Abstract
Photobacterium damselae subsp. piscicida is a bacterial fish pathogen that causes a disease known as pasteurellosis. Two transferable multiple-drug resistance (R) plasmids, pP99-018 (carrying resistance to kanamycin, chloramphenicol, tetracycline, and sulfonamide) and pP91278 (carrying resistance to tetracycline, trimethoprim, and sulfonamide), isolated from P. damselae subsp. piscicida strains from Japan (P99-018) and the United States (P91278), respectively, were completely sequenced and analyzed, along with the multiple-drug resistance regions of three other R plasmids also from P. damselae subsp. piscicida strains from Japan. The sequence structures of pP99-018 (150,057 bp) and pP91278 (131,520 bp) were highly conserved, with differences due to variation in the drug resistance and conjugative transfer regions. These plasmids, shown to be closely related to the IncJ element R391 (a conjugative, self-transmitting, integrating element, or constin), were divided into the conjugative transfer, replication, partition, and multiple-drug resistance regions. Each of the five multiple-drug resistance regions sequenced exhibited unique drug resistance marker composition and arrangement.
Antimicrobial drugs have historically been used to control bacterial and mycotic infections in fish farming (16). This practice has helped the industry improve production but at the same time allowed for the emergence of drug-resistant pathogens, which is known to be made possible by transferable R plasmids present in fish-pathogenic gram-positive and gram-negative bacteria (17). The R plasmids of Photobacterium damselae subsp. piscicida, a gram-negative pathogen, carry several drug resistance genes, also called drug resistance markers, that include a class A β-lactamase gene for ampicillin resistance (Apr), catI and catII for chloramphenicol resistance (Cpr), ppflo for florfenicol resistance (Ffr), aphA7 for kanamycin resistance (Kmr), sul2 for sulfonamide resistance (Sar), and tet(A) for tetracycline resistance (Tcr). Some of these drugs (ampicillin, florfenicol, sulfonamide, and tetracycline) are used in aquaculture in Japan, while others (chloramphenicol and kanamycin) are not (7).
P. damselae subsp. piscicida is the causative agent of pasteurellosis, a disease responsible for serious economic losses in fish farms across the world (17). However, even though the drug resistance patterns and genes contained in R plasmids from P. damselae subsp. piscicida strains in Japan, the United States, and Europe have been identified, the complete sequences of these R plasmids from P. damselae subsp. piscicida and the molecular mechanism responsible for the formation of resistance determinants in this species have not been well characterized.
In this study, we determined and analyzed the complete nucleotide sequences of R plasmids pP99-018 (Japan) and pP91278 (United States) from P. damselae subsp. piscicida, as well as the multiple-drug resistance regions of three other R plasmids (pSP98048, pSP98026, and pP9014) also collected in Japan. pP99-018 and pP91278 were shown to have a conserved backbone despite their geographic separation, differing only in the composition of their conjugation and multiple-drug resistance regions. The genes and regions that account for the mobility and drug resistance of the R plasmids studied here were similar to known genetic elements in other bacterial pathogens.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
Bacterial strains and plasmids and their sources are listed in Table 1. P. damselae subsp. piscicida strains were isolated from yellowtail (Seriola quinqueradiata) for the Japan samples and from moribund hybrid striped bass (Morone saxatilis × M. chrysops) for the U.S. samples. Heart infusion (Difco, Detroit, MI) supplemented with 1.5% sodium chloride was used for culture. Streptomycin-resistant (Smr) Escherichia coli HB101 cells were used as recipients of the R plasmids. Vector pBluescript II (Stratagene, CA) and E. coli JM109 were used to construct a random shotgun library. The medium for E. coli was 2× YT broth (1.6% tryptone, 1% yeast extract, and 0.5% NaCl). When necessary, the medium was supplemented with sulfamonomethoxine (a derivative of sulfonamide) (100 μg/ml), tetracycline (25 μg/ml), or ampicillin (100 μg/ml). The MICs of tetracycline, sulfamonomethoxine, and trimethoprim for strains carrying p91278 as determined by a standard twofold agar dilution method were 200, >800, and >800 μg/ml, respectively.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Description | Sourcea |
|---|---|---|
| Strains | ||
| E. coli | ||
| HB101 | supE44 Δ(mcrC-mrr) recA13 ara-14 proA2 lacY1 galK2 rpsL20 xyl-5 mtl-1 leuB6 thi-1 | Takara, Japan |
| JM109 | recA1 endA1 gyrA96 thi-1 hsdR17(rK− mK+) supE44 relA1 Δ(lac-proAB) [F′ traD36 proAB+lacIqlacZΔM15] | Takara, Japan |
| P. damselae subsp. piscicida | ||
| SP98048 | Wild type; Sar | This study (Ehime, 1998) |
| SP98026 | Wild type; Kmr Sar | This study (Ehime, 1998) |
| P9014 | Wild type; Apr Cpr Tcr | This study (Nagasaki, 1990) |
| P99-018 | Wild type; Kmr Cpr Tcr Sar | This study (Ehime, 1999) |
| P91278 | Wild type; Tcr Tmpr Sar | This study (United States, 1991) |
| Plasmids | ||
| pBluescript | Cloning vector (Apr) | Stratagene, United States |
| pGEM-T vector | Cloning vector (Apr) | Promega, United States |
Locations and years of isolation of P. damselae isolates are given in parentheses.
Plasmid DNA extraction and construction of random shotgun library.
The R plasmids of P. damselae subsp. piscicida were transferred into E. coli HB101 cells through mating experiments as previously described (7). The R plasmid DNA was extracted from the transconjugants by using alkaline lysis and purified by equilibrium centrifugation in a cesium chloride-ethidium bromide gradient. The purified plasmid DNA was fragmented by sonication. The sonicated DNA fragments were made blunt ended using a DNA blunting kit (Takara, Ohtsu, Japan) and fractionated by 1% agarose gel electrophoresis. DNA fragments 2 to 3 kb in length were isolated from gel. After dephosphorylation, fragments were ligated to the vector pBluecript II SK(−) and E. coli JM109 was transformed with the fragments by electrotransformation. White clones containing the inserts were randomly selected and amplified using a standard PCR method for DNA sequencing.
DNA sequencing.
pP99-018 and pP91278 clones were sequenced using a Thermosequenase sequencing kit (Amersham-Biotech, Piscataway, NJ) with automated DNA sequencer LC4200 (Li-Cor, Lincoln, NE). The final gaps were closed by direct sequencing of the products amplified by long accurate PCR with an LA PCR kit, version 2 (Takara, Ohtsu, Japan). Primers were designed from the ends of the assembled sequence. pGEM-T vector (Promega, Madison, WI) and JM109 were used for transformation. On the other hand, pSP98048, pSP98026, and pP9014 multiple-drug resistance region sequences were obtained by primer walking using primers designed from pP99-018. Plasmid DNA and deduced amino acid sequences were determined and analyzed with GENETYX version 7.0. The open reading frame (ORF) finder system (http://www.ncbi.nlm.nih.gov/gorf/gorf.html) was used to predict ORFs. The ORFs were compared to sequences in the public sequence databases of GenBank by using the BLAST algorithm.
Annotation and analysis of sequence data.
GENETYX version 7.0 (Software Development Co. Ltd., Tokyo, Japan) was used to align and assemble the sequences, while the predicted protein-encoding regions were initially defined by searching for ORFs longer than 50 codons by using the GAMBLER software (14). Searches of the protein databases for amino acid similarities were performed using the BLAST program. The G+C plot was calculated by again using GENETYX version 7.0, with the following parameters: a window of 1,000 nucleotides and a step size of 100 nucleotides.
Restriction analysis.
The extracted R plasmids pSP98048, pSP98026, pP9014, and pP99-018 were digested with PstI and electrophoresed in a 0.8% agarose gel containing ethidium bromide. Resulting bands were photographed with a densitometer (Atto).
Nucleotide sequence accession numbers.
The nucleotide sequences of pP99-018 and pP91278 have been deposited in the DDBJ, GenBank, and EMBL databases, under accession numbers AB277723 and AB277724, respectively, as well as those for the multiple-drug resistance regions of pSP98048 (AB305618), pSP98026 (AB305619), and pP9014 (AB305620).
RESULTS
Overview of pP99-018 from Japan.
The circular plasmid pP99-018 from a P. damselae subsp. piscicida isolate from Japan was 150,057 bp in length and contained 187 putative ORFs (see Fig. S1 and Table S1 in the supplemental material). A total of 187 ORFs initiated by 165 putative ATG, 11 GTG, and 11 TTG start codons were identified. The products of 78 of the ORFs (41.7%) showed significant similarity (80 to 100%) to protein sequences in the databases, while another 42 putative proteins (22.5%) were similar (<50%) to hypothetical proteins of unknown function. Putative genes of pP99-018 predicted to code for partition, replication, and conjugative transfer as backbone functions and Cpr, Kmr, Tcr, and Sar genes were identified, as were a number of transposable elements. Sixty-two putative ORFs had no significant identity (<50%) with sequences in GenBank. The percentage of unknown genes (35.0%) of pP99-018 was quite large compared to those of other previously described R plasmids. The overall G+C content was 51.4% (see Fig. S3 in the supplemental material).
Overview of pP91278 from the United States.
pP91278 was 130,520 bp in length, 19,537 bp smaller than pP99-018. The two plasmids were >99% identical if the additional resistance determinants and conjugative transfer region in pP99-018 were not considered (see Fig. S2 and Table S2 in the supplemental material). A total of 160 ORFs initiated by 140 ATG, 11 GTG, and 9 TTG putative start codons were identified (see Table S2 in the supplemental material). The products of 59 of these ORFs (36.9%) exhibited no significant similarity (80 to 100%) to protein sequences in the databases, while another 36 putative proteins (22.5%) had similarity (<50%) to hypothetical proteins of unknown function. This result suggests that pP91278, like pP99-018, encodes proteins with novel functions. Search results revealed putative genes which code for the backbone functions of pP91278 (replication, partition, and conjugative transfer) and the phenotypic traits of Sar, Tcr, and Tmpr. The overall G+C content of pP91278 was 51.7%, almost identical to the 51.4% of pP99-018 (see Fig. S3 in the supplemental material). From the G+C profiles, differences around the regions of the transposable elements and the conjugative transfer elements and in the multiple-drug resistance regions can be observed.
Regions of pP99-018 and pP91278.
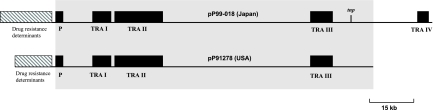
pP99-018 (Japan) and pP91278 (United States) sequences were almost identical, minus the additional resistance determinants, a 1-kb transposase gene (located at bases 126344 to 125250), and conjugative transfer region IV (TRA IV) in pP99-018 (Fig. 1).
FIG. 1.
Comparison of DNA sequences of R plasmids pP99-018 and pP91278. Regions of plasmids are indicated as drug resistance determinants (multiple-drug resistance regions); P (partition region); TRAs (transfer regions); and tnp (transposase gene), found only in pP99-018. Conserved regions are shown by gray shading.
(i) Replication.
Both plasmids consisted of an ORF (ORF140 in pP99-018 and ORF131 in pP91278) encoding a protein that was 98% homologous to the replication initiation, or RepA, protein of pRA1 from Aeromonas hydrophila. Downstream of the RepA gene was a DnaA box for the binding of host DnaA initiator protein. Another ORF (ORF123 in pP99-018 and ORF115 in pP91278) was found to be similar (44%) to a Ter binding protein gene, the Tus gene of Rts1 of Proteus vulgaris for replication termination.
(ii) Partitioning.
The product of one ORF (ORF29 in pP99-018 and ORF21 in pP91278) showed 33% similarity to partition protein A, or IncC, and the product of another ORF (ORF30 in pP99-018 and ORF22 in pP91278) showed 30% similarity to the transcriptional repressor protein encoded by korB of Aplysia punctata pFBAOT6. The incC and korB genes were homologous to genes reported to encode the nucleoprotein-forming complex ParA and ParB proteins. Another ORF (ORF124 in pP99-018 and ORF116 in pP91278) immediately downstream of the Ter binding protein gene was similar to the plasmid-partitioning kfrA gene of Pseudomonas sp. strain pADP-1.
(iii) Conjugative transfer.
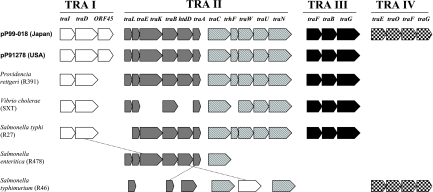
R plasmid pP99-018 contained four conjugative transfer regions (TRA I to IV) involving 21 transfer genes, while pP91278 had only three such regions (TRA I to III) involving 17 transfer genes (Fig. 2). The first cluster, TRA I, was about 6 kb and encoded three putative transfer proteins: TraI, with 51% homology to TraI of Providencia rettgeri; TraD, with 65% homology to TraD of Providencia rettgeri; and a putative conjugation factor that was 43% similar to the product of the SXT element of Vibrio cholerae. TRA II was separated from TRA I by four ORFs of unknown function and was about 20 kb. The TRA II cluster was composed of 11 ORFs in two subclusters separated only by one ORF for a hypothetical protein of unknown function (ORF64 in pP99-018 and ORF56 in pP91278) and an ORF similar to the dsbC gene of R391. The first subcluster was similar to the TRA II region of R391 in having traL, traE, traK, traB, htdD, and traA and to the TRA III region of R391, which has traC, trhF, traW, traU, and traN. The third cluster, TRA III, was about 6 kb and was located between the replication genes for Tus and RepA. TRA III included three genes which were homologous to traF, traH, and traG of R391 and R27. Interestingly, an additional transfer region (TRA IV) was present in pP99-018 compared to pP91278, separated from TRA III by a 7-kb region comprising 10 putative ORFs (ORF172 to ORF181), 9 of which encoded products with no significant homology to protein sequences in GenBank. TRA IV was composed of ORFs similar to the traE, traO, traF, and traG genes of R46 of Salmonella enterica serovar Typhimurium, with 97, 97, 98, and 100% similarity, respectively.
FIG. 2.
Relationship between the putative transfer genes of pP99-018, pP91278, and homologs present in related plasmid and conjugative mobile elements. The names of the transfer genes and transfer regions are indicated. Genes are transcribed in the direction indicated by the respective arrows. Dotted lines show homologous genes. Not drawn to scale.
(iv) Multiple-drug resistance.
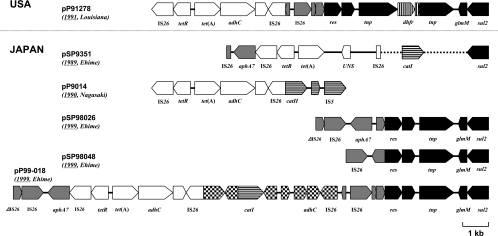
The region consisting of the resistance determinants of pP99-018, measuring about 19 kb, had a number of transposable elements carrying Kmr (ORF3), Tcr (ORF6), Cpr (ORF12), and Sar (ORF26) genes (arranged in the plasmid in this order), namely, aphA7, tet(A), catI, and sul2 genes, respectively (Fig. 3; see also Table S1 in the supplemental material). On the other hand, the pP91278 multiple-drug resistance region was about 13 kb and possessed ORFs for genes tet(A) (ORF3), dhfr (ORF14), and sul2 (ORF18), encoding Tcr, Tmpr, and Sar, respectively (see Table S2 in the supplemental material). The tet(A) and sul2 genes and their transposon-like structures were identical to those in pP99-018. However, dhfr was unique to pP91278 and had 53% similarity to a Pseudomonas syringae pv. syringae B728a chromosomal dihydrofolate reductase gene that encodes Tmpr. dhfr, unlike tet(A), was flanked by transposase genes rather than insertion sequence 26 (IS26).
FIG. 3.
Comparison of the multiple-drug resistance regions of R plasmids from Japan (pSP9351, pSP98048, pSP98026, pP9014, and pP99-018) and U.S. (pP91278) isolates. Broken lines represent unknown sequences.
Multiple-drug resistance regions of pSP98048, pSP98026, and pP9014.
The lengths of the multiple-drug resistance regions of pSP98048, pSP98026, and pP9014 were 6,493, 7,896, and 7,537 bp, respectively (Fig. 3; see also Tables S3 to S5 in the supplemental material). pSP98048 carries the Sar marker gene sul2 flanked by 6-bp direct repeats (DRs; 5′-GGACGC-3′). Between pSP98048 and pP99-018, there were two stretches of 100% sequence identity, bases 1 to 149701 and 12947 to 19001. In the pP99-018 sequence, the Cpr, Kmr, and Tcr determinants were located between 17-bp DRs (5′-GGCACTGTTGCAAATAGT-3′). pSP98026 is composed of Kmr and Sar genes, identified as aphA7 and sul2, respectively. The sequences of pSP98026 and pSP9808 were identical except for the Kmr region. The Kmr region was located between two regions homologous to those in pSP98048 and was bordered by two truncated genes and flanked by the 6-bp DRs (5′-CCCCTT). A complete copy of IS26 lay downstream of the Kmr determinant location. pP9014 comprised the Cpr and Tcr determinants, identified as catII and tet(D), respectively. catII was found at the 3′ end of IS5, downstream of the Tcr region. The supposed Apr determinant for this plasmid, the class A β-lactamase gene, was undetected in the sequenced region.
Phenotypes of R plasmids.
Of the R plasmids pSP98048, pSP98026, pP9014, and pP99-018, pP9014 showed the most different PstI restriction patterns (Fig. 4).
FIG. 4.
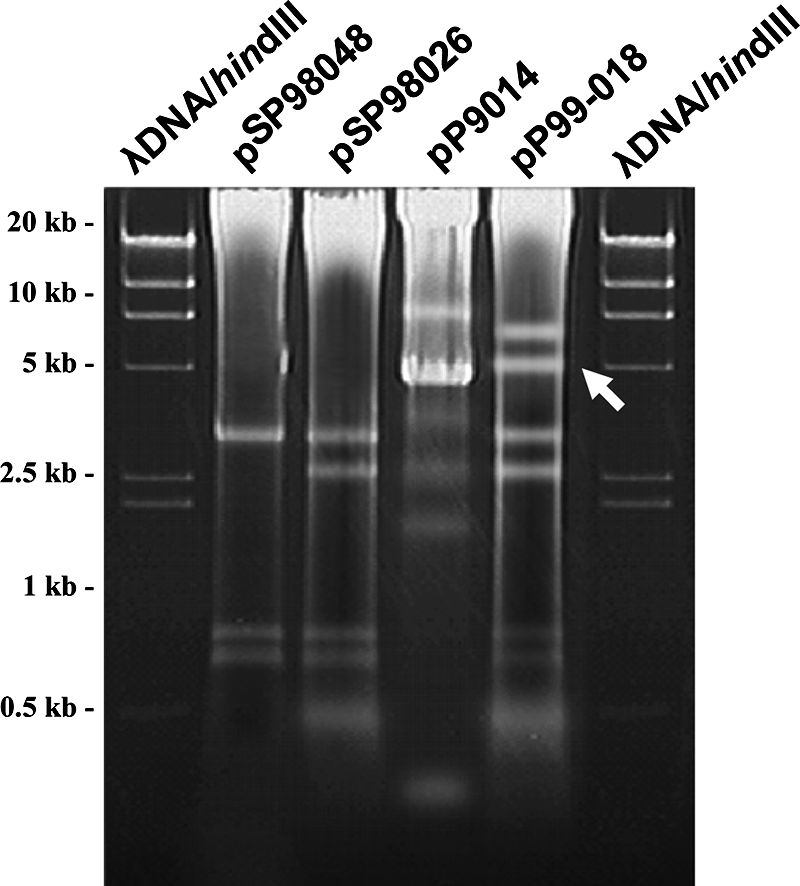
Electrophoretic profiles of four transferable R plasmids from P. damselae subsp. piscicida strains from Ehime and Kyushu, Japan, following PstI restriction. The white arrow points to the location of the multiple-drug resistance region in pP99-018.
DISCUSSION
P. damselae subsp. piscicida has been divided into two different clonal lineage groups, regardless of the host species: one group includes the U.S. and Japan strains, while the other group includes the European strains (5, 11, 12). The European and U.S. strains share the same 20- and 7-MDa plasmids, but the Japanese strains did not have these plasmids. Surveys done in Japan showed that the combination of Cpr, Kmr, Sar, and Tcr is the major resistance marker for this bacterial species (7).
The conjugative transfer regions of pP99-018 (from Japan), pP91278 (from the United States), R391 (from a South African Providencia rettgeri strain) (3), R27 (from a Salmonella enterica serovar Typhi strain from England) (15), SXT (from Vibrio cholerae) (1), and R478 (from Serratia marcescens) (4) were related based on their conserved backbone. This backbone, particularly its functional genes, has been shown to be responsible for conjugative transfers and the regulation and excision and integration of elements and, thus, allows for the horizontal transfer of DNA elements, making a wide range of bacteria adaptable to the changing environments. Based on nucleotide analysis, R391 is a conjugative integrating mosaic made up of elements from bacteriophages and plasmids and of transposable elements and is capable of site-specific integration into the bacterial chromosome by a phage-like integration system (3). Thus, R391 has been classified into a new group of constins (conjugative, self-transmitting, integrating elements), as opposed to being a conjugative transposon. Previously, R391 and other IncJ class mobile genetic elements were assumed to be plasmids, but this idea was later challenged because they do not possess a replicon for autonomous replication. R plasmids from P. damselae subsp. piscicida could be classified as a constins because of their high degree of similarity to R391, but the finding that they possess a replication region composed of genes responsible for DNA synthesis confirmed them to be plasmids. The classification of R plasmids in P. damselae subsp. piscicida is therefore in need of further confirmation. The absence of the long separating region between the subclusters of TRA II of pP99-018 and pP91278 compared to those of R391 is particularly interesting, and the reason for this difference is unclear.
The additional transfer region (TRA IV) in pP99-018 appeared to have been acquired through transposition because it was accompanied by a transposase gene, tnp. The presence of the extra region, consisting of four mobility-related genes and other unknown genes, may be a source of new plasmid function in pP99-018 and, therefore, warrants further studies.
In the multiple-drug resistance regions of the R plasmids of P. damselae subsp. piscicida, the Kmr, Tcr, and Cpr genes were flanked by IS26, making them class I transposons, while the Sar and Tmpr genes were bordered by transposase genes, suggesting that they are class II transposons. The genetic element of a similar, previously described plasmid, pSP9351 (with the structure IS26-Kmr gene-IS26-Tcr gene-IS26-Sar gene-IS26), is regarded as a complex transposon and exhibits a functional hybrid promoter made up of the transposable element IS26 (6, 8, 9). Direct evidence shows that IS26 provides part of the promoter of the antibiotic resistance operon (IAB) of pBWH77 and has an active role in the evolution and mobility of antibiotic resistance genes (10). Furthermore, insertion sequences, together with transposase, have been suggested to be responsible for a two-step, reactive junction pathway in bacterial transpositional recombination (2). Hence, it is likely that IS26 and the transposases promote DNA rearrangements in the R plasmid of P. damselae subsp. piscicida and that IS26 is the principal modulator of the evolution of drug resistance in this bacterial species. In addition, we found that the Tmpr gene dhfr, found in the U.S. isolates but not the Japan isolates, confirmed the Tmpr phenotype of the organism. Since pP91278 is resistant to trimethoprim, pP91278 can be classified as a type I R-plasmid dihydrofolate reductase instead of type II, which are practically insensitive to trimethoprim, methotrexate, and aminopterin (13, 18). Furthermore, the 746-bp segment containing dhfr had a lower G+C content (46.6%) than the overall G+C content of pP91278, suggesting that dhfr is not an indigenous pP91278 gene. These data suggest clearly that insertion sequence elements and transposase are deeply involved in the exchange or acquisition of resistance determinants in P. damselae subsp. piscicida.
Based on the restriction profiles, pP9014, isolated in Nagasaki, Japan, in 1990, appears to have a backbone structure different from that of pP99108, pSP98048, and pSP98026, all collected in 1999 in Ehime, Japan. The Apr determinant class A β-lactamase gene was also not detectable in the multiple-drug resistance region sequenced, even though it is clear that the strain is Apr based on results from transfer experiments (data not shown). Taken together, these data suggest that pP9014 is of a different R-plasmid type not related to the four other plasmids in this study.
With the complete DNA sequences of two R plasmids of P. damselae subsp. piscicida known, several downstream studies are possible to elucidate further the molecular interaction between P. damselae subsp. piscicida and the host. For example, DNA microarray chips could now be made to study the gene expression profile of the pathogen's R plasmid during infection.
Supplementary Material
Acknowledgments
This research was supported in part by the Research and Education Program for Seafood Safety Project: Special Fund for Education and Research from MEXT (Ministry of Education, Culture, Sports, Science and Technology of Japan) and grants-in-aid from the Food Safety Commission, Japan.
Footnotes
Published ahead of print on 10 December 2007.
Supplemental material for this article may be found at http://aac.asm.org/.
REFERENCES
- 1.Beaber, J. W., B. Hochhut, and M. K. Waldor. 2002. Genomic and functional analyses of SXT, an integrating antibiotic resistance gene transfer element derived from Vibrio cholerae. J. Bacteriol. 184:4259-4269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berger, B., and D. Haas. 2001. Transposase and cointegrase: specialized transposition proteins of the bacterial insertion sequence IS21 and related elements. Cell. Mol. Life Sci. 58:403-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Böltner, D., C. MacMahon, J. T. Pembroke, P. Strike, and A. M. Osborn. 2002. R391: a conjugative integrating mosaic comprised of phage, plasmid, and transposon elements. J. Bacteriol. 184:5158-5169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gilmour, M. W., N. R. Thomson, M. Sanders, J. Parkhill, and D. E. Taylor. 2004. The complete nucleotide sequence of the resistance plasmid R478: defining the backbone components of incompatibility group H conjugative plasmids through comparative genomics. Plasmid 52:182-202. [DOI] [PubMed] [Google Scholar]
- 5.Hawke, J. P., R. L. Thune, R. K. Cooper, E. Judice, and M. Kelly-Smith. 2003. Molecular and phylogenetic characterization of strains of Photobacterium damselae subsp. piscicida isolated from hybrid striped bass cultured in Louisiana, USA. J. Aquat. Anim. Health 5:189-201. [Google Scholar]
- 6.Kim, E. H., and T. Aoki. 1993. The structure of the chloramphenicol resistance gene on a transferable R plasmid from the fish pathogen, Pasteurella piscicida. Microbiol. Immunol. 37:705-712. [DOI] [PubMed] [Google Scholar]
- 7.Kim, E. H., and T. Aoki. 1993. Drug resistance and broad geographical distribution of identical R plasmids of Pasteurella piscicida isolated from cultured yellowtail in Japan. Microbiol. Immunol. 37:103-109. [DOI] [PubMed] [Google Scholar]
- 8.Kim, E. H., and T. Aoki. 1994. The transposon-like structure of IS26-tetracycline, and kanamycin resistance determinant derived from transferable R plasmid of fish pathogen, Pasteurella piscicida. Microbiol. Immunol. 38:31-38. [DOI] [PubMed] [Google Scholar]
- 9.Kim, E. H., and T. Aoki. 1996. Sulfonamide resistance gene in a transferable R plasmid of Pasteurella piscicida. Microbiol. Immunol. 40:397-399. [DOI] [PubMed] [Google Scholar]
- 10.Lee, K. Y., J. D. Hopkins, and M. Syvanen. 1990. Direct involvement of IS26 in an antibiotic resistance operon. J. Bacteriol. 172:3229-3236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Magariños, B., J. L. Romalde, I. Bandín, B. Fouz, and E. Toranzo. 1992. Phentoypic, antigenic and molecular characterization of Pasteurella piscicida strains isolated from fish. Appl. Environ. Microbiol. 58:3316-3322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Magariños, B., A. E. Toranzo, J. L. Barja, and J. L. Romalde. 2000. Existence of two geographically-linked clonal lineages in the bacterial fish pathogen Photobacterium damselae subsp. piscicida evidenced by random amplified polymorphic DNA analysis. Epidemiol. Infect. 125:213-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Patishall, K. H., J. Acar, J. J. Burchall, F. W. Goldstein, and R. J. Harvey. 1977. Two distinct types of trimethoprim-resistant dihydrofolate reductase specified by R-plasmids of different compatibility groups. J. Biol. Chem. 252:2319-2323. [PubMed] [Google Scholar]
- 14.Sakiyama, T., H. Takami, N. Ogasawara, S. Kuhara, T. Kozuki, K. Doga, A. Ohyama, and K. Horikoshi. 2000. An automated system for genome analysis to support microbial whole-genome shotgun sequencing. Biosci. Biotechnol. Biochem. 64:670-673. [DOI] [PubMed] [Google Scholar]
- 15.Sherburne, C. K., T. D. Lawley, M. W. Gilmour, F. R. Blattner, W. Burland, E. Grotbeck, D. J. Rose, and D. E. Taylor. 2000. The complete DNA sequence and analysis of R27, a large IncHI plasmid from Salmonella typhi that is temperature sensitive for transfer. Nucleic Acids Res. 28:2177-2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Siniesko, S. F. 1959. Antibiotics in fish diseases and fish nutrition. Antibiot. Chemother. 9:541-545. [PubMed] [Google Scholar]
- 17.Sørum, H. 2006. Antimicrobial drug resistance in fish pathogens. In F. M. Aarestrup (ed.), Antimicrobial resistance in bacteria of animal origin. ASM Press, Washington, DC.
- 18.Sundstrom, L., C. Jansson, K. Bremer, E. Heikkila, B. Olsson-Liljequist, and O. Skold. 1995. A new dhfrVIII trimethoprim-resistance gene, flanked by IS26, whose product is remote from other dihydrofolate reductases in parsimony analysis. Gene 154:7-14. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.