Abstract
Antimicrobial peptides are short, positively charged, amphipathic peptides that possess a wide spectrum of antimicrobial activity and have an important role in the host's innate immunity. Lack of, or dysfunctions in, antimicrobial peptides have been correlated with infectious diseases, including periodontitis. Porphyromonas gingivalis, a gram-negative anaerobe and a major pathogen associated with periodontal diseases, is resistant to antimicrobial peptides of human and nonhuman origin, a feature that likely contributes to its virulence. Expressing a robust proteolytic activity, P. gingivalis hydrolyzes antimicrobial peptides. In this study, P. gingivalis inactivated three antimicrobial peptides, while a d-enantiomer was resistant to degradation. P. gingivalis was resistant to the protease-resistant d-enantiomer peptide, and importantly, a protease-deficient P. gingivalis mutant was also resistant to the antimicrobial peptide. Finally, the binding of a fluorescently labeled antimicrobial peptide to protease-deficient P. gingivalis was much weaker than the binding of susceptible Escherichia coli. Our results suggest that the resistance of P. gingivalis ATCC 33277 to direct killing by antimicrobial peptides is protease independent and results (at least partially) from the low affinity of antimicrobial peptides to P. gingivalis.
Antimicrobial peptides are components of the innate immunity that mediate a broad range of antimicrobial activity. More than 800 cationic peptides have been described for insects, vertebrates, and humans (www.bbcm.units.it/∼tossi/pag1.htm). Though highly diverse, antimicrobial peptides share the features of a net positive charge and the ability to adopt an amphipathic structure in solution. The peptides are believed to kill bacteria through a multiple hit mechanism, whose targets can include the outer and inner membranes as well as cytoplasmic components (16). Thus, antimicrobial peptides seem to escape many of the bacterial drug resistance mechanisms and often show a synergistic effect with conventional antibiotics (44).
In mammals, antimicrobial peptides were also found to function as immunomodulators of the innate immune system that alter gene expression in host cells, induce or modulate chemokine and cytokine production, and elicit or inhibit a proinflammatory response (3, 38, 43). Since the recognition of their immunoregulatory functions, antimicrobial peptides are often referred to as host defense peptides.
In the human oral cavity, several kinds of antimicrobial peptides are found. These include α- and β-defensins, histatins, and LL-37 (6, 20, 34). Periodontal disease is a common bacterium-induced inflammatory disease (36) in which the tooth-supporting tissues are attacked. Deficiency of LL-37 in neutrophils and in saliva has been correlated with the occurrence of severe periodontal disease (34).
Porphyromonas gingivalis, the oral pathogen most associated with chronic periodontal disease (40), is resistant to many antimicrobial peptides (1, 15, 27). P. gingivalis is known for its robust proteolytic activity, in particular, the expression of extracellular Arg-gingipain and Lys-gingipain cysteine proteases that cleave at the C termini of arginine and lysine, respectively. Arginine and lysine, both being positively charged amino acids, are highly represented in cationic antimicrobial peptides. Therefore, it was rational to hypothesize that gingipains are important for the resistance of P. gingivalis to antimicrobial peptides. Our results, however, suggest that although P. gingivalis readily digests antimicrobial peptides, its ability to escape killing by antimicrobial peptides is independent of its proteolytic capacity.
Peptide synthesis is relatively straightforward; therefore, antimicrobial peptides present an appealing approach for controlling microbial pathogens. Better understanding of the mechanisms of bacterial resistance to antimicrobial peptides provides insight for the development of effective antibacterial therapies.
MATERIALS AND METHODS
Peptides.
The three antimicrobial peptides used in this study have been described by us before (1). Dhvar4 (KRLFKKLLFSLRKY-NH2; molecular weight [MW], 1,839.3) is a derivative of human histatin 5, a member of a family of antifungal and antibacterial histidine-rich proteins secreted from salivary glands (13, 17-19). K4-S4(1-15)a (ALWKTLLKKVLKAAA-NH2; MW, 1,653.1) is a short derivative of dermaseptin S4, a member of the dermaseptin family of antimicrobial peptides, isolated from the skin of tree frogs (11, 23). Human LL-37 (LLGDFFRKSKEKIGKEFKRIVQRIKDFLRNLVPRTES; MW, 4,493.3) is the only member of the cathelicidin family in humans and is cleaved and released from the C terminus of the cathelicidin hCAP18 propeptide secreted from granules of neutrophils (41) and from epithelial cells (2).
We synthesized the peptides by the solid-phase method (12), applying Fmoc (9-fluorenylmethyloxycarbonyl) active ester chemistry on an Applied Biosystems model 433A peptide synthesizer (Foster City, CA) as described before (25). The crude peptides were purified to more than 95% chromatographic homogeneity by reversed-phase high-performance liquid chromatography (Alliance-Waters, Milford, MA). The purified peptides were subjected to amino acid analysis and electrospray mass spectrometry (Micromass ZQ; Waters, Milford, MA) to confirm their composition and were stored as lyophilized powder at −20°C.
Bacterial strains and growth conditions.
Streptococcus mutans ATCC 27351 was cultured in brain heart infusion broth (BHI) (Difco, MD) at 37°C in an atmosphere enriched with 5% CO2. P. gingivalis ATCC 33277 was grown in Wilkin's broth (Oxoid, United Kingdom). P. gingivalis KDP136 (P. gingivalis ATCC 33277 mutant inactivated in rgpA, rgpB, and kgp), generously supplied by Koji Nakayama (39), was grown in enriched BHI broth (containing, per liter, 37 g of BHI [Difco], 5 g of yeast extract [Difco], 1 g of cysteine, 5 mg of hemin, and 1 mg of vitamin K1) supplemented with chloramphenicol, 20 μg/ml; erythromycin, 10 μg/ml; and tetracycline, 0.7 μg/ml. P. gingivalis strains were grown in jars containing an anaerobic atmosphere generation system (Oxoid, United Kingdom). Aggregatibacter (formerly Actinobacillus) actinomycetemcomitans ATCC 29523 (26) was cultured in 0.5% yeast extract, 1.5% Bacto tryptone, 0.75% d-glucose, 0.25% NaCl, 0.075% l-cysteine, 0.05% sodium thioglycolate, and 4% NaHCO3 at 37°C in 5% CO2. Escherichia coli ATCC 25922 was grown in BHI under aerobic conditions.
Effect of preincubation with bacterial culture supernatant on peptide antimicrobial activity.
Cultures of P. gingivalis or A. actinomycetemcomitans were grown to stationary phase and centrifuged for 10 min at 10,000 × g in a microcentrifuge (Eppendorf, Germany). Supernatants were collected and filter sterilized (0.2 μm; Whatman Schleicher & Schuell, Germany). Peptides K4-S4(1-15)a (100 μg/ml final concentration), Dhvar4a (800 μg/ml), LL-37 (2,000 μg/ml), and D-K4-S4(1-15)a (100 μg/ml) were incubated with 20% (final) P. gingivalis or A. actinomycetemcomitans filtrates for the indicated time.
Overnight cultures of bacteria were diluted 1:10 (for S. mutans) or 1:5,000 (for E. coli) in culture medium, sonicated for 1 min in a sonication bath (Transsonic T 640; ELMA, Germany), and distributed in aliquots of 190 μl into 96-well plates (Nunc, Denmark). Ten microliters of supernatant-treated peptides was added [K4-S4(1-15)a and Dhvar4a were added to S. mutans cultures, and LL-37 was added to an E. coli culture]. Plates were incubated at the appropriate growth conditions. Bacterial growth was determined by measurements of the optical density at 650 nm (OD650) using a ThermoMax microplate spectrophotometer (Molecular Devices). Percent inhibition of bacterial growth by the supernatant-treated peptides was calculated and compared to that for cells where fresh medium was added instead of peptides (no peptide, 0% growth inhibition) and controls where peptides were preincubated with fresh medium instead of culture supernatant (nontreated peptide, 100% growth inhibition). Data are means and standard deviations of three independent experiments performed in triplicate.
The MIC in planktonic bacteria was determined using a microdilution assay as described previously (1).
Fluorescent labeling of peptides.
Peptides (1.5 mg) were labeled using the Alexa Fluor 546 protein labeling kit (Molecular Probes, Oregon) and purified using Bio-Gel P-2 (Bio-Rad, Hercules, CA). The first 0.5-ml elution fraction collected (containing the majority of the labeled peptide) was used. Quantification of the labeled LL-37 was measured with a NanoDrop ND-1000 UV-Vis spectrophotometer at 226 nm (NanoDrop Technologies, Wilmington, DE). A calibration curve was made using known amounts of LL-37 at 226 nm to enable estimation of the concentration of the purified, fluorescently labeled LL-37.
Attachment of fluorescently labeled peptide to P. gingivalis and to E. coli.
Overnight bacterial cultures were diluted (1:5 for P. gingivalis KDP136 and 1:100 for E. coli ATCC 25922) and grown to an OD600 of approximately 1 (approximately 8 h for P. gingivalis and 3 h for E. coli). Cells were sedimented for 3 min at 10,000 × g and brought to an OD600 of 1 in phosphate-buffered saline (PBS). Labeled peptides (0 [negative control], 1, 4, or 16 μl [estimated concentration 0.8 mg/ml]) were added to 100 μl of bacterial suspension and incubated at room temperature for 10 min. Cells were washed in 0.5 ml PBS, resuspended in 0.1 ml PBS, and transferred to wells of a 96-well microtiter plates (Nunc, Denmark). Fluorescence of bacterium-attached labeled peptide was determined by using a fluorescence microplate reader (excitation, 544 nm; emission, 590 nm; FLUOstar+ Galaxy; BMG Laboratories, Offenburg, Germany). In order to test the integrity of peptide incubated with P. gingivalis KDP136, the supernatant of the labeling reaction mixture (containing unbound peptide) was used to react with E. coli ATCC 25922 as described above. Experiments were performed with minimal exposure to light. Means and standard deviations of three independent experiments are presented. Two microliters of each adherence reaction mixture was placed on a microscope slide and analyzed using an Axiovert 200, SensiCam Zeiss, PCO microscope (Zeiss, Germany).
RESULTS
Peptide antimicrobial activity diminishes following incubation with culture supernatant prepared from P. gingivalis.
Previous results by others (15, 21) and ourselves (1) demonstrated that most tested P. gingivalis strains showed resistance to antimicrobial peptides. The fact that P. gingivalis is a very proteolytic organism suggested that its resistance to antimicrobial peptides might stem from its ability to hydrolyze them. Proteolysis has been suggested before as a resistance mechanism against antimicrobial peptides in yeast (Candida albicans) (35) and bacteria (14, 37), including P. gingivalis (10).
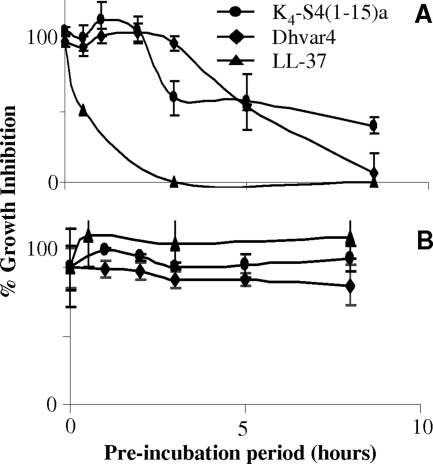
Previously, we reported that P. gingivalis is resistant to the antimicrobial peptides K4-S4(1-15)a, LL-37, and Dhvar4 (1). To test whether P. gingivalis can inactivate these three peptides, the growth inhibitions of S. mutans [susceptible to K4-S4(1-15)a and Dhvar4] and E. coli (susceptible to LL-37) (1) were tested with or without preincubation of the peptides with protease-containing P. gingivalis culture supernatant. As can be seen in Fig. 1A, the antimicrobial activity of all three antimicrobial peptides was diminished following preincubation with the culture supernatant prepared from P. gingivalis ATCC 33277 in a time-dependent manner. Culture supernatant prepared from the nonproteolytic periodontal pathogen A. actinomycetemcomitans in a manner similar to that described above did not reduce the antimicrobial activity of the peptides (Fig. 1B).
FIG. 1.
Effect of P. gingivalis ATCC 33277 or A. actinomycetemcomitans culture supernatant fluid on peptide antimicrobial activity: Peptides K4-S4(1-15)a, Dhvar4, and LL-37 were preincubated with supernatants prepared from P. gingivalis (A) or A. actinomycetemcomitans (B) for the indicated time and added to a growing culture of S. mutans [K4-S4(1-15)a (5 μg/ml final concentration), Dhvar4 (40 μg/ml final concentration)], or E. coli (LL-37, [100 μg/ml final concentration]). Percent bacterial growth inhibition by the supernatant-treated peptides was calculated as described in Materials and Methods. Data are means and standard deviations (error bars) of three independent experiments.
d-Enantiomer conformation of K4-S4(1-15)a renders resistance to inactivation by culture supernatants of P. gingivalis ATCC 33277.
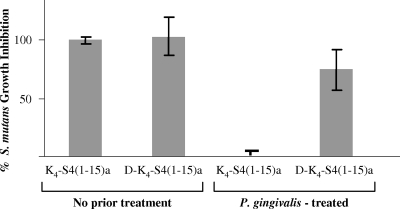
Peptides synthesized using d-amino acids acquire an increased resistance to proteolysis by bacterial proteases. Unlike the “parent” K4-S4(1-15)a peptide, the d-enantiomer retained anti-S. mutans activity following 8 h of incubation with culture supernatant prepared from P. gingivalis (Fig. 2). This observation suggested that the peptide in the d-conformation was resistant to degradation by P. gingivalis.
FIG. 2.
Effect of P. gingivalis supernatant on antibacterial activity of D-K4-S4(1-15)a, the d-enantiomer of K4-S4(1-15)a. Peptides K4-S4(1-15)a and D-K4-S4(1-15)a were treated (or not) for 8 h with supernatant prepared from P. gingivalis and added (5 μg/ml) to a growing culture of S. mutans. Percent inhibition was calculated and compared to those for cells grown without antimicrobial peptides (0% inhibition) and cells grown in the presence of untreated K4-S4(1-15)a (100% inhibition). Data are means and standard deviations (error bars) of three independent experiments.
P. gingivalis is resistant to the d-enantiomer of K4-S4(1-15)a.
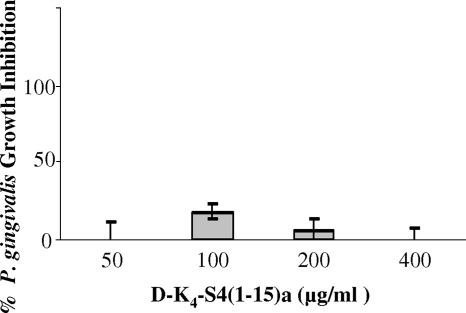
Although d-K4-S4(1-15)a remained effective against S. mutans, even after an 8-h preincubation with P. gingivalis supernatant, P. gingivalis ATCC 33277 remained resistant to D-K4-S4(1-15)a (Fig. 3). Some inhibition of D-K4-S4(1-15)a could be detected at 100 μg/ml; however, this effect was not observed at higher concentrations. This result suggested that the resistance of P. gingivalis ATCC 33277 to K4-S4(1-15)a is independent of its proteolytic capacity.
FIG. 3.
Effect of D-K4-S4(1-15)a on P. gingivalis growth. Increasing amounts of D-K4-S4(1-15)a were added to a growing culture of P. gingivalis. Percent inhibition was calculated according to that for nontreated cells (0%). Data are means and standard deviations (error bars) of three independent experiments.
Binding of fluorescently labeled peptides to P. gingivalis.
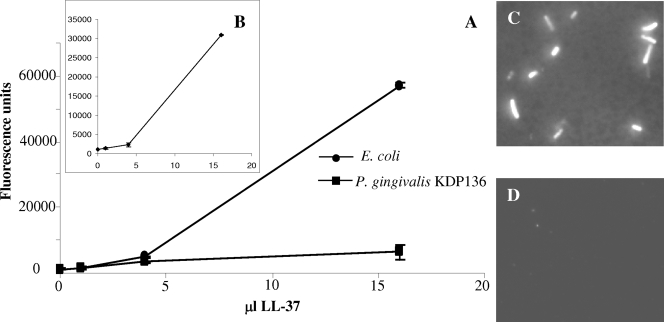
Fluorescent labeling of the K4-S4(1-15)a peptide abolished its bacterial binding capacity (data not shown), presumably because of steric interference by the chromophore, which has a significant molecular mass (410 Da) compared to that of the short K4-S4(1-15)a peptide (1,653 Da). However, fluorescently labeled LL-37 peptide bound E. coli ATCC 25922 in a dose-dependent manner (Fig. 4A and C). Fluorescently labeled LL-37 was immediately digested by P. gingivalis ATCC 33277 (data not shown). Therefore, a protease-deficient P. gingivalis strain (KDP136) (39) was used to determine the binding of LL-37 to P. gingivalis. Unlike E. coli, which exhibited strong binding to LL-37 (Fig. 4A and C), the protease-deficient P. gingivalis KDP136 strain demonstrated weak binding to the fluorescently labeled LL-37 peptide (Fig. 4A). Poor binding to P. gingivalis KDP136 (Fig. 4D) did not result from peptide degradation because the unbound peptide that remained in the incubation supernatant after removal of P. gingivalis KDP136 by centrifugation bound to E. coli with an affinity similar to that of the peptide not incubated with P. gingivalis KDP136 (Fig. 4B).
FIG. 4.
Binding of Alexa Fluor-labeled LL-37 to E. coli ATCC 25922 or P. gingivalis KDP136. (A) Total fluorescence of bacteria incubated with Alexa Fluor-labeled LL-37 (concentration is estimated at 0.8 mg/ml [see Materials and Methods]). (B) Total fluorescence of E. coli ATCC 25922 incubated with the supernatant of Alexa Fluor-labeled LL-37 that was preincubated with P. gingivalis KDP136 (see Results). Images in panels C and D show fluorescence of bacteria incubated with Alexa Fluor-labeled LL-37 recorded using identical instrument settings. Of the 13 E. coli cells present in the field of view presented in panel C, 12 were labeled. Out of the 51 P. gingivalis KDP136 cells present in the field of view presented in panel D, only two were labeled. Error bars indicate standard deviations.
Protease-deficient P. gingivalis KDP136 is resistant to LL-37.
Though not capable of degrading LL-37 (Fig. 4B), the protease-deficient P. gingivalis KDP136 mutant was found to be resistant to the LL-37 antimicrobial peptide (MIC higher than 200 μg/ml [data not shown]), supporting the above results indicating that the resistance of P. gingivalis to direct killing by antimicrobial peptides is protease independent.
DISCUSSION
P. gingivalis is a major pathogen involved in periodontal disease that flourishes in the gingival crevice, although it is bathed with antimicrobial peptides (6, 45). Indeed, in a recent study (1), P. gingivalis was found to be resistant to the three tested antimicrobial peptides (from human and nonhuman origins). Resistance to antimicrobial peptides is likely to contribute to the virulence of P. gingivalis. The high proteolytic capacity of P. gingivalis suggested that peptide degradation is the mechanism by which P. gingivalis resists host antimicrobial peptides. Indeed, in this study, wild-type P. gingivalis inactivated all three antimicrobial peptides in a time-dependent manner. Of the three antimicrobial peptides, the activity of human LL-37 was reduced the most and most rapidly (Fig. 1A). This might result from its increased length (37 amino acids) and its abundance of lysines (6) and arginines (5) that are the cleavage sites for the Arg- and Lys-gingipains of P. gingivalis. Dhvar4 (a derivative of the human salivary histatin 5 antimicrobial peptide) is 14 amino acids long and contains four lysines and two arginines and was more susceptible to proteolysis by P. gingivalis than was K4-S4(1-15)a (an analog of the amphibian dermaseptin S4 peptide), which is 15 amino acids long but contains only four lysines (Fig. 1A).
The fact that wild-type P. gingivalis ATCC 33277 is resistant to D-K4-S4(1-15)a (Fig. 3), the protease-resistant d-enantiomer of K4-S4(1-15)a, and that its protease-deficient mutant progeny P. gingivalis KDP136 retains resistance to LL-37, although it is incapable of degrading it, testify that the resistance of P. gingivalis ATCC 33277 to direct killing by these antimicrobial peptides is independent of protease activity. Rather, our results suggest that the resistance of P. gingivalis ATCC 33277 to antimicrobial peptides derives, at least in part, from its low affinity to these positively charged peptides (Fig. 4). Cationic antimicrobial peptides bind lipopolysaccharides (LPS) of gram-negative bacteria (32), and LPS structure has been shown to affect susceptibility or resistance to antimicrobial peptides (28). In accordance with our observations, it was suggested previously (9, 10) that the unique LPS structure of P. gingivalis that induces different host responses than those of most studied LPS (7, 24, 33) might contribute to its resistance to antimicrobial peptides. Alterations in LPS composition might also explain differences in antimicrobial susceptibility profiles observed among several strains of P. gingivalis (21). Modifications in LPS (9) or bacterial cell surfaces have previously been demonstrated to contribute to resistance to antimicrobial peptides in several bacterial pathogens, including Haemophilus influenzae (42), Staphylococcus aureus (8, 29, 30), and Pseudomonas aeruginosa (31). Treponema denticola is a periodontal pathogen that, in a manner similar to that of P. gingivalis, expresses robust proteolytic activity. The resistance of T. denticola to β-defensins was also shown to be associated with its unique cell surface structure that lacks LPS and demonstrated reduced defensin binding (4, 5).
In dental plaque, P. gingivalis is found in a closely packed interdigitated mixed-species biofilm. Though we demonstrated that the highly active gingipains of P. gingivalis are not essential for its resistance to direct killing by antimicrobial peptides, we propose (22) that the P. gingivalis proteases, by degrading local antimicrobial peptides, can provide protection to proximal, otherwise peptide-susceptible bacteria, such as Fusobacterium nucleatum (1) and A. actinomycetemcomitans (34). It also seems reasonable to speculate that in vivo, the degradation of local antimicrobial peptides by P. gingivalis disrupts the immunoregulatory functions of the peptides and assists P. gingivalis (and proximal plaque community species) in evading an otherwise orchestrated host immune response.
Acknowledgments
We thank Koji Nakayama for generously providing P. gingivalis KDP136 and Ronit Naor for her technical assistance.
This work was supported in part by the Israel Science Foundation (grant number 517/06) and, in part, by the United States-Israel Binational Science Foundation (grant number 2005084).
Footnotes
Published ahead of print on 17 December 2007.
REFERENCES
- 1.Altman, H., D. Steinberg, Y. Porat, A. Mor, D. Fridman, M. Friedman, and G. Bachrach. 2006. In vitro assessment of antimicrobial peptides as potential agents against several oral bacteria. J. Antimicrob. Chemother. 58:198-201. [DOI] [PubMed] [Google Scholar]
- 2.Bals, R., X. Wang, M. Zasloff, and J. M. Wilson. 1998. The peptide antibiotic LL-37/hCAP-18 is expressed in epithelia of the human lung where it has broad antimicrobial activity at the airway surface. Proc. Natl. Acad. Sci. USA 95:9541-9546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bowdish, D. M., D. J. Davidson, Y. E. Lau, K. Lee, M. G. Scott, and R. E. Hancock. 2005. Impact of LL-37 on anti-infective immunity. J. Leukoc. Biol. 77:451-459. [DOI] [PubMed] [Google Scholar]
- 4.Brissette, C. A., and S. A. Lukehart. 2007. Mechanisms of decreased susceptibility to β-defensins by Treponema denticola. Infect. Immun. 75:2307-2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brissette, C. A., and S. A. Lukehart. 2002. Treponema denticola is resistant to human β-defensins. Infect. Immun. 70:3982-3984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brogden, K. A., J. M. Guthmiller, M. Salzet, and M. Zasloff. 2005. The nervous system and innate immunity: the neuropeptide connection. Nat. Immunol. 6:558-564. [DOI] [PubMed] [Google Scholar]
- 7.Burns, E., G. Bachrach, L. Shapira, and G. Nussbaum. 2006. Cutting Edge: TLR2 is required for the innate response to Porphyromonas gingivalis: activation leads to bacterial persistence and TLR2 deficiency attenuates induced alveolar bone resorption. J. Immunol. 177:8296-8300. [DOI] [PubMed] [Google Scholar]
- 8.Collins, L. V., S. A. Kristian, C. Weidenmaier, M. Faigle, K. P. Van Kessel, J. A. Van Strijp, F. Gotz, B. Neumeister, and A. Peschel. 2002. Staphylococcus aureus strains lacking D-alanine modifications of teichoic acids are highly susceptible to human neutrophil killing and are virulence attenuated in mice. J. Infect. Dis. 186:214-219. [DOI] [PubMed] [Google Scholar]
- 9.Devine, D. A. 2003. Antimicrobial peptides in defence of the oral and respiratory tracts. Mol. Immunol. 40:431-443. [DOI] [PubMed] [Google Scholar]
- 10.Devine, D. A., P. D. Marsh, R. S. Percival, M. Rangarajan, and M. A. Curtis. 1999. Modulation of antibacterial peptide activity by products of Porphyromonas gingivalis and Prevotella spp. Microbiology 145:965-971. [DOI] [PubMed] [Google Scholar]
- 11.Feder, R., A. Dagan, and A. Mor. 2000. Structure-activity relationship study of antimicrobial dermaseptin S4 showing the consequences of peptide oligomerization on selective cytotoxicity. J. Biol. Chem. 275:4230-4238. [DOI] [PubMed] [Google Scholar]
- 12.Fields, G. B., and R. L. Noble. 1990. Solid phase peptide synthesis utilizing 9-fluorenylmethoxycarbonyl amino acids. Int. J. Pept. Protein Res. 35:161-214. [DOI] [PubMed] [Google Scholar]
- 13.Giacometti, A., O. Cirioni, W. Kamysz, G. D'Amato, C. Silvestri, M. S. Del Prete, A. Licci, A. Riva, J. Lukasiak, and G. Scalise. 2005. In vitro activity of the histatin derivative P-113 against multidrug-resistant pathogens responsible for pneumonia in immunocompromised patients. Antimicrob. Agents Chemother. 49:1249-1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Groenink, J., A. L. Ruissen, D. Lowies, W. van 't Hof, E. C. Veerman, and A. V. Nieuw Amerongen. 2003. Degradation of antimicrobial histatin-variant peptides in Staphylococcus aureus and Streptococcus mutans. J. Dent. Res. 82:753-757. [DOI] [PubMed] [Google Scholar]
- 15.Guthmiller, J. M., K. G. Vargas, R. Srikantha, L. L. Schomberg, P. L. Weistroffer, P. B. McCray, Jr., and B. F. Tack. 2001. Susceptibilities of oral bacteria and yeast to mammalian cathelicidins. Antimicrob. Agents Chemother. 45:3216-3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hancock, R. E., and H. G. Sahl. 2006. Antimicrobial and host-defense peptides as new anti-infective therapeutic strategies. Nat. Biotechnol. 24:1551-1557. [DOI] [PubMed] [Google Scholar]
- 17.Helmerhorst, E. J., I. M. Reijnders, W. van't Hof, I. Simoons-Smit, E. C. Veerman, and A. V. Amerongen. 1999. Amphotericin B- and fluconazole-resistant Candida spp., Aspergillus fumigatus, and other newly emerging pathogenic fungi are susceptible to basic antifungal peptides. Antimicrob. Agents Chemother. 43:702-704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Helmerhorst, E. J., W. van't Hof, P. Breeuwer, E. C. Veerman, T. Abee, R. F. Troxler, A. V. Amerongen, and F. G. Oppenheim. 2001. Characterization of histatin 5 with respect to amphipathicity, hydrophobicity, and effects on cell and mitochondrial membrane integrity excludes a candidacidal mechanism of pore formation. J. Biol. Chem. 276:5643-5649. [DOI] [PubMed] [Google Scholar]
- 19.Helmerhorst, E. J., W. Van't Hof, E. C. Veerman, I. Simoons-Smit, and A. V. Nieuw Amerongen. 1997. Synthetic histatin analogues with broad-spectrum antimicrobial activity. Biochem. J. 326:39-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hosokawa, I., Y. Hosokawa, H. Komatsuzawa, R. B. Goncalves, N. Karimbux, M. H. Napimoga, M. Seki, K. Ouhara, M. Sugai, M. A. Taubman, and T. Kawai. 2006. Innate immune peptide LL-37 displays distinct expression pattern from beta-defensins in inflamed gingival tissue. Clin. Exp. Immunol. 146:218-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Joly, S., C. Maze, P. B. McCray, Jr., and J. M. Guthmiller. 2004. Human beta-defensins 2 and 3 demonstrate strain-selective activity against oral microorganisms. J. Clin. Microbiol. 42:1024-1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kolenbrander, P. E., N. S. Jakubovics, N. I. Chalmers, and G. Bachrach. 2007. Coaggregation and distance-critical communication, p. 89-100. In K. A. Brogden, F. C. Minion, N. Cornick, T. B. Stanton, Q. Zhang, L. K. Nolan, and M. J. Wannemuehler (ed.), Virulence mechanisms of bacterial pathogens, 4th ed. ASM Press, Washington, DC.
- 23.Krugliak, M., R. Feder, V. Y. Zolotarev, L. Gaidukov, A. Dagan, H. Ginsburg, and A. Mor. 2000. Antimalarial activities of dermaseptin S4 derivatives. Antimicrob. Agents Chemother. 44:2442-2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martin, M., J. Katz, S. N. Vogel, and S. M. Michalek. 2001. Differential induction of endotoxin tolerance by lipopolysaccharides derived from Porphyromonas gingivalis and Escherichia coli. J. Immunol. 167:5278-5285. [DOI] [PubMed] [Google Scholar]
- 25.Marynka, K., S. Rotem, I. Portnaya, U. Cogan, and A. Mor. 2007. In vitro discriminative antipseudomonal properties resulting from acyl substitution of N-terminal sequence of dermaseptin S4 derivatives. Chem. Biol. 14:75-85. [DOI] [PubMed] [Google Scholar]
- 26.Norskov-Lauritsen, N., and M. Kilian. 2006. Reclassification of Actinobacillus actinomycetemcomitans, Haemophilus aphrophilus, Haemophilus paraphrophilus and Haemophilus segnis as Aggregatibacter actinomycetemcomitans gen. nov., comb. nov., Aggregatibacter aphrophilus comb. nov. and Aggregatibacter segnis comb. nov., and emended description of Aggregatibacter aphrophilus to include V factor-dependent and V factor-independent isolates. Int. J. Syst. Evol. Microbiol. 56:2135-2146. [DOI] [PubMed] [Google Scholar]
- 27.Ouhara, K., H. Komatsuzawa, S. Yamada, H. Shiba, T. Fujiwara, M. Ohara, K. Sayama, K. Hashimoto, H. Kurihara, and M. Sugai. 2005. Susceptibilities of periodontopathogenic and cariogenic bacteria to antibacterial peptides, β-defensins and LL37, produced by human epithelial cells. J. Antimicrob. Chemother. 55:888-896. [DOI] [PubMed] [Google Scholar]
- 28.Papo, N., and Y. Shai. 2005. A molecular mechanism for lipopolysaccharide protection of gram-negative bacteria from antimicrobial peptides. J. Biol. Chem. 280:10378-10387. [DOI] [PubMed] [Google Scholar]
- 29.Peschel, A., R. W. Jack, M. Otto, L. V. Collins, P. Staubitz, G. Nicholson, H. Kalbacher, W. F. Nieuwenhuizen, G. Jung, A. Tarkowski, K. P. van Kessel, and J. A. van Strijp. 2001. Staphylococcus aureus resistance to human defensins and evasion of neutrophil killing via the novel virulence factor MprF is based on modification of membrane lipids with l-lysine. J. Exp. Med. 193:1067-1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peschel, A., M. Otto, R. W. Jack, H. Kalbacher, G. Jung, and F. Gotz. 1999. Inactivation of the dlt operon in Staphylococcus aureus confers sensitivity to defensins, protegrins, and other antimicrobial peptides. J. Biol. Chem. 274:8405-8410. [DOI] [PubMed] [Google Scholar]
- 31.Pier, G. B. 2000. Role of the cystic fibrosis transmembrane conductance regulator in innate immunity to Pseudomonas aeruginosa infections. Proc. Natl. Acad. Sci. USA 97:8822-8828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Piers, K. L., M. H. Brown, and R. E. Hancock. 1994. Improvement of outer membrane-permeabilizing and lipopolysaccharide-binding activities of an antimicrobial cationic peptide by C-terminal modification. Antimicrob. Agents Chemother. 38:2311-2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pulendran, B., P. Kumar, C. W. Cutler, M. Mohamadzadeh, T. Van Dyke, and J. Banchereau. 2001. Lipopolysaccharides from distinct pathogens induce different classes of immune responses in vivo. J. Immunol. 167:5067-5076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pütsep, K., G. Carlsson, H. G. Boman, and M. Andersson. 2002. Deficiency of antibacterial peptides in patients with morbus Kostmann: an observation study. Lancet 360:1144-1149. [DOI] [PubMed] [Google Scholar]
- 35.Ruissen, A. L., J. Groenink, P. Krijtenberg, E. Walgreen-Weterings, W. van 't Hof, E. C. Veerman, and A. V. Nieuw Amerongen. 2003. Internalisation and degradation of histatin 5 by Candida albicans. Biol. Chem. 384:183-190. [DOI] [PubMed] [Google Scholar]
- 36.Satcher, D. S. 2000. Oral health in America: a report of the Surgeon General. [DOI] [PMC free article] [PubMed]
- 37.Schmidtchen, A., I. M. Frick, E. Andersson, H. Tapper, and L. Bjorck. 2002. Proteinases of common pathogenic bacteria degrade and inactivate the antibacterial peptide LL-37. Mol. Microbiol. 46:157-168. [DOI] [PubMed] [Google Scholar]
- 38.Scott, M. G., E. Dullaghan, N. Mookherjee, N. Glavas, M. Waldbrook, A. Thompson, A. Wang, K. Lee, S. Doria, P. Hamill, J. J. Yu, Y. Li, O. Donini, M. M. Guarna, B. B. Finlay, J. R. North, and R. E. Hancock. 2007. An anti-infective peptide that selectively modulates the innate immune response. Nat. Biotechnol. 25:465-472. [DOI] [PubMed] [Google Scholar]
- 39.Shi, Y., D. B. Ratnayake, K. Okamoto, N. Abe, K. Yamamoto, and K. Nakayama. 1999. Genetic analyses of proteolysis, hemoglobin binding, and hemagglutination of Porphyromonas gingivalis. Construction of mutants with a combination of rgpA, rgpB, kgp, and hagA. J. Biol. Chem. 274:17955-17960. [DOI] [PubMed] [Google Scholar]
- 40.Socransky, S. S., A. D. Haffajee, M. A. Cugini, C. Smith, and R. L. Kent, Jr. 1998. Microbial complexes in subgingival plaque. J. Clin. Periodontol. 25:134-144. [DOI] [PubMed] [Google Scholar]
- 41.Sørensen, O. E., P. Follin, A. H. Johnsen, J. Calafat, G. S. Tjabringa, P. S. Hiemstra, and N. Borregaard. 2001. Human cathelicidin, hCAP-18, is processed to the antimicrobial peptide LL-37 by extracellular cleavage with proteinase 3. Blood 97:3951-3959. [DOI] [PubMed] [Google Scholar]
- 42.Starner, T. D., W. E. Swords, M. A. Apicella, and P. B. McCray, Jr. 2002. Susceptibility of nontypeable Haemophilus influenzae to human β-defensins is influenced by lipooligosaccharide acylation. Infect. Immun. 70:5287-5289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang, D., A. Biragyn, D. M. Hoover, J. Lubkowski, and J. J. Oppenheim. 2004. Multiple roles of antimicrobial defensins, cathelicidins, and eosinophil-derived neurotoxin in host defense. Annu. Rev. Immunol. 22:181-215. [DOI] [PubMed] [Google Scholar]
- 44.Zasloff, M. 2002. Antimicrobial peptides of multicellular organisms. Nature 415:389-395. [DOI] [PubMed] [Google Scholar]
- 45.Zasloff, M. 2002. Innate immunity, antimicrobial peptides, and protection of the oral cavity. Lancet 360:1116-1117. [DOI] [PubMed] [Google Scholar]