Markowitz. 1998. Genotypic and phenotypic characterization of human immunodeficiency virus type 1 variants isolated from patients treated with the protease inhibitor nelfinavir. Antimicrob. Agents Chemother. 42:2637-2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rhee, S. Y., W. J. Fessel, A. R. Zolopa, L. Hurley, T. Liu, J. Taylor, D. P. Nguyen, S. Slome, D. Klein, M. Horberg, J. Flamm, S. Follansbee, J. M. Schapiro, and R. W. Shafer. 2005. HIV-1 Protease and reverse-transcriptase mutations: correlations with antiretroviral therapy in subtype B isolates and implications for drug-resistance surveillance. J. Infect. Dis. 192:456-465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rhee, S. Y., J. Taylor, G. Wadhera, A. Ben-Hur, D. L. Brutlag, and R. W. Shafer. 2006. Genotypic predictors of human immunodeficiency virus type 1 drug resistance. Proc. Natl. Acad. Sci. USA 103:17355-17360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Swanstrom, R., and J. Erona. 2000. Human immunodeficiency virus type-1 protease inhibitors: therapeutic successes and failures, suppression and resistance. Pharmacol. Ther. 86:145-170. [DOI] [PubMed] [Google Scholar]