Abstract
The frequency of alleles with triple mutations conferring sulfadoxine-pyrimethamine (SP) resistance in the Peruvian Amazon Basin has declined (16.9% for dhfr and 0% for dhps compared to 47% for both alleles in 1997) 5 years after SP was replaced as the first-line treatment for Plasmodium falciparum malaria. Microsatellite analysis showed that the dhfr and dhps alleles are of common origin.
After sulfadoxine-pyrimethamine (SP) resistance became widely prevalent in the Peruvian Amazon Basin, artesunate plus mefloquine was chosen as the recommended first-line treatment for uncomplicated Plasmodium falciparum malaria in 2001 (1, 4, 8). In the north coastal region of Peru, SP is used in combination with artesunate, as it remains efficacious against P. falciparum parasites in this area (7).
Resistance to pyrimethamine is attributed to mutations in dhfr codons 50, 51, 59, 108, and 164; and resistance to sulfadoxine is attributed to mutations in dhps codons 436, 437, 540, 581, and 613 (for a review, see reference 2). In Peru, the triple mutant N51I/S108N/I164L dhfr allele and the triple mutant A437G/K540E/A581G dhps allele have been correlated with high levels of SP resistance (4). A recent study with microsatellite markers has shown that highly resistant dhfr and dhps alleles with triple mutations in isolates from Venezuela have a common ancestral origin (9). It is not known whether Peruvian P. falciparum populations, which have a distinct dhfr allele with triple mutations, have a single origin or multiple origins.
The prevalence of the CQ-resistant pfcrt mutation K76T declined in Malawi and China after chloroquine (CQ) was removed from governmental treatment recommendations (5, 10). This decline correlated with the return of the clinical efficacy of CQ for the treatment of falciparum malaria (6). In Cambodia, however, alleles conferring CQ and SP resistance still occur at a high frequency two decades after these drugs were replaced (3). In Venezuela, we recently showed the complete fixation of mutant dhfr and dhps alleles 8 years after the withdrawal of SP use (9). Further studies are required to understand the roles of various ecological factors that determine the fate of resistance-conferring alleles following a change in the drug use.
In the present study, we attempted to determine if there were any changes in the frequencies of dhfr and dhps mutations in P. falciparum isolates from Iquitos, Peruvian Amazon Basin, after SP was removed from the national treatment recommendation in 2001. We analyzed 208 blood samples on filter paper (all P. falciparum positive by microscopy) collected between April 2005 and March 2006 during an evaluation of the efficacy and effectiveness of mefloquine and artesunate combination therapy for the treatment of uncomplicated malaria at Santa Clara and Morona Cocha health facilities, Iquitos, Peru. The study was approved by the Institutional Review Board of the Centers for Disease Control and Prevention, Atlanta, GA, and the Ministry of Health, Peru.
DNA was extracted from the filter paper strips by using a QIAamp DNA mini kit (Qiagen, Valencia, CA). Pyrosequencing was used to genotype mutations in dhfr at positions 50, 51, 59, 108, and 164 and mutations in dhps at positions 436, 437, 540, 581, and 613, as described previously (11). Microsatellite analysis was conducted as reported previously (9). A heminested PCR was performed for each locus. For the dhps microsatellite reactions, a number of new primers were created in order to conduct the heminested reactions. Microsatellite PCR primer sequences are provided in the supplemental material. Haplotypes were grouped as being different if they contained more than one different allele across the loci.
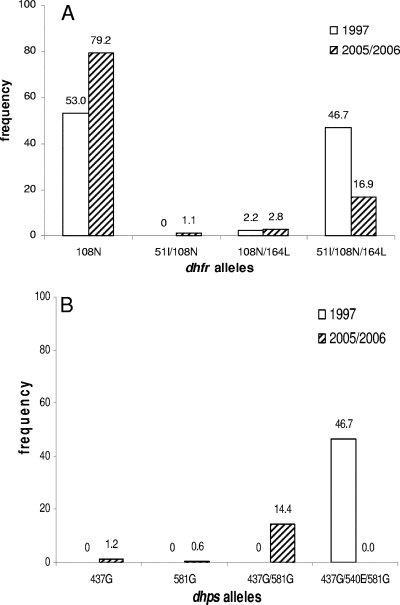
All isolates (100%) were found to have mutations in dhfr codon 108; 17.9% and 20.1% of the isolates had the N51I and I164L mutations, respectively. At codons 50 and 59, all the isolates were wild type. Figure 1A shows that isolates with the allele with the N51I/S108N/I164L triple mutations had declined significantly to 16.9%, whereas the previously reported rate from the same region was 47% (4). Other mutant alleles did not decline in their frequencies. Mutations at codons 437 and 581 were found in 15.3% of the isolates. Mutations at codons 436, 540, and 613 were absent. As shown in Fig. 1B, the dhps allele with the triple mutations A437G/K540E/A581G was completely absent in the samples from this study. Compared to the 47% prevalence in the previous data (4), this is a significant decline. The A437G/A581G allele was present in 14.4% of the isolates. Alleles with the A437G and A581G single mutations were each present in 1.2% and 0.6% of the isolates, respectively. A total of 83.8% isolates had wild-type dhps alleles.
FIG. 1.
Frequency of dhfr (A) and dhps (B) mutant alleles at two time points from the Iquitos region of Peru. The data for 2005 and 2006 came from this study; the data for 1997 were taken from the work of Kublin et al. (4). The difference in the frequencies of the dhfr and dhps alleles with triple mutations between the two points was highly significant (P < 0.0001), based on Fisher's exact test. The numbers of isolates were 141, 2, 5, and 30 for mutant dhfr alleles 108, 51/108, 108/164, and 51/108/164, respectively (a total of 178 samples were genotyped for dhfr). The numbers of isolates were 2, 1, 24, and 0 for mutant dhps alleles 437, 581, 437/581, and 437/540/581, respectively. The wild-type allele was present in 140 samples (a total of 167 samples were genotyped for dhps). One of the S108N/I164L allele isolates and all of the N51I/S108N/I164L allele isolates carried the Bolivia repeat.
Tables 1 and 2 show the dhfr and dhps haplotypes, respectively. Overall, the dhfr alleles showed five different haplotype groups. The dhfr alleles with triple mutations had at least three closely related haplotype groups, groups 1, 2, and 3. The allele with the N51I/S108N double mutation shared haplotype 3 and the S108N/I164L allele shared haplotype 1 with the allele with the triple mutations. The allele with the single mutation (S108N) was present in haplotype groups 3, 4, and 5 (Table 1). The dhps alleles also showed three haplotypes in this population (Table 2). The mutant alleles (A437G/A581G and A437G) had only one haplotype (group 1), while the wild-type alleles had three haplotype groups.
TABLE 1.
dhfr microsatellite haplotypes by mutant alleles, Iquitos, Peru
| Allele | Group |
dhfr haplotype at the following microsatellite locusa:
|
No. of isolates | ||||
|---|---|---|---|---|---|---|---|
| −5.3 | −3.87 | −0.3 | 0.52 | 5.87 | |||
| 50/108/164 | 1 | 204 | 215 | 101 | 97 | 122 | 9 |
| 50/108/164 | 1 | 224 | 215 | 101 | 97 | 122 | 7 |
| 50/108/164 | 1 | 204 | 194 | 101 | 97 | 122 | 6 |
| 50/108/164 | 1 | 204 | 202 | 101 | 97 | 122 | 1 |
| 50/108/164 | 2 | 224 | 202 | 101 | 97 | 122 | 3 |
| 50/108/164 | 2 | 224 | 194 | 101 | 97 | 122 | 2 |
| 50/108/164 | 3 | 204 | 194 | 101 | 97 | 108 | 5 |
| 50/108/164 | 3 | 204 | 101 | 97 | 108 | 1 | |
| 51/108 | 3 | 204 | 194 | 97 | 108 | 3 | |
| 108/164 | 1 | 204 | 215 | 97 | 1 | ||
| 108 | 3 | 204 | 194 | 103 | 108 | 11 | |
| 108 | 3 | 204 | 194 | 101 | 103 | 108 | 5 |
| 108 | 3 | 204 | 194 | 92 | 97 | 108 | 1 |
| 108 | 3 | 204 | 194 | 95 | 97 | 108 | 1 |
| 108 | 3 | 204 | 194 | 90 | 97 | 108 | 1 |
| 108 | 4 | 204 | 209 | 97 | 103 | 108 | 8 |
| 108 | 4 | 204 | 194 | 97 | 103 | 108 | 5 |
| 108 | 4 | 204 | 209 | 97 | 101 | 108 | 1 |
| 108 | 5 | 224 | 194 | 97 | 108 | 5 | |
| 108 | 5 | 224 | 194 | 92 | 97 | 108 | 4 |
| 108 | 5 | 224 | 194 | 78 | 97 | 108 | 3 |
| 108 | 5 | 224 | 194 | 74 | 97 | 108 | 2 |
| 108 | 5 | 224 | 194 | 97 | 97 | 108 | 1 |
| 108 | 5 | 224 | 194 | 100 | 97 | 108 | 1 |
| 108 | 5 | 224 | 194 | 103 | 97 | 108 | 1 |
A total of 88 isolates were tested. The numbers for the microsatellite loci are the location with respect to dhfr (in kb), where negative positions are 5′ to dhfr and nonnegative positions are 3′ to dhfr. Empty cells represent loci that did not amplify.
TABLE 2.
dhps microsatellite haplotypes by mutant alleles, Iquitos, Peru
| Allele | Group |
dhps haplotype at the following microsatellite locusa:
|
No. of isolates | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| −7.4 | −2.74 | −1.64 | −0.8 | 0.006 | 0.14 | 1.59 | 6.19 | 9.79 | 10.1 | |||
| 437/581 | 1 | 310 | 267 | 140 | 124 | 134 | 172 | 197 | 178 | 232 | 101 | 9 |
| 437/581 | 1 | 310 | 140 | 124 | 134 | 172 | 197 | 178 | 232 | 101 | 8 | |
| 437/581 | 1 | 310 | 140 | 124 | 134 | 172 | 197 | 178 | 232 | 1 | ||
| 437/581 | 1 | 310 | 267 | 140 | 124 | 172 | 197 | 178 | 232 | 101 | 1 | |
| 437/581 | 1 | 140 | 124 | 172 | 197 | 178 | 232 | 1 | ||||
| 437/581 | 1 | 140 | 124 | 134 | 172 | 197 | 178 | 101 | 1 | |||
| 437/581 | 1 | 140 | 124 | 134 | 172 | 178 | 232 | 1 | ||||
| 437 | 1 | 310 | 267 | 140 | 124 | 134 | 172 | 197 | 178 | 232 | 101 | 1 |
| 437 | 1 | 310 | 140 | 124 | 134 | 172 | 197 | 178 | 232 | 101 | 1 | |
| WTb | 1 | 310 | 267 | 140 | 124 | 134 | 172 | 197 | 180 | 232 | 101 | 12 |
| WT | 1 | 310 | 140 | 124 | 134 | 172 | 197 | 180 | 232 | 101 | 8 | |
| WT | 1 | 267 | 140 | 124 | 134 | 172 | 197 | 180 | 232 | 1 | ||
| WT | 1 | 140 | 124 | 172 | 197 | 180 | 232 | 101 | 1 | |||
| WT | 1 | 140 | 124 | 134 | 172 | 197 | 232 | 1 | ||||
| WT | 2 | 285 | 246 | 140 | 130 | 134 | 190 | 189 | 180 | 221 | 122 | 1 |
| WT | 2 | 285 | 246 | 140 | 130 | 134 | 189 | 180 | 221 | 122 | 1 | |
| WT | 2 | 246 | 140 | 130 | 134 | 189 | 221 | 1 | ||||
| WT | 2 | 140 | 130 | 134 | 189 | 180 | 122 | 1 | ||||
| WT | 2 | 140 | 132 | 134 | 172 | 180 | 1 | |||||
| WT | 3 | 292 | 270 | 140 | 136 | 134 | 166 | 181 | 172 | 221 | 122 | 1 |
A total of 53 isolates were tested. The numbers for the microsatellite loci are the location with respect to dhps (in kb), where negative positions are 5′ to dhps and nonnegative positions are 3′ to dhps. Empty cells represent loci that did not amplify.
WT, wild type.
To our knowledge, this may be a first report on the reduction in the frequency of alleles with triple mutations that confer high-level SP resistance after drug policy changes. It appears that the dhps allele with triple mutations is being replaced faster than the dhfr allele with triple mutations as the drug pressure is removed from the population. We do not know whether this is because the dhps allele with triple mutations is less biologically fit than the dhfr allele with triple mutations or is due to other reasons.
It is unclear why, in Cambodia and Venezuela, SP resistance alleles have remained at a high frequency after the replacement of SP (3, 9). The key difference seems to be that in Peru alleles that confer high levels of resistance have not yet reached fixation. Thus, there is an opportunity for wild-type alleles to compete with drug resistance-conferring alleles, which are assumed to carry a high fitness cost to the parasite in the absence of drug pressure, and eventually replace the resistance-conferring alleles (9). If this hypothesis is true, our findings may have significant impact on drug policy development and implementation.
We have also shown, using microsatellite markers, that both dhfr and dhps mutant alleles could have been derived from a common ancestor in this population. Although not all of the dhfr alleles with triple mutations had identical microsatellite haplotypes, they did appear to be closely related, and these additional haplotypes may have been generated by recombination and/or mutation. The dhps mutant and wild-type alleles all shared the same microsatellite haplotype, implying a common ancestor for these two mutant and wild-type alleles. In summary, molecular surveillance can be valuable in predicting the shifting trend in drug-resistant parasite populations following drug policy changes.
Supplementary Material
Acknowledgments
The financial support from the CDC Antimicrobial Resistance Working Group and the support of Atlanta Research Education Foundation are appreciated. S. M. Griffing is supported under a National Science Foundation Graduate Research Fellowship. We also thank the U.S. Agency for International Development for funding this project through the Amazon Malaria Initiative.
We thank the study team and the participating patients for their support. We thank Vincent Hill and Jothikumar Narayanan of the Parasitic Diseases Branch, Division of Parasitic Diseases, NCZVED, CDC, for allowing us to use the PSQ MA96 system for pyrosequencing. We thank John Barnwell and Larry Slutsker of the Malaria Branch for the support and encouragement of this project.
Footnotes
Published ahead of print on 19 November 2007.
Supplemental material for this article may be found at http://aac.asm.org/.
REFERENCES
- 1.Ayala, E., A. G. Lescano, R. H. Gilman, M. Calderon, V. V. Pinedo, H. Terry, L. Cabrera, and J. M. Vinetz. 2006. Polymerase chain reaction and molecular genotyping to monitor parasitological response to anti-malarial chemotherapy in the Peruvian Amazon. Am. J. Trop. Med. Hyg. 74:546-553. [PubMed] [Google Scholar]
- 2.Gregson, A., and C. V. Plowe. 2005. Mechanisms of resistance of malaria parasites to antifolates. Pharmacol. Rev. 57:117-145. [DOI] [PubMed] [Google Scholar]
- 3.Khim, N., C. Bouchier, M.-T. Ekala, S. Incardona, P. Lim, E. Legrand, R. Jambou, S. Doung, O. M. Puijalon, and T. Fandeur. 2005. Countrywide survey shows very high prevalence of Plasmodium falciparum multilocus resistance genotypes in Cambodia. Antimicrob. Agents Chemother. 49:3147-3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kublin, J. G., R. S. Witzig, A. H. Shankar, J. Q. Zurita, R. H. Gilma, J. A. Guarda, J. F. Cortese, and C. V. Plowe. 1998. Molecular assays for surveillance of antifolate-resistant malaria. Lancet 351:1629-1630. [DOI] [PubMed] [Google Scholar]
- 5.Kublin, J. G., J. F. Cortese, E. M. Njunjn, R. A. G. Mukadam, J. J. Wirima, P. N. Kazembe, A. A. Djimde, B. Kouriba, T. E. Talor, and C. V. Plowe. 2003. Reemergence of chloroquine-sensitive Plasmodium falciparum malaria after cessation of chloroquine use in Malawi. J. Infect. Dis. 187:1870-1875. [DOI] [PubMed] [Google Scholar]
- 6.Laufer, M. K., P. C. Thesing, N. D. Eddington, R. Masonga, F. K. Dzinjalamala, S. L. Takala, T. E. Taylor, and C. V. Plowe. 2006. Return of chloroquine antimalarial efficacy in Malawi. N. Engl. J. Med. 355:1959-1966. [DOI] [PubMed] [Google Scholar]
- 7.Marquino, W., J. R. Macarthur, L. M. Barat, F. E. Oblitas, M. Arrunategui, G. Garavito, M. L. Chafloque, B. Pardave, S. Gutierrez, N. Arrospide, C. Carrillo, C. Cabezas, and T. K. Ruebush II. 2003. Efficacy of chloroquine, sulfadoxine-pyrimethamine, and mefloquine for the treatment of uncomplicated Plasmodium falciparum malaria on the North coast of Peru. Am. J. Trop. Med. Hyg. 68:120-123. [PubMed] [Google Scholar]
- 8.Marquino, W., M. Huilca, C. Calampa, E. Falconi, C. Cabezas, R. Naupay, and T. K. Ruebush II. 2003. Efficacy of mefloquine and a mefoquine-artesunate combination therapy for the treatment of uncomplicated Plasmodium falciparum malaria in the Amazon Basin of Peru. Am. J. Trop. Med. Hyg. 68:608-612. [DOI] [PubMed] [Google Scholar]
- 9.McCollum, A. M., K. Mueller, L. Villegas, V. Udhayakumar, and A. A. Escalante. 2007. Common origin and fixation of Plasmodium falciparum dhfr and dhps mutations associated with sulfadoxine-pyrimethamine resistance in a low-transmission area in South America. Antimicrob. Agents Chemother. 51:2085-2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang, X., J. Mu, G. Li, P. Chen, X. Guo, L. Fu, L. Chen, X. Z. Su, and T. E. Wellems. 2005. Decreased prevalence of the Plasmodium falciparum chloroquine resistance transporter 76T marker associated with cessation of chloroquine use against P. falciparum malaria in Hainan, People's Republic of China. Am. J. Trop. Med. Hyg. 72:410-414. [PubMed] [Google Scholar]
- 11.Zhou, Z., A. C. Poe, J. Limor, K. K. Grady, I. Goldman, A. M. McCollum, A. A. Escalante, J. W. Barnwell, and V. Udhayakumar. 2006. Pyrosequencing, a high-throughput method for detecting single nucleotide polymorphisms in the dihydrofolate reductase and dihydropteroate synthetase gene of Plasmodium falciparum. J. Clin. Microbiol. 44:3900-3910. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.