Abstract
The acyclic nucleoside phosphonate drug (S)-9-[3-hydroxy-(2-phosphonomethoxy)propyl]adenine [(S)-HPMPA], is a broad-spectrum antiviral and antiparasitic agent. Previous work has shown that the active intracellular metabolite of this compound, (S)-HPMPA diphosphate [(S)-HPMPApp], is an analog of dATP and targets DNA polymerases. However, the mechanism by which (S)-HPMPA inhibits DNA polymerases remains elusive. Using vaccinia virus as a model system, we have previously shown that cidofovir diphosphate (CDVpp), an analog of dCTP and a related antiviral agent, is a poor substrate for the vaccinia virus DNA polymerase and acts to inhibit primer extension and block 3′-to-5′ proofreading exonuclease activity. Based on structural similarities and the greater antiviral efficacy of (S)-HPMPA, we predicted that (S)-HPMPApp would have a similar, but more pronounced effect on vaccinia polymerase than CDVpp. Interestingly, we found that (S)-HPMPApp is a good substrate for the viral enzyme, exhibiting Km and Vmax parameters comparable to those of dATP, and certainly not behaving like CDVpp as a functional chain terminator. Metabolic experiments indicated that (S)-HPMPA is converted to (S)-HPMPApp to a much greater extent than CDV is converted to CDVpp, although both drugs cause identical effects on virus DNA replication at their 50% effective concentration. Subsequent studies showed that both compounds can be faithfully incorporated into DNA, but when CDV and (S)-HPMPA are incorporated into the template strand, both strongly inhibit trans-lesion DNA synthesis. It thus appears that nucleoside phosphonate drugs exhibit at least two different effects on DNA polymerases depending upon in what form the enzyme encounters the drug.
(S)-9-[3-Hydroxy-(2-phosphonomethoxy)propyl]adenine [(S)- HPMPA] is an acyclic analog of dAMP and was the first of the nucleoside phosphonate drugs described in the research literature (11). (S)-HPMPA was first shown to exhibit activity against a range of DNA viruses, including herpesviruses, vaccinia virus, and adenovirus, as well as Moloney murine sarcoma retrovirus (11). Later work has shown that it is also effective against hepatitis B viruses (42, 43). Although the drug did not show any activity against human immunodeficiency virus in its original formulation (30), Hostetler et al. have recently shown that the alkoxyalkyl ester derivatives are active against this virus, as well as against mutant viruses resistant to azidothymidine, lamivudine, tenofovir, and nevirapine (14). The antimicrobial range of (S)-HPMPA also includes parasites such as trypanosomes (17, 18), Schistosoma mansoni (7), and Plasmodium falciparum and Plasmodium berghei (13, 34).
(S)-HPMPA is taken up into cells by endocytosis (29) and converted into the active metabolite (S)-HPMPA diphosphate [(S)-HPMPApp] by cellular kinases (25). (S)-HPMPApp is an analog of dATP, and different studies have described a variety of cytotoxic effects at higher drug doses, which are most likely caused by the inhibition of cellular DNA replication (8, 38, 39). Rat cell DNA polymerases α, δ, and ɛ can use (S)-HPMPApp as a substrate and incorporate two to four consecutive (S)-HPMPA molecules into a growing DNA strand (6, 21). More specifically (S)-HPMPApp is a strong inhibitor of DNA polymerase ɛ (Ki/Km = 0.07), a moderate inhibitor of DNA polymerase δ (Ki/Km = 0.25) and a weak inhibitor of DNA polymerase α (Ki/Km = 2.29) (21), but is not an inhibitor of DNA polymerase β (26). Interestingly, both DNA polymerases δ and ɛ can still excise (S)-HPMPA from a primer terminus, with polymerase ɛ showing more effective removal of the drug (6).
Similar studies have examined the effects of (S)-HPMPApp on parasitic and viral DNA synthesis. (S)-HPMPA inhibits the replication of human and duck hepatitis B viruses (42, 43) and herpes simplex virus type 1 (12). Herpes simplex virus DNA polymerase can use (S)-HPMPApp as a substitute for dATP, but curiously (S)-HPMPApp is a poor inhibitor of the enzyme itself (26). Adenovirus DNA polymerases are inhibited by (S)-HPMPApp, which causes a block in replication at the level of DNA elongation (28). Trypanosomal DNA replication is also inhibited by (S)-HPMPA (16), but although P. falciparum DNA polymerases α and γ are inhibited by (S)-HPMPApp, the in vivo target of the drug appears to be polymerase δ (13, 35).
These studies suggest that (S)-HPMPApp affects different polymerases in different ways, but the mechanism linking the effects on DNA synthesis to a cytotoxic, antiviral, or antiparasitic effect is not well understood. To address this question, we have chosen to examine the antiviral effects of nucleoside phosphonate drugs using vaccinia virus and vaccinia DNA polymerase as a model system. Orthopoxviruses are acutely sensitive to nucleoside phosphonate drugs, and it has been suggested they might prove useful for treating renascent smallpox (3, 19). Vaccinia polymerase is a B-family DNA polymerase (15) and possesses both 5′-to-3′ polymerase and 3′-to-5′ exonuclease activities (9). We have previously shown that when a related compound, cidofovir [(S)-1-(3-hydroxy-2-phosphonylmethoxypropyl)cytosine] [(S)-HPMPC] diphosphate (CDVpp), is incorporated into DNA it inhibits both primer extension and drug excision by vaccinia DNA polymerase (23). (S)-HPMPA and CDV differ only in the structure of the base moiety, so we hypothesized that the two compounds would have a similar mechanism of inhibition. (S)-HPMPA has been shown to be more toxic in mice than CDV (8), but subsequent work has yielded conflicting results on the relative cytotoxicity and therefore selectivity indices between the two drugs (3, 5, 19, 22, 36). The latter results are most likely due to differences in the sensitivity of the various cells lines used to (S)-HPMPA and CDV. (S)-HPMPA shows greater efficacy against orthopoxviruses in culture (12, 19, 22, 36) and the hexadecyloxypropyl (HDP) ester of (S)-HPMPA is 80-fold more active than the HDP-ester of CDV (5, 20). Therefore, we predicted that (S)-HPMPA would have a more profound effect on these aspects of enzyme activity than does CDV. Oddly, (S)- HPMPApp is not as good an inhibitor of primer extension as is CDVpp. We demonstrate that both drugs, when incorporated into the template strand, cause a profound block in DNA elongation. These results show that CDV and (S)-HPMPA are more complex drugs than has been previously recognized, affecting DNA elongation when in both the primer and template strands, and blocking 3′-to-5′ exonuclease activity when located in the primer strand.
MATERIALS AND METHODS
Chemicals.
(S)-HPMPApp and CDVpp were prepared by custom synthesis by TriLink Biotechnologies. (S)-HPMPA was a gift from G. Andrei, Rega Institute for Medical Research, Leuven, Belgium. CDV was obtained from Moravek Biochemicals. Radiolabeled cordycepin triphosphate ([α-32P]3′-deoxyATP) was purchased from Perkin-Elmer, [γ-32P]ATP and [α-32P]dATP were obtained from GE Healthcare, and the unlabeled deoxynucleoside triphosphates (dNTPs) were from Fermentas. Oligonucleotides were purchased from Sigma-Genosys or Integrated DNA Technologies. [2-14C]CDV (56 mCi/mmol), [8-14C]-(S)-HPMPA (57 mCi/mmol), and their alkoxyester derivatives HDP-[2-14C] CDV (50 mCi/mmol) and HDP-[8-14C] (S)-HPMPA (50 mCi/mmol) were synthesized by Moravek Biochemicals using unlabeled intermediates and methods provided by James R. Beadle as previously described (5).
Enzymes.
Vaccinia virus DNA polymerase was purified from cells coinfected with recombinant vaccinia vTMPOL and vTF7.5 viruses as described previously (24). The enzyme was freshly diluted in polymerase dilution buffer (25 mM potassium phosphate [pH 7.4], 5 mM β-mercaptoethanol, 1 mM EDTA, 10% [vol/vol] glycerol, 0.1 mg of bovine serum albumin/ml) prior to use. T4 polynucleotide kinase, the Klenow fragment of DNA polymerase I, uracil-DNA glycosylase (UDG), and terminal deoxynucleotidyl transferase (TdT) were purchased from Fermentas. Moloney murine leukemia virus (MMLV) reverse transcriptase was obtained from Invitrogen.
Cells and virus.
All cells and virus were obtained from the American Type Culture Collection. MRC-5 human lung fibroblasts were grown in minimal essential medium with Earle's salts containing 2% fetal bovine serum. BSC40 African green monkey kidney epithelial cells and vaccinia virus (Western Reserve [WR] strain) were cultured in minimal media containing 5% fetal calf serum, 1% amino acids, 1% l-glutamine, and 1% antibiotic/antimycotic. Cells were maintained at 37°C in a 5% CO2 atmosphere.
Plaque reduction assays.
Plaque reduction assays were performed in triplicate using 200 PFU of virus per 60-mm dish. Virus-infected BSC40 cells were cultured for 48 h and then stained with crystal violet. The 50% effective concentration (EC50) was calculated from a nonlinear curve fit by using Prism 4.0b software.
Slot blot hybridization.
BSC40 cells were infected with vaccinia virus at a multiplicity of infection of 10 in 60-mm dishes. The cells were then incubated at 37°C and harvested at the times indicated by scraping and centrifugation. [To examine the effects of drugs on virus replication, cells were preincubated with (S)-HPMPA or CDV for 24 h prior to infection, and then more drug was added after the virus was added.] The cell pellets were washed and resuspended in 1.5 ml of 10× saline sodium citrate containing 1 M ammonium acetate and stored at −80°C. The samples were frozen and thawed three times and clarified by centrifugation, and then 25-μl aliquots were mixed with an equal volume of 0.8 M NaOH plus 20 mM EDTA, boiled for 10 min, cooled on ice, and diluted with 125 μl of 0.4 M NaOH and 10 mM EDTA. The samples were applied in duplicate to a Zeta-Probe membrane (Bio-Rad) by using a vacuum manifold, washed, and immobilized with UV light. A 3.1-kb probe spanning the DNA polymerase gene was prepared by using the PCR (2, 41), purified, and labeled with [α-32P]dATP using a random priming labeling kit (Roche). The membrane was processed by using a Southern blot hybridization procedure (31) and label detected by using a Typhoon phosphorimager.
DNA polymerase and exonuclease assays.
Oligonucleotide primer-template pairs (Fig. 1) were used as substrates for DNA polymerase and exonuclease assays as previously described (23). The primers were first end labeled by using [γ-32P]ATP and T4 polynucleotide kinase. Reaction products were resolved on 10 to 15% polyacrylamide gels and also analyzed as described previously (23). To determine the Km and Vmax values for (S)-HPMPApp, we prepared 10-μl reaction mixtures containing ∼9 pmol of 32P-labeled primer P1, 35 pmol of template T11, 25 ng of vaccinia polymerase, 10 μM dGTP, various concentrations of dATP [or (S)-HPMPApp], and polymerase buffer [30 mM Tris-HCl (pH 7.9), 5 mM MgCl2, 70 mM NaCl, 1.8 mM dithiothreitol, 80 μg of bovine serum albumin/ml]. The reaction products were separated on 15% polyacrylamide gels and analyzed using phosphorimaging. To determine the fidelity of drug incorporation, 10-μl reactions were prepared containing 1 pmol of 5′-32P-end-labeled primer P1, 3 pmol of template DNA (templates T9 or T19 to T25, Fig. 1), 10 μM CDVpp or (S)-HPMPApp, buffer, and 25 ng of vaccinia virus DNA polymerase. In controls, dCTP and dATP were substituted for CDVpp and (S)-HPMPApp, respectively. Reactions were incubated for 1 min at 37°C for CDVpp and dCTP reactions and at 25°C for (S)-HPMPApp and dATP reactions and stopped by adding 5 μl of gel loading buffer.
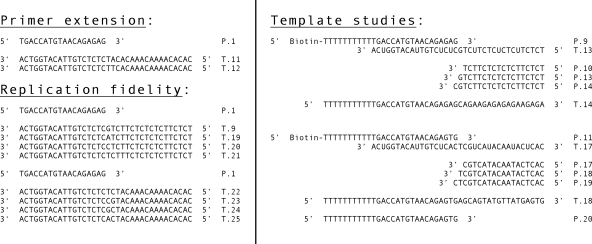
FIG. 1.
Oligonucleotide primer-template pairs used in the present study. Primer P1 was originally described by Xiong et al. (40). The primer DNA was 5′ end labeled with [γ-32P]ATP and polynucleotide kinase prior to annealing with the template DNA.
Preparation of CDV- and (S)-HPMPA-containing templates.
CDV-containing templates were prepared by using a two-step procedure. First, ∼25 pmol of primer P9 was annealed to a threefold excess of the deoxyuridine-containing template T13 (Fig. 1), followed by incubation at 37°C for 5 min in a reaction containing 25 μM (each) CDVpp (or dCTP), dATP, and dGTP and 2.5 ng vaccinia virus DNA polymerase/μl in polymerase buffer. The reactions were stopped by adding EDTA (to 45 μM), and the DNA was purified by using chloroform extraction and G-25 MicroSpin columns (GE Healthcare). The DNA was incubated at 37°C for 15 min with 1 U of UDG and then heated at 95°C for 10 min to cleave apyrimidinic sites. The reactions were extracted with phenol and chloroform, and the DNA was precipitated with ethanol. An aliquot of this DNA was also labeled using TdT and [α-32P]3′-deoxyATP to assess the extent of primer extension.
(S)-HPMPA-containing templates were prepared in a similar way except that (S)-HPMPA was incorporated using a different template plus one additional enzymatic step. About 30 pmol of primer P11 was mixed with a threefold excess of template T17 (Fig. 1), and the primer was then extended by two residues at 37°C for 1 h in a reaction containing 10 μM each (S)-HPMPApp and dGTP, 0.01 M dithiothreitol, first-strand buffer, and 8 U of MMLV reverse transcriptase/μl. The DNA was purified by using chloroform extraction and G-25 MicroSpin columns and precipitated with ethanol. This (S)-HPMPA-containing primer was then converted to a full-length extension product using vaccinia polymerase and all four dNTPs, purified, treated with UDG, purified again, and characterized as described above. A template containing dAMP [instead of (S)-HPMPA] was prepared the same way, except that the reverse transcriptase step was omitted.
DNA polymerase assays using CDV- or (S)-HPMPA-containing templates.
Enzyme substrates were prepared by annealing 5′-end-labeled primers to drug-containing templates in a ∼3:1 (template/primer) molar ratio. These DNAs were added to reactions containing the indicated dNTPs (50 μM each), polymerase buffer, and 2.5 ng of vaccinia DNA polymerase/μl and incubated at 37°C. The reactions were stopped by adding EDTA, and the biotinylated strands (and hybridized DNAs) were recovered by using M-280 streptavidin Dynabeads as directed by the manufacturer (Invitrogen). The products were then size fractioned and detected by phosphorimaging. Size standards were generated by using dideoxy-sequencing reactions and Klenow DNA polymerase plus the same 5′ end-labeled primers used in the vaccinia DNA polymerase assays annealed to templates T14 and T18 (Fig. 1).
Cell uptake and HPLC analysis of (S)-HPMPA and CDV metabolites.
[8-14C](S)-HPMPA and HDP-(S)-[8-14C]HPMPA (3 μM) were added to 24-well plates containing MRC-5 fibroblasts, and after 24 h at 37°C the media was removed, the cell monolayer was washed twice with cold phosphate-buffered saline, and the cell uptake of drug was assessed by liquid scintillation counting in quadruplicate as previously described (1). For measurement of (S)-HPMPA and HDP-(S)-HPMPA conversion to their mono- and diphosphates, radioactive drugs were added to 25-cm2 flasks of near-confluent MRC-5 cells (10 μM), followed by incubation for 24 h. The medium was removed, and the monolayer was washed twice with cold phosphate-buffered saline, followed by the addition of 0.6 ml of distilled water. The flasks were twice frozen and thawed and sonicated for 5 min in a cold sonicator bath, and the flask contents were scraped into a glass tube. Cold trichloroacetic acid was added to a final concentration of 8%, and the contents were vortex mixed and centrifuged for 10 min at 4°C. The supernatant was removed, an aliquot was subjected to liquid scintillation counting, and another aliquot (10,000 dpm) was subjected to high-pressure liquid chromatography analysis as previously described (1). The method used a Partisil 10 SAX column (4.6 by 15 cm), with SAX guard column, equilibrated with 20 mM potassium phosphate buffer (pH 5.8) and operating at a flow rate of 1 ml/min. The sample was applied to the column and, after 9 min of isocratic operation, eluted with a 20 to 700 mM potassium phosphate buffer gradient, over 20 min, followed by a 5-min terminal hold. Each 1-ml fraction was collected and analyzed by liquid scintillation counting using FloScint IV fluid. The retention times of [2-14C]CDVpp (25 to 27 min) (1) and (S)-[8-14C]HPMPApp (32 to 33 min) were identical to that of pure reference standards. The data shown are the average of two separate determinations.
RESULTS
Effect of (S)-HPMPA on vaccinia DNA replication.
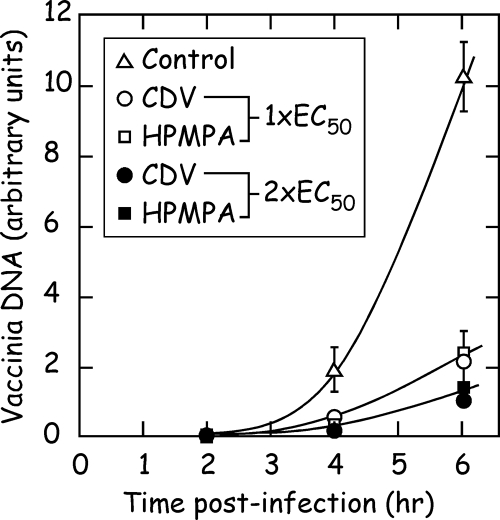
(S)-HPMPA inhibits viral DNA replication in vivo, but at lower concentrations than seen using CDV. A plaque reduction assay was first performed to determine the EC50 values for each drug against vaccinia virus strain WR in BSC40 cells. These values were determined to be 18 μM for (S)-HPMPA and 46 μM for CDV, confirming the greater efficacy of (S)-HPMPA against vaccinia virus (12, 19, 22, 36) and similar to values determined previously using vaccinia virus strain Copenhagen and in MRC-5 cells (Table 1). To determine the effect of (S)-HPMPA and CDV on virus DNA synthesis, we then tested two concentrations of each drug corresponding to the EC50 and twice the EC50. BSC40 cells were pretreated (or mock treated) with drug for 24 h prior to infection and then infected with virus while maintaining the drug selection pressure. The cells were harvested, and the amount of viral DNA accumulated was determined at different time points by slot blot hybridization using a 32P-labeled vaccinia gene probe. Figure 2 shows the results of this experiment. No viral DNA was detected in any of the mock-infected samples (data not shown) or in any of the infected cell samples at 2 h postinfection. In drug-free media, viral DNA began to be detected at 4 h postinfection, followed by a large accumulation at 6 h postinfection. As predicted, CDV and (S)-HPMPA cause a substantial reduction in DNA synthesis relative to what was detected in cells infected with virus in the absence of drug. Moreover, DNA replication is inhibited to a similar extent when cells are treated with concentrations of drug corresponding to the respective EC50s or to twice their respective EC50s. Thus, both drugs cause comparable effects on viral replication, but less (S)-HPMPA is required than CDV in the culture media.
TABLE 1.
Metabolic properties of CDV, (S)-HPMPA, and their hexadecyloxypropyl esters and antiviral activity against vaccinia virus strain Copenhagen in vitroa
| Drug | Cell uptake
|
Anabolic phosphorylation
|
Antiviral activity
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Drug concn (μM) | Mean uptake (pmol/well) ± SDb | Source or reference | Drug concn (μM) | N | Np | Npp | Source or reference | Mean EC50 (μM) ± SDb | Source or reference | |
| CDV | 3.0 | 3.60 ± 0.29 (3) | 1 | 10 | 274 | 1.0 | 1.3 | 1 | 46 ± 12 (2) | 20 |
| HDP-CDV | 3.0 | 187 ± 12.0 (2) | 1 | 10 | 697 | 63 | 132 | 1 | 0.80 ± 0.40 (2) | 20 |
| (S)-HPMPA | 3.0 | 2.77 ± 0.20 (4) | This study | 10 | 29 | 6.0 | 21 | This study | 2.70 ± 2.40 (2) | 5 |
| HDP-(S)-HPMPA | 3.0 | 155 ± 16 (4) | This study | 10 | 93 | 68 | 451 | This study | 0.01 ± 0.004 (2) | 5 |
Drug uptake was measured in MRC-5 fibroblasts in 24-well plates 24 h after each compound was added to the culture media. Anabolic phosphorylation was assessed in MRC-5 cells grown in T-75 flasks, 24 h after the addition of 14C-labeled versions of each compound. Abbreviations: N, nucleotide; Np, nucleotide monophosphate; Npp, nucleoside diphosphate.
Values in parentheses are the numbers of independent replicates.
FIG. 2.
Effect of (S)-HPMPA and CDV on vaccinia virus DNA synthesis in vivo. BSC40 cells were cultured for 24 h, with or without the indicated concentrations of drugs, and then infected with vaccinia WR at a multiplicity of infection of 10. Fresh culture medium was added, also containing (S)-HPMPA or CDV where indicated, and then all of the nucleic acids recovered at 2, 4, or 6 h postinfection. A slot blotting protocol, followed by phosphorimager analysis, was used to quantify the amount of accumulated viral DNA (in arbitrary units) using a 32P-labeled vaccinia DNA polymerase gene as a probe. The concentrations of each drug used correspond to the EC50 (18 and 45 μM) and twice the EC50 (36 and 90 μM) for (S)-HPMPA and CDV, respectively.
Cellular uptake and metabolism of (S)-HPMPA and CDV and their hexadecyloxypropyl esters.
To further evaluate the reasons for the much greater inhibitory effect of (S)-HPMPA and HDP-(S)-HPMPA versus CDV and HDP-CDV, we measured the uptake and conversion of these compounds to their diphosphates (the triphosphate equivalent and the active metabolite) as shown in Table 1. The cell uptake of CDV and (S)-HPMPA was comparable (4.0 versus 2.8 pmol/well), but the conversion of (S)-HPMPA to the diphosphate in 24 h was 16-fold greater than that of CDV (21 versus 1.3 pmol/plate). Esterification with hexadecyloxypropyl increased the cell uptake of both CDV and (S)-HPMPA by 47- and 55-fold, respectively. However, the amount of (S)-HPMPApp formed was still greater than CDVpp, (451 versus 132 pmol/flask; Table 1) (1). Thus, the intracellular level of CDVpp and (S)-HPMPApp correlate roughly with antiviral efficacy of the compounds shown in Table 1.
(S)-HPMPApp is a weak chain terminator.
These observations suggested that the greater bioactivity of (S)-HPMPA, relative to CDV, is at least partially explained by differences in the uptake and conversion of the two drugs to their diphosphoryl derivatives. Nevertheless, we decided to test the effect of (S)-HPMPApp on vaccinia DNA polymerase, to confirm that (S)-HPMPApp and CDVpp then have comparable effects on this enzyme. Curiously, they do not.
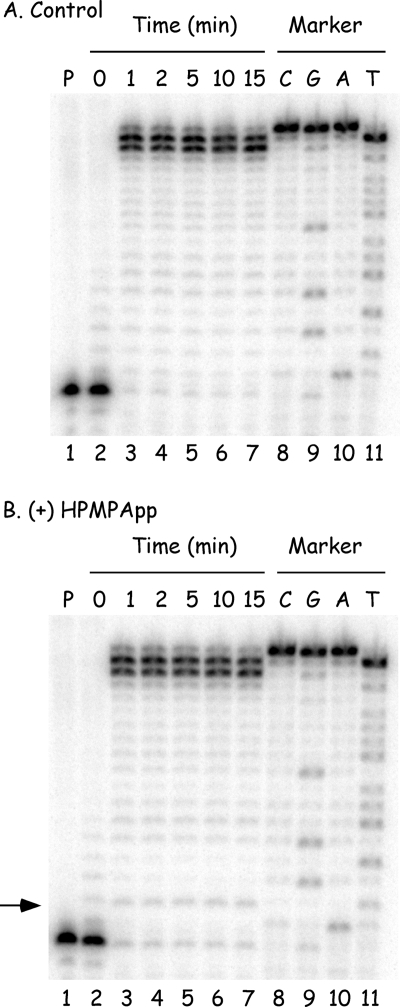
We first examined what happens when (S)-HPMPApp is added to a primer extension assay in a reaction containing an 18-nucleotide 32P-labeled primer annealed to a 36-nucleotide template (primer P1 and template T11, Fig. 1). The reactions also contained all four dNTPs at concentrations approximating those observed in vivo (5 μM dATP, 10 μM dCTP, 10 μM dGTP, and 10 μM dTTP) and were incubated at 37°C. When 10 μM (S)-HPMPApp was added to the reactions, it caused a very weak stop at the N+1 position, where N is the expected site of incorporation of (S)-HPMPA opposite a dTMP residue in the template (compare Fig. 3A and B). We also noted that adding (S)-HPMPApp caused a slight reduction in the production of molecules terminated at the N, N+2, and N+3 positions compared to the ladder of incomplete extension products seen in the control reaction. Since adding (S)-HPMPApp to the primer extension reaction produced only a weak stop, whereas adding CDVpp to a similar reaction mixture resulted in a strong stop one nucleotide after a template dG (23), we investigated whether the yield of premature termination products was affected by the (S)-HPMPApp concentration. No substantial differences in the intensity of the N+1 (or other) termination products were detected using (S)-HPMPApp concentrations varying from 0.1 to 100 μM (data not shown).
FIG. 3.
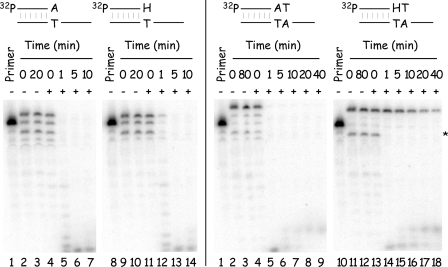
(S)-HPMPApp is a weak inhibitor of primer extension assays. The reactions contained 32P-labeled primer P1 annealed to template T11, four dNTPs (5 μM dATP, 10 μM dCTP, 10 μM dGTP, and 10 μM dTTP), 2.5 ng of vaccinia DNA/μl, and 0 μM (A) or 10 μM (B) (S)-HPMPApp. The mixture was incubated at 37°C, and samples were removed at the indicated time points and mixed with a formamide-containing stop/loading buffer. The reaction products were then separated on 10% denaturing polyacrylamide gels, and the radioactivity was detected by using a phosphorimager. The control reactions (lanes numbered 1) were incubated for 15 min with no added DNA polymerase. Size markers were prepared by using dideoxy sequencing reactions and Klenow DNA polymerase (lanes 8 to 11). The band corresponding to the (S)-HPMPA+1 extension product was seen (arrowed, panel B) but comprised ∼1.4% of the total label in each of lanes 3 to 7. The same band comprised ∼0.3% of the extension products in panel A.
Vaccinia DNA polymerase can use (S)-HPMPApp as a substrate and extend a primer containing (S)-HPMPA.
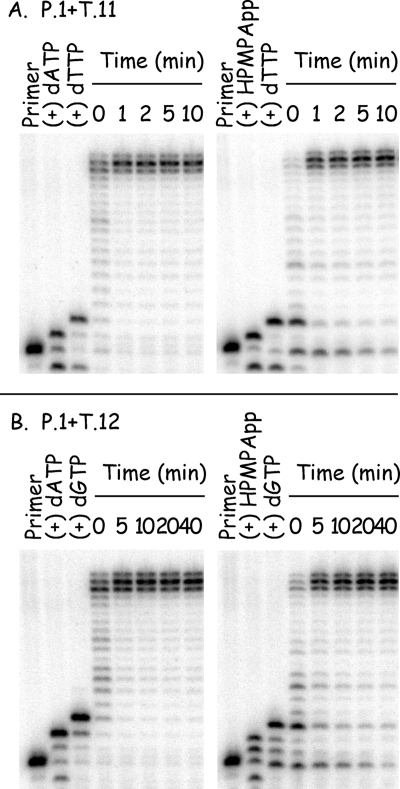
We next wanted to compare the substrate properties of (S)-HPMPApp versus dATP using the same P1-T11 primer-template combination. These experiments were complicated by the fact that, in the absence of other dNTPs, both compounds were incorporated into DNA and then sometimes excised so rapidly as to cause the complete degradation of the labeled primer strand in less than a minute at 37°C (data not shown). After some experimentation with reaction temperatures ranging from 0 to 25°C, we found that the problem can be avoided by incubating the reactions at 25°C (data not shown). This sufficiently slows the reaction to permit ready detection of both polymerase and exonuclease activities under steady-state conditions, and most of the subsequent experiments were performed at 25°C unless otherwise noted.
These assays were next used to determine the Km and Vmax for (S)-HPMPApp and for dATP. The 32P-labeled primer P1 was annealed to template T11, mixed with various concentrations of (S)-HPMPApp or dATP, followed by incubation at 37°C for 0, 1, 2, or 4 min with vaccinia DNA polymerase. We also added the nucleotide found at the 3′ end of the P1 strand to each reaction (dGTP, 10 μM) to minimize attack on that end by the 3′-to-5′ exonuclease and permit the use of a 37°C reaction temperature. The reaction products were separated by using a 15% polyacrylamide gel and detected by using a phosphorimager. The amount of primer extended by one nucleotide in each reaction was determined by using ImageQuant software, and the results were analyzed using one-phase exponential association, to determine an initial velocity, and the Michaelis-Menten equation. The Km of (S)-HPMPApp was calculated to be 3.8 ± 0.8 μM, and the Vmax was calculated at 2.1 ± 0.1 pmol/min. The Km and Vmax of dATP were determined to be 4.6 ± 0.5 μM and 2.0 ± 0.07 pmol/min, respectively. These data indicate that (S)-HPMPApp is as good a substrate for vaccinia DNA polymerase as is dATP.
Having shown that (S)-HPMPApp is a good substrate for vaccinia polymerase, we next examined what effect the presence of the drug near the primer terminus would have on chain extension. Of particular interest was the effect of (S)-HPMPA when incorporated into the penultimate position of the primer strand [“(S)-HPMPA+1”], since it is the CDV+1 structure that is less well used by vaccinia and herpes DNA polymerases. Figure 4A shows such a stepwise comparison of the substrate properties of dAMP versus (S)-HPMPA at 25°C. As noted above, (S)-HPMPA can be incorporated into DNA with kinetics resembling dATP. The next nucleotide (dTTP) was then added to the (S)-HPMPA-terminated primer in a manner also similar to that seen in the dAMP-terminated control reaction. Finally, when an equimolar mixture of dCTP, dGTP, and dTTP was added to each of these reactions (to 30 μM total concentration) the primer was rapidly extended out to the end of the template strand. We noted that there might be a slight lag in the production to the full-length products from a primer terminating in (S)-HPMPA plus dTMP compared to extension from a primer terminating in dAMP plus dTMP (Fig. 4A). Nevertheless, nearly all of the (S)-HPMPA-containing labeled primer was rapidly chased into higher-molecular-weight products within moments of starting the reactions.
FIG. 4.
(S)-HPMPA can be incorporated into DNA and extended by vaccinia DNA polymerase. (A) Primer P1 was labeled with 32P, annealed to template T11, and incubated for 1 min with 2.5 ng of vaccinia DNA polymerase/μl plus 10 μM dATP or (S)-HPMPApp at 25°C. A sample of the product was removed from each reaction and added to formamide stop buffer. dTTP was then added to each of the remaining mixtures, to a final concentration of 10 μM, and the incubation continued for another minute. A second sample was removed, followed by the addition of dCTP, dGTP, and dTTP (all to 10 μM final concentration), and the incubation was continued with periodic sampling. The reaction products were then size fractionated and detected by phosphorimaging. A slight lag may be seen in the extension of molecules terminated by (S)-HPMPA+dGMP, but the majority of the primer chases into a series of extension products in less than a minute at 25°C. (B) Experiment similar to that in panel A except that template T12 directs the incorporation of two consecutive molecules of dAMP or (S)-HPMPA.
Vaccinia DNA polymerase can use primers containing two consecutive (S)-HPMPA molecules.
CDV has been shown to greatly reduce the rate of primer extension by vaccinia virus and cytomegalovirus DNA polymerases whenever the template contains two consecutive dG residues (23, 40). These data have led to the suggestion that CDV might be especially prone to causing chain termination whenever it is incorporated into adjacent sites in the primer (10) and led us to test whether (S)-HPMPA might have similar effects on vaccinia DNA polymerase. For this experiment, a 32P-labeled primer P1 was annealed to template T12 (Fig. 1) and used as an alternative substrate. The P1-T12 primer-template was incubated with vaccinia DNA polymerase and with either 10 μM dATP or 10 μM (S)-HPMPApp (Fig. 4B) for 1 min. A sample was taken of each reaction, and the next nucleotide (dGTP) was added to the remainder to a final concentration of 10 μM. After another minute of incubation, a sample was taken, and the remaining reaction mixture was adjusted to include 10 μM each for dTTP and dCTP. The reaction was then sampled at different time points. We observed that vaccinia DNA polymerase can readily incorporate two consecutive molecules of dATP into DNA, extend this primer further by one nucleotide, and then rapidly extend this primer in the presence of all four dNTPs. Replacing dATP with (S)-HPMPApp produced similar results. Although there is again a slight lag on the initial production of fully extended primer products, as seen by the decreased intensity of the full-length bands in the first minute, the incorporation of two consecutive (S)-HPMPA molecules into the growing DNA strand did not cause a dramatic reduction in the rate of DNA elongation compared to the two molecules of CDV (23).
Effect of (S)-HPMPA on the 3′-to-5′ proofreading exonuclease activity.
To determine what effect (S)-HPMPA has on the 3′-to-5′ exonuclease activity of vaccinia DNA polymerase, the 5′-to-3′ polymerase activity was first used to incorporate (S)-HPMPA (or dAMP) into the terminus of 32P-labeled primer P1 annealed to template T11. We also added dTTP to some reactions to position the (S)-HPMPA/dAMP moiety at the penultimate location in the primer strand. Unincorporated nucleotides and (S)-HPMPApp were removed by gel filtration. The substrates were then incubated with fresh DNA polymerase, in the absence of dNTPs and at 25°C, to examine exonuclease activity in the absence of any polymerase activity. Figure 5 (left panel) shows the results obtained when the enzyme is presented with substrates incorporating dAMP or (S)-HPMPA at the 3′-primer terminus. A primer terminated with dAMP was completely converted to an array of smaller products by vaccinia DNA polymerase in less than a minute. A primer terminated with (S)-HPMPA was also rapidly degraded, although we noted that there was still a trace of the (S)-HPMPA-terminated primer band visible after 1 min of incubation (Fig. 5B, lane 12). A primer terminating in dAMP plus dTMP was also rapidly degraded by the vaccinia DNA polymerase (Fig. 5, right panel) with nearly complete conversion of this primer to smaller products seen after a minute of incubation. In contrast, a primer terminating in (S)-HPMPA plus dTMP was refractory to exonuclease activity. We estimate that the half-life of this product at 25°C is about 7 min and, although the exonuclease is more active if the reaction is incubated at 37°C, the half-life is still at least ∼4 min (data not shown). It should also be noted that these (S)-HPMPA-containing molecules are not irreversible inhibitors of the exonuclease activity. During the preparation of these substrates a small amount of a contaminating product is left behind that is one nucleotide smaller than the original primer P1 (Fig. 5, asterisk). This DNA should bear either a 3′-terminal dAMP or (S)-HPMPA residue and, even in the presence of the molecules terminated with (S)-HPMPA+dTMP, it is completely degraded within a minute of adding the enzyme.
FIG. 5.
Vaccinia DNA polymerase can excise (S)-HPMPA from the primer terminus but not if (S)-HPMPA is the penultimate 3′-nucleotide. Primer P1 was labeled with 32P, annealed to template T11, and incubated for 1 min with 2.5 ng of vaccinia DNA polymerase/μl at 25°C in the presence of 10 μM dATP, 10 μM (S)-HPMPA, 10 μM (each) dATP and dTTP or 10 μM (each) (S)-HPMPA and dTTP. This produced molecules bearing the 3′ structures indicated in the Figure [“H” = (S)-HPMPA]. The unincorporated nucleotides and (S)-HPMPApp were removed by gel filtration, and the purified substrates were incubated with fresh enzyme at 25°C. The reactions were sampled at the times indicated, mixed with formamide stop buffer, and size fractionated on 10% polyacrylamide gels, and the radioactivity was detected by phosphorimaging. Water was substituted for DNA polymerase in the “no polymerase” controls (indicated by “−” symbols). Primers terminated with (S)-HPMPA+dTMP are highly resistant to exonuclease attack (lanes 14 to 18, at right) but did not inhibit exonuclease attack on a trace of contaminating molecules terminated with dAMP or (S)-HPMPA (asterisk).
CDV and (S)-HPMPA are faithfully incorporated into DNA by vaccinia DNA polymerase.
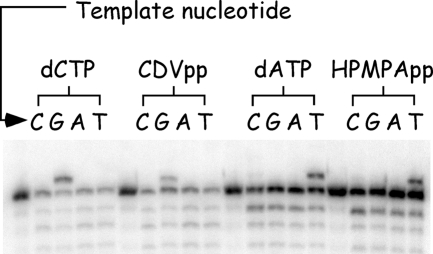
One mechanism that might account for the greater in vivo activity of (S)-HPMPA, relative to CDV, could be that (S)-HPMPA is a substrate, but one that is especially prone to misincorporation by vaccinia polymerase. This would cause mutations and deleterious effects on replication. To examine whether (S)-HPMPA is misincorporated into DNA by this enzyme, a simple primer extension analysis was performed. For these reactions, 32P-labeled primer P1 was annealed to different templates containing each of the four nucleotides located immediately after the primer terminus (Fig. 1). These primer-template pairs were incubated with vaccinia DNA polymerase and 10 μM CDVpp (or dCTP) or 10 μM (S)-HPMPApp (or dATP) for 1 min. The results are presented in Fig. 6, where it can be seen that CDV and dCMP are incorporated opposite only dG and that (S)-HPMPA and dAMP are incorporated opposite only dT. Thus, under the conditions of this experiment (which we estimate could detect 1 to 2% misincorporation), both CDV and (S)-HPMPA are faithfully incorporated into DNA by vaccinia polymerase.
FIG. 6.
CDV and (S)-HPMPA are faithfully incorporated into DNA by vaccinia DNA polymerase. The reaction mixtures contained 32P-end-labeled primer P1 annealed to different template strands and were incubated with the indicated nucleotides (each at 10 μM) and 2.5 ng of vaccinia DNA polymerase/μl at 37°C (dCTP and CDVpp) or 25°C [dATP and (S)-HPMPApp]. The nucleotide encoded by the template strand at the position immediately after the primer terminus is indicated on the figure. Each reaction was stopped after 1 min, and the products were separated by gel electrophoresis and detected by using a phosphorimager.
Preparation of oligonucleotide templates containing CDV or (S)-HPMPA.
When one considers the data outlined above, one factor that differentiates (S)-HPMPA from CDV is that (S)-HPMPA could be more readily incorporated into DNA than is CDV. This raises the interesting question of what effects the two drugs have on DNA synthesis when located in the template strand. Since there is currently no chemical method to synthesize these drug-containing DNAs, we used the enzymatic approach illustrated in Fig. 7 to produce these templates.
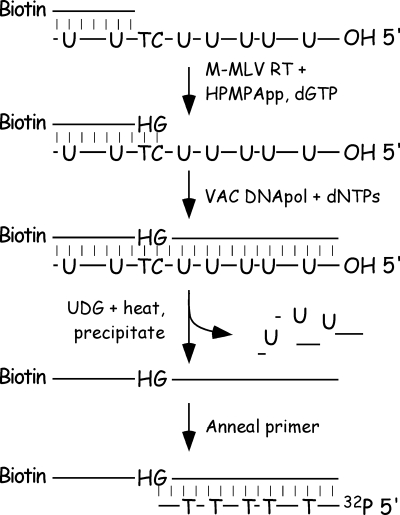
FIG. 7.
Scheme used to incorporate CDV and (S)-HPMPA into a template strand. The figure shows the method used to incorporate (S)-HPMPA into DNA. MMLV reverse transcriptase was used to add (S)-HPMPA (“H”) and dGMP to a DNA comprising primer P11 annealed to template T17 (Fig. 1). The products were purified and further extended using vaccinia DNA polymerase and four dNTPs. The T17 strand was then degraded with uracil glycosylase, and the (S)-HPMPA-containing strand was purified and annealed to 32P-labeled primers P17, P18, or P19 (Fig. 1). Molecules containing CDV, and control DNAs, were prepared the same way, except that we omitted the MMLV reverse transcriptase step. We also used different primers and templates to direct the incorporation of CDV and dCMP (Fig. 1).
In order to synthesize a CDV-containing template, the 5′ biotinylated primer, P9, was annealed to template T13 (Fig. 1) and extended using vaccinia DNA polymerase and CDVpp (or dCTP for control purposes) plus dATP and dGTP. The T13 template strand contains seven uracil residues that permitted its degradation using uracil-DNA glycosylase and heat. The UDG-resistant, CDV-containing (or dCMP-containing) DNAs were then purified for use as template strands. Aliquots of the UDG-resistant template DNA were also end labeled with 32P using TdT and cordycepin triphosphate, in order to assess the extent of P9 extension.
The preparation of an (S)-HPMPA-containing template followed a similar procedure, using 5′ biotinylated primer P11 annealed to template T17. However, this approach was complicated by the fact that the uracil in the template can promote the incorporation of many (S)-HPMPA molecules and attempts to add the drug in a stepwise manner, using vaccinia polymerase, were frustrated by exonuclease attack on the P11 primer. Therefore, we used MMLV reverse transcriptase to incorporate (S)-HPMPA plus the next nucleotide (dGMP) into the terminus of primer P11, purified the product, and generated a full-length copy using vaccinia polymerase and four dNTPs. The product was then processed as described above. We tried using the same method to prepare a dAMP-containing control template, but the extensive misincorporation of dAMP and/or dGMP by MMLV reverse transcriptase prevented us from doing so (Fig. 8, lanes 4 and 9). This problem can be avoided by limiting the time of incubation of the reverse transcriptase with (S)-HPMPApp and dGTP (Fig. 8, lane 10), but we found no good method to avoid it using dATP plus dGTP. As a result, this template was prepared like the CDV- and dCMP-containing templates, namely, by incubating the primer-template pair P11-T17 with vaccinia polymerase plus all four dNTPs, followed by UDG and heat treatment and ethanol precipitation.
FIG. 8.
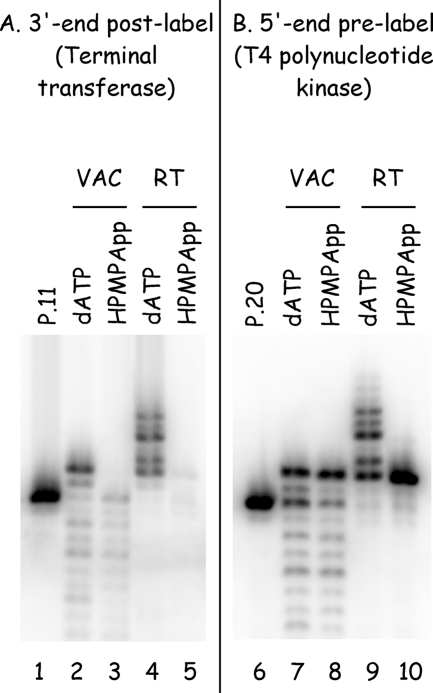
(S)-HPMPA inhibits labeling with TdT. (A) Primer P11 was annealed to strand T17 and incubated at 37°C with vaccinia DNA polymerase (1 min) or MMLV reverse transcriptase (1 h), in the presence of 10 μM dGTP and either 10 μM dATP or 10 μM (S)-HPMPApp. The T17 strand was degraded using UDG and heat, and the biotinylated P11 strand was recovered by using magnetic beads. The DNAs were labeled using terminal transferase and [α32P]3′- deoxyATP and subjected to electrophoresis, and the radioactivity was detected by using a phosphorimager. Primer P11 was purified and labeled the same way but not incubated with either polymerase (lane 1). (B) Primer P20 was 5′ end labeled and annealed to template T17 and then incubated with vaccinia polymerase or MMLV reverse transcriptase as described above. The reaction products were size fractionated by electrophoresis and detected by using a phosphorimaging. Both polymerases can incorporate (S)-HPMPA (and dGMP) into DNA, as judged by using prelabeled primers (lanes 8 and 10), but these N+1 products are not postlabeled with terminal transferase (lanes 3 and 5). The TdT is still active, as shown by the capacity to label any of the molecules encoding dAMP (lanes 2 and 4). VAC, vaccinia DNA polymerase; RT, reverse transcriptase.
During the preparation of these substrates, aliquots were taken of the reaction intermediates and labeled with 32P using terminal transferase to assess the extent of elongation of primer P11. We observed little or no labeling of the biotinylated-P11 products if they were expected to contain (S)-HPMPA (Fig. 8, lanes 3 and 5). We hypothesized that these might be poor substrates for TdT and confirmed this by performing control reactions using the nonbiotinylated primer P20 instead of P11 (Fig. 1). This permitted 5′ end labeling of the primer using T4 polynucleotide kinase and showed that the “absence” of an extension product (Fig. 8, lanes 3 and 5) was an artifact of the TdT not being able to extend (S)-HPMPA-containing primers.
CDV or (S)-HPMPA are faithfully copied by vaccinia DNA polymerase.
To test the coding properties of these drug lesions, the newly prepared templates were annealed to 5′-32P-end-labeled primers and incubated with vaccinia DNA polymerase plus one each of the four dNTPs at 50 μM. After a 1-min incubation, the products that had annealed to the template were retrieved using Dynabeads, size fractionated, and visualized by phosphorimaging. As shown in Fig. 9 (top panel), only dGMP is incorporated opposite dCMP and CDV and only dTMP is incorporated opposite dAMP and (S)-HPMPA (Fig. 9, bottom panel). These results show that CDV and (S)-HPMPA are both faithfully incorporated into DNA and then copied by vaccinia DNA polymerase.
FIG. 9.
CDV and (S)-HPMPA are faithfully copied by vaccinia DNA polymerase. The four template strands were prepared containing dCMP/CDV (upper panel) or dAMP/(S)-HPMPA (lower panel) and annealed to 32P-labeled primers P10 or P17 as indicated. These primer-template pairs were incubated with 2.5 ng of vaccinia DNA polymerase/μl at 37°C for 1 min in the presence of each of the indicated single dNTPs (50 μM), and the products were recovered using magnetic beads. The reaction products were then separated on a 10% polyacrylamide gel, and the radioactivity was visualized by phosphorimaging. CDV and (S)-HPMPA direct the incorporation of dGMP and dTMP, respectively. Each of the enzymatically prepared template strands was also separately labeled with terminal transferase to measure the length of the original extension products (template). Sequencing ladders were prepared using primer P10 annealed to template T14 (upper panel) or primer P17 annealed to template T18 (lower panel).
The presence of CDV or (S)-HPMPA in the template strand blocks primer extension.
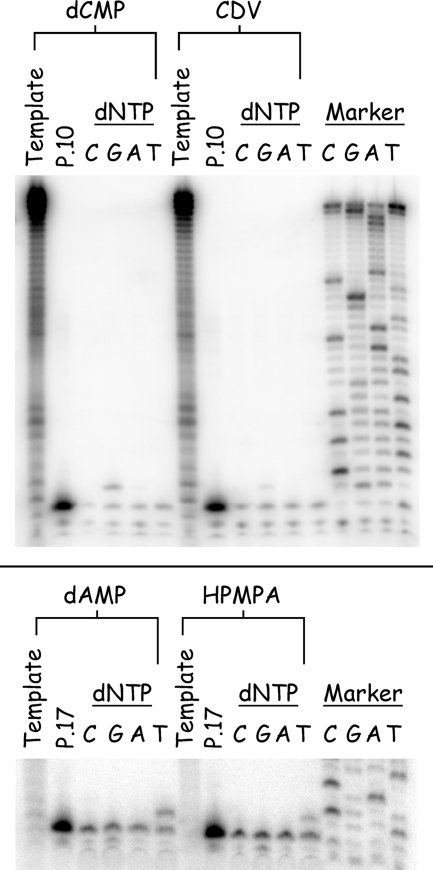
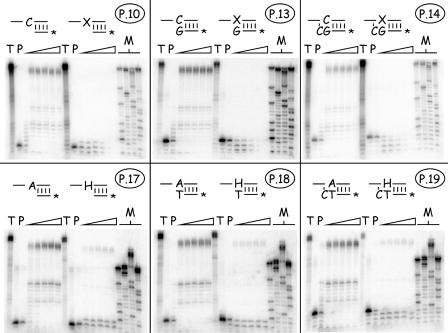
We next examined what effect these drug lesions had on primer extension. We synthesized three kinds of primers for the present study, designed so that the 3′ terminus was located one nucleotide preceding, next to, or one nucleotide past the site of incorporation of the drug residue (Fig. 1, primers P10 to P14 and P17 to P19). These strands were 32P end labeled, annealed to the prepared templates, and incubated at 37°C with vaccinia DNA polymerase and all four dNTPs at 50 μM each. The results of the present study are shown in Fig. 10. In all of these experiments, the primers annealed to control (i.e., drug-free) strands were all rapidly extended the full length of the enzymatically copied strand, regardless of where we positioned the 3′ end relative to the dAMP or dCMP template nucleotide. This demonstrated the integrity of templates produced in this manner. In contrast, the presence of CDV or (S)-HPMPA strongly inhibited primer extension.
FIG. 10.
Effects of templates bearing nucleoside phosphonate drugs on trans-lesion DNA synthesis. Templates were prepared bearing dCMP/CDV (upper panels) or dAMP/(S)-HPMPA (lower panels) and annealed to the six indicated 32P-labeled primer strands. These primers terminate at positions N+1 (P10 and P17), N (P13 and P18), and N−1 (P14 and P19), where N is the site of drug incorporation [“X” = CDV, “H” = (S)-HPMPA]. Each of these primer-template pairs was incubated with 2.5 ng of vaccinia DNA polymerase/μl at 37°C for 0, 1, 2, 5, 10, or 15 min (triangles) in the presence of all four dNTPs (50 μM each), and the products were recovered by using magnetic beads. The reaction products were then separated on a 10% polyacrylamide gel, and the radioactivity was visualized by phosphorimaging. All of the six primers were rapidly extended across control molecules bearing dCMP or dAMP residues. In contrast, P10 and P17 were extended only one nucleotide (left-hand column), and the drugs blocked net DNA synthesis from the other four primers. Each of the enzymatically prepared template strands was also separately labeled with terminal transferase and [α32P]3′-deoxyATP to measure the length of the original extension products (“T”). The electrophoretic properties of unmodified primer strands are illustrated in lanes marker “P.”
With CDV-containing templates (Fig. 10, top panel), primer P10 was first extended by just one nucleotide and then attacked and degraded by the 3′-to-5′ exonuclease. At no point did we ever see the complete conversion of the primer to the primer plus one nucleotide (N+1), instead a mix of molecules corresponding to N−1, N, and N+1 products was formed and then degraded (Fig. 10, top left image). No net DNA synthesis was seen with primers P13 and P14; instead, these DNAs were progressively degraded to a series of shorter molecules, with the most prominent products being the N−1 and N−2 bands (for P13) and N−2 and N−3 bands (for P14) (Fig. 10, top middle and top right images). A similar effect was seen with templates containing (S)-HPMPA. Primer P17 was converted to a mix of N and N+1 products, P18 was converted to a mix of N and N−1 products, and P19 was converted to a mix of N−1 and N−2 products (Fig. 10, bottom images). However, although both drugs had similar inhibitory effects on DNA extension, a dramatic difference was seen in the stability of the DNAs formed in these reactions. All of the substrates and products detected in reactions using the CDV-containing template were completely degraded in less than 15 min, whereas products that result from using the (S)-HPMPA-containing templates were stable throughout the entire reaction. This exonucleolytic attack on these strands occurs despite the use of high (200 μM) concentrations of dNTPs and suggests that the two different drugs, and resulting DNA structures, may have dramatically different effects on the rate of nucleotide turnover by vaccinia DNA polymerase.
DISCUSSION
The nucleoside phosphonate family of drugs exhibits a varying degree of activity against orthopoxviruses, with the purine analog, (S)-HPMPA, being more active than the pyrimidine analog, CDV, as judged by a 3- to 17-fold difference in EC50 in vivo (Table 1). Metabolic studies provide some insight into the reasons for this difference since cells exposed to these drugs also appear to more rapidly convert (S)-HPMPA into (S)-HPMPApp and provide intracellular concentrations of (S)-HPMPApp that are 16-fold higher than that of CDVpp with the unmodified nucleotides and 3.4-fold higher with HDP-(S)-HPMPA versus HDP-CDV (Table 1). A variety of data also clearly implicates the replication machinery, and DNA polymerase in particular, as being the primary target of both drugs. In particular, one selects for spontaneous mutations in the E9L (DNA polymerase) gene by continued passage of vaccinia virus in the presence of CDV, and such CDV-resistant mutations create cross-resistance to most other nucleoside phosphonate drugs, including (S)-HPMPA (2). When one exposes vaccinia virus-infected cells to biologically equivalent concentrations of both drugs (i.e., concentrations equal to the EC50), one sees nearly equal levels of inhibition of virus DNA replication, as judged by the amount of virus DNA synthesized (Fig. 2). However, what is not clear is what might be “wrong” with the substrate properties of (S)-HPMPApp, as judged by the primer extension assays used in the present study.
We have previously shown that the diphosphoryl derivative of CDV, CDVpp, can be used and incorporated into DNA by vaccinia DNA polymerase (23). CDVpp is a poor substrate for the vaccinia enzyme relative to the natural substrate, dCTP (Km, CDVpp = 23 ± 6 μM, Km, dCTP = 3.8 ± 0.7 μM [23]). Its incorporation results in only a slight decrease in the rate of chain extension after adding one CDV molecule but causes premature chain termination, as evidenced by the appearance of “pause sites,” after the addition of the next nucleotide (23). DNA synthesis is also profoundly inhibited when two consecutive molecules of CDV are incorporated into the 3′ end of the primer strand. These effects of CDVpp on vaccinia DNA polymerase are similar to the effects on cytomegalovirus DNA polymerase (40). Given that (S)-HPMPA is more biologically active than CDV but shares a similar chemical structure within the nucleoside phosphonate moiety, we hypothesized that (S)-HPMPApp would interact with vaccinia DNA polymerase in a manner similar to CDVpp but that the effects of the drug would simply be more exaggerated and perhaps detectable at lower concentrations. This is not what is observed.
The first experiment we performed was a simple primer extension assay containing all four dNTPs. When (S)-HPMPApp was added to this reaction, a weak stop at a position corresponding to two nucleotides longer than the original primer strand was detected (Fig. 3B). This is in contrast to that seen with CDVpp, where the addition of this compound at an equal concentration (10 μM) resulted in the formation of a strong stop at this N+1 position (23). We next examined the kinetics of incorporation of (S)-HPMPApp relative to dATP. These studies showed that (S)-HPMPApp is as good if not a better substrate for the polymerase than is dATP (Km, (S)−HPMPApp = 3.8 ± 0.8 μM versus Km, dATP = 4.6 ± 0.5 μM, with a comparable Vmax), and it is also very rapidly excised with kinetics comparable to dAMP from the 3′ end of the primer strand (Fig. 5). Our experiments also showed that when (S)-HPMPA is incorporated into DNA, it can then serve as a primer for further extension with kinetics similar to that seen in a control reaction, where dATP replaced (S)-HPMPApp (Fig. 4A). Finally, whereas molecules encoding CDV+1 nucleotide are poor primers, and molecules bearing two consecutive molecules of CDV are nearly inert in elongation reactions (23), the analogous (S)-HPMPA containing structures have not nearly as marked effects on the elongation rate (Fig. 4). The only striking difference between molecules bearing (S)-HPMPA and dAMP, as the penultimate 3′ nucleotide, is that the presence of (S)-HPMPA clearly inhibits the activity of the 3′-to-5′ exonuclease (Fig. 5). This is a property common to both (S)-HPMPA and CDV. Thus, these primer extension assays have identified no particular effect of (S)-HPMPApp on vaccinia polymerase that can directly account for the enhanced activity in vivo.
These observations suggested that (S)-HPMPA exerts its effects on vaccinia replication in a more indirect manner than CDV and in a way that is exacerbated by the relatively higher intracellular concentrations of (S)-HPMPApp versus CDVpp. A lower Km for (S)-HPMPApp (versus CDVpp), combined with a greater amounts of (S)-HPMPApp relative to dATP (compared to the CDVpp to dCTP ratio) and little effect of (S)-HPMPA on chain extension, would lead to relatively more incorporation of (S)-HPMPA than CDV into viral DNA. One possible consequence might then be that more (S)-HPMPA can be misincorporated into DNA than CDV. However, we examined incorporation of CDV and (S)-HPMPA opposite all four dNMPs and found that both drugs are faithfully incorporated opposite dGMP and dTMP, respectively (Fig. 6).
This led us to investigate what effects CDV and (S)-HPMPA have on DNA synthesis when located in the template strand. Most nucleoside and nucleotide analogues act as obligate chain terminators, whereas CDV, which still bears a 3′ hydroxyl, is generally classified as a nonobligate chain terminator. Since (S)-HPMPA can be incorporated into DNA without causing much chain termination (Fig. 4), some of the antiviral effects of this drug could be explained by what happens at the next round of replication. Since there are currently no chemical methods for the synthesis of CDV- and (S)-HPMPA-containing DNAs, we developed an enzymatic approach using vaccinia DNA polymerase, or vaccinia DNA polymerase in combination with MMLV reverse transcriptase (Fig. 7). A number of quality controls were conducted during the preparation of these substrates and one of the more interesting effects that we noted was that (S)-HPMPA-containing DNAs are also poorly labeled by TdT (Fig. 8). This suggests that DNAs bearing a nucleoside phosphonate drug, at the penultimate 3′ position, likely exhibit some structural feature that is broadly inhibitory to many different nucleotidyl transferases. Since TdT plays a key role in the development of immune diversity (37), this observation also raises some questions regarding what effect prolonged exposure to these drugs might have on immune responses to viral infection.
These new substrates were used to show that templates containing CDV and (S)-HPMPA cause a severe block in DNA extension. Although the polymerase can faithfully incorporate a nucleotide opposite either drug residue (Fig. 9), further elongation is inhibited (Fig. 10). Vaccinia DNA polymerase also rapidly degraded primers extending one nucleotide past the drug lesion in the presence of dNTPs, suggesting that such structures tend to be recognized as being mismatched by the 3′-to-5′ proofreading exonuclease. Although such effects have not been much studied, there have been a few reports of similar effects caused by other polymerase inhibitors and using other enzymes. For example Mikita and Beardsley (27) showed that arabinosylcytosine template residues partially blocked DNA elongation by Klenow, T4, and human α2 DNA polymerases, although it did not efficiently inhibit avian myeoloblastosis virus reverse transcriptase. Satake et al. (32, 33) also noted that 5-trifluoromethyl-2′-deoxyuridine caused a strong arrest one nucleotide before or after the lesion site using Klenow polymerase and human DNA polymerase α, respectively. Collectively, these results show that if nucleotide analogues are incorporated into the template strand, they can severely inhibit polymerase activity, much like some forms of DNA damage (4). This mechanism of action is not relevant for common DNA polymerase inhibitors, since most are obligate or de facto chain terminators. However, (S)-HPMPApp is a good substrate and a not very effective chain terminator (Fig. 4) and thus might well act more by inhibiting secondary rounds of DNA synthesis. Since both CDV and (S)-HPMPA block DNA synthesis to a similar extent, once incorporated into the template strand, the relatively greater efficacy of (S)-HPMPA is probably explained by a combination of factors related to higher intracellular levels of (S)-HPMPApp plus a greater likelihood that (S)-HPMPA would be incorporated into an irreparable DNA lesion.
Viewed from this perspective, these new insights into this mode of drug action can shed new light on the genetics of drug-resistant poxviruses. We have shown that CDV-resistant vaccinia viruses exhibit cross-resistance to (S)-HPMPA and acquire mutations in the E9L gene in both the DNA polymerase and the 3′-to-5′ exonuclease domains (2). Substitution mutations located in the exonuclease domain are probably the primary determinant of resistance and likely act to enhance drug excision from DNA (2). However, it is less clear how the substitution mutations located in the polymerase domain promote drug resistance. We have suggested that they might enhance the discrimination against CDVpp and (S)-HPMPApp during nucleotide selection (2). Based on the results of the present study, it is also possible that these mutations affect how mutant polymerases copy drug-containing (or otherwise damaged) templates and might help explain the weak mutator phenotype exhibited by virus encoding the A684V substitution mutation (2). Work is currently in progress to isolate these mutant polymerases, so they can be used in biochemical assays to investigate the role of these mutations on DNA polymerase activity.
Acknowledgments
We thank Chad Irwin and James Lin for the preparation of vaccinia virus DNA polymerase and G. Andrei for the gift of (S)-HPMPA.
This study was supported by CIHR and NSERC grants (D.H.E.) and by NIH grants AI-066499 and AI-064615 (K.Y.H.).
Footnotes
Published ahead of print on 3 December 2007.
REFERENCES
- 1.Aldern, K. A., S. L. Ciesla, K. L. Winegarden, and K. Y. Hostetler. 2003. Increased antiviral activity of 1-O-hexadecyloxypropyl-[2-14C]cidofovir in MRC-5 human lung fibroblasts is explained by unique cellular uptake and metabolism. Mol. Pharmacol. 63:678-681. [DOI] [PubMed] [Google Scholar]
- 2.Andrei, G., D. B. Gammon, P. Fiten, E. De Clercq, G. Opdenakker, R. Snoeck, and D. H. Evans. 2006. Cidofovir resistance in vaccinia virus is linked to diminished virulence in mice. J. Virol. 80:9391-9401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baker, R. O., M. Bray, and J. W. Huggins. 2003. Potential antiviral therapeutics for smallpox, monkeypox and other orthopoxvirus infections. Antivir. Res. 57:13-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baynton, K., and R. P. P. Fuchs. 2000. Lesions in DNA: hurdles for polymerases. Trends Biochem. Sci. 25:74-79. [DOI] [PubMed] [Google Scholar]
- 5.Beadle, J. R., W. B. Wan, S. L. Ciesla, K. A. Keith, C. Hartline, E. R. Kern, and K. Y. Hostetler. 2006. Synthesis and antiviral evaluation of alkoxyalkyl derivatives of 9-(S)-(3-hydroxy-2-phosphonomethoxypropyl)adenine against cytomegalovirus and orthopoxviruses. J. Med. Chem. 49:2010-2015. [DOI] [PubMed] [Google Scholar]
- 6.Birkus, G., D. Rejman, M. Otmar, I. Votruba, I. Rosenberg, and A. Holy. 2004. The substrate activity of (S)-9-[3-hydroxy-(2-phosphonomethoxy)propyl]adenine diphosphate toward DNA polymerases α, δ, and ɛ. Antiviral Chem. Chemother. 15:23-33. [DOI] [PubMed] [Google Scholar]
- 7.Botros, S., S. William, O. Hammam, Z. Zídek, and A. Holý. 2003. Activity of 9-(S)-[3-hydroxy-2-(phosphonomethoxy)propyl]adenine against Schistosomiasis mansoni in mice. Antimicrob. Agents Chemother. 47:3853-3858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bronson, J. J., I. Ghazzouli, M. J. M. Hitchcock, R. R. Webb II, E. R. Kern, and J. C. Martin. 1989. Synthesis and antiviral activity of nucleotide analogues bearing the (S)-(3-hydroxy-2-phosphonylmethoxy)propyl moiety attached to adenine, guanine, and cytosine, p. 88-102. In J. C. Martin (ed.), Nucleotide analogues as antiviral agents. American Chemical Society, Washington, DC.
- 9.Challberg, M. D., and P. T. Englund. 1979. Purification and properties of the deoxyribonucleic acid polymerase induced by vaccinia virus. J. Biol. Chem. 254:7812-7819. [PubMed] [Google Scholar]
- 10.De Clercq, E. 2004. Antivirals and antiviral strategies. Nat. Rev. Microbiol. 2:704-720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De Clercq, E., A. Holý, I. Rosenberg, T. Sakuma, J. Balzarini, and P. C. Maudgal. 1986. A novel selective broad-spectrum anti-DNA virus agent. Nature 323:464-467. [DOI] [PubMed] [Google Scholar]
- 12.De Clercq, E., T. Sakuma, M. Baba, R. Pauwels, J. Balzarini, I. Rosenberg, and A. Holý. 1987. Antiviral activity of phosphonylmethoxyalkyl derivatives of purine and pyrimidines. Antivir. Res. 8:261-272. [DOI] [PubMed] [Google Scholar]
- 13.de Vries, E., J. G. Stam, F. F. J. Franssen, H. Nieuwenhuijs, P. Chavalitshewinkoon, E. de Clercq, J. P. Overdulve, and P. C. van der Vliet. 1991. Inhibition of the growth of Plasmodium falciparum and Plasmodium berghei by the DNA polymerase inhibitor HPMPA. Mol. Biochem. Parasitol. 47:43-50. [DOI] [PubMed] [Google Scholar]
- 14.Hostetler, K. Y., K. A. Aldern, W. B. Wan, S. L. Ciesla, and J. R. Beadle. 2006. Alkoxyalkyl esters of (S)-9-[3-hydroxy-2-(phosphonomethoxy)propyl]adenine are potent inhibitors of the replication of wild-type and drug-resistant human immunodeficiency virus type 1 in vitro. Antimicrob. Agents Chemother. 50:2857-2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ito, J., and D. K. Braithwaite. 1991. Compilation and alignment of DNA polymerase sequences. Nucleic Acids Res. 19:4045-4057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaminsky, R., B. Nickel, and A. Holý. 1998. Arrest of Trypanosoma brucei rhodesiense and T. brucei brucei in the S-phase of the cell cycle by (S)-9-(3-hydroxy-2-phosphonylmethoxypropyl)adenine ((S)-HPMPA). Mol. Biochem. Parasitol. 93:91-100. [DOI] [PubMed] [Google Scholar]
- 17.Kaminsky, R., C. Schmid, Y. Grether, A. Holý, E. De Clercq, L. Naesens, and R. Brun. 1996. (S)-9-(3-hydroxy-2-phosphonylmethoxypropyl)adenine [(S)-HPMPA]: a purine analogue with trypanocidal activity in vitro and in vivo. Trop. Med. Int. Health 1:255-263. [DOI] [PubMed] [Google Scholar]
- 18.Kaminsky, R., E. Zweygarth, and E. De Clercq. 1994. Antitrypanosomal activity of phosphonylmethoxyalkylpurines. J. Parasitol. 80:1026-1030. [PubMed] [Google Scholar]
- 19.Keith, K. A., M. J. M. Hitchcock, W. A. Lee, A. Holý, and E. R. Kern. 2003. Evaluation of nucleoside phosphonates and their analogs and prodrugs for inhibition of orthopoxvirus replication. Antimicrob. Agents Chemother. 47:2193-2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kern, E. R., C. Hartline, E. Harden, K. Keith, N. Rodriguez, J. R. Beadle, and K. Y. Hostetler. 2002. Enhanced inhibition of orthopoxvirus replication in vitro by alkoxyalkyl esters of cidofovir and cyclic cidofovir. Antimicrob. Agents Chemother. 46:991-995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kramata, P., I. Votruba, B. Otová, and A. Holý. 1996. Different inhibitory potencies of acyclic phosphonomethoxyalkyl nucleotide analogs toward DNA polymerases α, δ, and ɛ. Mol. Pharmacol. 49:1005-1011. [PubMed] [Google Scholar]
- 22.Lebeau, I., G. Andrei, F. Dal Pozzo, J. R. Beadle, K. Y. Hostetler, E. De Clercq, J. van den Oord, and R. Snoeck. 2006. Activities of alkoxyalkyl esters of cidofovir (CDV), cyclic cidofovir, and (S)-9-(3-hydroxy-2-phosphonylmethoxypropyl)adenine against orthopoxviruses in cell monolayers and in organotypic cultures. Antimicrob. Agents Chemother. 50:2525-2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Magee, W. C., K. Y. Hostetler, and D. H. Evans. 2005. Mechanism of inhibition of vaccinia virus DNA polymerase by cidofovir diphosphate. Antimicrob. Agents Chemother. 49:3153-3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McDonald, W. F., and P. Traktman. 1994. Overexpression and purification of the vaccinia virus DNA polymerase. Protein Expr. Purif. 5:409-421. [DOI] [PubMed] [Google Scholar]
- 25.Merta, A., I. Votruba, J. Jindřich, A. Holý, T. Cihlář, I. Rosenberg, M. Otmar, and T. Y. Herve. 1992. Phosphorylation of 9-(2-phosphonomethoxyethyl)adenine and 9-(S)-(3-hydroxy-2-phosphonomethoxypropyl)adenine by AMP(dAMP) kinase from L1210 cells. Biochem. Pharmacol. 44:2067-2077. [DOI] [PubMed] [Google Scholar]
- 26.Merta, A., I. Votruba, I. Rosenberg, M. Otmar, H. Hřebabecký, R. Bernaerts, and A. Holý. 1990. Inhibition of herpes simplex virus DNA polymerase by diphosphates of acyclic phosphonylmethoxyalkyl nucleotide analogues. Antivir. Res. 13:209-218. [DOI] [PubMed] [Google Scholar]
- 27.Mikita, T., and G. P. Beardsley. 1988. Functional consequences of the arabinosylcytosine structural lesion in DNA. Biochemistry 27:4698-4705. [DOI] [PubMed] [Google Scholar]
- 28.Mul, Y. M., R. T. van Miltenburg, E. De Clercq, and P. C. van der Vliet. 1989. Mechanism of inhibition of adenovirus DNA replication by the acyclic nucleoside triphosphate analogue (S)-HPMPApp: influence of the adenovirus DNA binding protein. Nucleic Acids Res. 17:8917-8929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Palú, G., S. Stefanelli, M. Rassu, C. Parolin, J. Balzarini, and E. De Clercq. 1991. Cellular uptake of phosphonylmethoxyalkylpurine derivatives. Antivir. Res. 16:115-119. [DOI] [PubMed] [Google Scholar]
- 30.Pauwels, R., J. Balzarini, D. Schols, M. Baba, J. Desmyter, I. Rosenberg, A. Holy, and E. De Clercq. 1988. Phosphonomethoxyethyl purine derivatives, a new class of anti-human immunodeficiency virus agents. Antimicrob. Agents Chemother. 32:1025-1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 32.Satake, H., S. Takeda, A. Matsumura, M. Sasaki, N. Sugimoto, and Y. Wataya. 1992. Action of 5-trifluoromethyl-2′-deoxyuridine on DNA synthesis. Nucleic Acids Symp. Ser. 27:189-190. [PubMed] [Google Scholar]
- 33.Satake, H., S. Takeda, and Y. Wataya. 1991. Inhibition of in vitro DNA chain elongation of 5-trifluoromethyl-2′-deoxyuridine residue in the template. Nucleic Acids Symp. Ser. 25:37-38. [PubMed] [Google Scholar]
- 34.Smeijsters, L. J. J. W., F. F. J. Franssen, L. Naesens, E. de Vries, A. Holý, J. Balzarini, E. de Clercq, and J. P. Overdulve. 1999. Inhibition of the in vitro growth of Plasmodium falciparum by acyclic nucleoside phosphonates. Int. J. Antimicrob. Agents 12:53-61. [DOI] [PubMed] [Google Scholar]
- 35.Smeijsters, L. J. J. W., N. M. Zijlstra, J. Veenstra, B. E. Verstrepen, C. Heuvel, J. P. Overdulve, and E. de Vries. 2000. Plasmodium falciparum clones resistant to (S)-9-(3-hydroxy-2-phosphonylmethoxy-propyl)adenine carry amino acid substitutions in DNA polymerase δ. Mol. Biochem. Parasitol. 106:175-180. [DOI] [PubMed] [Google Scholar]
- 36.Snoeck, R., A. Holý, C. Dewolf-Peeters, J. Van Den Oord, E. De Clercq, and G. Andrei. 2002. Antivaccinia activities of acyclic nucleoside phosphonate derivatives in epithelial cells and organotypic cultures. Antimicrob. Agents Chemother. 46:3356-3361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thai, T.-H., and J. F. Kearney. 2005. Isoforms of terminal deoxynucleotidyltransferase: developmental aspects and function. Adv. Immunol. 86:113-136. [DOI] [PubMed] [Google Scholar]
- 38.Veselý, J., A. Merta, I. Votruba, I. Rosenberg, and A. Holý. 1990. The cytostatic effects and mechanism of action of antiviral acyclic adenine nucleotide analogs in L1210 mouse leukemia cells. Neoplasma 37:105-110. [PubMed] [Google Scholar]
- 39.Votruba, I., R. Bernaerts, T. Sakuma, E. De Clercq, A. Merta, I. Rosenberg, and A. Holý. 1987. Intracellular phosphorylation of broad-spectrum anti-DNA virus agent (S)-9-(3-hydroxy-2-phosphonylmethoxypropyl)adenine and inhibition of viral DNA synthesis. Mol. Pharmacol. 32:524-529. [PubMed] [Google Scholar]
- 40.Xiong, X., J. L. Smith, and M. S. Chen. 1997. Effect of incorporation of cidofovir into DNA by human cytomegalovirus DNA polymerase on DNA elongation. Antimicrob. Agents Chemother. 41:594-599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yao, X.-D., and D. H. Evans. 2004. Construction of recombinant vaccinia viruses using leporipoxvirus-catalyzed recombination and reactivation of orthopoxvirus DNA, p. 51-64. In S. N. Isaacs (ed.), Vaccinia virus and poxvirology: methods and protocols. Humana Press, Totowa, NJ. [DOI] [PubMed]
- 42.Yokota, T., K. Konno, E. Chonan, S. Mochizuki, K. Kojima, S. Shigeta, and E. De Clercq. 1990. Comparative activities of several nucleoside analogs against duck hepatitis B virus in vitro. Antimicrob. Agents Chemother. 34:1326-1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yokota, T., S. Mochizuki, K. Konno, S. Mori, S. Shigeta, and E. De Clercq. 1990. Phosphonylmethoxyalkyl derivatives of purine as inhibitors of human hepatitis B virus DNA synthesis. Nucleic Acids Symp. Ser. 1990:17-18. [PubMed] [Google Scholar]