Abstract
Five of the seven cases of vancomycin-resistant Staphylococcus aureus (VRSA) infection identified to date have occurred in southeastern Michigan. VRSA isolates from the four most recent cases (all from Michigan) were characterized. The vanA gene was localized to a single plasmid in each VRSA isolate. The pulsed-field gel electrophoresis patterns of chromosomal DNA and the restriction profile of the plasmid demonstrated that the four isolates were unique and differed from the first three VRSA isolates. Vancomycin-resistant Enterococcus (VRE) isolates, all of which were Enterococcus faecalis, were recovered from case patients 4 to 6. Each VRE isolate transferred vancomycin resistance to E. faecalis JH2-2 by conjugation. PCRs for vanA and the Inc18-like plasmid genes traA and repR confirmed the presence of an Inc18-like vanA plasmid in all VRE isolates and transconjugants. An Inc18-like vanA plasmid was identified in the VRSA isolate from case patient 7. These findings suggest a role of Inc18-like plasmids as vanA donors.
The first vanA-mediated, vancomycin-resistant Staphylococcus aureus (VRSA) strain was isolated in a Michigan hospital in 2002 (5, 7). Since then, six additional occurrences have been reported: one each in Pennsylvania and New York and four in southeastern Michigan (3, 6, 28a, 29). The vanA plasmid from the first VRSA isolate was extensively characterized (35). The vanA gene was localized to a 57.9-kb plasmid which consisted of a pSK41-like S. aureus plasmid with an insertion of a Tn1546-like element carrying the vanA operon. The researchers hypothesized that this S. aureus isolate acquired vanA from an isolate of vancomycin-resistant Enterococcus (VRE). The patient with the first case of VRSA infection was also infected with a vancomycin-resistant Enterococcus faecalis isolate at the time of VRSA isolation.
The vanA plasmid from this E. faecalis isolate was characterized by Flannagan et al. (15). In this isolate, vanA was localized to a plasmid belonging to the Inc18 plasmid family. These plasmids are broad-host-range conjugative plasmids; pIP501 and pAMβ1 are two well-characterized examples of Inc18 plasmids. Flannagan et al. (15) demonstrated that the E. faecalis vanA plasmid was conjugative and not pheromone responsive. That was the first report of vanA on an Inc18-like plasmid.
Details of the VRSA isolate from case patient 2 (VRSA-2; Pennsylvania) and VRSA-3 (New York) have been reported elsewhere (29, 36). Case patient 2 had a history of VRE infection, but no isolate was available for characterization. The VRE isolate was either coisolated from the same site along with the VRSA isolate or colonized the patient at another site. For VRSA-3 (New York), both Enterococcus faecium and E. faecalis were isolated from the patient, but only the E. faecium isolate contained the same vanA plasmid as the VRSA isolate and thus was hypothesized to be the donor of vanA to S. aureus (36).
The occurrence of vanA-mediated resistance in S. aureus is uncommon; however, a disproportionate number of isolates (five of seven) have occurred in a limited geographic area. In three of the four most recent cases from southeastern Michigan, a vancomycin-resistant E. faecalis isolate was recovered from the patient in association with VRSA; in the sixth case, a vancomycin-resistant Enterococcus avium isolate was also identified. Although several explanations for this observation are possible, a prominent hypothesis is that the VRE isolates recovered in Michigan have common characteristics that may facilitate the transfer of vanA-mediated resistance to S. aureus. The objectives of this study were to determine the genetic interrelatedness of the seven VRSA isolates recovered to date, identify the genetic factors shared by the Enterococcus isolates recovered from three of the latest four cases of VRSA infection from southeastern Michigan, and determine whether the Enterococcus vanA plasmids could also be identified in the corresponding VRSA isolates.
MATERIALS AND METHODS
Bacterial isolates.
All of the bacterial isolates analyzed in this study are listed in Table 1. To confirm the species identification of the isolates sent to the CDC, we used conventional biochemical analysis (2, 24).
TABLE 1.
VRSA strains and associated VRE isolates in this study
| Strain | Strain no. | Site | Mo/yr of isolation | MIC (μg/ml)a
|
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CC | DAP | DOX | ERY | GM | OXA | LEV | LNZ | PEN | Q/D | RIF | STR | TEC | VAN | ||||
| VRSA-4 | HIP14300 | Foot wound | 02/2005 | >16 | ≤0.5 | ≤0.25 | >8 | ≤2 | >16 | >16 | 2 | ≥2 | 0.5 | ≤0.5 | ND | 16 | 256 |
| E. faecalis | HIP14333 | Rectum | 02/2005 | ND | ND | 8 | >8 | >500 | ND | >8 | 2 | 4 | 16 | 1 | ≤1,000 | 128 | 1,024 |
| VRSA-5 | HIP15178 | Surgical wound | 10/2005 | >16 | ≤0.5 | ≤0.5 | >8 | ≤2 | >16 | >16 | 4 | >2 | 2 | ≤0.5 | ND | 8 | 512 |
| E. faecalis | HIP15179 | Co-isolated with VRSA | 10/2005 | ND | 1 | ≤2 | >8 | >500 | ND | 8 | 4 | 2 | 8 | 2 | >1,000 | 64 | 512 |
| VRSA-6 | AIS2006032 | Foot wound | 12/2005 | >16 | 1 | 1 | >8 | ≤2 | >16 | >16 | 2 | >2 | 1 | ≤0.5 | ND | 16 | 1,024 |
| E. faecalis | AIS2007003 | Rectum | 12/2005 | ND | 2 | 4 | >8 | >500 | ND | >8 | 2 | 2 | 16 | 1 | >1,000 | 64 | >512 |
| E. avium | AIS2007004 | Rectum | 12/2005 | ND | 1 | 8 | >8 | >500 | ND | 8 | 2 | 2 | ND | 1 | ≤1,000 | 8 | 256 |
| VRSA-7 | AIS2006049 | Arm wound | 10/2006 | >16 | 2 | ≤1 | >8 | ≤2 | >16 | 8 | 2 | >2 | 0.5 | ≤0.5 | ND | 16 | 512 |
Abbreviations: CC, clindamycin; DAP, daptomycin; DOX, doxycycline; ERY, erythromycin; GM, gentamicin; OXA, oxacillin, LEV, levofloxacin; LNZ, linezolid, PEN, penicillin; Q/D, quinupristin-dalfopristin; STR, streptomycin; TEC, teicoplanin, TET, tetracycline, VAN, vancomycin; ND, not determined.
Antimicrobial susceptibility testing.
Susceptibility to vancomycin and other antimicrobial agents was determined by the reference broth microdilution method by using in-house-prepared panels with cation-adjusted Mueller-Hinton broth (BD Biosciences, Sparks, MD). Susceptibility methods and interpretation were performed according to Clinical and Laboratory Standards Institute (formerly NCCLS) guidelines (10, 11). Quality control testing was performed daily by using strains S. aureus ATCC 29213 and E. faecalis ATCC 29212.
PFGE and MLST typing.
Pulsed-field gel electrophoresis (PFGE) was performed by digesting genomic DNA with the SmaI restriction enzyme and following the standard procedures for S. aureus (25) or enterococci (26). TIFF images of the pulsed-field gels were analyzed by using BioNumerics software (Applied Maths, Austin, TX) and Dice coefficients. The relatedness of the S. aureus strains was established by using previously published standards to identify the isolates as USA types (25). Multilocus sequence typing (MLST) of VRSA-6 was performed by amplification and sequence determination of seven genetic loci. The primers and PCR conditions were the same as those described for S. aureus at www.mlst.net (14). Both strands of each PCR product were sequenced by using the same primers. Sequencing was performed with a CEQ DTCS Quick Start kit and a CEQ 8000 genetic analysis system (Beckman Coulter, Fullerton, CA).
Isolation and manipulation of plasmid DNA.
Plasmids were prepared with a Qiagen Plasmid Midi or Maxi kit (Valencia, CA), according to the manufacturer's protocol, but with specific modifications for either Staphylococcus or Enterococcus. For S. aureus, the procedure was modified by the inclusion of lysostaphin (Sigma-Aldrich, St. Louis, MO) at a final concentration of 30 μg/ml in the cell lysis buffer. For the isolation of plasmids from Enterococcus species, lysozyme was added to a resuspension buffer at a final concentration of 5 mg/ml and the bacteria were incubated in the buffer at 37°C for 30 min before the lysis step. Also, the elution buffer was warmed to 65°C for the elution of plasmid DNA from the Qiagen cartridge.
PCR amplification of Inc18 and Tn1546 elements.
Six PCR primers were unique to this study (Table 2). PCRs for the detection of vanA, traA, and repR were prepared in a total volume of 50 μl, which consisted of 1.6 mM each deoxynucleoside triphosphate (Applied Biosystems, Foster City, CA), 400 μM each primer, 1× buffer, 1 mM MgCl2, 0.5 U of AmpliTaq Gold Enzyme (Applied Biosystems), and 2 μl of DNA extract (which was equal to 100 to 500 ng of DNA). PCRs were performed in a GeneAmp PCR system 9700 (Applied Biosystems) with the following reaction cycles: an initial denaturation step of 2 min at 94°C; 30 cycles of 15 s at 95°C, 30 s at 55°C, and 30 s at 72°C; and a final elongation at 72°C for 7 min. The PCR products were visualized on an agarose gel.
TABLE 2.
PCR Primers used in this study
| Primera | Sequence (5′ to 3′) | Fragment size (bp) |
|---|---|---|
| vanA F | CAT GAA TAG AAT AAA AGT TGC TGC AAT A | 1,032 |
| vanA R | CCC CTT TAA CGC TAA TAC GAT CAA | |
| traA F | TAA TCG CAA TGG CTT CTT ATC | 475 |
| traA R | TCT GCC CAA TCT TTA CGA AT | |
| repR F | GCT TCA TGA CGG CTT GTT A | 565 |
| repR R | TTG GCT GCT TTG ACA GAT TTA |
F, forward; R, reverse.
Primers P1 to P19 and the reaction conditions previously reported by Arthur et al. (1) were used to characterize the Tn1546-like elements in the VRSA and VRE isolates. In addition, Tn1546-like elements were characterized by using the long PCR protocol of Woodford and Stigter (38). The long PCR products were digested with EcoRI, EcoRV, HindIII, and XbaI.
The PCR primers and conditions for the detection of erythromycin-mediated resistance were the same as those described previously (27).
Conjugation.
Five filter mating experiments were performed in which the vanA donors were either E. faecalis HIP12467 (VRSA-1), HIP14333 (VRSA-4), HIP15179 (VRSA-5), or AIS2007003 (VRSA-6) or E. avium AIS2007004 (VRSA-6). The recipient strain was E. faecalis JH2-2. After overnight growth in brain heart infusion (BHI) broth with either 25 μg/ml vancomycin (donors) or 25 μg/ml fusidic acid (recipient), 100 μl of each culture was added to 5 ml of new BHI broth and incubated for 5 h. The mating mixtures were combined in a 20:1 ratio of donor to recipient (400 μl donor and 20 μl of recipient) and filtered through a Nalge 0.45-μm-pore-size filter under vacuum. The filter was removed from the filter unit and placed on a BHI agar plate, and the plate was incubated for 18 h. The filter with the overlying colonies was then removed from the agar and placed in 5 ml BHI broth, and the filter and broth were vortexed to remove the growth. Transconjugants were selected on BHI agar plates containing 25 μg/ml each of vancomycin and fusidic acid. The HindIII-digested plasmid DNA from the transconjugants was compared to the HindIII-digested plasmid DNA from the VRE donor to ensure that the transferred plasmid was the same as the donor plasmid.
Southern hybridization.
To determine the locations of the vanA and traA genes, plasmid DNA was digested with HindIII and examined by Southern blot analysis. The DNA fragments were separated on an agarose gel and transferred to a Zeta-probe GT genomic blotting membrane (Bio-Rad, Hercules, CA). The blot was probed with a 1,032-bp vanA PCR product (the PCR is described above) that was labeled with horseradish peroxidase by using an ECL direct nucleic acid labeling and detection systems kit (Amersham Biosciences, Piscataway, NJ). Hybridization occurred at 42°C overnight (12 h), and washes were performed according to the manufacturer's instructions. For the nick translation experiment, the HindIII-digested plasmid fragments from the VRSA strains and the VRE transconjugants were separated on an agarose gel and transferred to the blotting membrane. The blot was probed with the HindIII-digested plasmid fragments from the corresponding VRE transconjugant that were labeled by use of a NEBlot Phototope probe labeling kit (New England Biolabs). The hybridization was visualized with a Phototope-Star detection kit (New England BioLabs).
Clumping assay.
The clumping assays were performed by a microtiter serial twofold dilution method, as described previously (12). Briefly, a pheromone-containing supernatant was prepared from E. faecalis JH2-2. Cultures were prepared by diluting 1 ml of the overnight culture in 100 ml of fresh BHI broth and incubating the mixture at 37°C with shaking for 4 to 6 h. The culture was centrifuged at 8,000 × g for 10 min at 4°C, and the supernatant was then filtered through a 0.22-μm-pore-size filter unit (Nalgene, Rochester, NY) and boiled for 15 min (16). The ability of the supernatants to induce the clumping of Enterococcus strains was tested by mixing 50 μl of culture filtrate with 50 μl of fresh BHI broth and 25 μl of fresh log-phase Enterococcus cells. The mixtures were incubated for 2 to 3 h at 37°C with shaking and were examined for clumping. E. faecalis JH2-2 carrying pPD1 and E. faecalis JH2-2 carrying pAM373 were used as positive controls, and E. faecalis JH2-2 carrying pIP501 was used as a negative control.
RESULTS
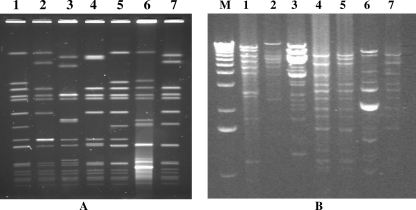
VRSA-4, VRSA-5, VRSA-6, and VRSA-7 were positive for vanA by PCR; and the vancomycin MICs ranged from 256 μg/ml to 1,024 μg/ml (Table 1). Typing of the VRSA isolates by PFGE indicated that the four most recent VRSA isolates were unique (Fig. 1A). Comparison of these patterns to the patterns of the VRSA isolates from the first three cases revealed that VRSA-5 and VRSA-1 had indistinguishable patterns. The PFGE patterns for VRSA-4 and VRSA-5 placed these isolates in the USA100 lineage, the most common MRSA lineage in the health care setting (25). The PFGE pattern of VRSA-6 was just outside of the 80% similarity range for USA100, but by MLST this isolate was sequence type 5 (ST5). This is the same ST as VRSA-4 and VRSA-5 (CDC, unpublished data) (Table 3).
FIG. 1.
PFGE analysis of VRSA genomic DNA (A) and restriction enzyme analysis of VRSA plasmid DNA (B). Each lane is labeled with the VRSA case number; lane M, 1-kb molecular marker. Genomic DNA was digested with SmaI, and plasmid DNA was digested with HindIII.
TABLE 3.
Summary of microbiological findings for seven VRSA cases
| Case no. | State | VRSA PFTa | Associated VRE (possible vanA donor) | Evidence of Inc18-like van A plasmid
|
Characterization of VRSA vanA plasmid | Reference(s) or source | |
|---|---|---|---|---|---|---|---|
| VRE | VRSA | ||||||
| 1 | MI | USA100 | E. faecalis | Yes | No | S. aureus plasmid with Tn1546 insertion | 15, 35 |
| 2 | PA | USA100 | Unknown | Not applicable | No | Uncharacterized | 29 |
| 3 | NY | USA800 | E. faecium | No | No | Maintained VRE vanA plasmid | 36 |
| 4 | MI | USA100 | E. faecalis | Yes | Yes | Maintained VRE vanA plasmid | This work |
| 5 | MI | USA100 | E. faecalis | Yes | Yes | Maintained VRE vanA plasmid | This work |
| 6 | MI | Not definedb | E. faecalis or E. avium | Yes | No | Probable S. aureus plasmid with Tn1546 insertion | This work |
| 7 | MI | USA100 | Unknown | Not applicable | Yes | Uncharacterized | This work |
PFT, pulsed-field type.
The PFGE pattern is related to USA100, but it is outside of the 80% similarity range.
We analyzed VRSA plasmid DNA by restriction analysis to determine if the VRSA isolates differed in their total plasmid DNA contents and by Southern blot analysis to locate the vanA gene. The HindIII plasmid restriction patterns for VRSA-1 and VRSA-5 were different (Fig. 1B, lanes 1 and 5, respectively). Therefore, we concluded that VRSA-1 and VRSA-5 are independent isolates. The restriction patterns of the plasmids from VRSA-4, VRSA-5, and VRSA-7 indicated that these plasmids are related (Fig. 1B, lanes 4, 5, and 7, respectively). The location of vanA on a plasmid was previously reported for VRSA-1 to VRSA-3 (29, 35, 36). By using the vanA sequences as a probe, the vanA gene was localized by Southern blot analysis to a 7-kb HindIII fragment of plasmid DNA in VRSA-4 to VRSA-7 (data not shown).
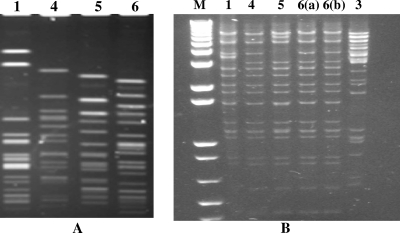
Since vancomycin-resistant E. faecalis strains were isolated from all but the most recent Michigan VRSA case, the isolates were typed by PFGE to determine if a single strain was a common donor for all of the Michigan VRSA cases. Each vancomycin-resistant E. faecalis isolate demonstrated a unique PFGE pattern (Fig. 2A). For all four VRE isolates (three E. faecalis isolates and one E. avium isolate) associated with the recent Michigan VRSA isolates, vanA-mediated vancomycin resistance was transferred to E. faecalis JH2-2 by conjugation. The transfer efficiency ranged from approximately 1.2 × 10−4 to 6.3 × 10−6 transconjugants per recipient cell. Analysis of uncut and HindIII-digested plasmid DNA from the transconjugant demonstrated that a single plasmid was present in each transconjugant, and these plasmids ranged in size from approximately 30 kb to 45 kb (data not shown). In each case, only vancomycin resistance (vanA mediated) and erythromycin resistance (ermB mediated) were transferred to E. faecalis JH2-2 by conjugation. Plasmids from all of the VRE transconjugants were similar by restriction pattern analysis with either HindIII (Fig. 2B) or EcoRI (data not shown). Plasmids from both the E. faecalis isolates and the E. avium isolate associated with VRSA-6 had a pattern that was indistinguishable from the pattern of the VRE plasmid from case 4 [Fig. 2B, lanes 4, 6(a), and 6(b)].
FIG. 2.
PFGE analysis of E. faecalis genomic DNA (A) and restriction enzyme analysis of VRE vanA plasmids from the transconjugants (B). Each lane is labeled with the VRSA case number; lane M, 1-kb molecular marker. For VRSA-6, VRE isolates of two species were recovered; lane 6(a) is the plasmid from E. faecalis and lane 6(b) is the plasmid from the E. avium. E. faecalis genomic DNA was digested with SmaI. Although E. faecalis was isolated from four VRSA-infected patients, the PFGE patterns of these indicated that these were different strains. Plasmid DNA from the transconjugants was digested with HindIII. The restriction patterns of plasmids from the Michigan VRE isolates were similar, but the plasmid from the VRE isolate from patient 3 (New York) was different.
Because the vanA VRE plasmid from the first VRSA case had previously been identified as an Inc18-like plasmid (15) and because this plasmid appeared to be related by restriction analysis to the VRE vanA plasmids associated with more recent Michigan VRSA cases, we used PCR analysis to further characterize the latter plasmids by detection of two Inc18-specific genes, traA and repR (30). The traA gene encodes a DNA nickase conjugative protein, and repR encodes a replication regulatory protein. Plasmids from both the native VRE isolates and the corresponding strain JH2-2 transconjugants were tested; and all were found to be positive for traA, repR, and vanA by PCR (Table 3). In addition, Southern blot analysis located the vanA and traA genes on the same plasmid in each transconjugant (data not shown).
The VRSA-associated VRE isolates (isolates HIP14333, HIP15179, AIS2007003, and AIS2007004) were tested for pheromone-responsive clumping when they were exposed to an E. faecalis JH2-2 filtrate. None of the VRE isolates demonstrated clumping in the presence of the filtrate, suggesting that their vanA plasmids are not pheromone responsive.
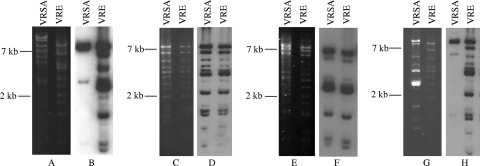
To determine how much of the VRE vanA plasmid was maintained in the VRSA isolates, we isolated the VRE vanA plasmids from the transconjugants, labeled them by nick translation, and performed a Southern blot using vanA plasmid DNA as the probe and HindIII-digested plasmid DNA from the corresponding VRSA as the target. The sequence of the first VRSA plasmid, pLW1043, has been determined (35). The S. aureus plasmid had acquired Tn1546, presumably from the VRE plasmid. This transposon was the only portion of the VRE vanA plasmid identified in the VRSA vanA plasmid. Our sequence results from the hybridization experiment with VRSA-1 were consistent with the previously published plasmid sequence. The HindIII digestion of pLW1043 was expected to result in a 7-kb fragment containing the Tn1546 sequence. This fragment was detected by Southern blot hybridization by using the corresponding VRE vanA plasmid as a probe (Fig. 3A). As a control, the VRE vanA plasmid probe was also used to detect HindIII fragments of the same plasmid from the VRE transconjugants, and all fragments were detected, indicating complete labeling of the plasmid DNA. The same hybridization experiment was performed with isolates from cases 4 to 6, as well as the corresponding VRE transconjugants. Hybridization of the VRSA-4 and VRSA-5 plasmids indicated that the VRSA isolates maintained the entire VRE vanA plasmid. Plasmids from VRSA-4 and VRSA-5 were positive for Inc18 traA and repR by PCR. In contrast, the VRE vanA plasmid probe hybridized to a 7-kb HindIII restriction fragment of VRSA-6 plasmid DNA. This HindIII fragment was the same 7-kb fragment that hybridized to the vanA probe described above. This result is similar to the results seen for VRSA-1, suggesting that in VRSA-6 Tn1546 inserted into an S. aureus plasmid and that no other VRE vanA plasmid DNA was maintained in the S. aureus isolate (Table 3). Plasmids from VRSA-1 and VRSA-2 were negative for traA and repR by PCR.
FIG. 3.
Detection of VRE vanA plasmid DNA in VRSA isolates by Southern blot analysis. Southern blot analyses of each VRSA plasmid DNA were performed by using a biotin-labeled probe prepared from the corresponding VRE vanA plasmid from the transconjugant. For each VRSA case, the left panel is an ethidium bromide gel and the right panel is the Southern blot. Molecular size standards are indicated at the left. (A and B) Isolates from VRSA-1; (C and D) isolates from VRSA-4; (E and F) isolates from VRSA-5; (G and H) the VRSA and the vancomycin-resistant E. faecalis isolates from VRSA-6.
For case 7, no VRE was isolated from sites of possible colonization (nares and rectum). Since some of the VRSA isolates maintained the VRE plasmid, we isolated plasmids from the VRSA isolates and used PCR to test for Inc18-specific genes traA and repR, as well as for vanA. One plasmid was identified in this isolate, and it was estimated to be 40 kb. The PCR for all three genes was positive, suggesting that an Inc18-like vanA plasmid was involved in this VRSA occurrence as well (Table 3).
Several different insertions and deletions within Tn1546 have been reported in VRE and VRSA isolates (8, 18, 36, 37). The transposon sequences from the first VRSA isolate and the corresponding VRE isolate were identical, and there were no insertions or deletions relative to the sequence of the prototype Tn1546 element (1, 35). To assess VRSA-4 to VRSA-7 and the corresponding VRE isolates from cases 4 to 6 for the presence of insertions or deletions within their Tn1546-like elements, we used a combination of primers to amplify 10 overlapping fragments of the entire element. The sizes of all PCR products were consistent with the size of the prototype Tn1546 element. Also, the results of restriction analyses of the amplified Tn1546-like elements in each isolate were consistent with those for the prototype Tn1546 element (data not shown). Therefore, all VRSA isolates from Michigan appeared to have the prototype transposon, whereas the transposon elements from the Pennsylvania and New York VRSA isolates each demonstrated unique insertions and deletions (8, 36).
DISCUSSION
We have identified an Inc18-like, vanA-encoding conjugative plasmid as a likely factor in the emergence of the five VRSA isolates that have been reported in southeastern Michigan since 2002. For each VRSA case, the isolate demonstrated a different restriction pattern for either the chromosomal or the plasmid DNA, so we conclude that each VRSA isolate was generated by a unique genetic event. However, each of the plasmids of the associated VRE isolates had similar restriction profiles, the same replication and conjugative proteins, and consistent Tn1546-like elements.
The Tn1546-like vanA element has been described on several different conjugative plasmids (13, 17, 19, 21-23, 28, 32), yet the transfer of vanA-mediated resistance to other bacteria, specifically, S. aureus, appears to be a rare event, with only seven VRSA isolates identified to date. It was anticipated that vanA, pheromone-responsive conjugative plasmids would be important for the occurrence of VRSA. S. aureus produces the pheromone cAM373, which can induce the clumping of Enterococcus carrying a cAM373-responsive plasmid, and the vanA gene was identified on such a plasmid in an isolate of E. faecalis (28). Also, vanA has been identified on other pheromone-responsive conjugative plasmids in both E. faecium and E. faecalis. It appears that all of the Michigan VRSA isolates described here occurred independently of pheromone-induced conjugation, since none of the vancomycin-resistant E. faecalis isolates associated with the Michigan VRSA isolates demonstrated pheromone-responsive clumping. Other environmental factors, e.g., the matrix of a biofilm, may play an important role in bringing the bacteria together in close proximity, where conjugation could take place.
The vanA gene has also been identified on non-pheromone-responsive conjugative plasmids. In E. faecium, vanA has been reported on pMG1-like plasmids (32). These plasmids demonstrate high-frequency conjugative transfer between enterococci (31, 32). The first report of a vanA Inc18-like plasmid was the characterization of the vanA plasmid in the E. faecalis isolate associated with the first VRSA case. Inc18-like conjugative plasmids (e.g., pIP501) characteristically demonstrate a broad host range. Conjugation between many different genera of gram-positive bacteria, in addition to at least one gram-negative species, Escherichia coli, has been demonstrated (9, 20, 33). The broad-host-range characteristic of Inc18-like plasmids is likely important for the conjugative transfer of vanA-mediated resistance from Enterococcus to S. aureus.
Little is known about the prevalence and geographical distributions of different vanA plasmids in VRE. If Inc18-like vanA plasmids are more common in Michigan than at other geographical locations, this may explain why five of the seven VRSA cases occurred in Michigan. It should be noted that the VRSA vanA plasmid in the isolate from the third VRSA case was identified in E. faecium (36). The E. faecium vanA plasmid is a conjugative plasmid, but it is negative for the Inc18 genes, traA and repR (W. Zhu, unpublished data). The nature of this plasmid is being investigated, but clearly, Inc18-like vanA plasmids are not the only plasmids that are capable of transferring vancomycin resistance from Enterococcus spp. to S. aureus.
S. aureus may have characteristics which allow some strains to be the more likely recipients of DNA transfer. All of the Michigan S. aureus recipients are part of the same clonal complex, ST5, and three of the four isolates clearly belong to the USA100 pulsed-field type. Even though they belong to a common lineage, there are differences in the PFGE patterns that identify each S. aureus recipient as unique. The association of isolates from the USA100, ST5 lineage with VRSA is likely because isolates within this lineage are the most common health care-associated MRSA strains (25). S. aureus strain RN4220 is often used in experimental studies because this strain can accept plasmid DNA from other species of bacteria by either conjugation or transformation. A study by Waldron and Lindsay demonstrated that a mutation in the hsdR gene, which encodes the Sau1 type I restriction-modification system, was responsible for RN4220's transformable phenotype (34). Whether the VRSA strains have similar mutations in the Sau1 type 1 restriction-modification system is being investigated.
The occurrence of VRSA appears to be either a one- or a two-step genetic event. The plasmid from the first VRSA isolate was sequenced and was found to be a previously recognized S. aureus plasmid containing a Tn1546 insertion (35). The proposed model of resistance transfer was a two-step genetic event in which the S. aureus isolate acquired the Enterococcus vanA plasmid, Tn1546 transferred from the Enterococcus plasmid to the S. aureus plasmid by transposition, and the Enterococcus plasmid was not maintained in the S. aureus recipient. In the third VRSA case, the entire Enterococcus vanA plasmid was maintained in the S. aureus recipient (36). This suggests a single genetic event in which the vanA plasmid is transferred from one isolate to the next, most likely by conjugation. In our analysis of the Michigan VRSA isolates, both outcomes were observed.
It is not clear how common VRSA will be in the future, but this is certainly a concerning antimicrobial resistance that should be prevented, if possible. An important aspect of VRSA prevention will be the control of VRE and methicillin-resistant S. aureus transmission. This is a challenging prospect, since both VRE and methicillin-resistant S. aureus are endemic in most health care settings in the United States (4). It may be prudent to implement the most rigorous control measures for patient populations and at locations where VRE isolates with Inc18-like plasmids occur.
Acknowledgments
We thank Fred C. Tenover, Linda M. Weigel, Roberta B. Carey, Brandi Limbago, James Rudrick, Dawn Sievert, and Marcus Zervos for their helpful comments regarding this work.
The findings and conclusions presented in this report are those of the authors and do not necessarily represent the views of the Centers for Disease Control and Prevention.
Footnotes
Published ahead of print on 3 December 2007.
REFERENCES
- 1.Arthur, M., C. Molinas, F. Depardieu, and P. Courvalin. 1993. Characterization of Tn1546, a Tn3-related transposon conferring glycopeptide resistance by synthesis of depsipeptide peptidoglycan precursors in Enterococcus faecium BM4147. J. Bacteriol. 175:117-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bannerman, T. L. 2003. Staphylococcus, Micrococcus, and other catalase-positive cocci that grow aerobically, p. 384-404. In P. R. Murray, E. R. Baron, J. H. Jorgensen, M. A. Pfaller, and R. H. Yolken (ed.), Manual of clinical microbiology, 8th ed. ASM Press, Washington, DC.
- 3.Centers for Disease Control and Prevention. 2004. Brief report: vancomycin-resistant Staphylococcus aureus—New York, 2004. Morb. Mortal. Wkly. Rep. 53:322-323. [PubMed] [Google Scholar]
- 4.Centers for Disease Control and Prevention. 2002. National Nosocomial Infections Surveillance (NNIS) System Report, data summary from January 1992 to June 2002, issued August 2002. Am. J. Infect. Control 30:458-475. [DOI] [PubMed] [Google Scholar]
- 5.Centers for Disease Control and Prevention. 2002. Staphylococcus aureus resistant to vancomycin—United States, 2002. Morb. Mortal. Wkly. Rep. 51:565-567. [PubMed] [Google Scholar]
- 6.Centers for Disease Control and Prevention. 2002. Vancomycin-resistant Staphylococcus aureus—Pennsylvania, 2002. Morb. Mortal. Wkly. Rep. 51:902. [PubMed] [Google Scholar]
- 7.Chang, S., D. M. Sievert, J. C. Hageman, M. L. Boulton, F. C. Tenover, F. P. Downes, S. Shah, J. T. Rudrik, G. R. Pupp, W. J. Brown, D. Cardo, and S. K. Fridkin. 2003. Infection with vancomycin-resistant Staphylococcus aureus containing the vanA resistance gene. N. Engl. J. Med. 348:1342-1347. [DOI] [PubMed] [Google Scholar]
- 8.Clark, N. C., L. M. Weigel, J. B. Patel, and F. C. Tenover. 2005. Comparison of Tn1546-like elements in vancomycin-resistant Staphylococcus aureus isolates from Michigan and Pennsylvania. Antimicrob. Agents Chemother. 49:470-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clewell, D. B., and M. V. Francia. 2004. Conjugation in gram-positive bacteria, p. 227-256. In B. E. Funnell and G. J. Phillips (ed.), Plasmid biology. ASM Press, Washington, DC.
- 10.Clinical and Laboratory Standards Institute. 2006. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 7th ed. Approved standard M7-A7. Clinical and Laboratory Standards Institute, Wayne, PA.
- 11.Clinical and Laboratory Standards Institute. 2006. Performance standards for antimicrobial susceptibility testing. Sixteenth informational supplement. M100-S16. Clinical and Laboratory Standards Institute, Wayne, PA.
- 12.Dunny, G. M., B. L. Brown, and D. B. Clewell. 1978. Induced cell aggregation and mating in Streptococcus faecalis: evidence for a bacterial sex pheromone. Proc. Natl. Acad. Sci. USA 75:3479-3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dutka-Malen, S., B. Blaimont, G. Wauters, and P. Courvalin. 1994. Emergence of high-level resistance to glycopeptides in Enterococcus gallinarum and Enterococcus casseliflavus. Antimicrob. Agents Chemother. 38:1675-1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Enright, M. C., N. P. Day, C. E. Davies, S. J. Peacock, and B. G. Spratt. 2000. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J. Clin. Microbiol. 38:1008-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Flannagan, S. E., J. W. Chow, S. M. Donabedian, W. J. Brown, M. B. Perri, M. J. Zervos, Y. Ozawa, and D. B. Clewell. 2003. Plasmid content of a vancomycin-resistant Enterococcus faecalis isolate from a patient also colonized by Staphylococcus aureus with a VanA phenotype. Antimicrob. Agents Chemother. 47:3954-3959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Flannagan, S. E., and D. B. Clewell. 2002. Identification and characterization of genes encoding sex pheromone cAM373 activity in Enterococcus faecalis and Staphylococcus aureus. Mol. Microbiol. 44:803-817. [DOI] [PubMed] [Google Scholar]
- 17.Handwerger, S., M. J. Pucci, and A. Kolokathis. 1990. Vancomycin resistance is encoded on a pheromone response plasmid in Enterococcus faecium 228. Antimicrob. Agents Chemother. 34:358-360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Handwerger, S., J. Skoble, L. F. Discotto, and M. J. Pucci. 1995. Heterogeneity of the vanA gene cluster in clinical isolates of enterococci from the northeastern United States. Antimicrob. Agents Chemother. 39:362-368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heaton, M. P., and S. Handwerger. 1995. Conjugative mobilization of a vancomycin resistance plasmid by a putative Enterococcus faecium sex pheromone response plasmid. Microb. Drug Resist. 1:177-183. [DOI] [PubMed] [Google Scholar]
- 20.Kurenbach, B., C. Bohn, J. Prabhu, M. Abudukerim, U. Szewzyk, and E. Grohmann. 2003. Intergeneric transfer of the Enterococcus faecalis plasmid pIP501 to Escherichia coli and Streptomyces lividans and sequence analysis of its tra region. Plasmid 50:86-93. [DOI] [PubMed] [Google Scholar]
- 21.Leclercq, R., E. Derlot, M. Weber, J. Duval, and P. Courvalin. 1989. Transferable vancomycin and teicoplanin resistance in Enterococcus faecium. Antimicrob. Agents Chemother. 33:10-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lim, S. K., K. Tanimoto, H. Tomita, and Y. Ike. 2006. Pheromone-responsive conjugative vancomycin resistance plasmids in Enterococcus faecalis isolates from humans and chicken feces. Appl. Environ. Microbiol. 72:6544-6553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Magi, G., R. Capretti, C. Paoletti, M. Pietrella, L. Ferrante, F. Biavasco, P. E. Varaldo, and B. Facinelli. 2003. Presence of a vanA-carrying pheromone response plasmid (pBRG1) in a clinical isolate of Enterococcus faecium. Antimicrob. Agents Chemother. 47:1571-1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martins Teixeira, L., and R. R. Facklam. 2003. Enterococcus, p. 422-433. In P. R. Murray, E. R. Baron, J. H. Jorgensen, M. A. Pfaller, and R. H. Yolken (ed.), Manual of clinical microbiology, 8th ed. ASM Press, Washington, DC.
- 25.McDougal, L. K., C. D. Steward, G. E. Killgore, J. M. Chaitram, S. K. McAllister, and F. C. Tenover. 2003. Pulsed-field gel electrophoresis typing of oxacillin-resistant Staphylococcus aureus isolates from the United States: establishing a national database. J. Clin. Microbiol. 41:5113-5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Murray, B. E., K. V. Singh, J. D. Heath, B. R. Sharma, and G. M. Weinstock. 1990. Comparison of genomic DNAs of different enterococcal isolates using restriction endonucleases with infrequent recognition sites. J. Clin. Microbiol. 28:2059-2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Raney, P. M., F. C. Tenover, R. B. Carey, J. E. McGowan, Jr., and J. B. Patel. 2006. Investigation of inducible clindamycin and telithromycin resistance in isolates of beta-hemolytic streptococci. Diagn. Microbiol. Infect. Dis. 55:213-218. [DOI] [PubMed] [Google Scholar]
- 28.Showsh, S. A., E. H. De Boever, and D. B. Clewell. 2001. Vancomycin resistance plasmid in Enterococcus faecalis that encodes sensitivity to a sex pheromone also produced by Staphylococcus aureus. Antimicrob. Agents Chemother. 45:2177-2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28a.Sievert, D. M., J. T. Rudrik, J. B. Patel, L. C. McDonald, M. J. Wilkins, and J. C. Hageman. Vancomycin-resistant Staphylococcus aureus in the United States, 2002-2006. Clin. Infect. Dis., in press. [DOI] [PubMed]
- 29.Tenover, F. C., L. M. Weigel, P. C. Appelbaum, L. K. McDougal, J. Chaitram, S. McAllister, N. Clark, G. Killgore, C. M. O'Hara, L. Jevitt, J. B. Patel, and B. Bozdogan. 2004. Vancomycin-resistant Staphylococcus aureus isolate from a patient in Pennsylvania. Antimicrob. Agents Chemother. 48:275-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thompson, J. K., and M. A. Collins. 2003. Completed sequence of plasmid pIP501 and origin of spontaneous deletion derivatives. Plasmid 50:28-35. [DOI] [PubMed] [Google Scholar]
- 31.Tomita, H., C. Pierson, S. K. Lim, D. B. Clewell, and Y. Ike. 2002. Possible connection between a widely disseminated conjugative gentamicin resistance (pMG1-like) plasmid and the emergence of vancomycin resistance in Enterococcus faecium. J. Clin. Microbiol. 40:3326-3333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tomita, H., K. Tanimoto, S. Hayakawa, K. Morinaga, K. Ezaki, H. Oshima, and Y. Ike. 2003. Highly conjugative pMG1-like plasmids carrying Tn1546-like transposons that encode vancomycin resistance in Enterococcus faecium. J. Bacteriol. 185:7024-7028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Trieu-Cuot, P., C. Carlier, and P. Courvalin. 1988. Conjugative plasmid transfer from Enterococcus faecalis to Escherichia coli. J. Bacteriol. 170:4388-4391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Waldron, D. E., and J. A. Lindsay. 2006. Sau1: a novel lineage-specific type I restriction-modification system that blocks horizontal gene transfer into Staphylococcus aureus and between S. aureus isolates of different lineages. J. Bacteriol. 188:5578-5585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weigel, L. M., D. B. Clewell, S. R. Gill, N. C. Clark, L. K. McDougal, S. E. Flannagan, J. F. Kolonay, J. Shetty, G. E. Killgore, and F. C. Tenover. 2003. Genetic analysis of a high-level vancomycin-resistant isolate of Staphylococcus aureus. Science 302:1569-1571. [DOI] [PubMed] [Google Scholar]
- 36.Weigel, L. M., R. M. Donlan, D. H. Shin, B. Jensen, N. C. Clark, L. K. McDougal, W. Zhu, K. A. Musser, J. Thompson, D. Kohlerschmidt, N. Dumas, R. J. Limberger, and J. B. Patel. 2007. High-level vancomycin-resistant Staphylococcus aureus isolates associated with a polymicrobial biofilm. Antimicrob. Agents Chemother. 51:231-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Woodford, N. 2001. Epidemiology of the genetic elements responsible for acquired glycopeptide resistance in enterococci. Microb. Drug Resist. 7:229-236. [DOI] [PubMed] [Google Scholar]
- 38.Woodford, N., and J. M. Stigter. 1998. Molecular investigation of glycopeptide resistance in gram-positive bacteria. Methods Mol. Med. 15:579-615. [DOI] [PubMed] [Google Scholar]