Abstract
Therapy with nucleoside reverse transcriptase inhibitors (NRTIs) can be associated with mitochondrial toxicity. In vitro studies have been used to predict the predisposition for and characterize the mechanisms causing mitochondrial toxicity. Entecavir (ETV) is an approved deoxyguanosine nucleoside for the treatment of chronic hepatitis B virus (HBV) infection that exhibits potent activity against viral reverse transcriptase. We assessed the potential for mitochondrial toxicity of ETV in long-term cultures of HepG2 hepatoma cells by measuring mitochondrial function (through lactate secretion), levels of mitochondrial DNA (mtDNA), and levels of mitochondrial proteins COX II and COX IV. Furthermore, we tested the activity of ETV-triphosphate (ETV-TP) against mitochondrial DNA polymerase γ (Pol γ) in vitro. ETV concentrations as high as 100 times the maximal clinical exposure (Cmax) did not affect cell proliferation, levels of lactate, mitochondrial DNA, or mitochondrial proteins throughout the 15-day culture. The lack of mitochondrial toxicity was consistent with the finding that ETV-TP was not recognized by mitochondrial DNA Pol γ and failed to be incorporated into DNA or inhibit the polymerase assay at the highest levels tested, 300 μM. Combinations of ETV with each of the other HBV NRTI antivirals, adefovir, tenofovir, and lamivudine at 10 times their respective Cmax levels also failed to result in cellular or mitochondrial toxicity. In summary, cell culture and enzymatic studies yielded no evidence that would predict mitochondrial toxicity of ETV at exposure levels in excess of those expected to be achieved clinically.
A subset of human immunodeficiency virus (HIV) nucleoside reverse transcriptase inhibitors (NRTIs) exhibit mitochondrial injury associated with long-term therapy (reviewed in references 4 and 28). Clinical manifestations of mitochondrial toxicity include various hematological disorders, peripheral neuropathy, skeletal and cardiac myopathy, pancreatitis, hepatic failure, and lactic acidosis. These diverse maladies can often be traced to a single origin, diminished capacity of the mitochondrial energy-generating system. Disruptions of the mitochondrial system leads to energy loss, leakage of electrons from the electron transport system, increased reactive oxygen species, and oxidative damage, as well as imbalances of the cellular redox state (increased NADH/NAD+ ratio) reversing the pyruvate/lactate balance in favor of increased lactate production (27).
A leading cause of mitochondrial dysfunction is through interactions with DNA polymerase γ (Pol γ), which is responsible for replicating mitochondrial DNA (mtDNA). Pol γ inhibition results in depletion of mtDNA, leading to decreased synthesis of mitochondrial proteins that maintain oxidative phosphorylation pathways. The triphosphates (TP) of many NRTIs (or diphosphates of phosphonate nucleotide analogs) have been shown to inhibit Pol γ in vitro. Lamivudine-TP (LVD-TP), adefovir-diphosphate (ADV-DP), and tenofovir-DP (TFV-DP), as well as zalcitabine-TP (ddCTP), zidovudine-TP (AZT-TP), and other HIV antivirals, inhibit Pol γ activity in vitro, although the extent of inhibition varies widely (4, 7). Pol γ can also incorporate NRTIs into mtDNA, which can lead to unfaithful replication or gene expression. Mitochondrial dysfunction associated with fialuridine (FIAU) therapy, which led to liver failure and death of patients in a clinical trial, has been proposed to originate from the incorporation of FIAU into mtDNA, impairing its ability to be faithfully transcribed and translated (10). Furthermore, incorporation of NRTIs into nuclear genes that encode mitochondrial proteins can also result in aberrant gene expression and mitochondrial toxicity, a mechanism proposed for AZT (34).
The propensity of nucleoside incorporation into mtDNA by Pol γ also varies and is generally higher with dideoxynucleotide-TPs (e.g., ddCTP) and lowest with LVD-TP and AZT-TP (29). The 3′-5′ exonuclease activity of Pol γ is also a variable; although Pol γ incorporates AZT-TP poorly, it is inefficiently proofread from the mtDNA, resulting in a lasting presence. In contrast, LVD-TP can function as both an inhibitor of Pol γ polymerization and a substrate for its 3′-5′ exonuclease activity, which results in less significant net incorporation into mtDNA (19). A number of studies have shown a correlation between clinical mitochondrial toxicity and in vitro mtDNA incorporation and failure to be exonucleolytically proofread, although clearly the clinical toxicities associated with AZT over long treatment periods are more severe than predicated by effects on Pol γ alone (21).
Long-term cell cultures and a battery of assays have been used to uncover mitochondrial effects for a number of HIV NRTIs (28, 34, 39). These studies typically measure mitochondrial biochemical function through extracellular levels of lactate and Pol γ effects through levels of mtDNA and proteins. Additional scrutiny has involved direct examination of Pol γ activity in the presence of the NRTI-triphosphates. In previous reports, LVD, ADV, and TFV, NRTIs used in long-term therapy of patients with chronic hepatitis B virus (HBV) infection, did not display mitochondrial toxicity in patients (15-17) or in HepG2 cell culture models (1, 2).
Entecavir (ETV; Baraclude, BMS-200475) is a deoxyguanosine nucleoside analog approved for the treatment of chronic HBV infection. ETV is a potent inhibitor of HBV polymerase (36). In a previous report, ETV exposures up to 75 μM for 4 days (2,500-fold the maximal clinical exposure [Cmax] at the 1.0-mg daily dosage) resulted in no appreciable reduction in mtDNA levels (20). In the present study we further explored the potential for mitochondrial toxicity of ETV. Human HepG2 hepatoma cells were cultured for 15 days in the presence of ETV at exposure levels up to 100 times the Cmax, in parallel with other HBV NRTIs at equivalent clinical exposure multiples. Specific mitochondrial and nonspecific cell toxicities were assessed at intervals during the course of the 15-day culture. In addition, combinations of ETV with the other anti-HBV NRTIs were also tested in culture. HepG2 cells were chosen for the present study because of their extensive use in the HBV field, including those that established inhibition of HBV replication and intracellular phosphorylation of the agents tested, as well as several key studies on the effects of NRTIs on mitochondrial metabolism (1, 2, 12, 34, 39). Finally, the ability of ETV-TP to be recognized by Pol γ was assessed in vitro. The results indicated no evidence for mitochondrial toxicity with ETV, either alone or in combination with other anti-HBV NRTIs.
MATERIALS AND METHODS
Cells, cell culture, and compounds.
Human HepG2 hepatoma cells were obtained from the American Type Culture Collection. Adherent cultures were maintained on collagen I (rat tail collagen)-coated tissue culture flasks in RPMI 1640 medium containing 10% heat-inactivated fetal bovine serum, 2 mM l-glutamine, 1 mM sodium pyruvate, and penicillin-streptomycin (100 U/ml and 100 μg/ml) and propagated at 37°C and 5% CO2, as described previously (20). All cell culture reagents were from Invitrogen. ETV was synthesized at Bristol-Myers Squibb (BMS). LVD (3TC) was purified from Epivir tablets (GlaxoSmithKline) at BMS. TFV (PMPA) and ADV (PMEA) were commercially available from Moravek Biochemicals, Inc., Brea, CA. 2,3-Dideoxycytidine (ddC) and dimethyl sulfoxide (DMSO) were obtained from Sigma. Cell numbers were determined by using trypan blue dye exclusion and hemacytometer counting.
Test compound exposures.
The effects of ETV and other NRTIs were assessed in HepG2 cell cultures essentially as described by Walker et al. (39). Cells (2.7 × 106) were seeded onto collagen I-coated T75 flasks at the start of each experiment. Compounds or DMSO vehicle control were added from DMSO stocks at 1,000 times the final exposure concentration. Final concentrations of test compounds corresponded to 1, 10, or 100 times the human steady-state peak plasma concentration (Cmax) achieved during HIV (LVD, TFV, and ddC) or HBV (ADV and ETV) therapy. Although ADV and TFV are administered as prodrugs, which provide increased oral adsorption in the gut, they are rapidly cleaved to the phosphonate by esterases. Thus, significant levels of the prodrug are not found in circulation (22, 35). We exposed cells to clinically relevant concentrations (Cmax) derived from measured levels of free-phosphonate in patients. For LVD, exposures were based on the 150-mg HIV dosage rather than the 100-mg HBV dosage to be consistent with previous studies (39). The Cmax values used were 8.3 μM for LVD (39), 1 μM for TFV (2), 177 nM for ddC (39), and 67 nM for ADV (1). For ETV, the Cmax used was 34 nM, which was 15% higher than the Cmax reported in healthy subjects (5). The DMSO concentrations in the vehicle-treated control cultures were 0.1% (vol/vol). Cultures were exposed to compounds for a total of 15 days, reseeding every 5 days to the original cell density. In addition, media and compounds were also replaced 3 days after seeding. This schedule resulted in replacement of media containing fresh compound on days 1, 3, 5, 8, 10, and 13. Cells and culture supernatants were harvested and assayed on days 5, 10, and 15.
Real-time PCR quantitation of mitochondrial and nDNA.
Total DNA was isolated from frozen HepG2 cell pellets containing 2.5 × 106 cells by using a Qiagen DNeasy tissue kit with RNase A treatment according to the manufacturer's instructions. A total DNA sample from DMSO control-treated HepG2 cells was utilized as the nuclear and mtDNA standard for all real-time PCR experiments. A real-time PCR method, as described by Gourlain et al. (18), was utilized to measure the levels of mtDNA, which measured the amount of the mitochondrial cytochrome b (cyt b) gene from total cellular DNA. The primers and labeled probe in the cyt b real-time PCR assay were forward primer cyt b-2s (5′-GCCTGCCTGATCCTCCAAAT-3′), reverse primer cyt b-2as (5′-AAGGTAGCGGATGATTCAGCC-3′), and probe cyt-p2 (5′-FAM-CACCAGACGCCTCAACCGCCTT-major groove binder-3′) (Applied Biosystems, Inc.). A primer-probe master mix was prepared that contained an optimized final concentration of 8 μM forward and reverse primers and 5 μM labeled probe. For quantitation of nuclear DNA (nDNA) in the total DNA sample, real-time PCR was used to determine the level of the chromosomal RNase P gene using a TaqMan RNase P control reagents kit (Applied Biosystems, Inc.), as described by Kim et al. (23). Real-time PCR was performed using the cyt b and RNase P primer/probes in parallel. Total DNA was diluted to a concentration of 1 ng/ml, and 5 μl was added to PCR wells containing 1 μl of primer-probe mix (cyt b or RNase P control kit), 4 μl of H2O, and 10 μl of TaqMan PCR Universal master mix (Applied Biosystems, Inc.). Real-time PCR was performed by using a TaqMan 7900HT instrument (Applied Biosystems, Inc.) and cycling conditions of 50°C for 2 min and 95°C for 10 min followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. Real-time PCR data were analyzed by using the 7900HT Sequence Detection System 2.2.1 software (Applied Biosystems, Inc.).
Lactate assay.
Extracellular lactate levels in culture supernatants were measured by using lactate reagent from Trinity Biotech (Wicklow, Ireland) after freezing and storage at −80°C. Assays used 10 μl of a 1:10 dilution of thawed supernatant mixed with 200 μl of lactate reagent in a 96-well assay plate. After a 10-min incubation at room temperature, plates were read spectrophotometrically at 540 nm. Lactate levels were determined by using linear regression analysis based upon standard curves prepared with commercial lactate standards (Trinity Biotech). Results, calculated as milligrams of lactate per 107 cells, based upon the total culture volume and number of cells per flask at the 5, 10, and 15 day harvest, were expressed as the percentage of DMSO vehicle-treated control cultures assayed in parallel.
Western blots.
HepG2 cells were seeded into collagen I-coated T75 flasks at a density of 2 × 106 cells/flask and cultured for 10 days in the presence of 3.4 μM ETV (100 times Cmax), 833 μM LVD (100 times Cmax), 1.8 μM ddC (10 times Cmax), or 0.1% DMSO (vehicle control). Medium and compounds were replenished after 3, 5, and 8 days in culture, and cells were reseeded at the initial plating density after 5 days. Cells were harvested after 10 days in culture, and mitochondria were isolated from 2 × 107 cells by using the Mitochondria Isolation Kit for Cultured Cells (Pierce, Rockford, IL) according to the manufacturer's protocol. Mitochondrial pellets were solubilized with Laemmli sample buffer (Bio-Rad, Hercules, CA) and separated by sodium dodecyl sulfate-10 to 20% polyacrylamide gel electrophoresis gels. Proteins were transferred to nitrocellulose according to the method of Towbin et al. (38), blocked with 5% nonfat dry milk in Tris-buffered saline containing 0.1% Tween 20, and then blotted with the following monoclonal antibodies: anti-Cytochrome c Oxidase (COX) subunit II (clone 12C4) at 2.2 μg/ml, anti-COX subunit IV (clone 10G8) at 5.5 μg/ml, and anti-mitochondrial porin (human) (clone 31HL) at 1 μg/ml (all from Invitrogen). Blots were developed by using the goat anti-mouse immunoglobulin G-horseradish peroxidase and ECL Western blotting system (Amersham) and autoradiographed. Autoradiographic images were scanned with a densitometer and analyzed by using Molecular Dynamics ImageQuant software (version 5.0).
Statistical analysis.
The absolute values for each endpoint measured (cell numbers, extracellular lactate, or mtDNA/nDNA ratio) were compared to the individual control values for each experiment, thus generating a percent control value for each treatment. Means and standard deviation (SD) or standard error of the mean (SEM) were determined for each treatment group (n = three or six replicates). Transformed results were analyzed by using one way analysis of variance with Dunnett's Post-Test (GraphPad Prism, version 3.0).
DNA Pol γ assays.
Pol γ primer extension kinetic assays were performed according to the method of Boosalis et al. (3). Pol γ (generously provided by Laurie Kaguni, Michigan State University) was purified from Drosophila embryos as a heterodimer of the catalytic and accessory subunits as described previously (40), and 4.4 U were used for experiments based on empirical determinations. Primer-templates consisted of a radiolabeled 19 base primer (5′-[32P]-AGGTCTTGTACTAGGAGGC-3′) annealed to a 40-base template (5′-ATTTAGTGCCCTGACCGATCGCCTCCTAGTACAAAGACCT-3′). DNA polymerase reactions were performed in a final volume of 10 μl of 50 mM Tris-HCl (pH 7.6), 100 mM KCl, 5 mM MgCl2, 10% glycerol, 1 mM dithiothreitol, and 100 μg of bovine serum albumin/ml and contained 0.16 pmol of 32P-labeled primer-template and 2 μM deoxynucleoside triphosphates. A total of 4.4 U of polymerase was used for reactions. Reactions were incubated for 30 min at 30°C, after which they were terminated by the addition of 5 μl of gel loading buffer (95% deionized formamide, 10 mM EDTA, 0.1% xylene cyanol, 0.1% bromophenol blue). Samples were heated at 80°C for 5 min and electrophoresed on 15% polyacrylamide-8 M urea gels in Tris-borate-EDTA buffer. Dried gels were quantitated by phosphorimager analysis using the Molecular Dynamics Storm 860 and ImageQuant Software (Molecular Dynamics). Kinetic inhibition (Ki) constants were calculated by the method of Cheng and Prusoff (6).
RESULTS
Effects of NRTIs on cell proliferation.
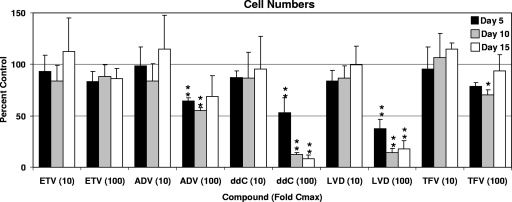
The initial evaluation in monitoring toxicity of ETV was to determine its potential for nonspecific cell cytotoxicity in culture. HepG2 cells were exposed to ETV and the other HBV NRTIs for various durations, and the cell numbers were determined. Compound exposure levels were based on multiples of observed clinical Cmax levels rather than on equivalent molar amounts simply based on the fact that the NRTIs exhibit a wide range of potencies and dosages that result in different patient exposures and because results based on clinical exposure levels are more likely to be relevant. HepG2 cells have been shown to readily phosphorylate these HBV NRTIs (13, 26, 35, 41). ETV, ADV, LVD, TFV, and ddC present at exposures that were 10 times their respective Cmax did not significantly affect cell numbers after 5, 10, or 15 days (Fig. 1). At 100 times the Cmax, however, several compounds adversely affected the cultures and resulted in decreased cell numbers. ddC significantly decreased the number of cells after 5 days, ultimately decreasing to <10% of the control values after 15 days. ADV at these concentrations resulted in a trend toward decreased cell numbers compared to control cultures, with 40 to 50% decreased cell numbers seen at days 5 through 15 of culture, although only the 5- and 10-day decreases reached statistical significance (P < 0.01). LVD treatment resulted in decreased cell numbers over the entire 15-day assay period (P < 0.01), with cell numbers nearly 60% lower than control cultures after 5 days and declining to <20% of control levels after 10 and 15 days of exposure. TFV displayed a 30% decrease at the 10-day time point (P < 0.05), but cells recovered by day 15. In contrast, ETV at exposures higher than 100 times the Cmax did not significantly affect HepG2 cell numbers at any time during the 15-day culture (Fig. 1). Subsequent assays were performed to determine whether the observed cytotoxicities were related to mitochondrial toxicity.
FIG. 1.
Effect of NRTI exposures on HepG2 cell numbers. Cell cultures were exposed to 10-fold or 100-fold the Cmax levels of the NRTIs listed, and cell numbers were determined after 5, 10, or 15 days. Values represent the percentage of 0.1% DMSO vehicle-only control cultures and represent the means ± the SD of three to six independent experiments. The duration of culture was 5 days (▪), 10 days (░⃞), and 15 days (□). *, P < 0.05; **, P < 0.001 (Dunnett's multiple-comparison test).
Mitochondrial function and lactate secretion.
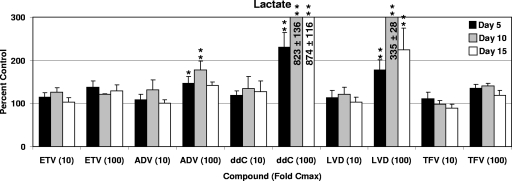
Increased secretion of extracellular lactate is most often a consequence of impaired mitochondrial function and as a result is used as an index of mitochondrial toxicity. Similar to the results obtained in cell proliferation assays, both ETV and TFV did not significantly affect the extracellular levels of lactate at the highest concentrations and longest durations tested (Fig. 2). Although ADV and LVD exposures at 10 times the Cmax did not change extracellular lactate levels, significant increases were observed when cells were incubated with 100-fold Cmax levels of ADV (5- and 10-day samples only) and LVD (P < 0.01 for 5-, 10-, and 15-day samples). These effects were consistent with decreased cell numbers at the higher concentrations of ADV and LVD. Consistent with previous reports, exposure of cells to 100-fold Cmax levels of ddC resulted in significant increases in extracellular lactate levels (P < 0.001), indicating impaired mitochondrial function.
FIG. 2.
Effect of NRTI exposures on extracellular lactate levels. HepG2 cell cultures were exposed to concentrations 10- or 100-fold higher than the Cmax levels of the NRTIs listed, and the levels of lactate in the media were determined after 5, 10, or 15 days. Lactate levels were normalized according to cell numbers. Values represent the percentage of 0.1% DMSO vehicle-only control cultures and represent the means ± the SEM of three to six independent experiments. The duration of culture was 5 days (▪), 10 days (░⃞), and 15 days (□). *, P < 0.05; **, P < 0.001 (Dunnett's multiple-comparison test). Levels greater than 300% were truncated to enhance presentation and were for, ddC-treated cells, 824 ± 136% and 875 ± 116% of the vehicle-treated control values for 10 and 100 times the Cmax, respectively, and for 100-fold the Cmax (for LVD) was 335% ± 28% of the vehicle-treated control.
mtDNA levels.
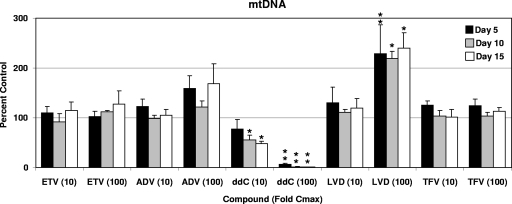
Directly monitoring mtDNA levels is a very sensitive method of measuring mitochondrial toxicity. The levels of HepG2 cell mtDNA were determined by measuring the mitochondrially encoded cyt b gene in total cellular DNA preparations by quantitative real-time PCR. Although equivalent quantities of total DNA (containing >99% nDNA) were analyzed, cyt b DNA levels were normalized using the nDNA levels found by quantitative PCR analysis of the same samples for the nuclear RNase P gene. This step controlled for possible reductions in cell numbers resulting from general cytotoxicity, as well as losses incurred during sample preparation. Exposure of HepG2 cells to ETV, ADV, or TFV did not result in any significant alterations in the level of mtDNA from DMSO-treated controls (Fig. 3). The slight elevation in mtDNA/nDNA ratio seen with ADV at concentrations 100-fold the Cmax, especially apparent after 5- and 15-day exposures, was found to be statistically insignificant. In contrast, exposures at 100 times the Cmax of LVD (833 μM) resulted in a significant increase in the mtDNA/nDNA ratio that was ca. 200% of DMSO-treated control levels at each of the 5-, 10-, and 15-day time points (P < 0.05). As previously reported, mtDNA was significantly reduced with exposure to ddC, with a noted decrease to 50% of control levels after 10- and 15-day exposures of ddC at concentrations only 10 times the Cmax (P < 0.05). At a ddC exposure of 100-fold, the Cmax, mtDNA was depleted by 93% after just 5 days of incubation, followed by nearly complete inhibition after 10 and 15 days of exposure.
FIG. 3.
Effect of NRTI exposures on mtDNA levels. HepG2 cell cultures were exposed to 10- or 100-fold the Cmax levels of the NRTIs listed, and the levels of intracellular mtDNA and nDNA were determined by real-time PCR. The ratios of mtDNA to nDNA were determined, and values representing the percentage of 0.1% DMSO vehicle-only control cultures are presented. Values represent the means ± the SEM of three to six independent experiments performed in duplicate. *, P < 0.05; **, P < 0.001 (Dunnett's multiple-comparison test).
Effects of ETV in combination with other NRTIs.
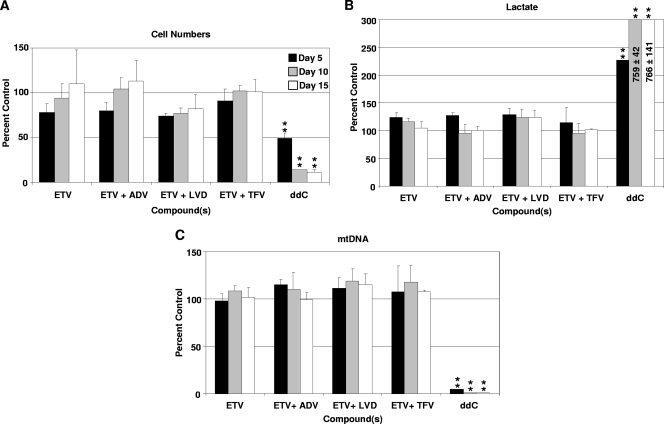
Recent reports have suggested that some HBV patients should be treated with combination therapy (14, 30). Before such combinations are used, the potential for additive or synergistic toxicities should be considered. Therefore, experiments were performed to determine whether exposure to ETV, combined with any of the other HBV NRTIs ADV, LVD, or TFV, increased the potential for mitochondrial toxicity in the 15-day HepG2 cell culture model. For these experiments, an intermediate concentration of 10 times the clinical Cmax was used for each agent because there was no evidence of toxicity at this exposure level in earlier assays using each of the NRTIs individually, and yet the exposures were still above levels expected to be achieved clinically. The combination of ETV with ADV, LVD, or TFV did not result in significant changes in cell numbers or lactate or mtDNA levels, with the overall results equivalent to those found with ETV alone (Fig. 4).
FIG. 4.
Effect of ETV-NRTI combinations. HepG2 cells were exposed to ETV at a concentration equivalent to 10-fold the Cmax either alone or in combination with ADV, LVD, or TFV also at 10-fold their respective Cmax. HepG2 cells exposed to 100-fold the Cmax of ddC was included as a positive control for mitochondrial toxicity. (A) Effect ETV-NRTI combinations on HepG2 cell numbers after exposure for 5 days (▪), 10 days (░⃞), and 15 days (□). The data represent the mean percentage of vehicle-treated control ± the SD of three independent experiments. (B) Levels of extracellular lactate normalized to cell number and compared to levels derived from vehicle-treated control. The data represent the means ± the SEM of three independent experiments. (C) Comparison of the effects of ETV-NRTI combinations on mtDNA levels in HepG2 cells exposed for 5, 10, and 15 days. The data represent the means ± the SEM of three independent experiments of mtDNA normalized to nDNA and compared as a percentage of vehicle-treated controls. **, P < 0.001 (Dunnett's multiple-comparison test).
Mitochondrial proteins.
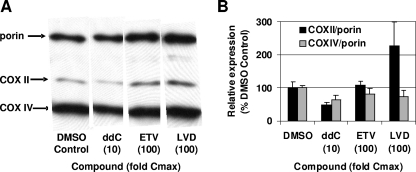
The steady-state levels of key mitochondrial proteins were monitored by Western blotting with monoclonal antibodies to mtDNA-encoded COX II protein and the nuclear encoded cyt c oxidase subunit IV protein (COX IV). The levels of COX II and COX IV were normalized to the levels of the mitochondrial protein porin, an outer mitochondrial membrane protein typically used to adjust for differences in the number of mitochondria present in each sample. In these experiments, ddC and LVD exposures at 10- and 100-fold the Cmax, respectively, were used as positive controls for mitochondrial toxicity. Figure 5 shows that ddC significantly decreased COX II levels relative to porin, although the levels of COX IV were also decreased, albeit to a lesser degree. Cells exposed to higher concentrations of ddC had insufficient mitochondria to be analyzed. LVD displayed a different pattern with an increase in the levels of COX II, but not the nuclear encoded COX IV protein. In contrast, HepG2 cells exposed to ETV at concentrations up to 100 times the Cmax showed control levels of COX II and COX IV. These results again confirmed that high levels of ETV did not affect mitochondrial proteins levels.
FIG. 5.
Western blot analysis of mitochondrial proteins. Cells were exposed to the 100-fold the Cmax values of ETV or LVD or 10-fold the Cmax of ddC for 10 days. Cells were processed and analyzed as described in Materials and Methods. (A) Western blot detection of mitochondrially encoded COX II or nuclear encoded COX IV and the mitochondrial porin protein. (B) The levels of COX II and COX IV proteins were normalized according to the levels of porin protein and plotted with respect to the levels in DMSO vehicle-treated cells. A representative Western blot is shown, and the average results of three independent experiments, performed in duplicate, are plotted as means ± the SEM.
mtDNA Pol γ.
Most often, mitochondrial toxicity caused by nucleoside analogs is presumed to arise through direct interaction with Pol γ. To characterize the interaction of ETV with Pol γ, polymerase primer extension reactions were performed with ETV-TP in place of dGTP as described in Materials and Methods. In experiments with all four dNTPs, Pol γ extended the primer template efficiently, resulting in the majority of products that were full-length extensions of the template. In contrast, when ETV-TP was used in place of dGTP with dATP, dCTP, and TTP, no extension of the primer template was observed, suggesting ETV-TP was not recognized by Pol γ. Table 1 shows the results of a kinetic analysis of the utilization of ETV-TP by Pol γ in the production of the +1 extended product as a function of dGTP or ETV-TP concentration. Both the Km for dGTP and the inhibition constant (Ki) for ETV-TP were determined. The results indicate that Pol γ did not appear to utilize or be inhibited by ETV-TP, even at the highest concentration used (300 μM). The ratio of Ki to Km indicates the preference of the enzyme for inhibitor over the natural substrate, with the lowest numbers indicating the greatest inhibitor selectivity. Although Pol γ obviously preferred dGTP over ETV-TP, the results for HBV RT (reported by Levine et al. [26]) indicate a dramatic preference for ETV-TP over dGTP. These results suggest that Pol γ did not bind or recognize ETV-TP, in concordance with the lack of mitochondrial toxicity in cell culture.
TABLE 1.
Kinetic analysis for ETV-TP
| Enzyme | dGTP Km (μM) | ETV-TP IC50 (μM) | ETV-TP Ki (μM) | Ki/Km |
|---|---|---|---|---|
| Pol γ | 0.12 | >300b | >160b | >1333 |
| HBV-RTa | 0.0013 | 0.00043 | 0.0002 | 0.15 |
Data reproduced from Levine et al. (26).
No incorporation was seen at 300 μM ETV-TP.
DISCUSSION
Mitochondrial toxicity is considered an inherent risk of therapies involving the nucleoside(tide) analog class, having been observed upon prolonged use of a subset of approved NRTIs. Therefore, a panel of in vitro assays has been established to determine the potential for mitochondrial toxicity. Previously, we demonstrated that a 4-day exposure of cells to 75 μM ETV (2,500 times the Cmax for the higher 1.0-mg dosage) did not reduce mtDNA levels (20). In the present study, we expand those studies by extending cell culture exposures, performing more assays for mitochondrial toxicity, and testing other approved NRTIs with anti-HBV activity in parallel. In addition, cells were exposed to concentrations of compounds based upon multiples of clinical exposures to provide more meaningful and relevant results. Evidence for toxicity was examined at multiple exposures and assay conditions to assess nonspecific mitochondrial dysfunction, steady-state levels of key mitochondrial proteins, inhibition of mtDNA Pol γ directly (in vitro) and indirectly (mtDNA levels), and nonspecific cytotoxicity. The results from all of these determinations was that ETV did not display any evidence of mitochondrial toxicity during the 15-day duration of the experiment at exposure levels >100-fold higher than those achieved in humans. These findings are supported by an absence of mitochondrial toxicity in woodchucks given ETV for 3 years (9) and in human clinical studies (31). In contrast, the clinical toxicity of FIAU (32) was accurately reflected in short-term toxicity in the woodchuck model (37).
In contrast to the findings with ETV, ddC, used as a positive control for mitochondrial toxicity, exhibited a significant decrease in mtDNA levels. The mechanism for mitochondrial toxicity of ddC lies in its efficient incorporation into mtDNA and its inefficient removal through polymerase proofreading (reviewed in reference 25). ADV, TFV, and LVD, with activity against both HIV and HBV, have been studied previously for mitochondrial toxicity. Although both LVD and TFV are utilized by Pol γ and incorporated into mtDNA in vitro, Pol γ displayed 2,800-fold and 11,400-fold preferences for the natural substrates dCTP and dATP, respectively (21). In contrast, Pol γ displayed only a 2.9-fold preference for dCTP over ddCTP. No evidence of in vitro mitochondrial toxicity has been reported for ADV (1), TFV (2), or LVD (39) at exposure multiples that were lower than those tested here. These agents had no effect in our assay at lower concentrations, although ADV and LVD resulted in decreased cell numbers and increased extracellular lactate at concentrations 100-fold higher than their respective Cmax levels. These results were not accompanied by decreased mtDNA levels. It is difficult to interpret the implications of the results obtained at these very high exposures, at which perturbations were observed with NRTIs that are not associated with mitochondrial toxicity in the clinical setting. However, the fact that ETV did not display any impact at these extremely high levels relative to clinical exposures strongly suggests an absence of relevant mitochondrial toxicity with ETV.
The results presented in Fig. 1 to 3 suggested that exposure of HepG2 cultures to LVD at 100 times the Cmax resulted in decreased cell numbers, increased extracellular lactate, and an increase in mtDNA. The toxicity of LVD at high concentrations was unexpected, given a lack of notable toxicity in the clinical setting. Additional effects were seen in the steady-state levels of mitochondrial proteins, with increased levels of both the nuclear encoded mitochondrial protein porin and the mitochondrion-encoded COX II (Fig. 5). These findings were accompanied by the observation that cells exposed to LVD at 100-fold its Cmax appeared larger microscopically after trypsin treatment (data not shown). Confirmation that these cells were indeed larger than those of DMSO-treated controls was provided by a comparison of total cellular protein/total DNA ratios, which revealed that cells treated with LVD at concentrations equivalent to 10-fold the Cmax were similar in size to DMSO-treated controls, whereas cells exposed to LVD at 100-fold the Cmax for 10 days or longer had a twofold increase in protein levels compared to DMSO-controls (P < 0.001) (data not shown). This apparent increase in cell size coincides with the measured twofold increase in mtDNA per cell, suggesting higher numbers of mitochondria per cell. Previous studies that did not show LVD mitochondrial toxicity in culture did not expose cells to concentrations of LVD as high as those tested in the present study (34, 39); the 833 μM concentration at which toxicity was observed after 15 days of exposure approach the 8-day 50% cytotoxicity concentration reported in HepG2 cells by Cihlar et al. (1,020 μM) (8). We hypothesize that cells may increase the synthesis of mitochondria and mtDNA in response to general cytotoxic effects to compensate for deficiencies in the cellular energy generating system, as evidenced by the increased secretion of lactate. These results further emphasize the extremely high exposures of all of the compounds tested, relative to the concentrations found in patients. Although the pathways for toxicities seen here with LVD and by Walker et al. (39) for AZT are beyond the scope of our study, Walker et al. discuss several possibilities. Also, Pol γ has less discrimination for LVD-TP than either TFV-TP or AZT-TP (21). More recently, de Baar et al. reported increased mtDNA in HepG2 cell cultures exposed to high concentrations (up to 300 μM) of LVD, as well as emtricitabine, zidovudine, and abacavir (12). Adverse events associated with LVD therapy that were consistent with mitochondrial toxicity have been reported on rare occasions (11, 33).
Recently, the idea of combination therapy for HBV has been proposed to address increasing resistance levels over time (14, 30). The potential for synergistic toxicities when different HBV NRTI combinations are used has not been studied. Here we saw no antagonism or potential for mitochondrial toxicity when ETV was combined with three currently approved NRTIs with anti-HBV activity, ADV, LVD, or TFV, at concentrations 10-fold the Cmax levels of each. These levels far exceed exposures reasonably expected clinically. The results of these in vitro studies using a single, established cell line may not entirely predict the levels of phosphorylated nucleotides found in various tissues throughout the body. However, basic liabilities targeting the mitochondria were able to be efficiently identified for the control compound ddC. In addition, the lack of toxicities seen are not due to insufficient levels of intracellular phosphorylation since HepG2 cells have been shown to readily phosphorylate the compounds and result in antiviral activity (13, 26, 35, 41).
In vitro assays for direct effects of ETV-TP on Pol γ completely explained the lack of mitochondrial toxicity even after exposure of cells to high concentrations of ETV. Pol γ failed to recognize the ETV-TP and incorporate it into DNA. ETV-TP did not inhibit Pol γ reactions containing dGTP. This result is in contrast to results obtained with HBV polymerase wherein ETV-TP was a potent inhibitor and a chain terminator in polymerase reactions (24, 26, 36).
In conclusion, long-term cell culture studies that examined adverse effects on mitochondria failed to uncover evidence of mitochondrial toxicity after ETV exposure at levels exceeding those reasonably expected during therapy. In contrast, some toxicity was seen for the other approved NRTIs with anti-HBV activity at the highest exposure multiples. Combinations of ETV with these other NRTIs, at 10 times their respective Cmax exposures, did not result in mitochondrial toxicity, yielding results that were similar to those obtained with ETV alone. These results support the concept that ETV has a very low potential for mitochondrial toxicity during therapy.
Acknowledgments
We gratefully acknowledge Bristol-Myers Squibb coworkers John Leet for providing lamivudine, Dominique Tersago for help in initial experimental design, Bruce Kreter for critical review of the manuscript, and Mary Jane Plym for expert technical assistance. We also gratefully acknowledge the generous gift of Pol γ from Laurie Kaguni.
Footnotes
Published ahead of print on 3 December 2007.
REFERENCES
- 1.Birkus, G., C. S. Gibbs, and T. Cihlar. 2003. Comparative effects of adefovir and selected nucleoside inhibitors of hepatitis B virus DNA polymerase on mitochondrial DNA in liver and skeletal muscle cells. J. Viral Hepat. 10:50-54. [DOI] [PubMed] [Google Scholar]
- 2.Birkus, G., M. J. Hitchcock, and T. Cihlar. 2002. Assessment of mitochondrial toxicity in human cells treated with tenofovir: comparison with other nucleoside reverse transcriptase inhibitors. Antimicrob. Agents Chemother. 46:716-723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boosalis, M. S., J. Petruska, and M. F. Goodman. 1987. DNA polymerase insertion fidelity. Gel assay for site-specific kinetics. J. Biol. Chem. 262:14689-14696. [PubMed] [Google Scholar]
- 4.Brinkman, K., H. J. ter Hofstede, D. M. Burger, J. A. Smeitink, and P. P. Koopmans. 1998. Adverse effects of reverse transcriptase inhibitors: mitochondrial toxicity as common pathway. Aids 12:1735-1744. [DOI] [PubMed] [Google Scholar]
- 5.Bristol-Myers Squibb. 2007. Baraclude prescribing information. [Online.] http://www.fda.gov/cder/foi/label/2007/021797s003,021798s003lbl.pdf.
- 6.Cheng, Y., and W. H. Prusoff. 1973. Relationship between the inhibition constant (Ki) and the concentration of inhibitor which causes 50 percent inhibition (lC50) of an enzymatic reaction. Biochem. Pharmacol. 22:3099-3108. [DOI] [PubMed] [Google Scholar]
- 7.Cherrington, J. M., S. J. Allen, B. H. McKee, and M. S. Chen. 1994. Kinetic analysis of the interaction between the diphosphate of (S)-1-(3-hydroxy-2-phosphonylmethoxypropyl)cytosine, ddCTP, AZTTP, and FIAUTP with human DNA polymerases beta and gamma. Biochem. Pharmacol. 48:1986-1988. [DOI] [PubMed] [Google Scholar]
- 8.Cihlar, T., G. Birkus, D. E. Greenwalt, and M. J. Hitchcock. 2002. Tenofovir exhibits low cytotoxicity in various human cell types: comparison with other nucleoside reverse transcriptase inhibitors. Antivir. Res. 54:37-45. [DOI] [PubMed] [Google Scholar]
- 9.Colonno, R. J., E. V. Genovesi, I. Medina, L. Lamb, S. K. Durham, M. L. Huang, L. Corey, M. Littlejohn, S. Locarnini, B. C. Tennant, B. Rose, and J. M. Clark. 2001. Long-term entecavir treatment results in sustained antiviral efficacy and prolonged life span in the woodchuck model of chronic hepatitis infection. J. Infect. Dis. 184:1236-1245. [DOI] [PubMed] [Google Scholar]
- 10.Cui, L., S. Yoon, R. F. Schinazi, and J. P. Sommadossi. 1995. Cellular and molecular events leading to mitochondrial toxicity of 1-(2-deoxy-2-fluoro-1-β-d-arabinofuranosyl)-5-iodouracil in human liver cells. J. Clin. Investig. 95:555-563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cupler, E. J., and M. C. Dalakas. 1995. Exacerbation of peripheral neuropathy by lamivudine. Lancet 345:460-461. [DOI] [PubMed] [Google Scholar]
- 12.de Baar, M. P., E. R. de Rooij, K. G. Smolders, H. B. van Schijndel, E. C. Timmermans, and R. Bethell. 2007. Effects of apricitabine and other nucleoside reverse transcriptase inhibitors on replication of mitochondrial DNA in HepG2 cells. Antivir. Res. 76:68-74. [DOI] [PubMed] [Google Scholar]
- 13.Delaney, W. E. t., A. S. Ray, H. Yang, X. Qi, S. Xiong, Y. Zhu, and M. D. Miller. 2006. Intracellular metabolism and in vitro activity of tenofovir against hepatitis B virus. Antimicrob. Agents Chemother. 50:2471-2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fung, S. K., and A. S. Lok. 2004. Management of hepatitis B patients with antiviral resistance. Antivir. Ther. 9:1013-1026. [PubMed] [Google Scholar]
- 15.GlaxoSmithKline. 2002. Epivir (lamivudine) prescribing information. [Online.] http://www.fda.gov/cder/foi/label/2002/20564s015and20596s016lbl.pdf.
- 16.Gilead Sciences, Inc. 2006. Hepsera Prescribing Information. [Online.] http://www.fda.gov/cder/foi/label/2006/021449s006lbl.pdf.
- 17.Gilead Sciences, Inc. 2003. Viread Prescribing information. [Online.] www.fda.gov/cder/foi/label/2003/21356slr005_viread_lbl.pdf.
- 18.Gourlain, K., B. Amellal, Z. Ait Arkoub, N. Dupin, C. Katlama, and V. Calvez. 2003. Quantitative analysis of human mitochondrial DNA using a real-time PCR assay. HIV Med. 4:287-292. [DOI] [PubMed] [Google Scholar]
- 19.Gray, N. M., C. L. Marr, C. R. Penn, J. M. Cameron, and R. C. Bethell. 1995. The intracellular phosphorylation of (−)-2′-deoxy-3′-thiacytidine (3TC) and the incorporation of 3TC 5′-monophosphate into DNA by HIV-1 reverse transcriptase and human DNA polymerase gamma. Biochem. Pharmacol. 50:1043-1051. [DOI] [PubMed] [Google Scholar]
- 20.Innaimo, S. F., M. Seifer, G. S. Bisacchi, D. N. Standring, R. Zahler, and R. J. Colonno. 1997. Identification of BMS-200475 as a potent and selective inhibitor of hepatitis B virus. Antimicrob. Agents Chemother. 41:1444-1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnson, A. A., A. S. Ray, J. Hanes, Z. Suo, J. M. Colacino, K. S. Anderson, and K. A. Johnson. 2001. Toxicity of antiviral nucleoside analogs and the human mitochondrial DNA polymerase. J. Biol. Chem. 276:40847-40857. [DOI] [PubMed] [Google Scholar]
- 22.Kearney, B. P., J. F. Flaherty, and J. Shah. 2004. Tenofovir disoproxil fumarate: clinical pharmacology and pharmacokinetics. Clin. Pharmacokinet. 43:595-612. [DOI] [PubMed] [Google Scholar]
- 23.Kim, J. Y., J. M. Hwang, H. S. Ko, M. W. Seong, B. J. Park, and S. S. Park. 2005. Mitochondrial DNA content is decreased in autosomal dominant optic atrophy. Neurology 64:966-972. [DOI] [PubMed] [Google Scholar]
- 24.Langley, D. R., A. W. Walsh, C. J. Baldick, B. J. Eggers, R. E. Rose, S. M. Levine, A. J. Kapur, R. J. Colonno, and D. J. Tenney. 2007. Inhibition of hepatitis B virus polymerase by entecavir. J. Virol. 81:3992-4001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee, H., J. Hanes, and K. A. Johnson. 2003. Toxicity of nucleoside analogues used to treat AIDS and the selectivity of the mitochondrial DNA polymerase. Biochemistry 42:14711-14719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Levine, S., D. Hernandez, G. Yamanaka, S. Zhang, R. Rose, S. Weinheimer, and R. J. Colonno. 2002. Efficacies of entecavir against lamivudine-resistant hepatitis B virus replication and recombinant polymerases in vitro. Antimicrob. Agents Chemother. 46:2525-2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lewis, W., and M. C. Dalakas. 1995. Mitochondrial toxicity of antiviral drugs. Nat. Med. 1:417-422. [DOI] [PubMed] [Google Scholar]
- 28.Lewis, W., B. J. Day, and W. C. Copeland. 2003. Mitochondrial toxicity of NRTI antiviral drugs: an integrated cellular perspective. Nat. Rev. Drug Discov. 2:812-822. [DOI] [PubMed] [Google Scholar]
- 29.Lim, S. E., and W. C. Copeland. 2001. Differential incorporation and removal of antiviral deoxynucleotides by human DNA polymerase gamma. J. Biol. Chem. 276:23616-23623. [DOI] [PubMed] [Google Scholar]
- 30.Locarnini, S., A. Hatzakis, J. Heathcote, E. B. Keeffe, T. J. Liang, D. Mutimer, J. M. Pawlotsky, and F. Zoulim. 2004. Management of antiviral resistance in patients with chronic hepatitis B. Antivir. Ther. 9:679-693. [PubMed] [Google Scholar]
- 31.Manns, M. P., M. Raptopoulou-Gigi, J. Sollano, R. G. Gish, T. T. Chang, M. Sherman, C. Yurdaydin, D. Shouval, A. S. Lok, E. Cooney, R. Hindes, J. Yang, A. Cross, and R. Wilber. 2005. Entecavir is well tolerated for the treatment of nucleoside-naive and lamivudine-refractory chronic hepatitis B: phase II/III safety results. J. Hepatol. 42:185. [Google Scholar]
- 32.McKenzie, R., M. W. Fried, R. Sallie, H. Conjeevaram, A. M. Di Bisceglie, Y. Park, B. Savarese, D. Kleiner, M. Tsokos, C. Luciano, et al. 1995. Hepatic failure and lactic acidosis due to fialuridine (FIAU), an investigational nucleoside analogue for chronic hepatitis B. N. Engl. J. Med. 333:1099-1105. [DOI] [PubMed] [Google Scholar]
- 33.Ojetti, V., A. Gasbarrini, A. Migneco, R. Flore, A. Santoliquido, D. De Martini, S. Agnes, N. Gentiloni Silveri, and P. Pola. 2002. Lamivudine-induced muscle mitochondrial toxicity. Dig. Liver Dis. 34:384-385. [DOI] [PubMed] [Google Scholar]
- 34.Pan-Zhou, X. R., L. Cui, X. J. Zhou, J. P. Sommadossi, and V. M. Darley-Usmar. 2000. Differential effects of antiretroviral nucleoside analogs on mitochondrial function in HepG2 cells. Antimicrob. Agents Chemother. 44:496-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ray, A. S., J. E. Vela, L. Olson, and A. Fridland. 2004. Effective metabolism and long intracellular half-life of the anti-hepatitis B agent adefovir in hepatic cells. Biochem. Pharmacol. 68:1825-1831. [DOI] [PubMed] [Google Scholar]
- 36.Seifer, M., R. K. Hamatake, R. J. Colonno, and D. N. Standring. 1998. In vitro inhibition of hepadnavirus polymerases by the triphosphates of BMS-200475 and lobucavir. Antimicrob. Agents Chemother. 42:3200-3208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tennant, B. C., B. H. Baldwin, L. A. Graham, M. A. Ascenzi, W. E. Hornbuckle, P. H. Rowland, I. A. Tochkov, A. E. Yeager, H. N. Erb, J. M. Colacino, C. Lopez, J. A. Engelhardt, R. R. Bowsher, F. C. Richardson, W. Lewis, P. J. Cote, B. E. Korba, and J. L. Gerin. 1998. Antiviral activity and toxicity of fialuridine in the woodchuck model of hepatitis B virus infection. Hepatology 28:179-191. [DOI] [PubMed] [Google Scholar]
- 38.Towbin, H., T. Staehelin, and J. Gordon. 1979. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl. Acad. Sci. USA 76:4350-4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Walker, U. A., B. Setzer, and N. Venhoff. 2002. Increased long-term mitochondrial toxicity in combinations of nucleoside analogue reverse-transcriptase inhibitors. Aids 16:2165-2173. [DOI] [PubMed] [Google Scholar]
- 40.Wernette, C. M., and L. S. Kaguni. 1986. A mitochondrial DNA polymerase from embryos of Drosophila melanogaster: purification, subunit structure, and partial characterization. J. Biol. Chem. 261:14764-14770. [PubMed] [Google Scholar]
- 41.Zhu, Y. L., D. E. Dutschman, S. H. Liu, E. G. Bridges, and Y. C. Cheng. 1998. Anti-hepatitis B virus activity and metabolism of 2′,3′-dideoxy-2′,3′-didehydro-β-l(−)-5-fluorocytidine. Antimicrob. Agents Chemother. 42:1805-1810. [DOI] [PMC free article] [PubMed] [Google Scholar]