Abstract
We previously reported that deficiency of the lytH gene, whose product is homologous to lytic enzymes, caused the elevation of methicillin resistance in Staphylococcus aureus strain SR17238, a strain of S. aureus with a low level of resistance to methicillin (low-level MRSA) (J. Bacteriol. 179:6294-6301, 1997). In this study, we demonstrated that deficiency of lytH caused the same phenomenon in four other clinical isolates of low-level MRSA, suggesting this deficiency to exist in clinical isolates. We therefore searched the region including lytH in 127 clinical isolates of MRSA by PCR and found one strain, SR17164 (methicillin MIC, 1,600 μg/ml), in which the lytH gene was inactivated by insertion sequence IS1182. lytH::IS1182 was replaced with intact lytH in this strain by integration and excision of the plasmid carrying the lytH region. Recombinants with intact lytH genes showed methicillin MICs of 800 μg/ml, twofold lower than those of the recombinants with lytH::IS1182 and the parent. In addition, S. aureus SR17164, which has a high level of methicillin resistance, had properties similar to those caused by lytH deficiency; that is, the resistance levels of strain SR17164 and lytH-deficient variants from strain SR17238 were not significantly affected by llm inactivation, which greatly lowered resistance levels in most other high-level MRSA strains. These findings suggest that lytH inactivation contributed, to some extent, to the resistance level of S. aureus SR17164. To the best of our knowledge, this strain is the first clinical isolate of MRSA for which the genetic base for high-level resistance has been clarified.
Methicillin-resistant Staphylococcus aureus (MRSA) is one of the noteworthy pathogens in nosocomial infections. MRSA has the mecA gene, encoding a low-affinity penicillin-binding protein, designated PBP2a or PBP2′, which is involved in cell wall synthesis and renders the bacteria methicillin resistant (9, 27, 28). Clinical isolates of MRSA show various resistance levels, with methicillin MICs ranging from 3.13 to more than 1,600 μg/ml. These differences in resistance levels observed for clinical isolates could not be correlated with the amount of PBP2a produced, and some other genetic factors have been demonstrated as being involved in the expression of methicillin resistance at high levels (21, 23). However, little is known about these factors.
Various genes affecting methicillin resistance levels have been identified by the transposon mutagenesis technique. Among these genes were fem genes, llm, fmtA, and mrp (10, 11, 12, 16, 19, 29). The inactivation of these genes caused a decrease in the methicillin resistance level of MRSA. On the other hand, we were able to identify a mutation of the lytH gene homologous to a gene for the lytic enzyme N-acetylmuramoyl-l-alanine amidase, which causes an increase in the methicillin resistance level of MRSA. The chromosomal DNA of Staphylococcus aureus strain SR17238 (a strain of S. aureus with a low level of resistance to methicillin [low-level MRSA]) (methicillin MIC, 6.3 μg/ml), and that of its high-level MRSA variant SRM1648 (methicillin MIC, 1,600 μg/ml) were analyzed by two-dimensional electrophoresis, and a deletion in the rel-orf1-lytH region was found to occur in the variant (6). On the basis of the facts that we simultaneously isolated the high-level MRSA variant SRM1663, which has a deletion within only the lytH gene, when strain SRM1648 was isolated and that LytH has homology to a cell wall-hydrolyzing enzyme, we considered that the inactivation of lytH was probably responsible for the increased resistance level. This variant was laboratory generated, however, and the question arose as to whether the lytH deletion observed in the laboratory would also occur in clinical settings.
In this paper, we describe a clinical isolate with a high level of methicillin resistance and with lytH disrupted by the transposition of an insertion sequence (IS).
MATERIALS AND METHODS
Bacteria and plasmids.
A total of 131 MRSA strains had been isolated from clinical specimens, such as sputum, blood, and skin, at 20 hospitals in Japan in 1992, 1994, 1995, and 2000. Four strains had low levels of resistance to methicillin, and the others had high levels of resistance. The S. aureus strains and plasmids used for transformation in this study are listed in Table 1.
TABLE 1.
S. aureus strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s)a | Source or reference |
|---|---|---|
| Strains | ||
| SR17238 | Low-level Mcr | 6 |
| SRM1648 | Variant (Δrel-lytH) from SR17238, high-level Mcr | 6 |
| SRM1663 | Variant (ΔlytH) from SR17238, high-level Mcr | 6 |
| SR17164 | Clinical isolate (lytH::IS1182), high-level Mcr | This study |
| SR20118 | Clinical isolate, low-level Mcr | This study |
| SR20137 | Clinical isolate, low-level Mcr | This study |
| SR20202 | Clinical isolate, low-level Mcr | This study |
| SR20280 | Clinical isolate, low-level Mcr | This study |
| SRM551 | High-level Mcr | 19 |
| SRM563 | Transformant (llm::Tn918) from SRM551, low-level Mcr | 19 |
| SRM1816 | Transformant (Δrel-lytH llm::Tn918) from SRM1648 | This study |
| SRM1825 | Transformant (ΔlytH llm::Tn918) from SRM1663 | This study |
| SRM1832 | Transformant (lytH::IS1182 llm::Tn918) from SR17164 | This study |
| RN4220 | Restriction-deficient strain | 13 |
| Plasmids | ||
| pCL52.1 | E. coli-S. aureus shuttle vector, temperature sensitive, Tetr | 14 |
| pSR100 | S. aureus plasmid, Cmr | 6 |
| pSR107 | pBluescript II with HindIII fragment including rel-hisS region from SR17238 | This study |
| pSR108 | pSR100 with HindIII fragment of rel-hisS region from SRM1648 | 6 |
| pSR428 | pCL52.1 with ScaI-PstI fragment of rel-hisS region from SR17238 | This study |
| pSR616 | pSR100 with HindIII fragment of rel-hisS region from SRM1663 | This study |
Low-level Mcr; low level of methicillin resistance; high-level Mcr, high level of methicillin resistance; Cm, chloramphenicol.
Susceptibility test.
MIC was determined by a serial twofold dilution method in tryptic soy agar (TSA) (Becton Dickinson, Sparks, MD). Overnight cultures of S. aureus strains in tryptic soy broth (TSB) (Becton Dickinson) were diluted to about 106 CFU/ml. Bacterial suspensions of 1 μl were spotted onto agar plates containing serial twofold dilutions of antibiotics and incubated for 24 h at 37°C before the MICs were scored.
Replacement of the mutated lytH gene with an intact one.
An S. aureus strain with an intact lytH gene in the chromosome was constructed using pCL52.1, a temperature-sensitive and tetracycline (Tet)-resistant Escherichia coli-S. aureus shuttle vector, by the method of Lin et al. (14). The ScaI-PstI fragment of the intact rel-hisS region (Fig. 1) from plasmid pSR107 (Table 1) was recloned into the SmaI-PstI site of pCL52.1, generating plasmid pSR428. This plasmid was electroporated into MRSA strain SR17164, having lytH::IS1182, via the restriction-deficient S. aureus strain RN4220 (13). The resultant transformant was grown in TSB containing 3 μg/ml of Tet at 30°C overnight. After dilution with fresh TSB containing Tet, the culture was incubated for 8 h at 42°C and then spread onto TSA plates containing Tet and the incubation was continued overnight at 42°C to select Tet-resistant cointegrants. To confirm integration of the plasmid into the rel-lytH region in chromosomal DNA, colonies grown at 42°C were checked by PCR with primers 1 and 2, described below. A single colony of cointegrants was grown in drug-free TSB at 30°C to excise the integrated plasmid from the chromosome. Cells from this culture were spread onto a drug-free TSA plate and incubated for 2 days at 30°C, and Tet-susceptible colonies were selected by the replica plate method.
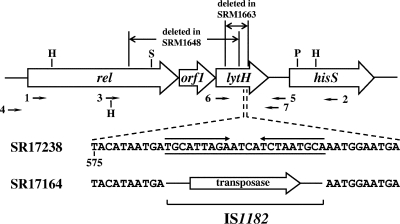
FIG. 1.
Scheme of open reading frame organization in the rel-hisS region, including lytH. The regions deleted in SRM1648 and SRM1663, reported in reference 6, are shown at the top of the figure. HindIII, ScaI, and PstI sites used for cloning experiments in this study are noted as H, S, and P, respectively. The arrows labeled 1 to 7 indicate the positions of the nucleotide sequences corresponding to PCR primers and the directions of the extension of DNA chain synthesized by PCR. Nucleotide sequences for a part of the lytH gene are shown below the site where the IS1182 insertion occurred. The upper and lower sequences are for strains SR17238 and SR17164, respectively. Inverted repeat sequences are shown with arrows above the sequence of SR17238. In SR17164, the sequence underlined was deleted and IS1182 containing the putative transposase gene was inserted instead.
PCR.
PCR was performed with ExTaq DNA polymerase (Takara Shuzo, Japan), as described previously (6), with modification of the extension reaction, which was performed at 66°C for 7 min. The sequences of the primers used were 5′GATTAGACGGACCGACGATTGTCGCAGGT for primer 1, 5′CTGGTTCAAAGTGTTTCACTAACGCTTCG for primer 2, 5′GATGGTTGAAGAAGCTTTAAAAGAGCAAGG for primer 3, 5′TATCGTCGACGGACGTATGATTGGTGTGGG for primer 4, 5′ACGAATATCCGTGTTGCCACCCTCATTGGT for primer 5, 5′AGTAGTGATTGCCTTTGTC for primer 6, and 5′TAAATAAACCCACTCAAAACG for primer 7. The nucleotide sequence underlined in primer 3 is the HindIII site. Their positions are shown schematically in Fig. 1.
Transformation.
The transformation of S. aureus strains with plasmids by electroporation was performed by a method described previously (6). The transformants were selected on the plates containing 3 μg/ml of Tet or 25 μg/ml of chloramphenicol. Transformation with chromosomal DNA of S. aureus was carried out using competent recipient cells prepared by treatment of the bacteria with helper phage 55 and 0.1 M CaCl2 (19).
Sequence analysis.
The nucleotide sequence was determined by the dideoxy chain termination method (24) with the automated sequencer ABI Prism 377 (PerkinElmer, MA).
RESULTS
Consequence of deletion of the rel-lytH region in clinical isolates.
In a previous report, we demonstrated that the deletion of lytH resulted in the elevation of the methicillin resistance level of the low-level MRSA strain SR17238 (6). To investigate whether these results could be extended to other clinical isolates, four MRSA strains, SR20118, SR20137, SR20202, and SR20280, with low levels of methicillin resistance (methicillin MICs, 12.5, 12.5, 25, and 6.3 μg/ml, respectively), were transformed by recombinant plasmid pSR108 (Table 1), which has the HindIII fragment of the rel-hisS region of the high-level MRSA strain SRM1648 with a deletion in the rel-lytH region (Fig. 1). Each transformant generated was cultivated in drug-free TSB at 37°C overnight, spread onto TSA plates containing 400 μg/ml of methicillin, and incubated at 37°C for 3 days. As described in our previous report (6), strain SRM1665, the transformant of SR17238 with pSR108, showed a frequency of colonies with high levels of resistance (growing on TSA containing methicillin) that was 103-fold higher than the frequencies of SR17238 and SRM1664, which is the transformant of SR17238 with the empty plasmid pSR100 (Table 2). Strains SRM2148, SRM2149, SRM2150, and SRM2151, the transformants of SR20118, SR20137, SR20202, and SR20280, respectively, also showed frequencies for colonies with high levels of resistance that were 3- to 84-fold higher than the frequencies for the parent strains, although the increases in frequency differed among the strains tested. In 24 colonies of each transformant, for which a high level of methicillin resistance was confirmed by regrowth on TSA plates containing 400 μg/ml of methicillin, we investigated a deletion in their chromosomal region from upstream of rel to downstream of lytH by PCR with primers 1 and 2. All tested colonies of SRM2148, SRM2149, and SRM2450, and 58.3% of tested colonies of SRM2151, gave PCR products of 2.8 kb, as strain SRM1665 did (Table 2). These results suggested that these colonies with high levels of resistance had undergone integration and excision of the plasmid at the rel-lytH region and consequently had undergone deletion within the rel-lytH region, as observed in SRM1648, and then acquired a high level of methicillin resistance. Therefore, clinical isolates of low-level MRSA other than SR17238 can also raise their resistance levels by a deletion in the rel-lytH region.
TABLE 2.
Frequency of high-level resistance in colonies of transformants of clinical strains of low-level MRSA with the plasmid harboring a deletion in the rel-lytH region
| Bacterial strain | Parent/plasmid | MIC of methicillin (μg/ml) | Frequency of resistant coloniesa | Frequency of deletion in the chromosomal rel-lytH regionb (%) |
|---|---|---|---|---|
| SR17238 | 6.3 | 1.7 × 10−6 | ||
| SRM1664 | SR17238/pSR100 | 6.3 | 1.1 × 10−6 | |
| SRM1665 | SR17238/pSR108 | 25 | 1.3 × 10−3 | 100 |
| SR20118 | 12.5 | 6.6 × 10−8 | ||
| SRM2148 | SR20118/pSR108 | 12.5 | 5.5 × 10−6 | 100 |
| SR20137 | 12.5 | 5.6 × 10−7 | ||
| SRM2149 | SR20137/pSR108 | 25 | 3.8 × 10−5 | 100 |
| SR20202 | 25 | 5.4 × 10−6 | ||
| SRM2150 | SR20202/pSR108 | 50 | 1.3 × 10−4 | 100 |
| SR20280 | 6.3 | 2.6 × 10−6 | ||
| SRM2151 | SR20280/pSR108 | 6.3 | 7.8 × 10−6 | 58.3 |
Frequency of colonies grown on the agar plates containing 400 μg/ml methicillin, which was spread with the overnight culture in methicillin-free broth and incubated for 3 days at 37°C.
In 24 colonies of each transformant, for which their high-level resistance was confirmed by regrowth on agar plates containing 400 μg/ml of methicillin, deletion in the chromosomal rel-lytH region was investigated by the PCR method with primers 1 and 2 as described in Materials and Methods.
To confirm that the deletion within the lytH gene causes an elevation in resistance, the rel-hisS region of the high-level methicillin-resistant variant SRM1663 with a 0.5-kb deletion in lytH was amplified by PCR with primers 2 and 3 and the HindIII fragment of this PCR product was cloned into the HindIII site of staphylococcal plasmid pSR100, generating pSR616 (Table 1 and Fig. 1). After the transformation of strain SR20280 with pSR616, the transformant was cultivated in drug-free TSB and bacterial cells were grown on TSA plates containing 400 μg/ml of methicillin as described above. The incidence of colonies with high levels of resistance was 4.8 × 10−6, which was twofold higher than that of the parent strain, SR20280, and 8.8% of 24 colonies tested were found to have a deletion in lytH, as observed for strain SRM1663. These results indicated that methicillin resistance was elevated by the inactivation of only lytH.
Detection of a lytH-deficient strain among clinical isolates.
Most clinical isolates of MRSA in Japan showed low levels of methicillin resistance until the 1980s (20); however, now they show high levels of resistance. We checked whether these strains resulted from a deficiency of lytH. By PCR with primers 4 and 5, we examined the rel-hisS region in 127 clinical isolates of high-level MRSA, of which 73% showed especially high methicillin MICs of 400 to 3,200 μg/ml. Although 126 out of 127 isolates gave the same 4.0-kb fragments as did low-level MRSA strain SR17238 (6), one strain, SR17164, with a methicillin MIC of 1,600 μg/ml, gave a product of 5.8 kb. This result indicated that a 1.8-kb fragment had been inserted into the rel-lytH region. Further PCR analysis with primers 6 and 7 revealed that the insertion site was located within lytH.
Sequence analysis of the DNA fragment of the lytH gene in strain SR17164 amplified by PCR with primers 6 and 7 revealed the presence of an IS, IS1182 (4), in lytH (Fig. 1). A pair of 9-bp inverted repeat sequences with a 4-bp spacer in the lytH gene of strain SR17238 (6) was deleted in strain SR17164, and IS1182 was inserted at this site instead. The translation of LytH in strain SR17164 was discontinued at TAG in the 5′ end of IS1182, and this alteration seemed to interfere with the expression of lytH. Sequence comparison with other IS1182s registered with GenBank (accession no. L43082 and L43098) indicated that IS1182 in lytH had an additional G and C at the 5′ and 3′ ends, respectively, and a substitution of G for T in a stop codon located in the middle of the transposase gene in the registered sequence. Consequently, this stop codon was replaced by a codon for glutamate and active transposase seemed to be produced in S. aureus SR17164. In the literature, a single copy of IS1182 in S. aureus BM3651 has also been reported to have the glutamate codon at the same position as seen that in strain SR17164 (4). This sequence is thought to encode normal transposase.
Association of lytH inactivation with high-level resistance in S. aureus SR17164.
To examine the association of the inactivation of lytH by IS1182 insertion with high-level methicillin resistance in S. aureus SR17164, the lytH::IS1182 region in its chromosome was replaced with intact lytH by Campbell-type integration and excision as described in Materials and Methods. When the cointegrant in which the plasmid pSR428 was integrated into the chromosome was incubated at 30°C to excise the integrated plasmid, it produced two kinds of recombinants, one with the intact lytH gene and the other with lytH::IS1182. MICs of methicillin and imipenem were determined for 10 strains each with the intact lytH gene or lytH::IS1182 in the chromosome. The susceptibility of the former to both antibiotics was found to be twofold higher than that of the latter (Table 3). Although the resistance level of strain SR17164 did not decrease significantly upon replacement of lytH::IS1182 with intact lytH, these results showed that the inactivation of lytH further raised the resistance level in S. aureus SR17164. Because the resistance level in SR17164 was unstable after several repeated cultivations, despite no alteration in lytH::IS1182, suggesting that some other factors are also involved with high level of resistance of this strain, competent cells of SR17164 in this experiment were prepared from cultures grown in the presence of 400 μg/ml of methicillin, which had little influence on the population of SR17164. This cultivation might raise the basal resistance level. Another possibility was that the putative clinical strain which produced strain SR17164 after the transposition of IS1182 into lytH had already possessed a high level of resistance.
TABLE 3.
Methicillin resistance of recombinants with or without deletion of the rel-lytH genes
| Bacterial straina | Genotype | MIC (μg/ml)
|
|
|---|---|---|---|
| Methicillin | Imipenem | ||
| SR17238 | Intact lytH | 6.3 | ≤0.1 |
| SRM1648 | Δrel-lytH | 1,600 | 50 |
| SR17164 | lytH::IS1182 | 1,600 | 100 |
| SR17164 recombinantb | Intact lytH | 800 | 50 |
| SR17164 recombinantb | lytH::IS1182 | 1,600 | 100 |
Details of each strain are listed in Table 1.
All 10 strains tested had the same MICs.
To further investigate the influence of lytH inactivation on high-level resistance, we employed the llm gene, which is similar to the teichoic acid linkage unit synthesis gene tagO from Bacillus subtilis and affects the methicillin resistance level (19, 26). Maki et al. demonstrated that a Tn918 insertional mutation of this gene caused a decrease in methicillin resistance in 16 out of 17 clinical strains of high-level MRSA. An exception was S. aureus SR17164 (19). Its high-level resistance was not greatly influenced by the insertion of Tn918 into llm. We next examined the influence of the insertional llm mutation on the resistance level of lytH mutants. S. aureus strains SRM1648, SRM1663, and SR17164 were transformed with chromosomal DNA of MRSA strain SRM563 with a Tn918 insertion in llm (19), producing the resultant transformants SRM1816, SRM1825, and SRM1832, respectively (Table 1). MICs of methicillin, imipenem, and cefazolin for the transformants indicated that they remained highly resistant, though their MICs decreased to some extent (Table 4). These results suggested that the exceptional properties of high-level MRSA SR17164 resulted from lytH inactivation.
TABLE 4.
Effect of llm inactivation on the methicillin resistance of lytH-deficient MRSA strains
| Bacterial straina | Genotypeb
|
MIC (μg/ml)
|
|||
|---|---|---|---|---|---|
| lytH | llm | Methicillin | Imipenem | Cefazolin | |
| SRM551 | + | + | 800 | 50 | 200 |
| SRM563 | + | − | 12.5 | 0.39 | 12.5 |
| SR17164 | − | + | 1,600 | 100 | 400 |
| SRM1832 | − | − | 800 | 50 | 200 |
| SRM1648 | − | + | 1,600 | 100 | 400 |
| SRM1816 | − | − | 100 | 25 | 100 |
| SRM1663 | − | + | 1,600 | 100 | 400 |
| SRM1825 | − | − | 400 | 50 | 200 |
Details of each strain are listed in Table 1.
+, the gene is active; −, the gene is inactivated by mutation.
DISCUSSION
Bacterial variants with high levels of methicillin resistance emerge spontaneously from strains with low levels of methicillin resistance at relatively high frequencies when selected by β-lactam antibiotics. The genetic factors involved in this phenomenon were long unknown. We succeeded for the first time in identifying lytH inactivation as one of the factors for MRSA strain SR17238 (6). In this study, we further demonstrated that a deletion in lytH caused an elevation in the resistance level even in other clinical isolates. These results indicated that an increase in resistance upon lytH inactivation would occur in many clinical isolates of MRSA and be a general phenomenon, not specific to strain SR17238 used in the previous study. Although we used the recombinant plasmid having a deletion in rel-lytH in some of the experiments, the following observations make it unlikely that an alteration of rel or orf1 other than lytH caused an increased resistance level. First, two high-level MRSA variants from SR17238, which are SRM1648 with a deletion in the rel-lytH region and SRM1663 with a deletion within the lytH gene, showed the same methicillin resistance levels (6). Second, the clinical isolate SR20280 with the plasmid harboring the deleted region from SRM1663 (within lytH) showed an increased frequency of colonies with high levels of resistance, as did the same isolate with the plasmid harboring the deleted region from SRM1648.
Finding the lytH-deficient strain SR17164 among 127 MRSA clinical isolates implies that the lytH mutation had actually occurred in clinical isolates and raised the methicillin resistance level. The contribution of lytH mutation to the resistance level was relatively small in this strain, suggesting that other unknown factors are also involved with the high level of resistance of this strain. Although it could not be clear which of lytH inactivation and other factors occurred first in SR17164, it is likely that the inactivation of lytH exerted at least some influence on the resistance level of SR17164, because lytH inactivation caused increased resistance level- in other clinical isolates of low-level MRSA. This consideration was also supported by the fact that the high-level methicillin resistance trait of SR17164 was characteristic of that caused by lytH deficiency, as shown in llm mutation experiments. That is, the inhibition of Llm decreased the resistance level in most clinical isolates of high-level MRSA, but this was not the case for SR17164 and the high-level MRSA mutants with the deletion of lytH (19). Because Llm is predicted to be involved in the synthesis of the teichoic acid linkage unit (26), the anionic cell wall polymer has some influence on high-level methicillin resistance but not on that caused by lytH deletion. The mechanisms underlying this phenomenon are not clear. To the best of our knowledge, strain SR17164 is the first clinical isolate of MRSA to have its genetic base for high-level methicillin resistance clarified. The frequency of detection of lytH-deficient strains among clinical isolates in the present study nearly agrees with the frequency of the deletion in the rel-lytH region reported previously, i.e., 0.74% (6). The high-level resistance of most clinical isolates, therefore, was not due to lytH inactivation. Mutations of several other genetic factors are likely involved in high-level methicillin resistance, thus suggesting the existence of multiple evolutional routes to high-level methicillin resistance.
It is worth noting that the mutation of lytH was caused by the transposition of IS1182. As the transposase of IS1182 within lytH of strain SR17164 was active, this IS is likely to be transposed into lytH and the resultant high-level MRSA strain SR17164 was selected in the presence of a high concentration of β-lactam in clinical settings. Several IS elements have been described for S. aureus (1, 3, 4, 8, 15). Hybridization patterns of IS elements varied among strains, as was shown for IS256 (5), and the transposition of some IS elements has been demonstrated to occur frequently in staphylococcal cells (2, 18, 22). IS elements were reported to be involved in the expression of resistance through the inactivation or activation of a gene. Examples of the former are IS1182 insertional inactivation of lytH as described in this paper and IS256 insertional inactivation of tcaA, which increased the glycopeptide resistance of S. aureus (17). Examples of the latter are the formation of hybrid promoters by IS256 transposition to enhance the expression of mutated llm (18) and IS257 transposition to direct the transcription of a tetracycline resistance gene, tetA (25). The IS element was also shown to control the synthesis of adhesins (30). In this way, IS elements seem to modulate the expression of various genes under the various environments encountered by the bacteria.
A comparison of the nucleotide sequences of the lytH genes of strains SR17164 and SR17238 showed that the inverted repeat sequence located in this gene was deleted at the insertion site of IS1182. Although these two strains were not isogenic, the deletion of the target sequence and the insertion of IS1182 probably occurred simultaneously. Generally, the insertion of IS elements often accompanies the duplication of several base pair sequences (7). Composite transposon Tn5405, with two copies of IS1182 at both ends, was also flanked by direct repeats of 8 bp (4). These observations, taken together with the insertion pattern observed for strain SR17164, indicate the likelihood of two ways of transposition for IS1182, i.e., concomitant deletion of inverted repeat sequences at the insertion site and duplication of the target sequence.
We previously reported that the deleted region in strain SRM1663 was flanked by 8-bp direct repeats at the site where the recombination and excision took place (6). Although the ways of mutation differ between strains SR17164 and SRM1663, peculiar nucleotide sequences, such as the direct repeat sequence and the inverted repeat sequence, are often locations that suffer some mutation. Therefore, the existence of such peculiar sequences in a genetic factor makes it easy for mutations to occur.
In conclusion, the insertional inactivation of lytH by IS1182 was demonstrated to cause an elevation of the methicillin resistance level in the clinical isolate of MRSA. Transposition of the IS element was suggested to contribute to the frequent emergence of high-level MRSA strains.
Footnotes
Published ahead of print on 10 December 2007.
REFERENCES
- 1.Barberis-Maino, L., B. Berger-Bächi, H. Weber, W. D. Beck, and F. H. Kayser. 1987. IS431, a staphylococcal insertion sequence-like element related to IS26 from Proteus vulgaris. Gene 59:107-113. [DOI] [PubMed] [Google Scholar]
- 2.Chesneau, O., R. Lailler, A. Derbise, and N. E. Solh. 1999. Transposition of IS1181 in the genomes of Staphylococcus and Listeria. FEMS Microbiol. Lett. 177:93-100. [DOI] [PubMed] [Google Scholar]
- 3.Derbise, A., K. G. H. Dyke, and N. E. Solh. 1994. Isolation and characterization of IS1181, an insertion sequence from Staphylococcus aureus. Plasmid 31:251-264. [DOI] [PubMed] [Google Scholar]
- 4.Derbise, A., K. G. H. Dyke, and N. E. Solh. 1996. Characterization of a Staphylococcus aureus transposon, Tn5405, located within Tn5404 and carrying the aminoglycoside resistance genes, aphA-3 and aadE. Plasmid 35:174-188. [DOI] [PubMed] [Google Scholar]
- 5.Dyke, K. G. H., S. Aubert, and N. E. Solh. 1992. Multiple copies of IS256 in staphylococci. Plasmid 28:235-246. [DOI] [PubMed] [Google Scholar]
- 6.Fujimura, T., and K. Murakami. 1997. Increase of methicillin resistance in Staphylococcus aureus caused by deletion of a gene whose product is homologous to lytic enzyme. J. Bacteriol. 179:6294-6301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Galas, D. J., and M. Chandler. 1989. Bacterial insertion sequences, p. 109-162. In D. E. Berg and M. M. Howe (ed.), Mobile DNA. American Society for Microbiology, Washington, DC.
- 8.Gillespie, L. T., B. R. Lyon, L. S. L. Loo, P. R. Matthews, P. R. Stewart, and R. A. Skurray. 1987. Homologous direct repeat sequences associated with mercury, methicillin, tetracycline and trimethoprim resistance determinants in Staphylococcus aureus. FEMS Microbiol. Lett. 43:165-171. [Google Scholar]
- 9.Hartman, B. J., and A. Tomasz. 1984. Low-affinity penicillin-binding protein associated with β-lactam resistance in Staphylococcus aureus. J. Bacteriol. 158:513-516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Henze, U., T. Sidow, J. Wecke, H. Labischinski, and B. Berger-Bächi. 1993. Influence of femB on methicillin resistance and peptidoglycan metabolism in Staphylococcus aureus. J. Bacteriol. 175:1612-1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jolly, L., S. W. Wu, J. van Heijenoort, H. de Lencastre, D. Megin-Lecreulx, and A. Tomasz. 1997. The femR315 gene from Staphylococcus aureus, the interruption of which results in reduced methicillin resistance, encodes a phosphoglucosamine mutase. J. Bacteriol. 179:5321-5325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Komatsuzawa, H., K. Ohta, H. Labischinski, M. Sugai, and H. Suginaka. 1999. Characterization of fmtA, a gene that modulates the expression of methicillin resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 43:2121-2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kreiswirth, B. N., S. Löfdahl, M. J. Betley, M. O'Reilly, P. M. Schlievert, M. S. Bergdoll, and R. P. Novick. 1983. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature 305:709-712. [DOI] [PubMed] [Google Scholar]
- 14.Lin, W. S., T. Cunneen, and C. Y. Lee. 1994. Sequence analysis and molecular characterization of genes required for the biosynthesis of type 1 capsular polysaccharide in Staphylococcus aureus. J. Bacteriol. 176:7005-7016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lyon, B. R., M. T. Gillespie, and R. A. Skurray. 1987. Detection and characterization of IS256, an insertion sequence in Staphylococcus aureus. J. Gen. Microbiol. 133:3031-3038. [DOI] [PubMed] [Google Scholar]
- 16.Maidhof, H., B. Reinicke, P. Blumel, B. Berger-Bächi, and H. Labischinski. 1991. femA, which encodes a factor essential for expression of methicillin resistance, affects glycine content of peptidoglycan in methicillin-resistant and methicillin-susceptible Staphylococcus aureus strains. J. Bacteriol. 173:3507-3513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maki, H., N. McCallum, M. Bischoff, A. Wada, and B. Berger-Bächi. 2004. tcaA inactivation increases glycopeptide resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 48:1953-1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maki, H., and K. Murakami. 1997. Formation of potent hybrid promoters of the mutant llm gene by IS256 transposition in methicillin-resistant Staphylococcus aureus. J. Bacteriol. 179:6944-6948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maki, H., T. Yamaguchi, and K. Murakami. 1994. Cloning and characterization of a gene affecting the methicillin resistance level and the autolysis rate in Staphylococcus aureus. J. Bacteriol. 176:4993-5000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Murakami, K., K. Nomura, M. Doi, and T. Yoshida. 1987. Production of low-affinity penicillin-binding protein by low- and high-resistance groups of methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 31:1307-1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Murakami, K., and A. Tomasz. 1989. Involvement of multiple genetic determinants in high-level methicillin resistance in Staphylococcus aureus. J. Bacteriol. 171:874-879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Needham, C., W. C. Noble, and K. G. H. Dyke. 1995. The staphylococcus insertion sequence IS257 is active. Plasmid 34:198-205. [DOI] [PubMed] [Google Scholar]
- 23.Ryffel, C., A. Strässle, F. H. Kayser, and B. Berger-Bächi. 1994. Mechanisms of heteroresistance in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 38:724-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Simpson, A. E., R. A. Skurray, and N. Firth. 2000. An IS257-derived hybrid promoter directs transcription of a tetA(K) tetracycline resistance gene in the Staphylococcus aureus chromosomal mec region. J. Bacteriol. 182:3345-3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Soldo, B., V. Lazarevic, and D. Karamata. 2002. tagO is involved in the synthesis of all anionic cell-wall polymers in Bacillus subtilis 168. Microbiology 148:2079-2087. [DOI] [PubMed] [Google Scholar]
- 27.Song, M. D., M. Wachi, M. Doi, F. Ishino, and M. Matsuhashi. 1987. Evolution of an inducible penicillin-target protein in methicillin-resistant Staphylococcus aureus by gene fusion. FEBS Lett. 221:167-171. [DOI] [PubMed] [Google Scholar]
- 28.Utsui, Y., and T. Yokota. 1985. Role of an altered penicillin-binding protein in methicillin- and cephem-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 28:397-403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu, S. W., and H. de Lencastre. 1999. Mrp—a new auxiliary gene essential for optimal expression of methicillin resistance in Staphylococcus aureus. Microb. Drug Res. 5:9-18. [DOI] [PubMed] [Google Scholar]
- 30.Ziebuhr, W., V. Krimmer, S. Rachid, I. Lossner, F. Gotz, and J. Hacker. 1999. A novel mechanism of phase variation of virulence in Staphylococcus epidermidis: evidence for control of the polysaccharide intercellular adhesin synthesis by alternating insertion and excision of the insertion sequence element IS256. Mol. Microbiol. 32:345-356. [DOI] [PubMed] [Google Scholar]