Abstract
Streptococcus uberis UCN 42, isolated from a case of bovine mastitis, was intermediately resistant to lincomycin (MIC = 2 μg/ml) while remaining susceptible to clindamycin (MIC = 0.06 μg/ml) and erythromycin. A 1.1-kb SacI fragment was cloned from S. uberis UCN 42 total DNA on plasmid pUC 18 and introduced into Escherichia coli AG100A, where it conferred resistance to both clindamycin and lincomycin. The sequence analysis of the fragment showed the presence of a new gene, named lnu(D), that encoded a 164-amino-acid protein with 53% identity with Lnu(C) previously reported to occur in Streptococcus agalactiae. Crude lysates of E. coli AG100A containing the cloned lnu(D) gene inactivated lincomycin and clindamycin in the presence of ATP and MgCl2. Mass spectrometry experiments demonstrated that the lnu(D) enzyme catalyzed adenylylation of clindamycin. A domain conserved in deduced sequences of lincosamide O-nucleotidyltransferases Lnu(A), Lnu(C), LinAN2, and Lin(D) and in the aminoglycoside nucleotidyltransferase ANT(2′′) was identified.
Streptococcus uberis is one of the most important causes of bovine clinical mastitis and subclinical intramammary infections. Previous studies have demonstrated that this microorganism has the ability to enter bovine mammary epithelial cells and may therefore escape antibiotic treatment (2, 21, 25). S. uberis is susceptible to β-lactams, which are the drugs of choice for therapy of infections due to this organism (12). However, these antibiotics do not penetrate into the cells. By contrast, macrolides and lincosamides are alternative antibiotics that are intracellularly active (11). In veterinary practice, two lincosamides, lincomycin and pirlimycin (26), are used to treat mastitis in lactating dairy cattle by intramammary infusion.
Lincosamides, including clindamycin, lincomycin, and pirlimycin, prevent protein synthesis by inhibiting the peptidyltransferase by binding to several nucleotides of 23S rRNA in the 50S subunit of the bacterial ribosome, including the key nucleotide A2058. Streptococci isolated from cows, in particular S. uberis and Streptococcus dysgalactiae subsp. dysgalactiae, have developed resistance to macrolides and/or lincosamides (11, 13, 15, 23). In streptococci, two major mechanisms are responsible for resistance to lincosamides: alteration of the antibiotic target site, which confers cross-resistance to macrolide, lincosamide, and streptogramin B-type antibiotics (the so-called MLSB phenotype), and enzymatic modification. The latter mechanism, initially reported by Dutta and Devriese to occur in streptococci isolated from bovine mastitis (9), confers resistance to lincomycin but not to erythromycin (the so-called L phenotype).
Enzymatic modification of lincosamides is due to nucleotidylation of the hydroxyl group at position 3 or 4 of lincosamides. Until now, two lincosamide O-nucleotidyltransferases encoded by the lnu(B) (4, 8, 20) and lnu(C) (1) genes have been reported to occur in streptococci and enterococci. Other lincosamide O-nucleotidyltransferase genes have been detected in staphylococci [lnu(A) and lnu(A′)] (5, 6, 19), in Bacteroides fragilis (linAN2) (27), and in enterobacteria (linF) (17).
Although the L phenotype of resistance to lincomycin has been reported to occur in S. uberis, the mechanism responsible for this resistance has not been investigated (15, 18). In this study, we describe a new gene, lnu(D), associated with the expression of borderline resistance to lincomycin in an S. uberis isolate.
MATERIALS AND METHODS
Bacterial strains and antimicrobial susceptibility testing.
S. uberis UCN42 was isolated from a case of clinical mastitis in a dairy cow. Preliminary identification of the microorganism was performed using Gram staining, catalase activity detection, and the ID32 strep system (bioMérieux, La Balme-les-Grottes, France). S. uberis was further distinguished from Streptococcus parauberis by PCR amplification and sequencing of the rrs gene (16S rRNA). Escherichia coli K-12 AG100A was used as a recipient strain in electrotransformation or mating experiments (22). E. coli AG100A is a mutant susceptible to lincosamides resulting from inactivation by transposon Tn903 harboring a kanamycin resistance gene of the AcrAB pump responsible for active efflux of lincosamides (22).
Antibiotic susceptibility was determined by the disk diffusion and agar dilution methods in Mueller-Hinton agar medium supplemented with 5% horse blood according to the recommendations of Comité de l'Antibiogramme de la Société Française de Microbiologie (CA-SFM) (7). Disks were from Bio-Rad (Marnes-la-Coquette, France). Clindamycin and lincomycin were purchased from Sigma (Saint Quentin Fallavier, France), and pirlimycin was provided by its manufacturer (Pfizer santé animale, Paris, France). Agar media were incubated at 37°C in 5% CO2. The reference strain Streptococcus pneumoniae ATCC 49619 and the susceptible isolate S. uberis 72 were included as controls. The CA-SFM MIC breakpoints for lincomycin and streptococci are ≤1 μg/ml (susceptible) and >8 μg/ml (resistant) (7).
Lincosamide inactivation.
The kinetics of lincomycin inactivation by resting cells of S. uberis UCN42 or E. coli K-12 AG100A transformants was tested in liquid medium as previously described (1). Bacterial cells were suspended in 0.01 M phosphate buffer (pH 7.0) containing 5 μg of clindamycin or lincomycin per ml and were incubated at 37°C for 24 h. The pH of the suspension was monitored and remained constant. Inactivation of clindamycin or lincomycin was followed by a bioassay with Micrococcus luteus ATCC 9341 as an indicator organism.
Modified clindamycin was prepared using conditions optimized for lincomycin. Cells of E. coli AG100A containing the cloned lnu(D) gene were lysed by sonication. Cell debris were removed by centrifugation at 40,000 × g for 45 min. Clindamycin (200 μg/ml) was added to the supernatant and incubated at 37°C for 18 h in the presence of ATP (2.5 mM) and MgCl2 (50 mM). Inactivation of the antibiotic was monitored as indicated above. Aliquots of inactivated clindamycin were freeze dried.
Samples were analyzed by using an electrospray ion trap mass spectrometer (MS) (LCQ Deca XP; Thermo Finnigan, San Jose, CA) coupled online with high-performance liquid chromatography (HPLC; Surveyor LC). They were separated by reverse-phase HPLC on a C18 capillary column (ThermoHyPurity C18; 150 by 0.18 mm). A linear gradient (flow rate, 5 μl/min) of 5 to 95% solution B, where solvent A was a 2 mM ammonium acetate aqueous solution and solution B was a 2 mM ammonium acetate solution in methanol, was used. The electrospray ionization parameters were as follows: spray voltage, 4.5 kV; spray current, 80 μA; sheath gas flow rate, 35; auxiliary gas flow rate, 10; capillary temperature, 250°C; capillary voltage, 10 V; and tube lens offset, −5 V. These parameters were issued from an optimization of the detection of lincomycin. Spectra were acquired in a mode that alternated a full MS scan (mass range, 200 to 1,000; three microscans; maximum ion time, 100 ms), followed by a collision-induced dissociation (CID)-MS2 and a CID-MS3 analysis (three microscans; maximum ion time, 400 ms; collision energy, 35%) of the most abundant ion detected in the previous spectra.
Cloning and sequencing of the lincosamide resistance determinant.
Total DNA from S. uberis UCN42 was digested with various restriction enzymes and ligated at 4°C to plasmid pUC18 DNA digested with the corresponding restriction enzymes. Recombinant plasmids were electrotransformed into E. coli AG100A cells, and transformants were selected on media containing lincomycin (5 μg/ml), ampicillin (100 μg/ml), and kanamycin (20 μg/ml). Both DNA strands were sequenced with a CEQ 8000 genetic analysis system sequencer (Beckman Coulter, Villepinte, France). Nucleotide and amino acid sequences were analyzed by using the BLAST and FASTA software available at the National Center for Biotechnology website (http://www.ncbi.nlm.nih.gov/). Multiple-sequence alignments, phylogenetic-tree construction, and a search for conserved motifs were performed with the ClustalW, PHYLIP, and PROSITE programs (available at the Institut Pasteur de Paris website, http://bioweb.pasteur.fr/).
Molecular techniques.
Detection of lnu(A), lnu(A′), lnu(B), lnu(C), linAN2, and linF was done by PCR as described previously (1). Strains of Staphylococcus haemolyticus BM4110 [lnu(A)] (5), Staphylococcus aureus BM411 [lnu(A′)] (6), E. coli DB10 pVMM26 [lnu(B)] (4), and Streptococcus agalactiae UCN36 [lnu(C)] (1) were used as controls.
Plasmid DNA was extracted from S. uberis UCN42, as described previously by Ehrenfeld and Clewell (10). Enterococcus faecalis containing plasmid pAD1 was used as a positive control for plasmid extraction (10).
Filter matings.
Transfer of lincomycin resistance from S. uberis to the recipient strain S. agalactiae BM132 (susceptible to lincomycin and resistant to rifampin and fusidic acid) (16) was attempted by filter mating, as previously described (4). Transconjugants were selected on brain heart infusion containing rifampin (20 μg/ml), fusidic acid (10 μg/ml), and lincomycin (2 μg/ml). Experiments were repeated three times.
Nucleotide sequence accession number.
The nucleotide sequence of the lnu(D) gene has been deposited in the GenBank data library under accession no. EF452177.
RESULTS AND DISCUSSION
Resistance to lincomycin in S. uberis UCN42.
By the disk diffusion method, S. uberis UCN42 was found to be intermediately resistant to lincomycin; susceptible to clindamycin, erythromycin, penicillin G, tetracyclines, trimethoprim-sulfamethoxazole, and vancomycin; and resistant to low levels of gentamicin. The MICs of erythromycin, clindamycin, lincomycin, and pirlimycin for S. uberis UCN42 were similar to those for the susceptible isolate S. uberis 72 except for that of lincomycin (2 μg/ml), which was 32 times higher (Table 1). This phenotype of borderline resistance to lincomycin but susceptibility to erythromycin and clindamycin is similar to that previously described for streptococcal or staphylococcal isolates that contain lnu genes (1, 5). However, we failed to detect by PCR any of the known lnu genes. A lincomycin inactivation bioassay showed that cells of S. uberis UCN42 inactivated clindamycin and lincomycin (data not shown). Therefore, the presence in our strain of a lincosamide resistance gene encoding an inactivating enzyme was presumed. The resistance was not transferable to S. agalactiae BM132 in mating experiments, and no plasmid DNA could be extracted from the strain.
TABLE 1.
MICs of erythromycin and lincosamides for S. uberis isolates and E. coli AG100A containing or not containing the lnu(D) gene
| Bacterial strain | Characteristic or plasmid | MIC (μg/ml) of:
|
|||
|---|---|---|---|---|---|
| Erythromycin | Clindamycin | Lincomycin | Pirlimycin | ||
| S. uberis 72 | Control | 0.06 | 0.03 | 0.06 | 0.06 |
| S. uberis UCN42 | lnu(D) | 0.12 | 0.06 | 2 | 0.12 |
| E. coli K-12 AG100A | pUC18 | 4 | 2 | 32 | 32 |
| E. coli K-12 AG100A | pUC18::lnu(D) | 4 | 32 | 128 | 64 |
Characterization of the lnu(D) gene.
DNA fragments from S. uberis UC8578 restricted by various enzymes were cloned in plasmid pUC18 digested similarly and introduced into E. coli AG100A. Transformants were selected with clindamycin or lincomycin. One of the transformants resistant to ampicillin, kanamycin, and clindamycin and harboring pUC18 with a 1.1-kb SacI insert was studied further. The transformant inactivated lincomycin and clindamycin according to the inactivation bioassay. The DNA insert was entirely sequenced. Analysis of the sequence revealed an open reading frame of 495 bp preceded by a GGAGG sequence similar to the ribosome binding site consensus sequence and separated by 6 bp from the ATG start codon. This open reading frame could possibly code for a 164-amino-acid protein. Comparison of the deduced sequence with those of proteins showed homology with various lincosamide and aminoglycoside nucleotidyltransferases. The highest levels of homology were with the lincosamide nucleotidyltransferases encoded by the lnu(C) gene from S. agalactiae (GenBank accession no. AY928180), lnu(A′) from S. aureus (GenBank accession no. J03947), lnu(A) from S. haemolyticus (GenBank accession no. M14039), and linAN2 from B. fragilis (GenBank accession no. AAF74724) (the identities are 53%, 29%, 33%, and 30%, respectively, and the similarities are 75%, 50%, 50%, and 50%, respectively). The lnu-related gene of S. uberis UCN42 was thus designated lnu(D).
Lnu(D) was also closely related to a putative aminoglycoside nucleotidyltransferase identified in silico in the genome of Bacillus cereus ATCC 10987 (81% identity) and displayed significant homology with putative aminoglycoside nucleotidyltransferases of Alkaliphilus metalliredigenes QYMF (32% identity), Mycobacterium flavescens PYR-GCK (32% identity), and Streptomyces coelicolor A3(2) (36% identity). So far, neither the functions nor the substrates (lincosamides or aminoglycosides) of these putative enzymes have been characterized.
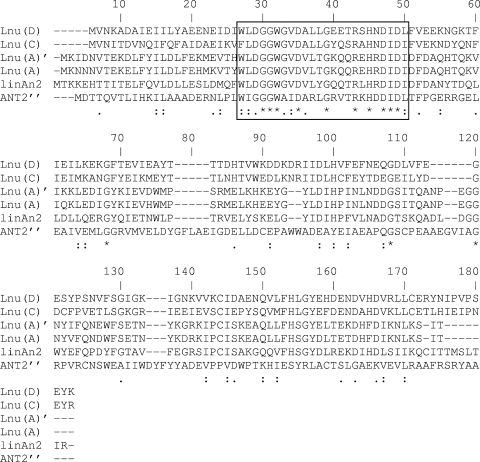
Furthermore, the N terminus of the Lnu(D) protein was homologous to that of the aminoglycoside (2′′) nucleotidyltransferase [ANT(2′′)] spread in enterobacteria and other gram-negative bacilli (24). Alignment of nucleotidyltransferases revealed the presence of a conserved domain in the N termini of the closely related Lnu and ANT(2′′) enzymes (Fig. 1). No specific function could be predicted for this domain (GGWXXDXXXGXXXRXHXDID, where X is any amino acid).
FIG. 1.
Partial alignment of the N termini of the deduced sequences of lincosamide O-nucleotidyltransferases Lnu(A) (GenBank accession no. M14039), Lnu(C) (GenBank accession no. AY928180), LinAN2 (GenBank accession no. AAF74724), and Lnu(D) (GenBank accession no. EF452177) and aminoglycoside nucleotidyltransferase ANT(2′′) (GenBank accession no. CAA87464). The conserved domain is boxed. *, identical residues; :, conserved substitutions; ., semiconserved substitutions.
No significant homology was found with lincosamide nucleotidyltransferases Lnu(B) and LinF from Enterococcus faecium and E. coli, respectively (GenBank accession no. AF110130 and AJ561197, respectively) or any aminoglycoside nucleotidyltransferase other than ANT(2′′) [aminoglycoside (4′) nucleotidyltransferases, aminoglycoside (6) nucleotidyltransferases, streptomycin (3) nucleotidyltransferase, aminoglycoside (3′′)(9) nucleotidyltransferase, and aminoglycoside (9) nucleotidyltransferase].
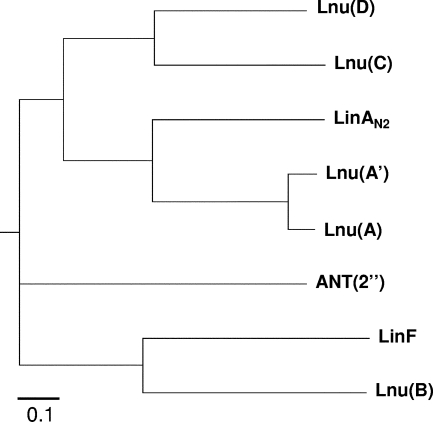
Alignment of nucleotidyltransferases was used to construct a phylogenetic tree (14). The Lnu enzymes from streptococci [Lnu(C) and Lnu(D)], staphylococci [Lnu(A) and Lnu(A′)], and B. fragilis (LinAN2) and ANT(2′′) formed distinct groups (Fig. 2). Other lincosamide and aminoglycoside nucleotidyltransferases were phylogenetically remote. Among lincosamide O-nucleotidyltransferases, Lnu(B) and LinF formed a group, distinct from the other enzymes in particular by their sizes [267 and 273 amino acids, respectively, versus 161, 164, and 164 amino acids for Lnu(A), Lnu(C), and Lnu(D), respectively]. In addition, LnuB modifies a hydroxyl group at position 3 in both clindamycin and lincomycin whereas the Lnu(A) nucleotidyltransferase modifies a hydroxyl group of clindamycin and lincomycin at positions 3 and 4, respectively. The precise sites of nucleotidylation of lincomycin and clindamycin by the other enzymes have not been characterized.
FIG. 2.
Phylogenetic relationship among lincosamide and aminoglycoside nucleotidyltransferases. The tree was constructed using the neighbor-joining method with the closely related amino acid sequences from S. uberis [Lnu(D)] (GenBank accession no. EF452177), S. haemolyticus [Lnu(A)] (GenBank accession no. M14039), S. aureus [Lnu(A′)] (GenBank accession no. J03947), S. agalactiae [Lnu(C)] (GenBank accession no. AY928180), and B. fragilis (LinAN2) (GenBank accession no. AAF74724). More distantly related sequences from Enterococcus faecium [Lnu(B)] (GenBank accession no. AF110130), E. coli (LinF) (GenBank accession no. AJ561197), and E. coli [ANT(2′′)] (GenBank accession no. CAA87464) were also included.
Mechanism of resistance.
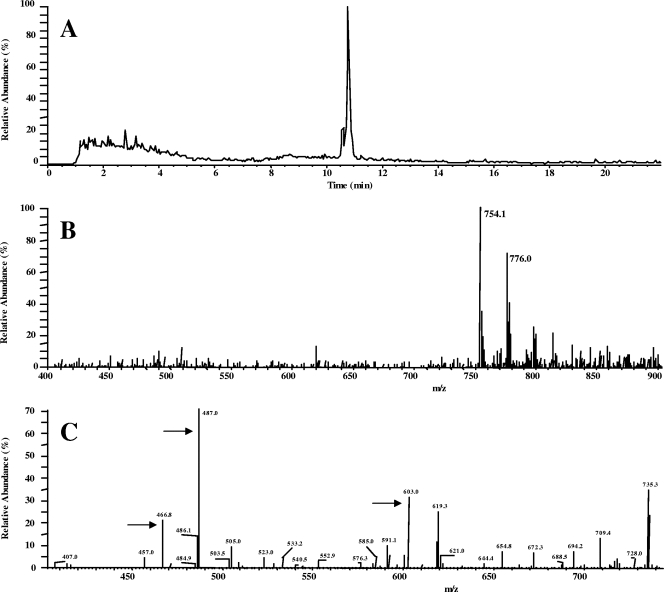
Inactivation of 200 μg of clindamycin per ml was obtained when crude extracts of E. coli AG100A/pUC18::lnu(D) were incubated with ATP and MgCl2 but not when cells were incubated in the absence of ATP. An HPLC-electrospray ionization chromatogram of the treated samples revealed a major peak eluted at 10.8 min (Fig. 3). MS analysis of this fraction revealed the dominance of a 754.1-atomic-mass-unit (amu) compound and its Na adduct (+22 amu), which is 329 Da heavier than the original clindamycin, in agreement with adenylylation of clindamycin. Adenylylation of clindamycin was ascertained by the CID-MS2 analysis. The precise site of nucleotidylation of clindamycin was not characterized.
FIG. 3.
HPLC-electrospray ionization MS analysis of inactivated clindamycin. (A) HPLC chromatogram showing a major peak eluting at 10.8 min. (B) Full MS analysis of the 10.8-min-eluted fraction reveals the dominance of a 754.1-amu compound and its Na adduct (+22 amu), which is 329 Da heavier than the original clindamycin. (C) Adenylylation of clindamycin was ascertained by the CID-MS2 analysis. Results are shown for fragments observed after CID. Fragments indicated by arrows resulted from leakage of the adenine or of the adenosine moiety of the compound.
Expression of lincosamide resistance.
The expression of the multicopy lnu(D) gene in E. coli AG100A resulted in high levels of resistance to both clindamycin and lincomycin (Table 1). In the original gram-positive host and in the E. coli transformant, both lincomycin and clindamycin were inactivated. However, resistance to lincomycin only was expressed in S. uberis and resistance to both lincomycin and clindamycin was expressed in E. coli. The reason for the difference in the resistance phenotypes according to the background remains unexplained. Hypothetically, the difference, which was also reported for the lnu(A) and lnu(C) genes (1, 6), might be related to differences in the relative affinities of clindamycin and lincomycin for the ribosomes of gram-positive and gram-negative organisms and for the Lnu(D) enzyme: clindamycin might have better affinity for the gram-positive ribosomes than for Lnu(D). It should be noted that the level of resistance to lincomycin conferred by lnu(D) in S. uberis is much lower than those conferred by lnu(A) in S. aureus and lnu(C) in S. agalactiae (MICs of lincomycin = 16 to 32 μg/ml). From a practical point of view, this may lead to difficulties in the detection of lincomycin resistance which is borderline. Furthermore, lincomycin resistance is misidentified when only clindamycin is tested. This is of particular importance for veterinary laboratories since lincomycin is used whereas clindamycin has not been approved for cows. The spread of lnu genes that confer resistance to lincomycin but not to clindamycin and pirlimycin in animal streptococci would lead to establishment of interpretive criteria for the testing of this antibiotic in veterinary laboratories.
The L phenotype has already spread in animal streptococci. In a previous study, Guérin-Faublée et al. demonstrated that 42% of S. uberis strains isolated from clinical mastitis in cows expressed resistance to lincomycin and that, among them, 33% were resistant to lincomycin while remaining susceptible to erythromycin (15). Further epidemiological studies, including detection of resistance genes, should be conducted to assess more accurately the prevalence of the various mechanisms of resistance to macrolides and lincosamides in streptococci isolated from animal infections.
Acknowledgments
We thank Michel Auzou for excellent technical assistance.
Footnotes
Published ahead of print on 17 December 2007.
REFERENCES
- 1.Achard, A., C. Villers, V. Pichereau, and R. Leclercq. 2005. New lnu(C) gene conferring resistance to lincomycin by nucleotidylation in Streptococcus agalactiae UCN36. Antimicrob. Agents Chemother. 49:2716-2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Almeida, R. A., and S. P. Oliver. 2006. Trafficking of Streptococcus uberis in bovine mammary epithelial cells. Microb. Pathog. 41:80-89. [DOI] [PubMed] [Google Scholar]
- 3.Reference deleted.
- 4.Bozdogan, B., L. Berrezouga, M. Kuo, D. Yurek, K. Farley, B. Stockman, and R. Leclercq. 1999. A new resistance gene, linB, conferring resistance to lincosamides by nucleotidylation in Enterococcus faecium HM1025. Antimicrob. Agents Chemother. 43:925-929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brisson-Noël, A., and P. Courvalin. 1986. Nucleotide sequence of gene linA encoding resistance to lincosamide in Staphylococcus haemolyticus. Gene 43:247-253. [DOI] [PubMed] [Google Scholar]
- 6.Brisson-Noël, A., P. Delrieu, D. Samain, and P. Courvalin. 1988. Inactivation of lincosaminide antibiotics in Staphylococcus. Identification of lincosaminide O-nucleotidyltransferases and comparison of the corresponding resistance genes. J. Biol. Chem. 263:15880-15887. [PubMed] [Google Scholar]
- 7.Comité de l'Antibiogramme de la Société Française de Microbiologie. February posting date. Communiqué 2006. http://www.sfm.asso.fr/.
- 8.de Azavedo, J. C., M. McGavin, C. Duncan, D. E. Low, and A. McGeer. 2001. Prevalence and mechanisms of macrolide resistance in invasive and noninvasive group B streptococcus isolates from Ontario, Canada. Antimicrob. Agents Chemother. 45:3504-3508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dutta, G. N., and L. A. Devriese. 1982. Resistance to macrolide, lincosamide and streptogramin antibiotics and degradation of lincosamide antibiotics in streptococci from bovine mastitis. J. Antimicrob. Chemother. 10:403-408. [DOI] [PubMed] [Google Scholar]
- 10.Ehrenfeld, E. E., and D. B. Clewell. 1987. Transfer functions of the Streptococcus faecalis plasmid pAD1: organization of plasmid DNA encoding response to sex pheromone. J. Bacteriol. 169:3473-3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Erskine, R. J., J. Cullor, M. Schaellibaum, B. Yancey, and A. Zecconi. 2004. Bovine mastitis pathogens and trends in resistance to antibacterial drugs. National Mastitis Council Research Committee Report. National Mastitis Council. http://www.nmconline.org/docs/ResPaper.pdf.
- 12.Erskine, R. J., S. Wagner, and F. J. de Graves. 2003. Mastitis therapy and pharmacology. Vet. Clin. North Am. Food Anim. Pract. 19:109-138. [DOI] [PubMed] [Google Scholar]
- 13.Erskine, R. J., R. D. Walker, C. A. Bolin, P. C. Bartlett, and D. G. White. 2002. Trends in antibacterial susceptibility of mastitis pathogens during a seven-year period. J. Dairy Sci. 85:1111-1118. [DOI] [PubMed] [Google Scholar]
- 14.Felsenstein, J. 1988. Phylogenies from molecular sequences: inference and reliability. Annu. Rev. Genet. 22:521-565. [DOI] [PubMed] [Google Scholar]
- 15.Guérin-Faublée, V., F. Tardy, C. Bouveron, and G. Carret. 2002. Antimicrobial susceptibility of Streptococcus species isolated from clinical mastitis in dairy cows. Int. J. Antimicrob. Agents 19:219-226. [DOI] [PubMed] [Google Scholar]
- 16.Heir, E., B. A. Lindstedt, T. M. Leegaard, E. Gjernes, and G. Kapperud. 2004. Prevalence and characterisation of integrons in blood culture Enterobacteriaceae and gastrointestinal Escherichia coli in Norway and reporting of a novel class 1 integron-located lincosamide resistance gene. Ann. Clin. Microbiol. Antimicrob. 3:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Horodniceanu, T., L. Bougueleret, N. El-Solh, D. H. Bouanchaud, and Y. A. Chabbert.. 1979. Conjugative R plasmids in Streptococcus agalactiae (group B). Plasmid 2:197-206. [DOI] [PubMed] [Google Scholar]
- 18.Loch, I. M., K. Glenn, and R. N. Zadoks. 2005. Macrolide and lincosamide resistance genes of environmental streptococci from bovine milk. Vet. Microbiol. 111:133-138. [DOI] [PubMed] [Google Scholar]
- 19.Luthje, P., and S. Schwarz.. 2006. Antimicrobial resistance of coagulase-negative staphylococci from bovine subclinical mastitis with particular reference to macrolide-lincosamide resistance phenotypes and genotypes. J. Antimicrob. Chemother. 57:966-969. [DOI] [PubMed] [Google Scholar]
- 20.Luthje, P., and S. Schwarz.. 2007. Molecular basis of resistance to macrolides and lincosamides among staphylococci and streptococci from various animal sources collected in the resistance monitoring program BfT-GermVet. Int. J. Antimicrob. Agents 29:528-535. [DOI] [PubMed] [Google Scholar]
- 21.Matthews, K. R., R. A. Almeida, and S. P. Oliver. 1994. Bovine mammary epithelial cell invasion by Streptococcus uberis. Infect. Immun. 62:5641-5646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Okusu, H., D. Ma, and H. Nikaido. 1996. AcrAB efflux pump plays a major role in the antibiotic resistance phenotype of Escherichia coli multiple-antibiotic-resistance (Mar) mutants. J. Bacteriol. 178:306-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rossitto, P. V., L. Ruiz, Y. Kikuchi, K. Glenn, K. Luiz, J. L. Watts, and J. S. Cullor. 2002. Antibiotic susceptibility patterns for environmental streptococci isolated from bovine mastitis in central California dairies. J. Dairy Sci. 85:132-138. [DOI] [PubMed] [Google Scholar]
- 24.Shaw, K. J., P. N. Rather, R. S. Hare, and G. H. Miller. 1993. Molecular genetics of aminoglycoside resistance genes and familial relationships of the aminoglycoside-modifying enzymes. Microbiol. Rev. 57:138-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tamilselvam, B., R. A. Almeida, J. R. Dunla, and S. P. Oliver. 2006. Streptococcus uberis internalizes and persists in bovine mammary epithelial cells. Microb. Pathog. 40:279-285. [DOI] [PubMed] [Google Scholar]
- 26.Thornsberry, C., J. K. Marler, J. L. Watts, and R. J. Yancey. 1993. Activity of pirlimycin against pathogens from cows with mastitis and recommendations for disk diffusion tests. Antimicrob. Agents Chemother. 37:1122-1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang, J., N. Shoemaker, G. R. Wang, and A. Salyers. 2000. Characterization of a Bacteroides mobilizable transposon, NBU2, which carries a functional lincomycin resistance gene. J. Bacteriol. 182:3559-3571. [DOI] [PMC free article] [PubMed] [Google Scholar]