Abstract
The antibiotics lactonamycin and lactonamycin Z provide attractive leads for antibacterial drug development. Both antibiotics contain a novel aglycone core called lactonamycinone. To gain insight into lactonamycinone biosynthesis, cloning and precursor incorporation experiments were undertaken. The lactonamycin gene cluster was initially cloned from Streptomyces rishiriensis. Sequencing of ca. 61 kb of S. rishiriensis DNA revealed the presence of 57 open reading frames. These included genes coding for the biosynthesis of l-rhodinose, the sugar found in lactonamycin, and genes similar to those in the tetracenomycin biosynthetic gene cluster. Since lactonamycin production by S. rishiriensis could not be sustained, additional proof for the identity of the S. rishiriensis cluster was obtained by cloning the lactonamycin Z gene cluster from Streptomyces sanglieri. Partial sequencing of the S. sanglieri cluster revealed 15 genes that exhibited a very high degree of similarity to genes within the lactonamycin cluster, as well as an identical organization. Double-crossover disruption of one gene in the S. sanglieri cluster abolished lactonamycin Z production, and production was restored by complementation. These results confirm the identity of the genetic locus cloned from S. sanglieri and indicate that the highly similar locus in S. rishiriensis encodes lactonamycin biosynthetic genes. Precursor incorporation experiments with S. sanglieri revealed that lactonamycinone is biosynthesized in an unusual manner whereby glycine or a glycine derivative serves as a starter unit that is extended by nine acetate units. Analysis of the gene clusters and of the precursor incorporation data suggested a hypothetical scheme for lactonamycinone biosynthesis.
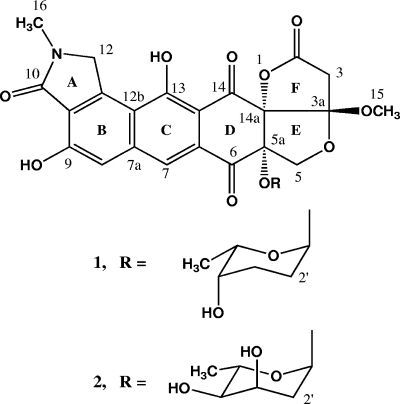
Lactonamycin (compound 1 [Fig. 1]) is an antibiotic of novel structure isolated from Streptomyces rishiriensis in a screen for new antibiotics active against drug-resistant bacterial strains (29, 30). Lactonamycin exhibits potent antimicrobial activity against methicillin-resistant Staphylococcus aureus and vancomycin-resistant Enterococcus as well as strong antitumor activity (29). Lactonamycin Z (compound 2 [Fig. 1]), a closely related antibiotic isolated from Streptomyces sanglieri, displays significant antitumor activity, although it exhibits weaker antibacterial activity than lactonamycin (18). Both antibiotics possess an aglycone core, lactonamycinone, whose structure is unlike that of any other antibiotic. Because of the unique structural core present in these antibiotics, they may possess an unusual mode of action that could be exploited for anticancer and antibacterial drug discovery. The presence of a carbohydrate residue in both antibiotics may also allow the creation of lactonamycinone derivatives with a new spectrum of bioactivity by changing the glycosylation pattern using combinatorial biosynthesis or glycorandomization. Both of these two techniques have been successfully employed to create libraries of glycosylated natural product variants (7, 25). The structure of lactonamycinone suggests that these antibiotics are polyketides related to the naphthacenequinone antibiotics tetracenomycin C (TcmC) and elloramycin. The biosynthetic gene clusters for TcmC and elloramycin have both been cloned, and the functions of many of the genes in the Tcm cluster have been elucidated (11, 19, 38). Detailed investigations of TcmC biosynthesis have shown that novel oxygenase chemistry is associated with the formation of the angular cis-1,2-diol function found in this antibiotic (37, 54). The presence of an analogous cis-1,2-diol in lactonamycinone and the presence of the novel, oxygen-rich E-F ring system suggest that unusual oxygenase chemistry probably plays a role in the biosynthesis of these antibiotics. Manipulation of these post-polyketide synthase (post-PKS) tailoring enzymes may also provide access to new lactonamycinone variants (40). For these reasons, we initiated investigations to clone and characterize the lactonamycin and lactonamycin Z gene clusters.
FIG. 1.
Structures of lactonamycin (compound 1) and lactonamycin Z (compound 2).
MATERIALS AND METHODS
General methods, bacterial strains, and cloning vectors.
Bacterial strains and plasmids used in this study are listed in Table 1. Escherichia coli strains were grown and transformed as described by Sambrook et al. (41). Culture media were obtained from Becton Dickinson and Co. (Sparks, MD) unless otherwise noted. Chemical reagents were purchased from Sigma-Aldrich Co. (Milwaukee, WI). Isotopically labeled compounds were obtained from Cambridge Isotopes (Andover, MA). Streptomyces rishiriensis MJ773-88K4 was obtained from Tomio Takeuchi at the Institute of Microbial Chemistry, Tokyo, Japan, while S. sanglieri AK 623 was obtained from the culture collection at the University of Newcastle. S. sanglieri AK 623 was cultured at 27°C in a medium containing 1% starch, 1% glucose, 1% corn steep powder, 0.5% casein-peptone, 0.2% yeast extract, 0.1% NaCl, pH 7.0, in tap water. S. rishiriensis was cultured under reported conditions (29) and under the conditions used for S. sanglieri AK 623. S. rishiriensis and S. sanglieri were maintained on ISP 2 (4 g/liter yeast extract, 10 g/liter malt extract, 4 g/liter dextrose, 20 g/liter agar) and ISP 3 agar (20 g/liter oatmeal extract, 1 ml/liter trace elements solution [in 100 ml H2O, 0.1 g FeSO4·7H2O, 0.1 g MnCl2·4H2O, 0.1 g ZnSO4·7H2O]), respectively.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s)a | Reference or source |
|---|---|---|
| Bacterial strains | ||
| Streptomyces rishiriensis MJ773-88K4 | Producer of lactonamycin | 29 |
| Streptomyces sanglieri AK 623 | Producer of lactonamycin Z | 18 |
| S. sanglieri AK 623 Δlcz36 | S. sanglieri AK 623 with nt 40 to 1281 of lcz36 replaced by pIJ773 aac(3)IV-oriT cassette; Apr | This work |
| Escherichia coli DH10B | General cloning host | Invitrogen |
| E. coli 12567 | dam-13::Tn9 dcm-6 hsdM | 27 |
| E. coli BW25113 | K-12 derivative; ΔaraBAD ΔrhaBAD | 10 |
| Plasmids | ||
| pBluescript II SK(+) | E. coli cloning vector; Amr | Stratagene |
| pSP72 | E. coli cloning vector; Amr | Promega |
| pGEM-T Easy | PCR cloning vector; Amr | Promega |
| pOJ446 | E. coli-Streptomyces shuttle cosmid for conjugal transfer; Apr | 5 |
| pIJ773 | pBluescript II SK(+) containing an aac(3)IV-oriT cassette | 16 |
| pIJ790 | λ Red (gam bet exo) cat araC rep101ts Cmr | 16 |
| pIJ702 | pIJ101 derivative; rep101 Tsr | 24 |
| pUZ8002 | tra neo RP4 | 36 |
| SuperCos1 | E. coli cosmid cloning vector; Carbr Kanr | Stratagene |
| pGEM-T Easy DH35 | 495-bp PCR internal lct45 fragment cloned into pGEM-T Easy | This work |
| pBS-VII-41F | pGEM-3zf containing ermEp* promoter as 0.45-kb EcoRI/SacI fragment | B. Shen, University of Wisconsin |
| pSET152 | E. coli-Streptomyces | 5 |
| Shuttle plasmid; oriT intφC31 Amr | ||
| pXZ152 | pSET152 derivative with tsr gene; Apr Tsr | This work |
| pOE152 | pXZ152 derivative containing ermEp* promoter; Apr Tsr | This work |
| pOElcz36 | pOE152 with lct36 cloned downstream of the ermE* promoter; Apr Tsr | This work |
| pKOlcz36 | Cosmid pCAS2 with nt 40 to 1281 of lcz36 replaced by pIJ773 aac(3)IV-oriT cassette | This work |
| pCAK7 | pOJ446 cosmid clone of S. sanglieri AK 623, hybridizing to lac36 | This work |
| pCAS2 | SuperCos1 cosmid clone of S. sanglieri AK 623, hybridizing to lct36 | This work |
| pCSR5 | pOJ446 cosmid clone of S. rishiriensis MJ773-88K4 hybridizing to the pGEM-T Easy DH35 lct36 insert | This work |
| pCSR8 | pOJ446 cosmid clone of S. rishiriensis MJ773-88K4 hybridizing to the pGEM-T Easy DH35 lct36 insert | This work |
| pCSR13 | pOJ446 cosmid clone of S. rishiriensis MJ773-88K4 hybridizing to lct10 | This work |
Drug resistance markers used are ampicillin (Am), apramycin (Ap), carbacillin (Carb), chloramphenicol (Cm), kanamycin (Kan), and thiostrepton (Ts).
DNA manipulations and sequence analysis.
Plasmid and cosmid DNA were purified with a QIAprep Spin Miniprep kit (Qiagen, Valencia, CA). PCR products were separated on agarose gels and purified from the gels using a Geneclean Spin kit (MP Biomedicals, Solon, OH). Digestion with restriction endonucleases and ligation experiments were carried out by standard procedures under conditions recommended by the manufacturers. Automated DNA sequencing was performed using universal and synthetic oligonucleotide primers. Cosmids were sequenced by release of the inserts with XbaI and SpeI followed by further digestion with BamHI or PstI to generate a pool of fragments that were then cloned into pBluescript II SK(+) or pSP72 plasmid vectors. The clones were then sequenced with universal primers followed by primer walking with designed primers. DNA gaps between subclones were filled in by direct sequencing of the parent cosmid. Sequences were determined by complete sequencing of both DNA strands with multiple sequencing of some regions.
PCR amplification of an NDP-hexose 2,3-dehydratase from S. rishiriensis.
The presence of an l-rhodinose moiety in lactonamycin suggested that a nucleoside diphosphate (NDP)-hexose 2,3-dehydratase gene should reside within the lactonamycin biosynthetic gene cluster. Accordingly, several sets of degenerate oligonucleotide primers were designed to amplify an internal region of an NDP-hexose 2,3-dehydratase gene from the lactonamycin producer S. rishiriensis MJ 773-88K4. The sequences of these primers were derived from conserved domains of NDP-hexose 2,3-dehydratase genes found in antibiotic biosynthetic gene clusters of Streptomyces violaceoruber Tu22 (gra ORF27) (13), Streptomyces peucetius (dnmT) (43), Saccharopolyspora erythraea (eryBVI) (14, 49), and Streptomyces nogalater (snogH) (53). One set of degenerate primers led to the successful amplification of a 495-bp NDP-hexose 2,3-dehydratase gene fragment. The degenerate primers were N-terminal primer DHF (5′-GGSCGSTTCTTCTCSGTSGAG-3′) and C-terminal primer DHR (5′-ACSGTSCGSGHGTCCATGTT-3′, where S is C or G).
Cosmid library construction and screening.
S. rishiriensis and S. sanglieri were grown in yeast extract-malt extract (YEME) medium (24), and genomic DNA was extracted by standard methods (24). Cosmid libraries of S. rishiensis and S. sanglieri DNA in cosmid vector pOJ446 (5) were constructed in a manner previously described (57). In a similar way, a cosmid library of S. sanglieri DNA in cosmid vector SuperCos1 was also constructed. Screening of the cosmid libraries by Southern hybridizations was carried out as previously described (57). The S. rishiriensis pOJ446 cosmid library was screened with the 495-bp NDP-hexose 2,3-dehydratase fragment amplified from S. rishiriensis by PCR, while the two S. sanglieri cosmid libraries were screened with a 1,320-bp PCR product obtained by PCR amplification of the S. rishiriensis lct36 gene.
Fermentation of S. rishiriensis MJ773-88K4.
Fermentation of S. rishiriensis MJ773-88K4 was carried out according to published methods (29) and also under the conditions employed for S. sanglieri AK 623 (see below). The levels of lactonamycin production were low in both media, and reproducible antibiotic production was difficult to maintain. Analytical high-performance liquid chromatography (HPLC) detection of lactonamycin was carried out on a 4.6- by 250-mm YMC-Pack ODA-A column as described below for lactonamycin Z. The retention time for lactonamycin in this system was ca. 27 min. Small amounts of lactonamycin could be isolated from the fermentation broth by preparative HPLC using a 10- by 250-mm YMC-Pack ODS-A column and the conditions described for preparative HPLC of lactonamycin Z (see below).
Fermentation of S. sanglieri and isolation of lactonamycin Z.
Fermentation of S. sanglieri AK 623 was carried out at 27°C and 120 rpm in the medium described above. The medium (100 ml) was inoculated from a spore suspension and grown at 27°C for 48 h. Fresh medium (five 200-ml portions) was then inoculated with five 10-ml samples of the seed culture, and fermentation was allowed to proceed for 96 h. The pH of the medium was readjusted to 4.5, and the mixture was centrifuged to separate the fermentation broth from the mycelium. The supernatant was extracted three times with ethyl acetate, while the mycelium was suspended in excess 50% aqueous acetone and stirred for 1 hour. The aqueous acetone extract was filtered, and the acetone was removed in vacuo. The aqueous residue was extracted three times with ethyl acetate, and the resulting extracts were combined with the ethyl acetate extracts of the fermentation broth. The total ethyl acetate extract was dried over anhydrous sodium sulfate and filtered, and the ethyl acetate was removed in vacuo. The residue was dissolved in chloroform and filtered to remove insoluble material. The filtered chloroform extract was taken to dryness in vacuo, and lactonamycin Z was isolated from the residue by preparative HPLC as described below. The estimated titer of lactonamycin Z in the fermentation broth is ca. 5 mg per liter.
HPLC analysis of lactonamycin Z.
HPLC analysis was carried out with a Varian ProStar 325 UV-visible detector set at 300 nm and a YMC-Pack ODS-A column (4.6 by 250 mm). Elution was carried out by running a linear gradient over 30 min from 40:60 methanol-water containing 0.5% formic acid to 80:20 methanol-water containing 0.5% formic acid with a flow rate of 1 ml/min. Under these conditions, lactonamycin Z exhibited a retention time of ca. 21 min. For preparative applications, the same gradient was applied to a YMC-Pack ODS-A column (10 by 250 mm) with a flow rate of 3 ml/min. Approximately 1 to 2 mg of purified lactonamycin Z could be obtained from 1 liter of fermentation broth.
Preparation of lactonamycinone from lactonamycin Z.
Lactonamycinone was prepared from lactonamycin Z by a modification of the procedure used to convert lactonamycin into lactonamycinone (30). Lactonamycin Z (3.5 mg) was dissolved in 0.5 ml of spectral grade tetrahydrofuran, and 0.2 ml of 1 N HCl was added. The resulting solution was stirred at room temperature and monitored periodically by HPLC. After 67 h, the tetrahydrofuran was removed in vacuo at room temperature, and the remaining aqueous solution was diluted with 1 ml of water and extracted three times with chloroform. The combined chloroform extracts were dried, and the solvent was removed in vacuo to yield ca. 1.0 mg of lactonamycinone, which exhibited an HPLC retention time of 19 min under the same conditions used for analytical HPLC analysis of lactonamycin and lactonamycin Z. Nuclear magnetic resonance (NMR) analysis confirmed the assigned structure: 1H NMR (500 MHz, CDCl3, δ values), 2.99 (1H, d, J = 17.0 Hz, H-3), 3.09 (1H, d, J = 17.0 Hz, H-3), 3.20 (3H, s, OCH3), 3.29 (3H, s, NCH3), 4.10 (1H, d, J = 9.6 Hz, H-5), 4.91 (1H, d, J = 9.6 Hz, H-5), 4.98 (2H, s, H-12), 7.31 (1H, s, H-8), 8.07 (1H, s, H-7), 9.36 (1H, s, very broad, C-9 OH), 13.70 (1H, s, broad, C-13 OH); 13C NMR (125 MHz, CDCl3), 29.2 (C-16, NCH3), 37.7 (C-3), 52.7 (C-15, OCH3), 55.0 (C-12), 73.8 (C-5), 82.6 (C-5a), 90.6 (C-14a), 109.4 (C-13a), 112.4 (C-3a), 112.9 (C-8), 116.6 (C-12b), 121.6 (C-7), 129.8 (C-6a), 141.8 (C-7a), 142.7 (C-12a), 157.4 (C-9), 164.0 (C-13), 168.9 (C-10), 170.6 (C-2), 189.5 (C-6), 192.2 (C-14). NMR spectral data for lactonamycinone have not been previously reported.
Inactivation of lactonamycin Z production by double-crossover disruption of the glycosyltransferase gene (lcz36).
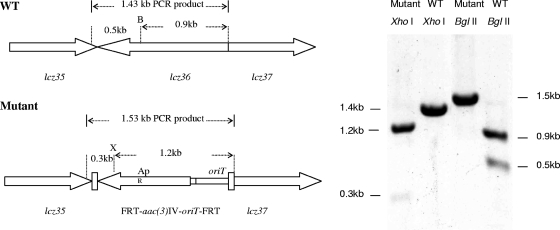
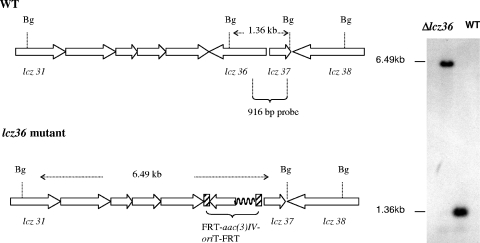
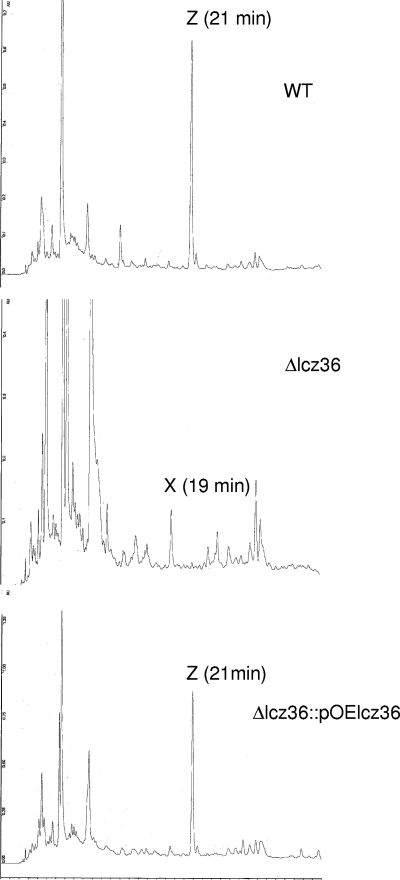
Double-crossover disruption of the lactonamycin Z glycosyltransferase gene (lcz36) was carried out by using the lambda Red-mediated recombination methodology (15, 16). The PCR product required for disruption of the lcz36 gene was obtained by amplification of the 1,383-bp aac(3)IV-oriT sequence using pIJ773 as a template with primer pairs containing 39-nucleotide (nt) extensions derived from the 5′ and 3′ ends of the lcz36 gene. The forward and reverse primers were ZGLTF (5′-ATGTGTTCGCTCCGGGAAAAGGATTGGAGAGCATGGATGATTCCGGGGATCCGTCACC-3′) and ZGLTR (5′-GGGAC CGGCCCGCCGACGCAGATGCCTGACGATTTGTCATGTAGGCTGGAGC TGCTTC-3′, respectively, where the underlined portions show the 39-nt extensions). Electrocompetent cells of E. coli BW25113 (10) containing pIJ790 (Cmr) and the SuperCos1-derived cosmid pCAS2 (Carbr Kanr) containing lcz36 were prepared using the method of Gust et al. (15). The electrocompetent cells were transformed with purified PCR product using a Bio-Rad Gene Pulser II set at 200 Ω, 25 μF, and 2.5 kV. The shocked cells were incubated in 1 ml of LB broth at 37 οC for 1 h and then selected on LB agar containing ampicillin (100 μg/ml), kanamycin (50 μg/ml), and apramycin (50 μg/ml). Transformants carrying the desired cosmid, pKOlcz36, were identified by restriction digestion of the isolated cosmids. Plasmid pKOlcz36 was then introduced into the nonmethylating E. coli ET12567 (27) containing the RP4 derivative pUZ8002 (36) and transferred into Streptomyces sanglieri AK 623 by intergeneric conjugation. For intergeneric conjugation, spores of Streptomyces sanglieri AK 623 were heat shocked at 50°C for 10 min, mixed with the E. coli cells prepared according to a standard protocol (24), and plated out on AS1 medium (1). After incubation at 30°C for 20 h, the plates were then overlaid with 3 ml of 0.7% soft tryptic soy agar containing 0.5 mg nalidixic acid and 1.25 mg apramycin. Incubation was then continued at 30°C for 5 to 7 days. Exconjugants were selected, and their antibiotic resistance was evaluated. Cosmid DNA was isolated from Kans Apr colonies, and the presence of the expected double-crossover disruption was verified by PCR amplification of the disrupted region using the following primer set: Flcz36 (5′-GGTCTTGTATCGGGAAAG-3′) and Rlcz36 (5′-CCAGTACGGCAGTATGCG-3′). Sequencing of the PCR product (data not shown) and restriction digests of the amplified fragment (Fig. 2) confirmed the presence of the predicted disruption. In addition, genomic DNA was extracted from the Δlcz36 disruptant, digested with BglII, and analyzed by Southern hybridization using a 916-bp probe amplified from the lcz36-lcz37 region of wild-type S. sanglieri using the following primer set: Lcz36F (5′-CGGTCACCTTCGCGTCCC-3′) and Lcz36R (5′-CTCCCAGTTGCGGGCGAAG-3′). The hybridization pattern exhibited by the Δlcz36 disruptant was completely consistent with the genetic alteration expected from the double-crossover disruption (Fig. 3). Fermentation of the Δlcz36 disruptant and analysis of the combined organic extracts of the mycelium and fermentation broth by HPLC showed that lactonamycin Z production had been abolished. A small peak (compound X) that exhibited approximately the same retention time as the aglycone lactonamycinone was also present in the extracts, but mass spectrometry analysis indicated that compound X is not lactonamycinone (Fig. 4).
FIG. 2.
Verification of S. sanglieri Δlcz36 mutant by restriction digestion of PCR products from wild-type (WT) and mutant strains. B, BglII; X, XhoI.
FIG. 3.
Southern hybridization analysis of S. sanglieri Δlcz36 mutant and wild-type S. sanglieri (WT) using a 916-bp probe derived from lcz36 and lcz37. In the Δlcz36 mutant, lcz36 is replaced by the FLP recombination target (FRT)-aac(3)IV-oriT-FRT cassette from pIJ773. Bg, BglII.
FIG. 4.
HPLC analysis of fermentation broth from wild-type S. sanglieri (WT), the Δlcz36 mutant, and the complemented Δlcz36 mutant using HPLC conditions described in Materials and Methods. Z, lactonamycin Z; X, unknown.
Construction of plasmid pOElcz36.
To complement S. sanglieri AK 623 Δlcz36, a derivative of the integrating plasmid pSET152 (5) was constructed. First, the thiostrepton resistance gene (tsr) from plasmid pIJ702 was amplified by PCR using the following forward and reverse primers: Tsr-XbaIF (AAAAATCTAGATCACTGACGAATCGAGGTCGAGGAAC) and Tsr-NheIR (AAAAAGCTAGCAGGCGAATACTTCATATGCGGGGATC, where the underlined bases correspond to XbaI and NheI sites, respectively). The resulting PCR fragment was digested with XbaI and NheI and then cloned into the NheI site of pSET152. A construct with the tsr gene in the same orientation as aac(3)IV of pSET152 was then identified by restriction digestion and designated pXZ152. The 0.45-kb ermEp* promoter cassette of plasmid pBS-VII-41F was PCR amplified using the following forward and reverse primers: ErmMfeI (5′-AAAAACAATTGAGCCCGACCCGAGCACGC-3′) and ErmEcoRI (5′-AAAAAGAATTCCCATGGGGTCGCACCCACCGACGAC-3′, where the underlined nucleotides correspond to MfeI and EcoRI sites, respectively).
The PCR product was digested with MfeI and EcoRI and then cloned into the EcoRI site of pXZ152. A construct with ermEp* promoter in the same orientation as the lacZα gene of pSET152 was identified by restriction digestion and designated pOE152 (data and map not shown). To complement the Δlcz36 mutant, the PCR primers 5′-AAAAAGAATTCATGAGAGTTCTGTTTGCT-3′ and 5′-AAAAATCTAGATCAGGACCGGTGCCGGGC-3′ were used to amplify the 1,284-bp lcz36 gene from S. sanglieri (EcoRI and XbaI sites of primers underlined). The purified PCR product was digested with EcoRI and XbaI and then cloned into the EcoRI-XbaI sites of pOE152 downstream of the ermEp* promoter, therefore generating the expression plasmid pOElcz36.
Complementation of the S. sanglieri AK 623 Δlcz36 mutant.
Plasmid pOElcz36 was introduced into E. coli ET12567/pUZ8002 and intergeneric conjugation was carried out with the Δlcz36 mutant in the manner described above. Exconjugants were selected using thiostrepton at 50 μg/ml. The integration of pOElcz36 into the chromosome of mutant Δlcz36 was verified by PCR (data not shown). Fermentation of S. sanglieri Δlcz36 (pOElcz36) was carried out in the usual manner, and the combined organic extracts of the mycelium and fermentation broth were analyzed for the presence of lactonamycin Z by HPLC. The analysis showed that lactonamycin Z production had been restored (Fig. 4). This was confirmed by isolation and NMR characterization of lactonamycin Z from the complemented strain (data not shown). The level of lactonamycin Z production by the complemented strain was estimated to be ca. one-third that of the wild-type strain.
Administration of isotopically labeled precursors to S. sanglieri AK 623.
Sterile S. sanglieri AK 623 culture medium (eight 200-ml portions) was inoculated with eight 10-ml samples of a 48-h culture of S. sanglieri grown under standard conditions. After incubation at 27°C, 120 rpm, for 48 h, a filter-sterilized aqueous solution containing 1.0 g of the labeled precursor was distributed equally to each of the eight shaking flasks, and the fermentation was allowed to continue for an additional 48 h. At the end of this time, lactonamycin Z was isolated from the fermentation broth by preparative HPLC as described above. All precursors were 99% enriched with 13C. The [2-13C, 15N]glycine was 98% enriched with 15N.
Nucleotide sequence accession numbers.
The nucleotide sequences were deposited in the GenBank database and given accession numbers EU147298 and EU147299.
RESULTS
Precursor incorporation experiments with S. sanglieri.
The structures of lactonamycin and lactonamycin Z suggest that the aglycone portion of these antibiotics, lactonamycinone, is biosynthetically related to the decaketide TcmC. This hypothesis was evaluated by administration of sodium [1-13C]-, [2-13C]-, and [1,2-13C2]acetate to the S. sanglieri fermentation. The results of these experiments are summarized in Table 2 . Several conclusions can be drawn from these data. First, the labeling patterns produced by each of these precursors are consistent with the head-to-tail incorporation of nine acetate units into lactonamycinone. Second, the data indicate that the 13C chemical shift assignments reported for C-3 and C-2′ of lactonamycin Z (18) need to be reversed. This conclusion follows from the labeling patterns observed with [2-13C]- and [1,2-13C2]acetate and is consistent with the 13C chemical shift assigned to C-3 of lactonamycin (30) (see supplemental material for more detail). Third, the labeling pattern is consistent with a mechanism for formation of the E/F ring system that involves oxidative cleavage of ring D of a naphthacenequinone intermediate similar to TcmD3 (Fig. 5). Last, the labeling patterns show that C-12 and C-12a are not derived from acetate, since none of the forms of labeled acetate produced enrichment in these carbon atoms. The absence of labeling on C-12 and C-12a suggested that an alternative starter unit is employed for polyketide assembly. A likely candidate for a starter unit appeared to be glycine. This possibility was examined by administration of [1,2-13C2]-, [1-13C]-, and [2-13C, 15N]glycine to S. sanglieri AK 623. The results of these experiments (Table 2) demonstrate that C-12a and C-12 of lactonamycinone are derived from C-1 and C-2 of glycine, respectively, and that the nitrogen atom in the δ-lactam ring of lactonamycinone is derived from the nitrogen atom of glycine. In addition to labeling some of the carbon atoms of the δ-lactam ring, both [1,2-13C2]- and [2-13C, 15N]glycine led to efficient labeling of the O-methyl and N-methyl groups of the aglycone (C-15 and C-16). However, no labeling of the O- and N-methyl groups was observed when [1-13C]glycine was administered to the fermentation. Labeling of the O-methyl and N-methyl groups by [1,2-13C2]- and [2-13C, 15N]glycine can be explained if C-2 of glycine is entering the C1 pool via metabolism of the labeled amino acid by the glycine cleavage system. The glycine cleavage system is a multienzyme complex composed of four proteins that catalyzes the oxidative decarboxylation of glycine to carbon dioxide, ammonia, and a C1 unit that is incorporated into 5,10-methylenetetrahydrofolate (34, 35). A hypothetical biosynthetic pathway for lactonamycinone based upon the results of the precursor incorporation experiments is shown in Fig. 5. In this pathway, the postulated formation of the 1,2-cis diol moiety of lactonamycinone by the formal cis ring opening of an epoxide is based upon an analogous process in TcmC biosynthesis (37), while the timing for formation of the δ-lactam ring of lactonamycinone is arbitrarily shown to occur at a late stage in the pathway.
TABLE 2.
Incorporation of labeled precursors into lactonamycin Z
| Precursor | Labeling pattern | One-bond coupling(s) (Hz) |
|---|---|---|
| Sodium [1-13C] acetate | C-2, C-3a, C-5a, C-6a, C-7a, C-9, C-10, C-13, C-14 | None observed |
| Sodium [2-13C] acetate | C-3, C-5, C-6, C-7, C-8, C-9a, C-12b, C-13a, C-14a | None observed |
| Sodium [1,2-13C2] | C-2, C-3, C-3a, C-5, | C-2, C-3 = 52.7 |
| acetate | C-5a, C-6, C-6a, C-7, | C-5, C-5a = 40.8 |
| C-7a, C-8, C-9, C-9a, | C-6, C-6a = 55.3 | |
| C-10, C-12b, C-13, | C-7, C-7a = 55.2 | |
| C-13a, C-14, C-14a | C-8, C-9 = 73.0 | |
| C-9a, C-10 = 64.1 | ||
| C-12b, C-13 = 67.3 | ||
| C-13a, C-14 = 59.8 | ||
| C-14a, C-3a = 37.1 | ||
| [1,2-13C2]glycine | C-12, C-12a, C-15, C-16 | C-12, C-12a = 41.6 |
| [1-13C]glycine | C-12a | None observed |
| [2-13C, 15N]glycine | C-12, C-15, C-16, N-11 | C-12, 15N = 11.3 |
FIG. 5.
Hypothetical biosynthetic pathway for lactonamycinone.
Cloning of the lactonamycin gene cluster from S. rishiriensis.
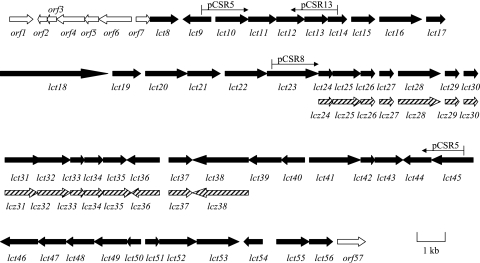
Initial efforts to clone the lactonamycin gene cluster were focused upon the lactonamycin producer S. rishiriensis. The presence of an l-rhodinose moiety in the antibiotic suggested that cloning could be accomplished by using deoxyhexose biosynthetic genes as probes. Using the conserved domains of NDP-hexose 2,3-dehydratases associated with several antibiotic biosynthetic pathways in Streptomyces species, degenerate primers were designed to amplify an NDP-hexose 2,3-dehydratase gene from the genomic DNA of S. rishiriensis. One set of primers led to the amplification of a 495-bp PCR product, which was verified to be part of a NDP-hexose 2,3-dehydratase gene by sequencing. The 495-bp fragment of an NDP-hexose 2,3-dehydratase gene was then used to probe an S. rishiriensis cosmid library constructed in cosmid vector pOJ446, yielding a total of 46 positive clones from which 2, pCSR5 and pCSR8, were chosen for sequencing based upon their restriction maps. Sequencing of these two overlapping cosmids revealed a total of 49 ORFs spanning ca. 56 kb of DNA. In addition, cosmid pCSR13, which overlaps the 5′ end of pCSR5, was isolated by using the lct10 gene as a probe. Partial sequencing of pCSR13 revealed eight ORFs that are located immediately upstream of the lct9 gene.
DNA sequence analysis of lactonamycin cluster from S. rishiriensis.
Sequencing a total of ca. 61 kb of DNA contained within cosmids pCSR5, pCSR8, and pCSR13 revealed the presence of 57 open reading frames (ORFs) (Fig. 6). The 57 sequenced genes and the actual or putative functions assigned to them are listed in Table 3. The borders of the lct cluster could not be defined by gene disruption experiments, since lactonamycin production could not be sustained in our laboratory. However, tentative borders can be assigned on the basis of the sequence analysis. The upstream end of the lct cluster probably begins with the 4′-phosphopantetheinyltransferase homolog lct8, since the upstream ORF (orf7) exhibits strong similarities to S-adenosylmethionine decarboxylase, an enzyme that is unlikely to be required for the lactonamycin biosynthetic pathway. The downstream border of the lct cluster is probably defined by orf57, which exhibits the signature of a transposase. The genes contained within the region from lct8 to lct56 can be divided into five groups as follows.
FIG. 6.
Genetic organization of the lct gene cluster from S. rishiriensis and the partial lcz gene cluster from S. sanglieri.
TABLE 3.
Organization and analysis of lactonamycin gene cluster from S. rishiriensis
| Gene designation | Size (aa)a | Protein homolog(s) [GenBank accession number(s)], % similarity/% identity | Possible function(s) |
|---|---|---|---|
| orf1 | 288 | SCC61A.21c (CAB892266), 88/79 | Tyrosinase |
| orf2 | 98 | SteO22.21c (CAI59990), 86/78 | Putative small membrane protein |
| orf3 | 92 | Dace_2175 (EAT15876), 58/32 | Acyl carrier protein |
| orf4 | 360 | RHA1_ro02212 (ABG94019), 51/38 | Phosphatase, acyltransferase |
| orf5 | 191 | EAV71565 (CcelDRAFT_0530), 56/35 | N-Acetyltransferase |
| orf6 | 409 | RPD_3346 (ABE40570), 55/41 | Hypothetical protein, methyltransferase? |
| orf7 | 126 | CYA_0574 (ABC98790), 61/47 | S-Adenosylmethionine decarboxylase |
| lct8 | 298 | AAP09275 (BC_2311) 48/34 | 4′-Phosphopantetheinyltransferase |
| lct9 | 314 | Orf85 (BAC76543), 82/78 | AfsA homolog, β-ketoacyltransferase, γ-butyrolactone biosynthesis |
| lct10 | 442 | Orf84 (BAC76542), 75/62 | Cytochrome P450, γ-butyrolactone biosynthesis? |
| lct11 | 254 | Orf83 (BAC76541), 75/66 | Phosphatase, γ-butyrolactone biosynthesis |
| lct12 | 249 | SngB (AAX97701), 72/61 | Ketoacyl acyl carrier protein/CoA reductase, γ-butyrolactone biosynthesis |
| lct13 | 222 | TarA (AAF06961), 82/72; TylP (AAD40801), 78/68 | γ-Butyrolactone binding protein |
| lct14 | 160 | TylQ (AAD40803), 63/50 | Transcriptional regulator |
| lct15 | 276 | Orf75 (NP_851497), 79/69; TylS (AAD40804), 75/65 | Streptomyces antibiotic regulatory protein (SARP) |
| lct16 | 500 | SAV3380 (NP_824557), 64/52 | FAD-dependent monooxygenase |
| lct17 | 327 | Orf21 (BAC79034), 73/66 | Carbohydrate kinase (glucose kinase) |
| lct18 | 1,133 | Orf49 (BAC76507), 92/87 | 5-Methyltetrahydrofolate:homocysteine S-methyltransferase |
| lct19 | 285 | Orf (+9) (AAN85549), 87/81 | 5,10-Methylenetetrahydrofolate reductase |
| lct20 | 469 | MtmH (CAK50773), 97/93 | Adenosylhomocysteinase |
| lct21 | 220 | Orf71 (BAC76569), 64/53 | SARP |
| lct22 | 509 | SAV3632 (BAC71344), 53/42 | AfsR-like regulatory protein |
| lct23 | 563 | SnorA (CAA12016), 51/38 | AfsR-like regulatory protein |
| lct24 | 92 | Sim4 (AAL15582), 69/52 | Acyl carrier protein |
| lct25 | 349 | TcmN (P16559), 66/50; FdmN (AAQ08925), 58/40 | O-Methyltransferase; aligns with the C-terminal region of TcmN |
| lct26 | 83 | RPC_2301 (ABD87855), 67/47 | Acyl carrier protein |
| lct27 | 152 | FdmI (AAQ08919), 73/59; TcmN (P16559), 68/53 | Polyketide cyclase; aligns with the N-terminal region of TcmN |
| lct28 | 510 | SAV1250 (BAC68960), 63/52 | Carboxylic acid adenylation |
| lct29 | 109 | ElmI (AAF73052), 63/42; TcmI (P39890), 63/43 | Polyketide cyclase |
| lct30 | 138 | TcmJ (P16558), 70/49; ElmJ (AAF73053), 65/48 | Polyketide cyclase |
| lct31 | 421 | TcmK (P16538), 80/69 | KSα |
| lct32 | 406 | TcmL (P16539), 74/64 | KSβ |
| lct33 | 115 | TcmH (P39889), 66/52; ElmH (AAF73051), 64/42 | Monooxygenase |
| lct34 | 248 | G111934 (BAC89875), 51/35 | Thioesterase |
| lct35 | 322 | TcmO (P39896), 61/50 | O-Methyltransferase |
| lct36 | 429 | UrdGT1a (AAF00214), 66/51 | Glycosyltransferase |
| lct37 | 139 | G113466 (BAC91407), 62/40 | Hypothetical protein, ester cyclase |
| lct38 | 493 | SAV1847 (BAC69558), 68/49 | Membrane efflux protein |
| lct39 | 332 | SCO6554 (CAA22358), 61/45 | Transcriptional regulatory protein |
| lct40 | 252 | SCO7810 (CAC03628), 70/55 | Oxidoreductase |
| lct41 | 558 | TcmG (P39888), 63/46; ElmG (AAF73050), 61/46 | Dioxygenase |
| lct42 | 108 | ElmH (AAF73051), 60/38; TcmH (P39889), 57/44 | Monooxygenase |
| lct43 | 308 | SAV4759 (BAC72471), 55/43 | Oxidoreductase |
| lct44 | 337 | LanT (ADD13550), 74/63 | NDP-hexose 3-ketoreductase |
| lct45 | 468 | LanS (AAD13549), 76/70; UrdS (AAF72552), 76/68 | NDP-hexose 2,3-dehydratase |
| lct46 | 427 | UrdQ (AAF72550), 91/80; LanQ (AAD13547), 90/80 | NDP-hexose 3-dehydratase |
| lct47 | 310 | LanZ3 (AAD13561), 56/44; UrdZ3 (AFF72549), 53/44 | NDP-hexose 4-ketoreductase |
| lct48 | 326 | LanH (AAD13546), 84/73; UrdH (AAF00211), 80/67 | NDP-hexose 4,6-dehydratase |
| lct49 | 355 | LanG (AAD13545), 77/65; UrdG (AAF00210), 76/63 | NDP-hexose synthetase |
| lct50 | 161 | LanZ1 (AAD13558), 80/69, UrdZ1 (AAF00208), 71/59 | NDP-hexose 3,5-epimerase |
| lct51 | 158 | Glr0227 (BAC88168), 55/38 | Hypothetical protein, ester cyclase |
| lct52 | 405 | MetK (AAD22464), 91/84 | S-Adenosylmethionine synthetase |
| lct53 | 542 | SCO6284 (CAC37885), 88/80 | Probable acyl-CoA decarboxylase |
| lct54 | 194 | SAV5143 (BAC72855), 69/49 | ArsR regulatory protein |
| lct55 | 378 | SCO3305 (CAB45339), 69/52 | Integral membrane acyltransferase |
| lct56 | 297 | SAV4467 (BAC72179), 90/85 | rRNA methyltransferase, lactonamycin resistance? |
| orf57 | 322 | SAV4466 (BAC72178), 91/82 | Putative IS1650 family transposase |
aa, amino acids.
(i) Genes encoding the PKS and associated enzymes.
The lactonamycin minimal PKS appears to be composed of Lct31 (KSα), Lct32 (KSβ), and Lct24 (acyl carrier protein [ACP]). Lct31 and Lct32 exhibit 69% and 64% identity to TcmK and TcmL, respectively, the KSα and KSβ from the TcmC biosynthetic gene cluster, respectively (19). Lct31 contains the highly conserved sequence STGCTSGLD that is found around the ketosynthase active site cysteine residue (55). The lct cluster appears to encode two ACPs, Lct24 and Lct26. Neither resides in close proximity to the ketosynthase components of the minimal PKS. Lct24 exhibits 52% identity to the PKS ACP Sim4 from the simocyclinone gene cluster as well as a high degree of similarity to ACPs from other type II PKS gene clusters. These similarities suggest that Lct24 is part of the lactonamycin minimal PKS. Lct26 exhibits the highest degree of similarity to ACPs from Rhodopseudomonas and Thermoanaerobacter sp. It is conceivable that Lct26 plays a role in the loading of glycine or a glycine derivative onto Lct31 (see Discussion). Both ACPs display the conserved LG(x)DS motif containing the serine residue that is the attachment site for a 4′-phosphopantetheine prosthetic group (21). The lct8 gene appears to encode a 4′-phosphopantetheinyltransferase. The function of this enzyme may be to convert Lct24 and Lct26 from the apo form to the holo form.
Three putative polyketide cyclases, Lct27, Lct29, and Lct30, reside within the lct cluster. Lct27 exhibits 59% identity to FdmI, a cyclase from the fredericamycin gene cluster (55). Lct27 also shows high similarity to the N-terminal cyclase domain of the bifunctional protein TcmN from the Tcm cluster (50). Lct27 may catalyze cyclizations leading to the formation of an anthrone intermediate resembling TcmF2 (Fig. 5). Lct29 exhibits 43% identity to the cyclase TcmI (46), suggesting that Lct29 probably catalyzes a cyclization leading to an intermediate similar to TcmF1. Lct30 displays 49% identity to TcmJ. TcmJ has been shown to increase the production of TcmF2 in vitro, although the mode of action of TcmJ is unclear (2). An unusual feature of the lct cluster is the presence of the Lct34 thioesterase. Thioesterases are commonly found in type I PKS gene clusters and nonribosomal peptide gene clusters, but they are only rarely associated with type II PKS genes. A thioesterase gene has been reported to occur in a type II PKS gene cluster that resides on the linear pSLA2-L plasmid of Streptomyces rochei (32).
(ii) Genes encoding tailoring enzymes or proteins of unknown function.
The lactonamycin cluster encodes a relatively large number of oxygenases and oxidoreductases. Potential functions can be assigned to only two of the oxygenases. Lct41 displays 46% identity to TcmG and 46% identity to ElmG. The latter enzymes have been shown to catalyze the triple hydroxylation of TcmA2 to TcmC, a reaction leading to formation of a cis-1,2-diol function (37, 47, 54). Lct41 may be involved in the formation of the similar cis-1,2-diol moiety found in lactonamycin. Lct33 and Lct42 exhibit strong similarities to TcmH, a monooxygenase that catalyzes the oxidation of the naphthacenone TcmF1 to the naphthacenequinone TcmD3 (45). Structure determination of the related enzyme ActVA-ORF6 led to the characterization of an “antibiotic biosynthesis monooxygenase” (ABM) domain found in this family of proteins and the identification of three active site residues (Asn, Trp, and Arg) (42). Both Lct33 and Lct42 exhibit many of the conserved residues displayed by the ABM domain, but only Lct33 contains all three active site residues. This suggests that Lct33 plays a role in lactonamycin biosynthesis similar to that of TcmH in TcmC biosynthesis, while the function of Lct42 is unclear. An additional oxidative enzyme is encoded by lct10, whose translation product exhibits strong similarities to cytochrome P450 enzymes. Since lct10 is situated between genes that appear to be involved in the biosynthesis and binding of a γ-butyrolactone autoregulator, it is likely that lct10 plays a role in the biosynthesis of a γ-butyrolactone. The lct cluster contains one glycosyltransferase, Lct36. Lct36 is presumed to catalyze the attachment of l-rhodinose to lactonamycinone.
Two genes encoding methyltransferases are present in the lct cluster. Lct25 aligns with the C-terminal methyltransferase domain of the bifunctional protein TcmN. TcmN methylates the C-11 phenolic hydroxyl group of TcmD3 (44). It is possible that Lct25 methylates the C-13 hydroxyl group of a TcmD3 analog on the pathway to lactonamycin. The second methyltransferase is Lct35. This protein shows 50% identity to TcmO, an enzyme that methylates the C-3 phenolic hydroxyl of TcmB3 (50). Since the corresponding C-3 hydroxyl group of lactonamycin is unmethylated, it is possible that Lct35 is required for formation of the N-methyl group of lactonamycin.
Eight additional ORFs within the lct cluster encode proteins whose functions cannot be predicted from sequence analysis. The translation product of the lct16 gene exhibits strong similarity to FAD-dependent monooxygenases that catalyze the hydroxylation of aromatic rings, while lct40 and lct43 encode oxidoreductases. All three of these genes might play a role in the formation of the lactonamycin E/F ring system. The protein encoded by lct28 shows high similarity to acyl coenzyme A (acyl-CoA) synthetases and carboxylic acid adenylation enzymes. Because of the specific incorporation of glycine into lactonamycin Z, it is conceivable that Lct28 might transform glycine or a glycine derivative into the corresponding adenylate for use as the starter unit in polyketide assembly (see Discussion). Lct37 and Lct51 exhibit similarities to ester cyclases, while Lct55 exhibits high similarity to a family of membrane-bound acyltransferases. Last, Lct53 appears to encode an acyl CoA decarboxylase. No clear function can be assigned to any of these proteins.
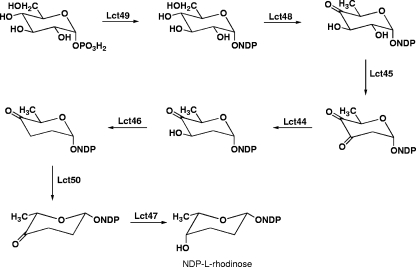
(iii) Genes encoding enzymes for NDP-l-rhodinose biosynthesis.
Lactonamycin contains an l-rhodinose moiety, which is attached to the C-5a hydroxyl group of the aglycone. Investigations of the biosynthesis of the aromatic polyketide urdamycin have identified seven genes that are required for NDP-l-rhodinose biosynthesis in Streptomyces fradiae (17). Homologs of each of these genes are present in the lactonamycin gene cluster (Table 3). The potential roles played by these genes in the formation of NDP-l-rhodinose are shown in Fig. 7.
FIG. 7.
Potential role played by lactonamycin genes in biosynthesis of NDP-l-rhodinose.
(iv) Genes encoding potential regulatory and resistance proteins.
The lactonamycin gene cluster contains an unusually large number of genes that display signatures for regulatory proteins. Potential functions can be assigned to three of these proteins, Lct13, Lct14, and Lct15. These three proteins exhibit similarities of 78%, 63%, and 75% to TylP, TylQ, and TylS, respectively. The latter three proteins are part of a regulatory network that controls production of the macrolide antibiotic tylosin in S. fradiae (4). In this network, both TylP and TylQ act as repressors, while TylS functions as an activator. The repressor activity of TylP is regulated by the binding of a γ-butyrolactone signaling molecule. A similar function would be anticipated for Lct13. The role assigned to Lct13 is supported by the fact that the proteins encoded by lct9, lct11, and lct12 exhibit strong similarities to proteins required for γ-butyrolactone biosynthesis (22, 23). Both Lct15 and TylS show strong similarities to the family of activator proteins, such as ActII-ORF4 and DnrI that are known as SARPs (Streptomyces antibiotic regulatory proteins) (56).
The lct gene cluster contains two genes whose encoded proteins might provide S. rishiriensis with lactonamycin resistance. The first of these proteins is Lct38, which appears to be a member of the major facilitator superfamily of membrane efflux proteins. A protein in this superfamily has been found to confer resistance to the azoxy antibiotic valanimycin on the producing organism, Streptomyces viridifaciens (26). The second candidate for a lactonamycin resistance protein is encoded by lct56. Lct56 exhibits high similarity to rRNA methyltransferases. Methylation of specific 23 S rRNA nucleotides is a well-established mechanism for bacterial resistance to macrolide antibiotics (12).
(v) Genes encoding proteins associated with primary metabolism.
There are four genes embedded within the lct cluster that encode proteins associated with primary metabolism. The proteins encoded by these genes, Lct18, Lct19, Lct20, and Lct52, exhibit high similarities to proteins that are required to regenerate S-adenosyl-l-methionine (AdoMet) from S-adenosylhomocysteine after the transfer of a methyl group from AdoMet. The function of these genes is presumably to supply AdoMet to the methyltransferases Lct25 and Lct35. A similar set of AdoMet biosynthetic genes has been found within the biosynthetic gene cluster of the type I polyketide antibiotic lankamycin (32).
Cloning of the lactonamycin Z biosynthetic gene cluster from S. sanglieri.
Investigations with the lactonamycin producer S. rishiriensis revealed that consistent production of lactonamycin could not be maintained. Fermentations derived either from S. rishiriensis spore stocks stored at −80°C or from lyophilized spore stocks gave erratic results. Furthermore, lactonamycin production levels were negatively affected by protoplast formation and by the manipulations required to produce exconjugants. Consequently, investigations of S. sanglieri AK 623 were commenced. The production of lactonamycin Z by this organism was found to be more robust than the production of lactonamycin by S. rishiriensis. Southern hybridization of the genomic DNA of the S. sanglieri AK 623 with probes derived from the lct33, lct36, and lct53 genes showed that unique homologs of these genes exist in S. sanglieri AK 623, suggesting the existence of a similar gene cluster for lactonamycin Z (data not shown). Therefore, the lct36 gene of S. rishiriensis was amplified by PCR, labeled with 32P, and used to screen S. sanglieri cosmid libraries constructed in pOJ446 and SuperCos1. A total of 38 positive pOJ446-derived cosmids and 17 positive SuperCos1-derived cosmids were isolated. One pOJ446-derived cosmid, pCAK7, was initially chosen for sequencing, since it also exhibited strong hybridization to probes derived from the lct33 and lct53 genes (data not shown). Sequencing of ca. 13 kb of DNA from the insert in cosmid pCAK7 disclosed the presence of 15 ORFs. These ORFs are listed in Table 4. All 15 ORFs in the sequenced region of S. sanglieri DNA exhibit high similarities of genes found in the lct cluster. Furthermore, the organization of these genes is identical to that found in the lct cluster. These findings provide strong evidence that both the S. rishiriensis and S. sanglieri clusters are involved in the production of similar metabolites. The functions of the 15 lcz genes are expected to be similar or identical to those of the corresponding lct genes.
TABLE 4.
Organization and analysis of partial lactonamycin Z gene cluster from S. sanglieri AK 623
| Gene designation | Size (aa)a | Protein homolog(s) [GenBank accession number(s)], % similarity/% identity | Possible function, % identity to S. rishiriensis homolog |
|---|---|---|---|
| lcz24 | 93 | Sim4 (AAL15582), 71/52 | Acyl carrier protein, 81 |
| lcz25 | 335 | TcmN (P16599), 66/49 | O-Methyltransferase, 83 |
| lcz26 | 83 | RPC_2301 (ABD87855), 70/44 | Acyl carrier protein, 69 |
| lcz27 | 153 | RubF (AAG03070), 70/58 | Polyketide cyclase, 87 |
| lcz28 | 524 | SAV1250 (BAC68960), 63/51 | Carboxylic acid adenylation, 75 |
| lcz29 | 109 | ElmI (AAF73052), 64/46 | Polyketide cyclase, 83 |
| lcz30 | 145 | TcmJ (P16558), 70/50 | Polyketide cyclase, 90 |
| lcz31 | 421 | TcmK (P16538), 80/67 | KSα, 90 |
| lcz32 | 406 | TcmL (P16539), 74/65 | KSβ, 87 |
| lcz33 | 115 | TcmH (P39889), 67/52; ElmH (AAF73051), 64/43 | Monooxygenase, 80 |
| lcz34 | 246 | G111934 (BAC89875), 52/37 | Thioesterase, 70 |
| lcz35 | 323 | TcmO (P39896), 62/49 | O-Methyltransferase, 84 |
| lcz36 | 427 | UrdGT1a (AAF00214), 66/51 | Glycosyltransferase, 83 |
| lcz37 | 137 | SAV3997 (BAC71709), 50/34 | Hypothetical protein, ester cyclase, 81 |
| lcz38 | 491 | SAV1847 (BAC69558), 65/47 | Membrane efflux protein, 81 |
aa, amino acids.
Confirmation that the cloned locus from S. sanglieri is required for lactonamycin Z production.
To confirm that the cloned locus from S. sanglieri contains genes that are required for lactonamycin Z production, a double-crossover gene disruption was created in the glycosyltransferase gene lcz36. The disruption cosmid, pKOlcz36, was created by replacement of nt 40 to 1281 of lcz36 in cosmid pCAS2 with the aac(3)IV-oriT cassette of pIJ773 using lambda Red-mediated recombination (15, 16). Introduction of cosmid pKOlcz36 into S. sanglieri by intergeneric conjugation led to the desired disruptant, as judged by PCR amplification of the disrupted region and restriction digests of the resulting PCR product (Fig. 2) as well as by Southern hybridization experiments (Fig. 3). The Δlcz36 mutant was cultured under standard conditions, and the combined organic extracts of the broth and mycelium were analyzed by HPLC. The HPLC analysis (Fig. 4) showed the absence of lactonamycin Z. A complementation construct, pOElcz36, was generated by placing the ermEp* promoter in front of the lcz36 gene cloned into plasmid pXZ152. Plasmid pXZ152 was derived from the integrating plasmid pSET152 by introduction of the thiostrepton resistance gene of pIJ702. The complementation construct was introduced into the Δlcz36 mutant by intergeneric conjugation. Restoration of lactonamycin Z production was confirmed by HPLC analysis (Fig. 4) and by isolation and NMR characterization of lactonamycin Z from the complemented strain (data not shown).
DISCUSSION
PCR primers designed to conserved regions of NDP-hexose 2,3-dehydratase genes were used to amplify a 2,3-dehydratase DNA probe from the lactonamycin producer S. rishiriensis. This probe was used to clone a gene cluster from S. rishiriensis whose sequence analysis suggested it was likely to be the lactonamycin biosynthetic gene cluster of this organism. Unfortunately, lactonamycin production by this organism was readily lost during the course of genetic manipulations. This behavior created major obstacles to the interpretation of gene disruption experiments. For this reason, investigations of the lactonamycin Z producer, S. sanglieri, were initiated. The lactonamycin Z gene cluster was cloned from this organism by using the glycosyltransferase gene lct36 from the lactonamycin gene cluster as a probe. Sequencing of ca. 13 kb of the cloned S. sanglieri DNA disclosed the presence of 15 ORFs. All 15 of these genes exhibit high similarity to genes within the S. rishiriensis cluster as well as an identical organization. This suggests that both gene clusters are coding for the production of similar metabolites in both organisms. Unequivocal evidence that the cloned S. sanglieri genes are required for lactonamycin Z biosynthesis was obtained by inactivation of the glycosyltransferase gene lcz36 by means of a double-crossover gene disruption and by complementation of the resulting mutation. HPLC analysis of the spectrum of metabolites produced by the lcz36 disruptant revealed the absence of lactonamycin Z (Fig. 4). Introduction of an intact copy of lcz36 under the control of the ermEp* promoter into the lcz36 disruptant restored production of lactonamycin Z (Fig. 4). These results confirm that the genetic locus cloned from S. sanglieri is required for lactonamycin Z production, and they support the hypothesis that the highly similar locus cloned from S. rishiriensis is necessary for lactonamycin production. Although the entire lactonamycin Z gene cluster of S. sanglieri has not been fully sequenced, the more extensive sequencing of the S. rishiriensis gene cluster appears to have reached the lactonamycin cluster boundaries. The 5′ end of the lactonamycin gene cluster probably begins with lct8, since the ORF immediately upstream appears to encode S-adenosylmethionine decarboxylase. The 3′ end of the lactonamycin cluster is probably defined by lct56, since the ORF immediately downstream exhibits a high degree of similarity to the IS1650 family of transposases.
Important insights into the biosynthesis of the aglycone core, lactonamycinone, were obtained from precursor incorporation experiments. Administration of sodium [1-13C]-, [2-13C]-, and [1,2-13C2]acetate to S. sanglieri produced labeling patterns that are consistent with the head-to-tail incorporation of nine acetate units into lactonamycinone (Table 2). The 13C-labeling pattern observed for rings E and F of lactonamycinone derived from the labeled forms of acetate can be rationalized by the oxidative cleavage of ring D of a napthacenequinone intermediate similar to TcmD3 (Fig. 5). Most significantly, no labeling of C-12 and C-12a was produced in any of these incorporation experiments. This observation suggested that the polyketide assembly process is initiated by use of an alternative starter unit. This hypothesis was confirmed by administration of [1,2-13C2]-, [1-13C]-, and [2-13C, 15N]glycine to S. sanglieri. The labeling patterns observed in lactonamycin Z derived from these precursors indicate that the nitrogen atom and both carbon atoms of glycine serve as the source of the polyketide starter unit (Table 2). The use of alternate starter units in type II aromatic polyketide assembly is rare, but not unprecedented. For example, propionate is used as the starter unit for daunorubicin biosynthesis in S. peucetius (3, 39), several short-chain carboxylic acids serve as starter units for the anthroquinones R1128A-D produced by Streptomyces sp. strain R1128 (28), and benzoate functions as the starter unit for enterocin in Streptomyces maritimus (20). The mechanism involved in the incorporation of alternative starter units varies in each of these examples (33). The lactonamycin and lactonamycin Z gene clusters exhibit several novel features that may provide a clue to the mechanism of glycine incorporation. One of these features is the presence of genes (lct28 and lcz28) that encode a protein with high similarities to acyl-CoA synthetases and carboxyl adenylation enzymes. Recent studies have identified the amino acid residues that determine the substrate specificity of enzymes or domains that catalyze the adenylation of α-amino acids (8, 48). Analysis of Lct28 and Lcz28 reveals that the specificity codes for these proteins are DLFVLCLL and DLFILCLL, respectively. Since the consensus specificity code for glycine is DILQ(L/V)G(L/M/V)I (9, 31), it appears unlikely that glycine serves as a substrate for either Lct28 or Lcz28. An alternative substrate could be sarcosine (N-methylglycine), for which a specificity code has not been clearly defined. Another novel feature of the lct and lcz gene clusters is the presence of two genes in each cluster that encode ACPs. Of these ACPs, only Lct24 and Lcz24 exhibit strong similarity to the ACPs of other type II PKSs. This suggests that the other ACPs, Lct26 and Lcz26, may play a role in the initiation of lactonamycinone biosynthesis with a glycine derivative. This hypothesis is supported by the fact that both the R1128 and frenolicin gene clusters contain an additional ACP that functions to initiate polyketide synthesis with an alternative starter unit (52). Therefore, it appears that initial stages of lactonamycinone biosynthesis may involve activation of a glycine derivative by Lct28 or Lcz28, attachment of the activated amino acid to Lct26 or Lcz26 as a thioester, and transfer of the amino acid from Lct26 or Lcz26 to the ketosynthase Lct31 or Lcz31 by thioester exchange.
A third unusual feature of the lactonamycin gene clusters is the presence of a thioesterase (Lct34 and Lcz34) in each cluster. Although thioesterase domains and stand-alone thioesterases are often found in association with type I polyketide synthases, the occurrence of thioesterases within type II polyketide gene clusters is very rare (32). Recent studies of the acyltransferase homolog ZhuC found within the R1128 PKS have shown that ZhuC exhibits thioesterase activity and selectively hydrolyzes acetyl-ACPs formed by malonyl-ACP decarboxylation (51). ZhuC therefore appears to function as an editing enzyme to remove acetate starter units that might otherwise compete with the alkyl acyl starter units. Investigations by Bisang et al. (6) have shown that the decarboxylation of malonyl-ACPs is catalyzed by the KSβ (chain length factor) of the minimal PKS and to a lesser extent by the KSα. The ability of the KSβ protein to catalyze malonyl-ACP decarboxylation is dependent upon the presence of a conserved glutamine residue. It appears likely that the KSβ proteins in the lct and lcz clusters can catalyze malonyl-ACP decarboxylation, since the conserved glutamine residue is present in the amino acid sequences of both proteins. For this reason, it is conceivable that the thioesterase encoded in each cluster may function in a similar manner to ZhuC in order to prevent initiation of lactonamycinone biosynthesis with acetate. Additional investigations will clearly be required to elucidate the details of glycine incorporation into lactonamycinone.
Supplementary Material
Acknowledgments
Support of this work by the National Institutes of Health (GM053818 and AI058045) and by the Robert A. Welch Foundation (C-0729) is gratefully acknowledged. We also acknowledge the National Science Foundation (grant CHE-9708978) for partial funding of the Chemistry Department 500-MHz NMR spectrometer.
Thanks are also due to Ben Shen for plasmid pBS-VII-41F, Tomio Takeuchi for a culture of S. rishiriensis, and S. Yates and X. Qian for assistance with mass spectral measurements. We also acknowledge Youlin Xia for 800-MHz NMR spectra obtained at the University of Houston.
Footnotes
Published ahead of print on 10 December 2007.
Supplemental material for this article may be found at http://aac.asm.org/.
REFERENCES
- 1.Baltz, R. H. 1980. Genetic recombination in Streptomyces fradiae by protoplast fusion and cell regeneration. Dev. Ind. Microbiol. 21:43-54. [DOI] [PubMed] [Google Scholar]
- 2.Bao, W., E. Wendt-Pienkowski, and C. R. Hutchinson. 1998. Reconstitution of the iterative type II polyketide synthase for tetracenomycin F2 biosynthesis. Biochemistry 37:8132-8138. [DOI] [PubMed] [Google Scholar]
- 3.Bao, W. L., P. J. Sheldon, E. Wendt-Pienkowsi, and C. R. Hutchinson. 1999. The Streptomyces peucetius dpsC gene determines the choice of starter unit in biosynthesis of the daunorubicin polyketide. J. Bacteriol. 181:4690-4695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bate, N., D. R. Bignell, and E. Cundliffe. 2006. Regulation of tylosin biosynthesis involving ‘SARP-helper’ activity. Mol. Microbiol. 62:148-156. [DOI] [PubMed] [Google Scholar]
- 5.Bierman, M., R. Logan, K. O'Brien, E. T. Seno, R. N. Rao, and B. E. Schoner. 1992. Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene 116:43-49. [DOI] [PubMed] [Google Scholar]
- 6.Bisang, C., P. F. Long, J. Cortes, J. Westcott, J. Crosby, A. L. Matharu, R. J. Cox, T. J. Simpson, J. Staunton, and P. F. Leadlay. 1999. A chain initiation factor common to both modular and aromatic polyketide synthases. Nature 401:502-505. [DOI] [PubMed] [Google Scholar]
- 7.Blanchard, S., and J. S. Thorson. 2006. Enzymatic tools for engineering natural product glycosylation. Curr. Opin. Chem. Biol. 10:263-271. [DOI] [PubMed] [Google Scholar]
- 8.Challis, G. L., J. Ravel, and C. A. Townsend. 2000. Predictive, structure-based model of amino acid recognition by nonribosomal peptide synthetase adenylation domains. Chem. Biol. 7:211-224. [DOI] [PubMed] [Google Scholar]
- 9.Chen, X. H., J. Vater, J. Piel, P. Franke, R. Scholz, K. Schneider, A. Koumoutsi, G. Hitzeroth, N. Grammel, A. W. Strittmatter, G. Gottschalk, R. D. Süssmuth, and R. Borriss. 2006. Structural and functional characterization of three polyketide synthase gene clusters in Bacillus amyloliquefaciens FZB 42. J. Bacteriol. 188:4024-4036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Decker, H., J. Rohr, H. Motamedi, H. Zähner, and C. R. Hutchinson. 1995. Identification of Streptomyces olivaceus Tü 2353 genes involved in the production of the polyketide elloramycin. Gene 166:121-126. [DOI] [PubMed] [Google Scholar]
- 12.Douthwaite, S., P. F. Crain, M. Liu, and J. Poehlsgaard. 2004. The tylosin-resistance methyltransferase RlmA(II) (TlrB) modifies the N-1 position of 23S rRNA nucleotide G748. J. Mol. Biol. 337:1073-1077. [DOI] [PubMed] [Google Scholar]
- 13.Draeger, G., S.-H. Park, and H. G. Floss. 1999. Mechanism of the 2-deoxygenation step in the biosynthesis of the deoxyhexose moieties of the antibiotics granaticin and oleandomycin. J. Am. Chem. Soc. 121:2611-2612. [Google Scholar]
- 14.Gaisser, S., G. A. Bohm, J. Cortes, and P. F. Leadlay. 1997. Analysis of seven genes from the eryAI-eryK region of the erythromycin biosynthetic gene cluster in Saccharopolyspora erythraea. Mol. Gen. Genet. 256:239-251. [DOI] [PubMed] [Google Scholar]
- 15.Gust, B., G. L. Challis, K. Fowler, T. Kieser, and K. F. Chater. 2003. PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc. Natl. Acad. Sci. USA 100:1541-1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gust, B., G. Chandra, D. Jakimowicz, T. Yuqing, C. J. Bruton, and K. F. Chater. 2004. Lambda red-mediated genetic manipulation of antibiotic-producing Streptomyces. Adv. Appl. Microbiol. 54:107-128. [DOI] [PubMed] [Google Scholar]
- 17.Hoffmeister, D., K. Ichinose, S. Domann, B. Faust, A. Trefzer, G. Drager, A. Kirschning, C. Fischer, E. Kunzel, D. Bearden, J. Rohr, and A. Bechthold. 2000. The NDP-sugar co-substrate concentration and the enzyme expression level influence the substrate specificity of glycosyltransferases: cloning and characterization of deoxysugar biosynthetic genes of the urdamycin biosynthetic gene cluster. Chem. Biol. 7:821-831. [DOI] [PubMed] [Google Scholar]
- 18.Höltzel, A., A. Dieter, D. G. Schmid, R. Brown, M. Goodfellow, W. Beil, G. Jung, and H. P. Fiedler. 2003. Lactonamycin Z, an antibiotic and antitumor compound produced by Streptomyces sanglieri strain AK 623. J. Antibiot. (Tokyo) 56:1058-1061. [DOI] [PubMed] [Google Scholar]
- 19.Hutchinson, C. R. 1997. Biosynthetic studies of daunorubicin and tetracenomycin C. Chem. Rev. 97:2525-2535. [DOI] [PubMed] [Google Scholar]
- 20.Izumikawa, M., Q. Cheng, and B. S. Moore. 2006. Priming type II polyketide synthases via a type II nonribosomal peptide synthetase mechanism. J. Am. Chem. Soc. 128:1428-1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jiralerspong, S., V. Rangaswamy, C. L. Bender, and R. J. Parry. 2001. Analysis of the enzymatic domains in the modular portion of the coronafacic acid polyketide synthase. Gene 270:191-200. [DOI] [PubMed] [Google Scholar]
- 22.Kato, J. Y., N. Funa, H. Watanabe, Y. Ohnishi, and S. Horinouchi. 2007. Biosynthesis of γ-butyrolactone autoregulators that switch on secondary metabolism and morphological development in Streptomyces. Proc. Natl. Acad. Sci. USA 104:2378-2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kawachi, R., T. Akashi, Y. Kamitani, A. Sy, U. Wangchaisoonthorn, T. Nihira, and Y. Yamada. 2000. Identification of an AfsA homologue (BarX) from Streptomyces virginiae as a pleiotropic regulator controlling autoregulator biosynthesis, virginiamycin biosynthesis and virginiamycin M1 resistance. Mol. Microbiol. 36:302-313. [DOI] [PubMed] [Google Scholar]
- 24.Kieser, T., M. J. Bibb, M. J. Buttner, K. F. Chater, and D. A. Hopwood. 2000. Practical Streptomyces genetics. The John Innes Foundation, Norwich, United Kingdom.
- 25.Langenhan, J. M., B. R. Griffith, and J. S. Thorson. 2005. Neoglycorandomization and chemoenzymatic glycorandomization: two complementary tools for natural product diversification. J. Nat. Prod. 68:1696-1711. [DOI] [PubMed] [Google Scholar]
- 26.Ma, Y., J. Patel, and R. J. Parry. 2000. A novel valanimycin-resistance determinant (vlmF) from Streptomyces viridifaciens MG456-hF10. Microbiology 146:345-352. [DOI] [PubMed] [Google Scholar]
- 27.MacNeil, D. J., K. M. Gewain, C. L. Ruby, G. Dezeny, P. H. Gibbons, and T. MacNeil. 1992. Analysis of Streptomyces avermitilis genes required for avermectin biosynthesis utilizing a novel integration vector. Gene 111:61-68. [DOI] [PubMed] [Google Scholar]
- 28.Marti, T., Z. H. Hu, N. L. Pohl, A. N. Shah, and C. Khosla. 2000. Cloning, nucleotide sequence, and heterologous expression of the biosynthetic gene cluster for R1128, a non-steroidal estrogen receptor antagonist—insights into an unusual priming mechanism. J. Biol. Chem. 275:33443-33448. [DOI] [PubMed] [Google Scholar]
- 29.Matsumoto, N., T. Tsuchida, M. Maruyama, N. Kinoshita, Y. Homma, H. Iinuma, T. Sawa, M. Hamada, T. Takeuchi, N. Heida, and T. Yoshioka. 1999. Lactonamycin, a new antimicrobial antibiotic produced by Streptomyces rishiriensis MJ773-88K4. I. Taxonomy, fermentation, isolation, physico-chemical properties and biological activities. J. Antibiot. 52:269-275. [DOI] [PubMed] [Google Scholar]
- 30.Matsumoto, N., T. Tsuchida, H. Nakamura, R. Sawa, Y. Takahashi, H. Naganawa, H. Iinuma, T. Sawa, T. Takeuchi, and M. Shiro. 1999. Lactonamycin, a new antimicrobial antibiotic produced by Streptomyces rishiriensis MJ773-88K4. II. Structure determination. J. Antibiot. 52:276-280. [DOI] [PubMed] [Google Scholar]
- 31.Miao, V., M. F. Coeffet-Legal, P. Brian, R. Brost, J. Penn, A. Whiting, S. Martin, R. Ford, I. Parr, M. Bouchard, C. J. Silva, S. K. Wrigley, and R. H. Baltz. 2005. Daptomycin biosynthesis in Streptomyces roseosporus: cloning and analysis of the gene cluster and revision of peptide stereochemistry. Microbiology 151:1507-1523. [DOI] [PubMed] [Google Scholar]
- 32.Mochizuki, S., K. Hiratsu, M. Suwa, T. Ishii, F. Sugino, K. Yamada, and H. Kinashi. 2003. The large linear plasmid pSLA2-L of Streptomyces rochei has an unusually condensed gene organization for secondary metabolism. Mol. Microbiol. 48:1501-1510. [DOI] [PubMed] [Google Scholar]
- 33.Moore, B. S., and C. Hertweck. 2002. Biosynthesis and attachment of novel bacterial polyketide synthase starter units. Nat. Prod. Rep. 19:70-99. [DOI] [PubMed] [Google Scholar]
- 34.Nakai, T., N. Nakagawa, N. Maoka, R. Masui, S. Kuramitsu, and N. Kamiya. 2003. Coexpression, purification, crystallization and preliminary X-ray characterization of glycine decarboxylase (P-protein) of the glycine-cleavage system from Thermus thermophilus HB8. Acta Crystallogr. Sect. D 59:554-557. [DOI] [PubMed] [Google Scholar]
- 35.Okamura-Ikeda, K., N. Kameoka, K. Fujiwara, and Y. Motokawa. 2003. Probing the H-protein-induced conformational change and the function of the N-terminal region of Escherichia coli T-protein of the glycine cleavage system by limited proteolysis. J. Biol. Chem. 278:10067-10072. [DOI] [PubMed] [Google Scholar]
- 36.Paget, M. S., L. Chamberlin, A. Atrih, S. J. Foster, and M. J. Buttner. 1999. Evidence that the extracytoplasmic function sigma factor σE is required for normal cell wall structure in Streptomyces coelicolor A3(2). J. Bacteriol. 181:204-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rafanan, E. R., Jr., C. R. Hutchinson, and B. Shen. 2000. Triple hydroxylation of tetracenomycin A2 to tetracenomycin C involving two molecules of O2 and one molecule of H2O. Org. Lett. 2:3225-3227. [DOI] [PubMed] [Google Scholar]
- 38.Rafanan, E. R., Jr., L. Le, L. Zhao, H. Decker, and B. Shen. 2001. Cloning, sequencing, and heterologous expression of the elmGHIJ genes involved in the biosynthesis of the polyketide antibiotic elloramycin from Streptomyces olivaceus Tu2353. J. Nat. Prod. 64:444-449. [DOI] [PubMed] [Google Scholar]
- 39.Rajgarhia, V. B., N. D. Priestley, and W. R. Strohl. 2001. The product of dpsC confers starter unit fidelity upon the daunorubicin polyketide synthase of Streptomyces sp. strain C5. Metab. Eng. 3:49-63. [DOI] [PubMed] [Google Scholar]
- 40.Rix, U., C. Fischer, L. L. Remsing, and J. Rohr. 2002. Modification of post-PKS tailoring steps through combinatorial biosynthesis. Nat. Prod. Rep. 19:542-580. [DOI] [PubMed] [Google Scholar]
- 41.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 42.Sciara, G., S. G. Kendrew, A. E. Miele, N. G. Marsh, L. Federici, F. Malatesta, G. Schimperna, C. Savino, and B. Vallone. 2003. The structure of ActVA-Orf6, a novel type of monooxygenase involved in actinorhodin biosynthesis. EMBO J. 22:205-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Scotti, C., and C. R. Hutchinson. 1996. Enhanced antibiotic production by manipulation of the Streptomyces peucetius dnrH and dnmT genes involved in doxorubicin (adriamycin) biosynthesis. J. Bacteriol. 178:7316-7321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shen, B., and C. R. Hutchinson. 1996. Deciphering the mechanism for the assembly of aromatic polyketides by a bacterial polyketide synthase. Proc. Natl. Acad. Sci. USA 93:6600-6604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shen, B., and C. R. Hutchinson. 1993. Tetracenomycin F1 monooxygenase: oxidation of a naphthacenone to a naphthacenequinone in the biosynthesis of tetracenomycin C in Streptomyces glaucescens. Biochemistry 32:6656-6663. [DOI] [PubMed] [Google Scholar]
- 46.Shen, B., and C. R. Hutchinson. 1993. Tetracenomycin F2 cyclase: intramolecular aldol condensation in the biosynthesis of tetracenomycin C in Streptomyces glaucescens. Biochemistry 32:11149-11154. [DOI] [PubMed] [Google Scholar]
- 47.Shen, B., and C. R. Hutchinson. 1994. Triple hydroxylation of tetracenomycin A2 to tetracenomycin C in Streptomyces glaucescens. Overexpression of the tcmG gene in Streptomyces lividans and characterization of the tetracenomycin A2 oxygenase. J. Biol. Chem. 269:30726-30733. [PubMed] [Google Scholar]
- 48.Stachelhaus, T., H. D. Mootz, and M. A. Marahiel. 1999. The specificity-conferring code of adenylation domains in nonribosomal peptide synthetases. Chem. Biol. 6:493-505. [DOI] [PubMed] [Google Scholar]
- 49.Summers, R. G., S. Donadio, M. J. Staver, E. Wendt-Pienkowski, C. R. Hutchinson, and L. Katz. 1997. Sequencing and mutagenesis of genes from the erythromycin biosynthetic gene cluster of Saccharopolyspora erythraea that are involved in L-mycarose and D-desosamine production. Microbiology 143:3251-3262. [DOI] [PubMed] [Google Scholar]
- 50.Summers, R. G., E. Wendt-Pienkowski, H. Motamedi, and C. R. Hutchinson. 1992. Nucleotide sequence of the tcmII-tcmIV region of the tetracenomycin C biosynthetic gene cluster of Streptomyces glaucescens and evidence that the tcmN gene encodes a multifunctional cyclase-dehydratase-O-methyl transferase. J. Bacteriol. 174:1810-1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tang, Y., A. T. Koppisch, and C. Khosla. 2004. The acyltransferase homologue from the initiation module of the R1128 polyketide synthase is an acyl-ACP thioesterase that edits acetyl primer units. Biochemistry 43:9546-9555. [DOI] [PubMed] [Google Scholar]
- 52.Tang, Y., T. S. Lee, S. Kobayashi, and C. Khosla. 2003. Ketosynthases in the initiation and elongation modules of aromatic polyketide synthases have orthogonal acyl carrier protein specificity. Biochemistry 42:6588-6595. [DOI] [PubMed] [Google Scholar]
- 53.Torkkell, S., K. Ylihonko, J. Hakala, M. Skurnik, and P. Mantsala. 1997. Characterization of Streptomyces nogalater genes encoding enzymes involved in glycosylation steps in nogalamycin biosynthesis. Mol. Gen. Genet. 256:203-209. [DOI] [PubMed] [Google Scholar]
- 54.Udvarnoki, G., C. Wagner, R. Machinek, and J. Rohr. 1995. Biosynthetic origin of the oxygen atoms of tetracenomycin C. Angew. Chem. Int. Ed. Engl. 34:565-567. [Google Scholar]
- 55.Wendt-Pienkowski, E., Y. Huang, J. Zhang, B. Li, H. Jiang, H. Kwon, C. R. Hutchinson, and B. Shen. 2005. Cloning, sequencing, analysis, and heterologous expression of the fredericamycin biosynthetic gene cluster from Streptomyces griseus. J. Am. Chem. Soc. 127:16442-16452. [DOI] [PubMed] [Google Scholar]
- 56.Wietzorrek, A., and M. Bibb. 1997. A novel family of proteins that regulates antibiotic production in Streptomycetes appears to contain an OmpR-like DNA-binding fold. Mol. Microbiol. 25:1181-1184. [DOI] [PubMed] [Google Scholar]
- 57.Zhang, X., and R. J. Parry. 2007. Cloning and characterization of the pyrrolomycin biosynthetic gene clusters from Actinosporangium vitaminophilum ATCC 31673 and Streptomyces sp. strain UC 11065. Antimicrob. Agents Chemother. 51:946-957. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.