Abstract
A major obstacle in hepatitis C virus (HCV) research has been the lack of a permissive cell culture system that produces infectious viral particles. Significant breakthroughs have been achieved lately in establishing such culture systems. Yet to date, there are no reports of the applications of any of these systems in HCV drug screening. Here, we report the generation of two monocistronic, chimeric genotype 1 full-length HCV genome molecules. These molecules, C33J-Y835C-UBI and C33J-Y835C-FMDV2A, both contain the structural protein region from genotype 1 (subtype 1b, Con1) and the remaining region from the genotype 2a (JFH1) clone. Both contain the humanized Renilla luciferase reporter gene which is separated from the rest of the HCV open reading frame by two different cleavage sites. The viral RNAs replicated efficiently in transfected cells. Viral particles produced were infectious in naïve Huh7.5 cells, and the infectivity could be blocked by monoclonal antibody against a putative HCV entry cofactor, CD81. A pilot high-throughput screen of 900 unknown compounds was executed by both the genotype 2a subgenomic replicon system and the infectious system. Thirty-one compounds were identified as hits by both systems, whereas 78 compounds were identified as hits only for the infectious system, suggesting that the infectious system is capable of identifying inhibitors targeting the viral structural proteins and steps involving them in the viral life cycle. The infectious HCV system developed here provides a useful and versatile tool which should greatly facilitate the identification of HCV inhibitors currently not identified by the subgenomic replicon system.
Hepatitis C virus (HCV) infection is one of the leading causes of liver disease in the United States. In the United States, nearly 4 million people are positive for HCV, accounting for 1.8% of the total population (2). Approximately 170 million people are infected by HCV worldwide (27). Severe liver diseases, including cirrhosis and hepatocellular carcinoma, are the most common consequences of chronic HCV infection. Tremendous progress has been made in the search for specific anti-HCV therapies, as exemplified by the recent phase II clinical trials of VX-950 from Vertex and SCH-503034 from Schering Plough as well as previous efforts by Wyeth/Viropharma for HCV-796 and Boehringer Ingelheim for BILN-2061. As of yet, no specific anti-HCV drug has been approved by the FDA. The standard of care for treating HCV infection is the combination of pegylated alpha interferon and ribavirin, with only about 50 to 60% of patients responding to this therapy (11, 15). More effective therapies are greatly needed for the treatment of disease caused by HCV infection.
The emergence of HCV subgenomic replicon systems represents major progress in HCV research and drug discovery in recent years. The HCV subgenomic replicons (18), however, contain only nonstructural (NS) regions, either NS2-NS5B or NS3-NS5B. The study of the remaining components of the open reading frame (C-E1-E2-P7) for antiviral therapeutic intervention is not possible using the subgenomic replicon system. The subgenomic replicon system does not produce infectious viral particles either, which makes it impossible to use this system as a tool for the study of several critical steps in the viral life cycle, including viral attachment, entry, and infection. There is clearly a need to create a cell culture system which can produce infectious HCV in vitro to overcome the limitations of the subgenomic replicon-based systems.
In 2005, significant breakthroughs in establishing the infectious HCV cell culture system were achieved. Three individual groups reported the production of infectious HCV particles upon transfection of Huh7 or its derivative cell lines with genotype (GT) 2a (JFH1) genomes (17, 26, 29). However, GT 1a and 1b HCVs are the most prevalent HCV genotypes in infected patients in the United States and most of the rest of the world. Moreover, GT 1a and 1b HCVs are relatively more resistant to interferon treatment and are more typically associated with the development of cirrhosis and liver cancer. As such, Lemon's group generated a cell culture system (28) which produced infectious GT 1a (Hutchinson strain) HCV particles, but the titer of this cell culture-grown HCV was 100- to 1,000-fold lower than that of a GT 2a system described by others (17, 26, 29). Moreover, no cell-based drug screening system using infectious GT 1 HCV has been reported thus far.
To develop a robust screening tool that targets the structural proteins of GT 1, we generated GT 1b/2a based chimeric monocistronic luciferase reporter viruses. These viruses encoded the structural proteins and the amino terminus of NS2 from GT 1b (Con1) and the remaining sequence from GT 2a (JFH1). These constructs involved the introduction of a short, codon-optimized luciferase reporter gene, i.e., humanized Renilla luciferase (hRLuc), separated from the viral structural proteins by the sequence encoding a cleavable peptide, either from the foot-and-mouth disease virus 2A (FMDV 2A) or a ubiquitin monomer (UBI). The inclusion of a reporter gene allowed us to avoid the time-consuming and labor-intensive quantitative reverse transcription-PCR methods used traditionally in screening. The antiviral activity of the screened compounds could be easily monitored through measurement of the luciferase activity. This facilitated the development of a high-throughput screen (HTS) targeting the steps in the HCV life cycle that are not accessible with the subgenomic replicon system. More interestingly, when we were testing the two reporter viruses with different cleavage sites, we found that the reporter virus containing the FMDV 2A cleavage site consistently showed higher luciferase activity than the one with the UBI cleavage site. This unique feature makes the reporter virus containing FMDV 2A a better candidate for antiviral activity screening. To validate the newly developed infectious system regarding its ability to identify compounds with antiviral activity, we tested several known HCV inhibitors using both the 1b/2a infectious system and the 2a subgenomic replicon system in parallel. As expected, the 50% effective concentrations (EC50s) obtained from the two assay systems were comparable. Furthermore, we screened 900 unknown Pfizer compounds by employing the above two systems. Thirty-one hits were identified by both systems whereas 78 hits were identified only by the infectious 1b/2a system but not the 2a replicon system. This indicates that the newly developed 1b/2a infectious HCV system, in addition to being able to identify the inhibitors targeting the RNA replication machinery, is also capable of identifying inhibitors specifically targeting the viral structural protein regions of GT 1b and/or important steps of the viral life cycle that are not accessible with the subgenomic replicon system, such as viral particle attachment and entry.
MATERIALS AND METHODS
Inhibitors.
Compounds A (5), BILN-2061 (16), and Pfizer compound plates were synthesized at Pfizer Global Research and Development (La Jolla, CA).
Cell culture.
The Huh7.5 cell line was obtained from Apath LLC (St. Louis, MO). The FT3-7 cell line is a clonal derivative of Huh7 cells obtained following transformation with a Toll-like receptor 3 expression vector, kindly provided by Stanley Lemon (The University of Texas Medical Branch at Galveston, Galveston, Texas) (28). The 2a replicon stable cell line was made in house by introducing the JFH1 subgenomic replicon (12) into Huh7.5 cells. All cells were propagated in Dulbecco's modified Eagle medium (DMEM; Invitrogen, Carlsbad, CA) containing 10% fetal bovine serum (HyClone, Logan, UT), 100 IU/ml of penicillin, 100 μg/ml of streptomycin sulfate (Invitrogen), 2 mM l-glutamine (Invitrogen), and 0.1 mM nonessential amino acids (Invitrogen) at 37°C and 5% CO2. The culture medium for the FT3-7 cell line and 2a replicon cell line was further supplemented with 2 μg/ml of blasticidin (Calbiochem, San Diego, CA) and 200 μg/ml geneticin (Invitrogen), respectively.
Construction of plasmids. (i) RB10-hR-SM.
The hRLuc gene and the sequence of the 2A protease of FMDV from pBB7M4hRLuc (9) were introduced into monocistronic subgenomic HCV replicon RB10 (I389/hyg-ubi/NS3-3′/5.1) (8) through an AscI site to generate RB10-hR. The fragment containing four single mutations on pBB7M4hRLuc (9) was used to replace the corresponding fragment on RB10-hR, resulting in RB10-hR-SM.
(ii) 1b-2a-hR.
The sequences of all oligonucleotides used for PCRs are listed in a table in the supplemental material. Five PCRs were employed to generate construct 1b-2a. In the first PCR oligonucleotides EcoRI-2a(+) and 1b-PmeI-2a-Core(−) were used to amplify the plasmid pSG-JFH1. In the second PCR, oligonucleotides 2a-PmeI-1b-Core(+) and 2a-NS3-1b-NS2(−) were used to amplify RB9 (I389/core-3′-5.1) (21). For the third PCR, the PCR products from first and second PCR were used as the templates, and oligonucleotides EcoRI-2a(+) and 2a-NS3-1b-NS2(−) were used as primers. The resulting PCR product contained the 2a 5′ nontranslated region, the first 51 nucleotides of the 2a core region, and the C-E1-E2-P7-NS2 region of 1b. In the fourth PCR, oligonucleotides 1b-NS2-2a-NS3(+) and SpeI-2a(−) were used to amplify pSG-JFH1. For the fifth PCR, the PCR products from the third and fourth PCRs were used as the templates, and oligonucleotides EcoRI-2a(+) and SpeI-2a(−) were used as primers. The final PCR product was then introduced into pSG-JFH1 through EcoRI and SpeI sites, resulting in the 1b-2a chimera. RB10-hR-SM was then used as a template, and PCR was carried out with primers PmeI-hR(+) and PmeI-FMDV2A(−). The resulting PCR fragment and 1b-2a chimera were both digested with PmeI and then ligated to generate 1b-2a-hR.
(iii) 1b-2a-hR-UBI.
To replace the FMDV 2A fragment of 1b-2a-hR with a UBI fragment, five PCRs were performed. In the first PCR, oligonucleotides hRLuc-421-450(+) and UBI-hR(−) were used to amplify the RB10-hR-SM. In the second PCR, oligonucleotides hR-UBI(+) and 2a-Core-UBI(−) were used to amplify the RB10-hR-SM. For the third PCR, the PCR products from the first and second PCRs were used as the templates, and oligonucleotides hRLuc-421-450(+) and 1b-Core-Ubi(−) were used as primers. In the fourth PCR, oligonucleotides Ubi-1b-Core(+) and Con1-MluI(−) were used to amplify 1b-2a-hR. For the fifth PCR, the PCR products from the third and fourth PCRs were used as the templates, and oligonucleotides hRLuc-421-450(+) and Con1-MluI(−) were used as primers. The final PCR product was then introduced into 1b-2a-hR through NruI and MluI sites, resulting in the construct 1b-2a-hR-UBI.
(iv) Con1-C3-JFH1.
To make the 1b-2a junction at the first 30 amino acids of NS2, six PCRs were performed. In the first PCR, oligonucleotides JFH1 2377-2423(+) and JFH1 3452-3408(−) were used to amplify the JFH1 region of nucleotides 2399 to 3430 (GenBank accession number AB047639). In the second PCR, oligonucleotides JFH1 3408-3452(+) and JFH1 4147-4118(−) were used to amplify the 1b-2a construct. For the third PCR, the PCR products from the first and second PCRs were used as the templates, and oligonucleotides JFH1 2377-2423(+) and JFH1 4147-4118(−) were used as primers. In the fourth PCR, oligonucleotides Con1-MluI(+) and JFH1-Con1-NS2(−) were used to amplify 1b-2a-hR. For the fifth PCR, oligonucleotides Con1-JFH1-NS2(+) and JFH1 4147-4118(−) were used to amplify the PCR product from the third PCR. In the sixth PCR, the PCR products from fourth and fifth PCRs were used as the templates, and oligonucleotides Con1-MluI(+) and JFH1 4147-4118(−) were used as primers. The final PCR product was then introduced into the 1b-2a-hR-UBI construct through MluI and SpeI sites, resulting in Con1-C3-JFH1.
(v) C33J-UBI.
To make the 1b-2a junction at the first 33 amino acids of NS2, three PCRs were performed. In the first PCR, oligonucleotides Con1-MluI(+) and Con1-NS2-33-JFH1(−) were used to amplify Con1-C3-JFH1. In the second PCR, oligonucleotides Con1-NS2-33-JFH1(+) and JFH1 4147-4118(−) were used to amplify the Con1-C3-JFH1 construct. For the third PCR, the PCR products from first and second PCRs were used as the templates, and oligonucleotides Con1-MluI(+) and JFH1 4147-4118(−) were used as primers. Con1-C3-JFH1 and the PCR product of the third PCR were digested with MluI and SpeI and then ligated to generate C33J-UBI.
(vi) C33J-Y835C-UBI.
To make the Y835C mutation, a mutation of TAT to TGT was generated through three PCRs. In the first PCR, oligonucleotides C33J-MluI-F and C33J-4059-R were used to amplify C33J-UBI. In the second PCR, oligonucleotides C33J-4059-F and C33J-4340-R were used to amplify C33J-UBI. Both C33J-4059-F and C33J-4059-R contain a mutation of A to G. In the third PCR, the PCR products from the first and second PCRs were used as the templates, and oligonucleotides C33J-MluI-F and C33J-4340-R were used as primers. The resulting PCR product was used to replace the corresponding fragment in C33J-UBI through MluI and AflII digestion followed by ligation.
(vii) C33J-Y835C-FMDV2A.
To replace UBI with FMDV2A, three PCRs were performed. In the first PCR, oligonucleotides 2a-FL-NruI-F and 1b-FMDV-R were used to amplify 2a-FL-FMDV2A. In the second PCR, oligonucleotides 1b-FMDV-F and C33J-AfeI-R were used to amplify C33J-Y835C-UBI. In the third PCR, PCR products of the first and second PCR were amplified by 2a-FL-NruI-F and C33J-AfeI-R. C33J-Y835C-UBI and the third PCR product were both cut with NruI and AfeI and then ligated.
(viii) C33J-GND.
To make a D-to-N mutation, a mutation of GAT to AAT was introduced by replacing the HpaI/XbaI fragment of C33J-UBI with the corresponding fragment from pJFH1-GND (12).
(ix) C33J-ΔE1-E2.
To delete the E1-E2 region in C33J-UBI, three PCRs were performed. In the first PCR, oligonucleotides C33J-PmeI-F and C33J-2129/3793-R were used to amplify C33J-UBI. In the second PCR, oligonucleotides C33J-2129/3793-F and C33J-AflII-R were used to amplify C33J-UBI. For the third PCR, products of the first and second PCR were amplified by C33J-PmeI-F and C33J-AflII-R. C33J-UBI and the third PCR product were digested by PmeI and AflII and then ligated. All constructs were confirmed by sequencing.
HCV RNA transfection and virus production.
HCV RNAs were transcribed in vitro with a T7 Megascript kit (Ambion, Austin, TX) following the manufacturer's protocol. Trypsinized FT3-7 cells were washed twice with phosphate-buffered saline (PBS) without Ca2+ or Mg2+ (Invitrogen) and then resuspended at 2 × 107 cells/ml. Ten micrograms of in vitro transcripts was mixed with 0.4 ml of the cell suspension in a 4-mm cuvette, and electroporation was performed using a Bio-Rad Gene Pulser system set to deliver a single pulse at 220 V and 950 μF. Immediately after electroporation, cells were resuspended with complete DMEM and seeded in 96-well plates for hRLuc activity measurement and in 25-cm2 flasks for virus production. For virus production, transfected cells were transferred from 25-cm2 flasks into 75-cm2 flasks 48 h after transfection. Cells were passaged with a 1:3 split every 2 days for the indicated period. Cell culture supernatants were collected upon each passage and stored at 4°C with the addition of 1% HEPES.
Infectivity assay.
Huh7.5 cells were seeded in black-walled, clear-bottom 96-well plates at a density of 1.5 × 104 cells/well. After overnight incubation, 50 μl of cell culture supernatants collected at indicated time periods (see Fig. 3A) from transfected cells was added into each well. After being incubated for 2 days, the cell culture medium was completely removed from each well, and cells were lysed in 20 μl of lysis buffer (Promega, Madison, WI). hRLuc activity was measured with Renilla luciferase assay buffer and substrate (Promega, Madison, WI), following the manufacturer's instructions, using a 1450 Trilux MicroBeta Jet (Perkin-Elmer, Wellesley, MA). For the neutralization assay, Huh7.5 cells in 96-well plates were incubated with serial dilutions of normal mouse immunoglobulin G (IgG) (Upstate, Temecula, CA) or CD81 monoclonal antibody (MAb) JS-81 (BD Pharmingen, San Jose, CA) at 37°C for 1 h before being inoculated with virus stock containing 100 focus-forming units (FFU) of virus. hRLuc activity was measured 2 days later, as described above.
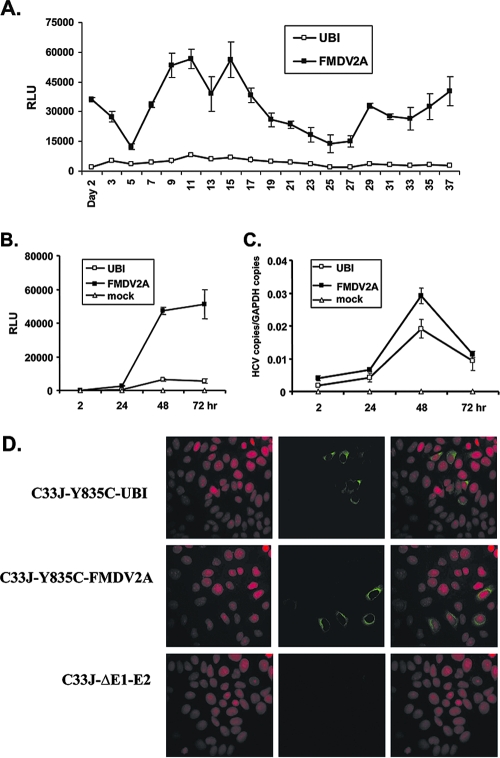
FIG. 3.
Infectious viral particles were produced from cells transfected with both reporter constructs. (A) Naïve Huh7.5 cells were infected with culture supernatants collected at indicated time points from cells transfected with C33J-Y835C-UBI (UBI) and C33J-Y835C-FMDV2A (FMDV2A). hRLuc activity of infected cells was measured 48 h after infection. Results are expressed as mean relative light units (RLU) ± standard deviations for three wells of a 96-well plate. (B) Culture supernatants collected on day 11 posttransfection were used to infect Huh7.5 cells, and hRLuc activity was measured 2 h, 24 h, 48 h, and 72 h postinfection. Culture supernatant of nontransfected cells served as a mock control. (C) In parallel, HCV RNA copies were measured by TaqMan assay. RNA copies of GAPDH were used to normalize the input. (D) Culture supernatants collected from cells transfected with C33J-Y835C-UBI, C33J-Y835C-FMDV2A, and the E1-E2 deletion mutant C33J-ΔE1-E2 were used for infecting naïve Huh7.5 cells. Forty-eight hours after inoculation, immunofluorescent staining against HCV core antigen was performed. Cell nuclei were counterstained by propidium iodide.
TaqMan assay of RNA replication.
Huh7.5 cells were seeded in 96-well plates at a density of 1.5 × 104 cells/well. After overnight incubation, 50 μl of cell culture supernatants collected at day 11 from cells transfected with both reporter viruses was added into each well. After incubation for 2, 24, 48, and 72 h, cell culture medium was removed, and the cells were washed gently with PBS. Total RNA was extracted using a Qiagen RNeasy 96 kit (Valencia, CA). Twenty microliters of the RNA was subjected to reverse transcription, using random hexamers to generate the cDNA. Quantitative PCR was carried out for HCV and glyceraldehyde-3-phosphate dehydrogenase (GAPDH), an internal control, according to the manufacturer's instructions (ABI, Foster City, CA), using the following primers and probes: HCV forward primer, 5′-GTCTGCGGAACCGGTGAGTAC-3′; HCV reverse primer, 5′-CTACGAGACCTCCCGGGGCAC-3′; HCV probe, 5′-FAM-GCGAAAGGCCTTGTGGTACTGC-BHQ1-3′; GAPDH forward primer, 5′-GAAGGTGAAGGTCGGAGTC-3′; GAPDH reverse primer, 5′-GAAGATGGTGATGGGATTTC-3′; GAPDH probe, 5′-FAM-CAAGCTTCCCGTTCTCAGCC-TAMRA-3′ (where FAM is 6-carboxyfluorescein, TAMRA is 6-carboxytetramethylrhodamine, and BHQ is Black Hole quencher). HCV and GAPDH RNA quantifications were based on a standard curve run on the same plate as the experimental samples. The signal of HCV RNA was normalized to that of GAPDH mRNA at each time point.
Immunofluorescent staining of core antigen and viral titration.
Huh7.5 cells were seeded in eight-well chamber slides (Nalge Nunc, Rochester, NY) 24 h before inoculation with 100 μl of supernatants. Forty-eight hours after inoculation, cells were fixed with methanol:acetone (1:1) at room temperature for 9 min and blocked with 2% bovine serum albumin at 37°C for 30 min. Cells were then incubated with primary antibody against HCV core antigen (Affinity Bioreagents, Golden, CO) at 37°C for 2 h, followed by washing with PBS and incubation with fluorescein isothiocyanate-conjugated goat anti-mouse IgG secondary antibody (Southern Biotech, Birmingham, AL) at 37°C for 1 h. Nuclei were counterstained by propidium iodide. Slides were examined under a Nikon TS100 microscope. A cluster of infected cells was considered as one single FFU. All cell clusters of infected cells were counted in each well, and virus titers were calculated accordingly in terms of FFU/ml.
In vitro antiviral and cytotoxicity assays.
Antiviral and cytotoxicity assays were carried out in black-walled, clear-bottom 96-well plates. Huh7.5 cells or 2a replicon cells were seeded at a density of 1.5 × 104 cells/well in 100 μl of DMEM culture medium and incubated overnight. For the antiviral activity assay, cell culture supernatants containing 100 FFU of C33J-Y835C-FMDV2A reporter virus were added into the wells and incubated for 8 h. Before the compounds were added, all medium was removed. Eight threefold serial dilutions of inhibitors or controls were prepared in DMEM and added to the appropriate wells, yielding final concentrations of 1,000 to 0.05 μM for compound A, 1,000 to 0.05 nM for BILN-2061, and 2.5 × 10−4 to 1.3 × 10−4 μg/ml for JS-81 antibody in 200 μl of DMEM. Dimethyl sulfoxide (DMSO; at a concentration of 1%) was used as the compound control, and PBS was used as the control for the antibody. Alternatively, the same amounts of the reporter viruses and the diluted compounds were added onto the cells at the same time. For the cytotoxicity assay, only compounds or 1% DMSO was added but not the cell culture supernatants containing virus. Following 2 days of incubation, hRLuc activity was measured as described above, representing the antiviral activity of the compounds. Cytotoxicity was measured using a CellTiter-Glo (CTG) Luminescent Cell Viability assay kit (Promega), following the manufacturer's instructions. The percent inhibition was determined from hRLuc and CTG values, using the following formulas: for luciferase activity, (1 − hRLuc signal of compound well/mean hRLuc signal of DMSO well) × 100; for CTG values, (1 − CTG signal of compound well/mean CTG signal of DMSO well) × 100.
The percent inhibition was plotted against the concentration of compound using the XLFit program (IDBS, Emeryville, CA), and the EC50 and 50% cytotoxicity concentration (CC50) were calculated.
Pilot HTS using C33J-Y835C-FMDV2A reporter virus and 2a replicon cells.
A total of 15,000 Huh7.5 cells or 2a replicon cells were seeded into 10 96-well plates, using a Multidrop system (ThermoLabsystems, Franklin, MA). After overnight incubation, 900 compounds from 10 96-well Pfizer compound plates from the Pfizer file were added onto 2a replicon cells at a final concentration of 10 μM. The same amounts of compounds together with 100 FFU of C33J-Y835C-FMDV2A reporter virus were added onto Huh7.5 cells. In all plates, an antiviral control (10 μM AG-21541) was added to well D8, and a cytotoxicity control (100 μM 5,6-dichloro-1-beta-D-ribofuranosyl-benzimidazole) was added to well D9. The wells D1 to D3 and D7 were used as DMSO-only controls. For the cytotoxicity assay, the same set of compounds was added onto 2a replicon cells. After 48 h of incubation, hRLuc activity and CTG values were measured, and percent antiviral inhibition and cytotoxicity were calculated as described above.
RESULTS
Construction of monocistronic GT 1b/2a reporter viruses.
In this study, we constructed two HCV GT 1b/2a chimeras, C33J-Y835C-UBI and C33J-Y835C-FMDV 2A, that carry a hRLuc reporter gene inserted 5′ of the coding sequence of the structural proteins (Fig. 1). These reporter viruses permit easy monitoring of viral infectivity by the simple measurement of luciferase activity. To ensure the separation and subsequently the proper functioning of the luciferase enzyme, the hRLuc reporter was separated from the viral structural proteins by a cleavable peptide encoding either the UBI monomer or the FMDV 2A protease sequence. Our experience with the full-length hRLuc-containing HCV GT 2a (JFH1) clone indicated that a reporter virus containing the FMDV 2A cleavage site generally showed more robust luciferase activity than the one with the UBI cleavage site (data not shown). In order to achieve the most efficient virus production, the region of the GT 1b (Con1) core to NS2 was fused with GT 2a (JFH1) at the site located right after the first transmembrane domain of NS2 (the first 33 amino acids were derived from Con1 and the rest of the sequence was from JFH1) (20). The cell culture-adaptive mutation Y835C (13) was introduced, which further augmented virus production (data not shown).
FIG. 1.
Schematic structures of 1b/2a chimeric reporter viruses with UBI and FMDV 2A cleavage sites. (A) C33J-Y835C-UBI. The HCV IRES drives the translation of both the hRLuc reporter gene and the viral genome. An ubiquitin sequence (UBI) is placed between the hRLuc and viral proteins to allow proteolytic processing. Polyproteins derived from GT 1b (Con1) and GT 2a (JFH1) are represented by open boxes and gray boxes, respectively, with the intergenotypic junction located in NS2. The adaptive mutation Y835C located in Con1 NS2 is indicated by an asterisk. (B) C33J-Y835C-FMDV2A. The structure is identical to that shown in panel A, except that the hRLuc is separated from the viral genome by the FMDV 2A cleavage sequence.
Replication competence of both reporter viruses.
In vitro transcripts of both reporter viruses, together with the transcript of an RNA replication-deficient mutant containing a Gly-Asn-Asp (GND) substitution for the conserved Gly-Asp-Asp (GDD) motif in the NS5B polymerase active site, were electroporated into the replication-permissive cell line FT3-7. RNA replication was monitored by the measurement of hRLuc activity 4 h and 72 h postelectroporation. The hRLuc activity measured at 4 h after transfection represents the electroporation efficiency because RNA replication had not started at that time and all the luciferase was translated from the input RNA (14). The ratio of hRLuc activity at 72 h to hRLuc activity at 4 h was considered as the relative increase in the level of RNA replication. As shown in Fig. 2, transfection of RNAs of both reporter viruses supported efficient RNA replication. Input RNAs replicated 13- or 10-fold in cells transfected with C33J-Y835C-UBI or C33J-Y835C-FMDV2A, respectively. Interestingly, although the RNA replication rates were comparable in both types of transfected cells, the hRLuc signals of FMDV 2A-transfected cells was four- to fivefold higher than that of UBI-transfected cells at either 4 h or 72 h. As expected, the RNA of the replication deficient mutant C33J-GND did not replicate over the 72-h period (Fig. 2).
FIG. 2.
Transfection of both reporter constructs supported efficient RNA replication. C33J-Y835C-UBI, C33J-Y835C-FMDV2A, and C33J-GND RNAs were transfected into FT3-7 cells. hRLuc activities were measured 4 h and 72 h posttransfection. Results are expressed as mean relative light units (RLU ± standard deviations) for three wells of a 96-well plate. The ratio of the RLU at 72 h to the RLU at 4 h is considered as the relative change in RNA replication.
Production of infectious viral particles in reporter viral RNA-transfected cells.
After transfection, cells were passaged every 2 days for up to 37 days, when the experiment was terminated. The medium was collected before each passage and was centrifuged. The supernatant was collected and stored at each time point. The collected supernatants were used to infect naïve Huh7.5 cells, and hRLuc activity of infected cells was measured 48 h postinfection. As shown in Fig. 3A, transfection of both reporter viral RNAs successfully produced fully infectious viral particles in the culture supernatants, which could then be used to effectively infect naïve cells, generating hRLuc signals. For C33J-Y835C-FMDV2A-transfected cells, production of infectious virus was evident as early as 2 days after electroporation and was sustained up to day 37, with the viral particle production peaking on day 9 to day 15. The hRLuc signals from cells infected with the C33J-Y835C-UBI virus were much lower than the ones infected with the C33J-Y835C-FMDV2A virus.
To understand the kinetics of viral infection and RNA replication through hRLuc production, day 11 supernatants collected from cells transfected with each of the viral RNAs were used to infect naïve Huh7.5 cells. Cells were lysed at 2 h, 24 h, 48 h, and 72 h postinfection, and hRLuc activity (Fig. 3B) was measured at each time point. In parallel, the HCV RNA replication in infected cells was monitored by quantitative PCR using a TaqMan assay (Fig. 3C). Twenty-four hours after infection, detectable hRLuc signals were generated efficiently in C33J-Y835C-FMDV2A-infected cells; the signals increased sharply to up to 48 h and remained high at 72 h (Fig. 3B). Interestingly, hRLuc signals of C33J-Y835C-UBI supernatant-infected cells were generated much less efficiently, despite comparable RNA replication levels (Fig. 3B and C). Moreover, we observed that after 48 h postinfection, the RNA replication levels of both viruses started to decrease, whereas the hRLuc signals remained high, suggesting that the hRLuc protein may be accumulated in the cells and turned over gradually at the 72 h time point. However, the strength of the hRLuc signal mirrored the HCV RNA levels quite well up to the 48 h time point, which was hence used as a time point for developing the antiviral assays. The reduction of intracellular RNA as detected by the TaqMan assay at the 72-h time point could be due to packaging and release of the RNA as virus particles.
A total of 100 μl of supernatants collected from C33J-Y835C-UBI and C33J-Y835C-FMDV 2A viral RNA-transfected cells as well as supernatant collected from cells transfected with a deletion encompassing the E1-E2 region in C33J-Y835C-UBI (C33J-ΔE1-E2) was used for inoculating naïve Huh7.5 cells. Forty-eight hours after inoculation, immunofluorescent staining against HCV core antigen was performed. As shown in Fig. 3D, core antigen was expressed in cells inoculated with supernatants collected from C33J-Y835C-UBI- and C33J-Y835C-FMDV2A-transfected cells, suggesting that both reporter viral RNAs upon transfection could produce and secrete infectious viruses into the culture supernatants. As a control, viral RNA from the construct C33J-ΔE1-E2 lost the ability to produce infectious viral particles. No infectious virus was released into the corresponding supernatant; as a result, no core antigen was detected in the cells inoculated with this supernatant (Fig. 3D, lowest panel). Interestingly, similar numbers of FFU were observed in cells inoculated with UBI and FMDV 2A supernatants, i.e., 2 × 103 to 3 × 103 FFU per milliliter of supernatant observed at 72 h posttransfection. This suggested that the two reporter viruses produced comparable numbers of infectious viral particles.
Neutralization of viral infectivity by CD81 MAb.
The cell surface protein CD81 has been shown to bind to HCV envelope protein E2 and is thus thought to assist in mediating the entry of virus particles (1, 22). A recombinant MAb against CD81, JS-81, at final concentrations of 0, 0.02, 0.1, 0.5, and 2.5 μg/ml was preincubated with Huh7.5 cells at 37°C for 1 h before inoculation with C33J-Y835C-UBI and C33J-Y835C-FMDV2A supernatants. Forty-eight hours after infection, hRLuc activity was measured. We found that the anti-CD81 MAb blocked the infection of C33J-Y835C-UBI and C33J-Y835C-FMDV2A viruses efficiently in a dose-dependent manner, whereas the control normal mouse IgG had little effect on viral infectivity (Fig. 4A and B). These data suggest that both C33J-Y835C-UBI and C33J-Y835C-FMDV2A viral particles bind to and enter the cells through a mechanism involving the CD81 cell surface protein. As shown in Fig. 4A and B, the infectivity of reporter viruses was almost completely blocked by CD81 MAb with a concentration of 2.5 μg/ml.
FIG. 4.
CD81 MAb neutralized infectivity of two reporter viruses. Naïve Huh7.5 cells were preincubated with CD81 MAb (JS-81) at the indicated concentrations for 1 h before inoculation with culture supernatants of C33J-Y835C-UBI (A) and C33J-Y835C-FMDV2A (B). hRLuc activity of infected cells was measured 48 h postinfection. Normal mouse IgG served as a negative control. Data are expressed as the percent infectivity of supernatant alone from two independent experiments.
Antiviral and cytotoxicity assays using infectious C33J-Y835C-FMDV2A reporter virus.
Based on the fact that cells infected by C33J-Y835C-FMDV2A virus consistently generated higher hRLuc signals than the ones infected by C33J-Y835C-UBI virus, C33J-Y835C-FMDV2A was used in the development of an antiviral assay for anti-HCV compound screening. To validate the C33J-Y835C-FMDV2A infectious virus system, the antiviral activities of three known HCV inhibitors, including a protease inhibitor, BILN-2061 (16), a polymerase inhibitor, compound A (5), and an entry inhibitor, CD81 MAb (JS-81), were tested in antiviral assays with the use of both the infectious C33J-Y835C-FMDV2A and a GT 2a (JFH1) subgenomic replicon cell line. For the C33J-Y835C-FMDV2A virus system, two assay conditions were tested, i.e., adding the virus and compound at the same time or preincubating the cells with virus for 8 h before adding the compounds. The EC50 values obtained from the two assay conditions were similar for the replication inhibitors, BILN-2061 and compound A. The EC50 values for BILN-2061 were 152 ± 14 nM using an 8 h-preincubation and 109 ± 7 nM using the simultaneous addition condition; for compound A the values were 17 ± 5 μM and 15 ± 4 μM, respectively (Table 1). More importantly, all the EC50 values obtained using the infectious C33J-Y835C-FMDV2A virus system were comparable to the ones obtained using the 2a subgenomic replicon cell line, which were 265 ± 20 nM for BILN-2061 and 12 ± 1 μM for compound A. As an entry inhibitor, CD81 antibody did not block viral infectivity if added 8 h after virus addition to the cells. However, when added at the same time with virus, CD81 antibody efficiently neutralized viral infectivity, with the EC50 at 0.1 μg/ml. Taken together, these data suggest that the newly developed C33J-Y835C-FMDV2A reporter virus system is sensitive to replication inhibitors and capable of identifying HCV entry inhibitors.
TABLE 1.
Determination of antiviral activity and cytotoxicity of control inhibitors by using reporter virus and a subgenomic replicon cell line
| Inhibitor | Category | EC50a
|
CC50b | ||
|---|---|---|---|---|---|
| FMDV 2A virus
|
2a replicon cells | ||||
| 8 h | 0 h | ||||
| BILN-2061 (nM) | Protease inhibitor | 152 ± 14 | 109 ± 7 | 256 ± 20 | >1,000 |
| Compound A (μM) | Polymerase inhibitor | 17 ± 5 | 15 ± 4 | 12 ± 1 | 491 |
| JS-81 MAb (μg/ml) | Entry inhibitor | >2.5 | 0.1 ± 0 | ND | >2.5 |
EC50 values were determined using C33J-Y835C-FMDV2A infection assay or 2a replicon cell line as described in Materials and Methods. Cells were preincubated with reporter virus for 8 h before the addition of the corresponding compounds (8 h), or reporter virus and the corresponding compounds were added to the cells at the same time (0 h). EC50 values are expressed as mean ± standard deviation from three independent experiments. ND, not determined.
CC50, 50% cytotoxic concentration. Values were determined using the CTG assay as described in Materials and Methods.
Characterization of infectious C33J-Y835C-FMDV2A reporter virus in an HTS format.
To further evaluate the capability of the C33J-Y835C-FMDV2A reporter virus system in identifying compound hits in an HTS format, Huh7.5 cells were plated in 96-well plates, and C33J-Y835C-FMDV2A supernatants were added into all wells. In the same plate, compounds BILN-2061 and compound A were added into three wells at three different plate locations. Each compound was added at a concentration equal to three times their respective EC50 values. DMSO (1%) was added into the remaining wells to be used as a control. Table 2 represents a plate with raw percentage inhibition readout. The average hRLuc count of the DMSO control was 18,224. The percent inhibition of all nine wells across the three different positions was 99.52% ± 0.37% and 90.77% ± 3.87% for BILN-2061 and compound A, respectively. In addition, no cytotoxicity was observed at these concentrations, consistent with the previous data (Table 1). These data suggest that the infectious C33J-Y835C-FMDV2A reporter virus system could reproducibly identify HCV inhibitors in an HTS format, regardless of the position in the 96-well plate.
TABLE 2.
Evaluation of the capability of C33J-Y835C-FMDV 2A to identify hits in an HTS formata
| Row | % Inhibition relative to DMSO controls at the indicated well position
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | |
| A | 90 | 89 | 94 | 8 | −11 | −10 | 9 | 9 | −20 | 99 | 99 | 99 |
| B | 0 | 26 | −14 | 28 | 2 | 6 | −18 | −26 | −5 | 44 | −3 | −25 |
| C | 15 | −5 | 29 | 36 | −31 | −36 | 16 | −20 | 23 | −20 | 22 | −14 |
| D | 3 | 25 | 27 | 95 | 97 | 93 | 100 | 100 | 99 | 35 | 12 | 28 |
| E | −12 | 3 | −5 | 29 | −3 | 14 | 0 | 13 | −1 | −12 | 34 | −44 |
| F | 11 | −14 | −8 | 33 | 9 | 30 | 15 | −14 | 10 | −18 | 18 | −43 |
| G | −18 | −13 | −16 | −8 | −7 | −15 | 13 | 21 | −19 | −1 | 18 | −76 |
| H | 100 | 100 | 100 | 13 | −24 | 18 | −23 | −16 | −36 | 86 | 90 | 84 |
A representative 96-well plate was displayed. Compound A and BILN-2061 were added at a concentration of 3× EC50. Results are expressed as percent inhibition of the DMSO (1%) control wells. Italicized boldface values indicate the addition of compound A, and boldface values indicate the addition of BILN-2061; all other wells represent the DMSO controls. The average 1% DMSO hRLuc count was 18,224.
Pilot HTS run with C33J-Y835C-FMDV2A reporter virus system.
To further validate the feasibility of using the infectious C33J-Y835C-FMDV2A reporter virus system to identify HCV compounds with specific antiviral activity, 900 compounds from the Pfizer compound archive were tested at a concentration of 10 μM in an HTS format, using 10 96-well plates. In parallel, the same set of compounds was also tested in the GT 2a (JFH1) subgenomic replicon cell line. A “hit” was defined as a compound having antiviral inhibition of ≥60% and cytotoxicity of ≤40% compared to values of the 1% DMSO controls. As shown in Table 3, using these criteria, a hit rate of 12.1% was obtained with the C33J-Y835C-FMDV2A reporter virus system, and 4.2% was obtained using the GT 2a subgenomic replicon system. More interestingly, 31 hits were identified by both systems, whereas 78 compound hits were identified only by the C33J-Y835C-FMDV2A reporter virus but not the GT 2a replicon system. These results indicate that the former group of compound hits may target the replication machinery of the GT 2a genome, whereas the latter group of compound hits is potentially targeting steps in the virus life cycle, such as the viral attachment, entry, disassembly, and morphogenesis steps.
TABLE 3.
Pilot HTS with Pfizer's compound platesa
| Plate no. | No. of hits identified
|
||||
|---|---|---|---|---|---|
| 2a replicon system | FMDV2A system | 2a replicon- FMDV2A overlap | 2a replicon system only | FMDV2A system only | |
| 1 | 1 | 16 | 0 | 1 | 16 |
| 2 | 7 | 21 | 5 | 2 | 16 |
| 3 | 0 | 5 | 0 | 0 | 5 |
| 4 | 2 | 11 | 2 | 0 | 9 |
| 5 | 0 | 3 | 0 | 0 | 3 |
| 6 | 0 | 5 | 0 | 0 | 5 |
| 7 | 1 | 10 | 0 | 1 | 10 |
| 8 | 3 | 5 | 2 | 1 | 3 |
| 9 | 16 | 25 | 15 | 1 | 10 |
| 10 | 8 | 8 | 7 | 1 | 1 |
| Total | 38 | 109 | 31 | 7 | 78 |
Ten 96-well compound plates were screened, as described in Materials and Methods. Numbers of hits are given for each system separately and for overlapping hits as well as for exclusive hits (2a replicon only and FMDV2A only). FMDV2A, C33J-Y835C-FMDV2A system.
DISCUSSION
While the development of infectious HCV cell culture systems reached several milestones in the past 2 years (17, 26, 29), these systems have not yet become a truly useful and robust tool for antiviral drug screening, especially in a high-throughput format. In this study, we have constructed two monocistronic GT 1b/2a chimeric viruses, which encode a short codon-optimized reporter gene. These constructs efficiently replicate their viral RNA and produce infectious viral particles. Importantly, this novel infectious system is capable of producing a high level of luciferase signal for monitoring the antiviral activity of compounds. Therefore, it allows us to use the full-length viruses for the high-throughput anti-HCV compound screening and identify novel inhibitors not captured by using a replicon system.
To separate the fused hRLuc and the viral proteins, two cleavage peptides were employed, UBI and FMDV 2A protease. UBI is a small protein composed of 76 amino acids that are fused between hRLuc and the 1b core protein in C33J-Y835C-UBI. After the fusion protein is synthesized, UBI is cleaved at the C-terminal end by the cellular enzyme ubiquitin C-terminal hydrolase, thus releasing the hRLuc-UBI complex from the rest of the viral proteins (7, 10). The 2A protease derived from FMDV is a short polypeptide of 20 amino acids that cleaves the polyprotein of FMDV at the 2A/2B junction cotranslationally. The self-processing activity in FMDV 2A leads to cleavage between the terminal glycine of the 2A product and the initial proline of 2B (24). In C33J-Y835C-FMDV2A, the cleavage of the polyprotein product occurs at the C-terminal end of the 2A coding region, leaving this peptide fused to the upstream hRLuc protein and releasing the downstream viral protein intact with the addition of an N-terminal proline. In our study we observed several things. (i) After transfection of viral RNAs, although the RNA replication efficiency was comparable for C33J-Y835C-UBI and C33J-Y835C-FMDV2A viral RNAs, the hRLuc signals generated from C33J-Y835C-FMDV2A RNA-transfected cells were four- to fivefold higher than signals of C33J-Y835C-UBI-transfected cells, at either 4 h or 72 h (Fig. 2). (ii) Similarly, after infection of naïve cells, infectious C33J-Y835C-FMDV2A reporter virus produced much higher hRLuc signals than the C33J-Y835C-UBI virus (Fig. 3A and B), despite comparable RNA replication levels (Fig. 3C) and similar viral titers (data not shown). Several possibilities may account for the findings. First, FMDV 2A sequence holds the ability to self-cleave, unlike UBI which requires an additional cellular enzyme, i.e., ubiquitin C-terminal hydrolase, for its cleavage. This self-processing ability may grant C33J-Y835C-FMDV2A a more efficient cleavage upon translation and therefore release the hRLuc-FMDV 2A complex more efficiently. Second, the addition of extrinsic peptide to the upstream gene product, in our case the hRLuc protein, might interfere with its function. In our experience, addition of the 2A peptide has little to no effect on the function of hRLuc, as revealed by the high hRLuc counts detected. This is consistent with earlier reports showing that the addition of 2A peptide did not impair the functions or localizations of other proteins such as O6-methylguanine-DNA methyltransferase and homeobox transcription factor HOXB4 (6, 19). It is still unclear to what extent the addition of UBI would interfere with the function of the associated hRLuc protein. Taken together, the unique feature of generating a high hRLuc signal makes C33J-Y835C-FMDV2A a better candidate for antiviral assay development.
Compared to the widely used dicistronic reporter constructs which contain both HCV and encephalomyocarditis virus (EMCV) internal ribosome entry sites (IRESs), the monocistronic reporter viruses we constructed in the current study have several potential advantages. First, the additional EMCV IRES increases the length and complexity of the viral genome, thereby delaying viral RNA replication. Indeed, a recent report by the Bartenschlager group found that replication of dicistronic genomes was significantly delayed compared to monocistronic genomes (25). Second, the monocistronic construct design eliminates the “false-positive” selection of the compounds targeting the EMCV IRES, thereby simplifying and increasing the specificity of anti-HCV compound screening. Third, the monocistronic design reduces the possibility that after multiple rounds of virus culture, the hRLuc reporter gene is deleted (except in the case of in-frame deletions), thereby losing the reporter signal for monitoring replication.
The use of the subgenomic replicon system represents a milestone in HCV drug discovery. The replicons replicate autonomously to high levels in human hepatoma cell lines, which enables the screening of inhibitors targeting the traditional targets, such as NS3 protease and NS5B polymerase (3). However, without the inclusion of viral structural regions, these replicon systems are not able to identify inhibitors targeting important steps of the viral life cycle, such as viral attachment, entry, disassembly, and morphogenesis. Our newly developed reporter virus system, by generating full-length infectious viral particles, overcomes these drawbacks of the replicon system. Similar to the replicon systems, it identifies the nonstructural protein inhibitors, e.g., the protease inhibitor BILN-2061 and the polymerase inhibitor compound A. More importantly, it is also capable of identifying the inhibitors targeting other life cycle steps, e.g., CD81 antibody which targets the viral entry (Table 1). This feature of the infectious virus system promises more opportunities in discovering novel HCV inhibitors. Furthermore, this system represents an authentic HCV particle generation system where budding probably occurs at the endoplasmic reticulum (23). This could potentially be an advance compared to the HCV pseudoparticle system for screening entry inhibitors once the glycosylation pattern of particles from the infectious system is examined. In the retroviral pseudoparticle system, a modification forces the expression of envelope proteins on the cell surface, and glycosylation patterns may thus be altered from patterns that arise when envelope proteins are retained in the endoplasmic reticulum (4). Using such a system might miss certain inhibitors that operate by recognition of the true conformations on the envelope protein generated by a glycosylation pattern in the endoplasmic reticulum. Therefore, investigation of the glycosylation patterns of our cell culture-derived infectious HCVs would be of great interest for future studies. Altogether, using culture-derived HCV reporter viruses as a screening tool advances anti-HCV drug discovery. However, one thing of note is that culture-derived HCV does not completely mimic the HCV present in human serum, which normally is associated with very-low-density lipoprotein, low-density lipoprotein, high-density lipoprotein, and IgG, which may modify the efficacy of inhibitors. Therefore, modifications to screening assay conditions using the infectious reporter virus reported here with components of human plasma and serum could potentially mimic conditions that the virus experiences in nature.
After being validated with three known inhibitors, BILN-2061, compound A, and CD81 antibody, this new system was tested for its ability to identify compound hits in an HTS format. As expected, it successfully discovered known inhibitors in 96-well plates, regardless of the position of the inhibitors on the 96 well-plate (Table 2). To further test its feasibility in identifying unknown compound hits in an HTS format, the infectious system was used in parallel with the GT 2a replicon system for screening 900 unknown compounds. This resulted in 31 compound hits discovered by both systems, while 78 hits were identified only by the new infectious system. It is reasonable to speculate that the overlapping 31 hits identified are the ones targeting the replication and translation components of GT 2a. In contrast, the 78 hits (rate of 8.7%) identified by only the 1b/2a chimeric infectious virus system are the ones potentially targeting the structural regions of the GT 1b virus and/or targets/steps involved in attachment, entry, disassembly, and assembly components of the viral life cycle. The high hit rate could possibly be due to the inhibition of the FMDV 2A protease. Employing an FMDV 2A inhibition assay in secondary assays could help the identification and elimination of false positives identified in the infectious system. We also observed seven hits that were identified only by the GT 2a replicon system but not by the 1b/2a infectious system. This difference may arise because the subgenomic replicon assay has a higher signal-to-noise ratio and potentially higher sensitivity than the infectious virus assay. Additionally, the 2a replicon is a dicistronic replicon that contains an EMCV IRES, which could also be the source of false negatives identified only by the 2a replicon system since the infectious constructs lack the EMCV IRES. Based on these findings, this pilot HTS supports the feasibility of using the newly established infectious viral system in screening inhibitors targeting all of the HCV viral proteins and the steps of the whole HCV life cycle in a large-scale HTS.
In summary, here we report the development of a novel HCV reporter virus system which generates infectious HCV particles in cell culture and can be used as a robust screening tool for HCV drug discovery. The most notable advantage of this system over the current replicon systems is that it broadens target range, which maximizes the identification of compounds inhibiting HCV through specific mechanisms that target all events of the viral life cycle. To our knowledge, this is the first cell-based infectious HCV system that is suitable for drug screening on a large scale.
Supplementary Material
Acknowledgments
This work was made possible by support from Amy Patick and Zuzana Hostomska, who supported this proposal for the Pfizer postdoctoral program. This work was carried out in the Department of Virology, Pfizer, La Jolla.
We thank Tihua Chen for technical support during the pilot HTS and Stephanie Shi for technical help with the immunofluorescent staining experiment. We are also indebted to all of the former colleagues from the Department of Virology for their encouragement during the study.
Footnotes
Published ahead of print on 26 November 2007.
Supplemental material for this article may be found at http://aac.asm.org/.
REFERENCES
- 1.Akazawa, D., T. Date, K. Morikawa, A. Murayama, M. Miyamoto, M. Kaga, H. Barth, T. F. Baumert, J. Dubuisson, and T. Wakita. 2007. CD81 expression is important for the permissiveness of Huh7 cell clones for heterogeneous hepatitis C virus infection. J. Virol. 81:5036-5045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alter, M. J., D. Kruszon-Moran, O. V. Nainan, G. M. McQuillan, F. Gao, L. A. Moyer, R. A. Kaslow, and H. S. Margolis. 1999. The prevalence of hepatitis C virus infection in the United States, 1988 through 1994. N. Engl. J. Med. 341:556-562. [DOI] [PubMed] [Google Scholar]
- 3.Bartenschlager, R. 2002. Hepatitis C virus replicons: potential role for drug development. Nat. Rev. Drug Discov. 1:911-916. [DOI] [PubMed] [Google Scholar]
- 4.Bartosch, B., J. Dubuisson, and F. L. Cosset. 2003. Infectious hepatitis C virus pseudo-particles containing functional E1-E2 envelope protein complexes. J. Exp. Med. 197:633-642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Camp, D., C. Matthews, S. Neville, M. Rouns, R. Scott, and T. Truong. 2006. Development of a synthetic process towards a hepatitis C polymerase inhibitor. Org. Process Res. 10:814-821. [Google Scholar]
- 6.Chinnasamy, D., M. D. Milsom, J. Shaffer, J. Neuenfeldt, A. F. Shaaban, G. P. Margison, L. J. Fairbairn, and N. Chinnasamy. 2006. Multicistronic lentiviral vectors containing the FMDV 2A cleavage factor demonstrate robust expression of encoded genes at limiting MOI. Virol. J. 3:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Finley, D., and V. Chau. 1991. Ubiquitination. Annu. Rev. Cell Biol. 7:25-69. [DOI] [PubMed] [Google Scholar]
- 8.Frese, M., K. Barth, A. Kaul, V. Lohmann, V. Schwarzle, and R. Bartenschlager. 2003. Hepatitis C virus RNA replication is resistant to tumour necrosis factor-alpha. J. Gen. Virol. 84:1253-1259. [DOI] [PubMed] [Google Scholar]
- 9.Hao, W., K. J. Herlihy, N. J. Zhang, S. A. Fuhrman, C. Doan, A. K. Patick, and R. Duggal. 2007. Development of a novel dicistronic reporter-selectable hepatitis C virus replicon suitable for high-throughput inhibitor screening. Antimicrob. Agents Chemother. 51:95-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hershko, A. 1991. The ubiquitin pathway for protein degradation. Trends Biochem. Sci. 16:265-268. [DOI] [PubMed] [Google Scholar]
- 11.Hugle, T., and A. Cerny. 2003. Current therapy and new molecular approaches to antiviral treatment and prevention of hepatitis C. Rev. Med. Virol. 13:361-371. [DOI] [PubMed] [Google Scholar]
- 12.Kato, T., T. Date, M. Miyamoto, A. Furusaka, K. Tokushige, M. Mizokami, and T. Wakita. 2003. Efficient replication of the genotype 2a hepatitis C virus subgenomic replicon. Gastroenterology 125:1808-1817. [DOI] [PubMed] [Google Scholar]
- 13.Kaul, A., I. Worz, T. Pietschmann, A. Shavinskaya, and R. Bartenschlager. 2006. Adaptation of authentic and chimeric hepatitis C virus to a human hepatoma cell line, abstr. 219. Abstr. 13th International Meeting on Hepatitis C Virus and Related Viruses, Cairns, Queensland, Australia, 27 to 31 August, 2006.
- 14.Krieger, N., V. Lohmann, and R. Bartenschlager. 2001. Enhancement of hepatitis C virus RNA replication by cell culture-adaptive mutations. J. Virol. 75:4614-4624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lake-Bakaar, G. 2003. Current and future therapy for chronic hepatitis C virus liver disease. Curr. Drug Targets Infect. Disord. 3:247-253. [DOI] [PubMed] [Google Scholar]
- 16.Lamarre, D., P. C. Anderson, M. Bailey, P. Beaulieu, G. Bolger, P. Bonneau, M. Bos, D. R. Cameron, M. Cartier, M. G. Cordingley, A. M. Faucher, N. Goudreau, S. H. Kawai, G. Kukolj, L. Lagace, S. R. LaPlante, H. Narjes, M. A. Poupart, J. Rancourt, R. E. Sentjens, R. St George, B. Simoneau, G. Steinmann, D. Thibeault, Y. S. Tsantrizos, S. M. Weldon, C. L. Yong, and M. Llinas-Brunet. 2003. An NS3 protease inhibitor with antiviral effects in humans infected with hepatitis C virus. Nature 426:186-189. [DOI] [PubMed] [Google Scholar]
- 17.Lindenbach, B. D., M. J. Evans, A. J. Syder, B. Wolk, T. L. Tellinghuisen, C. C. Liu, T. Maruyama, R. O. Hynes, D. R. Burton, J. A. McKeating, and C. M. Rice. 2005. Complete replication of hepatitis C virus in cell culture. Science 309:623-626. [DOI] [PubMed] [Google Scholar]
- 18.Lohmann, V., F. Korner, J. Koch, U. Herian, L. Theilmann, and R. Bartenschlager. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285:110-113. [DOI] [PubMed] [Google Scholar]
- 19.Milsom, M. D., L. B. Woolford, G. P. Margison, R. K. Humphries, and L. J. Fairbairn. 2004. Enhanced in vivo selection of bone marrow cells by retroviral-mediated coexpression of mutant O6-methylguanine-DNA-methyltransferase and HOXB4. Mol. Ther. 10:862-873. [DOI] [PubMed] [Google Scholar]
- 20.Pietschmann, T., A. Kaul, G. Koutsoudakis, A. Shavinskaya, S. Kallis, E. Steinmann, K. Abid, F. Negro, M. Dreux, F. L. Cosset, and R. Bartenschlager. 2006. Construction and characterization of infectious intragenotypic and intergenotypic hepatitis C virus chimeras. Proc. Natl. Acad. Sci. USA 103:7408-7413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pietschmann, T., V. Lohmann, A. Kaul, N. Krieger, G. Rinck, G. Rutter, D. Strand, and R. Bartenschlager. 2002. Persistent and transient replication of full-length hepatitis C virus genomes in cell culture. J. Virol. 76:4008-4021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pileri, P., Y. Uematsu, S. Campagnoli, G. Galli, F. Falugi, R. Petracca, A. J. Weiner, M. Houghton, D. Rosa, G. Grandi, and S. Abrignani. 1998. Binding of hepatitis C virus to CD81. Science 282:938-941. [DOI] [PubMed] [Google Scholar]
- 23.Rouille, Y., F. Helle, D. Delgrange, P. Roingeard, C. Voisset, E. Blanchard, S. Belouzard, J. McKeating, A. H. Patel, G. Maertens, T. Wakita, C. Wychowski, and J. Dubuisson. 2006. Subcellular localization of hepatitis C virus structural proteins in a cell culture system that efficiently replicates the virus. J. Virol. 80:2832-2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ryan, M. D., A. M. King, and G. P. Thomas. 1991. Cleavage of foot-and-mouth disease virus polyprotein is mediated by residues located within a 19 amino acid sequence. J. Gen. Virol. 72:2727-2732. [DOI] [PubMed] [Google Scholar]
- 25.Schaller, T., N. Appel, G. Koutsoudakis, S. Kallis, V. Lohmann, T. Pietschmann, and R. Bartenschlager. 2007. Analysis of hepatitis C virus superinfection exclusion by using novel fluorochrome gene-tagged viral genomes. J. Virol. 81:4591-4603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wakita, T., T. Pietschmann, T. Kato, T. Date, M. Miyamoto, Z. Zhao, K. Murthy, A. Habermann, H. G. Krausslich, M. Mizokami, R. Bartenschlager, and T. J. Liang. 2005. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat. Med. 11:791-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.World Health Organization. 2000. Hepatitis C fact sheet 164. http://www.who.int/mediacentre/factsheets/fs164/en.
- 28.Yi, M., Y. Ma, J. Yates, and S. M. Lemon. 2007. Compensatory mutations in E1, p7, NS2, and NS3 enhance yields of cell culture-infectious intergenotypic chimeric hepatitis C virus. J. Virol. 81:629-638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhong, J., P. Gastaminza, G. Cheng, S. Kapadia, T. Kato, D. R. Burton, S. F. Wieland, S. L. Uprichard, T. Wakita, and F. V. Chisari. 2005. Robust hepatitis C virus infection in vitro. Proc. Natl. Acad. Sci. USA 102:9294-9299. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.