Abstract
β-d-2′-Deoxy-2′-fluoro-2′-C-methylcytidine (PSI-6130) is a potent inhibitor of hepatitis C virus (HCV) RNA replication in an HCV replicon assay. The 5′-triphosphate of PSI-6130 is a competitive inhibitor of the HCV RNA-dependent RNA polymerase (RdRp) and acts as a nonobligate chain terminator. Recently, it has been shown that the metabolism of PSI-6130 also results in the formation of the 5′-triphosphate of the uridine congener, β-d-2′-deoxy-2′-fluoro-2′-C-methyluridine (PSI-6206; RO2433). Here we show that the formation of the 5′-triphosphate of RO2433 (RO2433-TP) requires the deamination of PSI-6130 monophosphate and that RO2433 monophosphate is subsequently phosphorylated to the corresponding di- and triphosphates by cellular UMP-CMP kinase and nucleoside diphosphate kinase, respectively. RO2433-TP is a potent inhibitor of the HCV RdRp; however, both enzymatic and cell-based assays show that PSI-6130 triphosphate is a more potent inhibitor of the HCV RdRp than RO2433-TP.
Hepatitis C virus (HCV), a member of the Flaviviridae family of viruses, is one of the major causes of liver disease. Nearly 2% of the U.S. population and an estimated 170 million people worldwide are believed to be infected with HCV (2, 20). Approximately 80% of infected individuals develop a chronic infection, and long-term chronic HCV infection can lead to liver cirrhosis and hepatocellular carcinoma (7, 20, 24). The current standard of care is a combination of pegylated interferon alpha and ribavirin (2, 6, 8, 21), which produces viral response rates in approximately 50% of patients infected with genotype 1 virus. Due to the adverse effects associated with both interferon and ribavirin therapy and the lack of an optimal sustained viral response in the majority of patients infected with HCV, there is a need for more potent anti-HCV compounds with fewer adverse effects.
The HCV RNA-dependent RNA polymerase (RdRp; the NS5B protein) is essential for virus RNA replication and therefore represents an attractive target for therapy (3, 11, 14, 15, 25). Since nucleoside analogs form the cornerstone of therapy against human immunodeficiency virus, hepatitis B virus, and herpesviruses, such an approach to the treatment of HCV infection should prove equally effective. Recently, several nucleoside analogs with modifications at either the 2′ or the 4′ position have demonstrated good activity against HCV in vitro and in vivo (4, 12, 18).
The discovery and development of nucleoside analogs require an understanding of the pathways and enzymes involved in the anabolism of an analog to the active triphosphate form. We have shown that β-d-2′-deoxy-2′-fluoro-2′-C-methylcytidine (PSI-6130) is a potent and selective inhibitor of HCV RNA synthesis in an HCV replicon assay (5, 19, 22). Recently, we reported that PSI-6130 is anabolized to the 5′-triphosphate by enzymes involved in the deoxycytidine salvage pathway and that 2′-fluoro-2′-C-methylcytidine 5′-triphosphate (PSI-6130-TP) is an inhibitor of the HCV RdRp (19). In vitro metabolism studies have now shown that in addition to the formation of PSI-6130-TP, the 5′-triphosphate of the uridine congener, β-d-2′-deoxy-2′-fluoro-2′-C-methyluridine 5′-triphosphate (RO2433-TP), is formed in cells incubated with PSI-6130 (16). In this report we describe the enzymes involved in the metabolism of PSI-6130 to RO2433-TP and the kinetics of inhibition of the HCV RdRp by the UTP metabolite. In addition, we explore the roles of both PSI-6130-TP and RO2433-TP in the overall activity of PSI-6130 against the wild-type clone A replicon and the clone A S282T mutant replicon.
MATERIALS AND METHODS
Enzymes.
Human deoxycytidylate deaminase (DCTD) was cloned from HepG2 cells. Total RNA was isolated from cells grown to 75 to 85% confluence. Poly(A) RNA was isolated from total RNA by using a Poly(A) Purist MAG kit (Ambion, Austin, TX), and reverse transcription was performed with 1/10 of the poly(A) RNA by using a First Strand cDNA kit (Roche Applied Sciences, Indianapolis, IN) and the reverse primer from the first round of PCR. The target gene was amplified by a nested PCR procedure. The first round of PCR consisted of 25 cycles with the outer primer pair. A further 35 cycles utilized 2 μl of the PCR product from the first amplification as the template and the inner primer pair. The inner primer pair also introduced restriction enzyme sites that allowed directional cloning. The final PCR product was digested and cloned into the Escherichia coli expression vector pQE-60 (Qiagen, Valencia, CA), which introduced a six-histidine tail for affinity purification at the carboxyl terminus of the protein. The resulting product was verified by sequencing, and the construct was cotransformed into XL1-Blue MRF′ competent E. coli cells (Stratagene, La Jolla, CA) along with plasmid pRep4, which codes for the lac repressor protein. The enzyme was purified by metal affinity chromatography with Talon resin (Clontech, Mountain View, CA). Human cytidine deaminase (CDA) was cloned from Huh-7 cells. RNA isolation, cDNA preparation, amplification, cloning, expression, and purification were carried out exactly as described above for DCTD, except that the appropriate primer pairs were used. The 21-amino-acid carboxy-terminal truncated NS5B protein was cloned in our laboratory previously (19, 23). However, the clone expressed very low amounts of the protein, and the polymerase activity was also low. DNA sequencing analysis showed that the resulting enzyme contained four different amino acid residues compared to the consensus wild-type sequence, namely, Arg20, Phe182, Thr215, and Gly402. These residues were corrected to the consensus residues Lys, Leu, Met, and Arg, respectively, by using a QuikChange mutagenesis kit (Stratagene). The gene was inserted into a pET28a vector (Novagen, Inc., Madison, WI) between the NcoI and HindIII sites. The resulting plasmid, pET28a/NS5B_d21, was verified by sequencing and was transformed into BL21-gold(DE3) competent cells (Stratagene) driven by T7 RNA polymerase for protein expression. The cells were grown at 37°C until the optical density at 600 nm reached 0.6. Protein expression was induced by 0.5 mM isopropyl-β-d-thiogalactopyranoside. The cells were further grown at 20°C overnight. The S282T mutant RdRp was created by using the QuikChange mutagenesis kit (Stratagene). The sequences of both the wild type and the S282T clone were verified by DNA sequencing. The enzyme was purified by metal affinity chromatography with Talon resin (Clontech). Deoxycytidine kinase (dCK), uridine-cytidine kinase 1 (UCK-1), UMP-CMP kinase (YMPK), and nucleoside diphosphate kinase (NDPK) were cloned, expressed, and purified as described previously (19). Human creatine kinases (isoforms BB, MB, and MM), lactate dehydrogenase (LDH), and yeast 3-phosphoglycerate kinase were purchased from Sigma (St. Louis, MO). Rabbit muscle pyruvate kinase (PK) was obtained from MP Biomedicals (Solon, OH).
Other materials.
PSI-6130 and 2′-fluoro-2′-deoxycytidine (2′-F-2′-deoxycytidine) were synthesized in-house. 2′-Fluoro-2′-C-methylcytidine monophosphate (PSI-6130-MP), PSI-6130-TP, 2′-fluoro-2′-C-methyluridine (PSI-6206), 2′-fluoro-2′-C-methyluridine monophosphate (PSI-6206-MP), 2′-fluoro-2′-C-methyluridine diphosphate (PSI-6206-DP), and RO2433-TP were synthesized by ChemCyte (San Diego, CA). 2′-C-Methylcytidine was synthesized by Expicor, (Hyderabad, India). Other natural nucleosides/nucleotides were purchased from Sigma. Zebularine [1-(β-d-ribofuranosyl)pyrimidin-2-one] was purchased from Berry and Associates (Dexter, MI). [α-32P]UTP was purchased from Perkin-Elmer (Waltham, MA), and [3H]PSI-6130 was synthesized by Moravek Biochemicals (Berea, CA). HCV subgenomic replicon RNA-containing Huh-7 cells (clone A cells) were obtained from Apath, LLC (St. Louis, MO).
Preparation of 2′-deoxy-2′-fluoro-2′-C-methyluridine 5′-(phenyl methoxyalanyl phosphate).
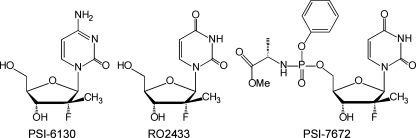
Phenyl methoxyalaninyl phosphorochloridate (1 g, 6.5 eq) dissolved in 3 ml of tetrahydrofuran was added to a mixture of 2′-deoxy-2′-fluoro-2′-C-methyluridine (0.15 g, 1 eq) and N-methylimidazole (0.3 g, 8 eq) in 3 ml tetrahydrofuran with vigorous stirring at room temperature, and then the reaction mixture was stirred overnight. The solvent was removed by the use of reduced pressure. The resulting crude product was dissolved in methanol purified by preparatory high-pressure liquid chromatography (HPLC) on a YMC column (25 by 30 by 2 mm) with a water-acetonitrile gradient elution mobile phase. The acetonitrile and water were removed under reduced pressure to give the desired product, PSI-7672 (50.1 mg, 15.6%) (Fig. 1). 1H nuclear magnetic resonance (dimethyl sulfoxide-d6) δ 1.20 to 1.27 (m, 6H), 3.58 (d, J = 16.0 Hz, 3H), 3.75 to 3.92 (m, 2H), 4.015 to 4.379 (m, 2H), 5.54 (t, J = 10.2 Hz, 1H), 5.83 to 5.91 (m, 1H), 6.00 to 6.16 (m, 1H), 7.18 (d, J = 8.0 Hz, 2H), 7.22 (s, 1H), 7.35 (t, J = 4.4 Hz, 2H), 7.55 (s, 1H), 11.52 (s, 1H); mass spectrometry, m/e 502 (M + 1)+.
FIG. 1.
Chemical structures of PSI-6130, RO2433, and PSI-7672.
Enzyme assays.
CDA assays were performed with a Perkin-Elmer (Waltham, MA) Lambda 35 spectrophotometer by monitoring the spectrum change from cytidine to uridine. All assays were performed at 30°C in a reaction mixture (1 ml) containing 50 mM Tris-HCl (pH 7.5), various concentrations of nucleoside substrate, and enzyme. The reactions were started by the addition of enzyme. The final enzyme concentrations used with each substrate were 118 nM for cytidine and deoxycytidine, 5.9 μM for PSI-6130, 6.5 μM for 2′-C-methylcytidine, and 65 nM for 2′-F-2′-deoxycytidine. When cytidine and deoxycytidine were used as the substrates, deamination was measured at a single wavelength at 282 nm (change in ɛ [Δɛ] = −3,600 M−1 cm−1). When PSI-6130, 2′-fluorocytidine, and 2′-C-methylcytidine were used as the substrates, the reaction was measured at various wavelengths, depending upon the substrate concentration. The Δɛ value was determined at each wavelength: for PSI-6130, up to 100 μM, 282 nm (Δɛ = −3,300); up to 1 mM, 291 nm (Δɛ = −1,330); up to 2 mM, 293.5 nm (Δɛ = −680); and up to 5 mM, 297 nm (Δɛ = −219) After each assay, the rate of decrease in the absorbance per minute was determined from the linear portion of the kinetic trace. The rate of product formation was calculated on a molar basis by dividing the change in absorbance by the appropriate Δɛ value, according to Beer's law (A = ɛ·C·L, where A is the absorbance, C is the concentration, and L is the light path length). The data were then entered into the GraFit program, version 5 (Erithacus Software, Horley, Surrey, United Kingdom), where Vmax and Km were determined by fitting to the Michaelis-Menten equation. The turnover number (kcat) was calculated from the molar enzyme concentration and Vmax, and the catalytic efficiency was calculated by dividing kcat by Km.
The DCTD assays were performed by using a spectrophotometric assay for the natural substrate, dCMP, and an HPLC assay for PSI-6130-MP. The spectrophotometric assay was essentially the same as the CDA assay described above. The reactions were performed at 37°C, and the change in absorbance at 290 nm (Δɛ = 1,694 M−1 cm−1) was monitored. The final enzyme concentration was 9.3 nM. The reactions for the HPLC assay were initiated by adding 0.56 μg of enzyme to a reaction mixture (500 μl) containing 50 mM Tris-HCl, pH 7.5, 2 mM MgCl2, 20 mM β-mercaptoethanol, and the substrate at the indicated concentrations. The reaction mixtures were incubated at 37°C; and at 5, 10, and 20 min, 100 μl of the reaction mixture was removed and quenched with 3.5 to 3.8% (wt/wt) HCl. Prior to HPLC analysis, each quenched sample was passed through a YM-10 Microcon centrifugal filter unit (Millipore), stored at −20°C, and analyzed within 24 h with a Perkin-Elmer (Waltham, MA) series 200 HPLC system. Strong-anion-exchange HPLC was performed on a Whatman 10-μm Partisil SAX column (Whatman, Maidstone, England) which was preequilibrated with buffer A (0.02 M KH2PO4, pH 3.5). Elution of the nucleotides was accomplished with a 10-min linear gradient with from 0 to 10% buffer B (1 M KH2PO4, pH 3.5) at a flow rate of 1.0 ml/min. The nucleotide substrate and product were detected at 254 nm and were quantified on the basis of a standard curve of the peak area versus the concentration.
All the kinase assays employed a coupled system with PK and LDH, and the oxidation of NADH was monitored at 340 nm with a Lambda 35 UV/visible spectrometer (Perkin-Elmer, Waltham, MA). Assays were performed at 25°C in a 1-ml reaction mixture containing 64 mM Tris-HCl, pH 7.5, 3.8 mM EDTA, 180 mM KCl, 12.8 mM MgCl2, 24 mM (NH4)2SO4, 1 mM ATP, 0.5 mM phosphoenolpyruvate, 0.1 mM NADH, 5 IU/ml PK, 13.8 IU/ml LDH, nucleoside substrate, and kinase. The reaction rate with different concentrations of the nucleoside substrate was determined for each enzyme, and steady-state parameters were calculated by using the GraFit program, version 5 (Erithacus Software).
RdRp assays were performed by monitoring the de novo synthesis of RNA by using a previously described method, with some modifications (19). A steady-state reaction was performed at 27°C in a total volume of 140 μl containing 2.8 μg of unprimed minus internal ribosomal entry site RNA template, 140 U of anti-RNase (Ambion), 2.8 μg of NS5B, various concentrations of natural and modified nucleotides, 5 mM MgCl2, and 2 mM dithiothreitol in 50 mM HEPES buffer, pH 7.5. In order to determine the Ki for PSI-6130-TP, the concentrations of UTP, GTP, and ATP were fixed at 10, 100, and 100 μM, respectively, and the 32P-labeled CTP and PSI-6130-TP concentrations were varied. For the experiment with RO2433-TP, the CTP, ATP, and GTP concentrations were fixed at 10, 100, and 100 μM, respectively, and the 32P-labled UTP and RO2433-TP concentrations were varied. At the desired times, 20-μl aliquots were taken and the reaction was quenched by mixing 80 μl of stop solution containing 12.5 mM EDTA, 2.25 M NaCl, and 225 mM sodium citrate into the reaction mixture. Quantification of the product and data analysis were performed as described previously (19).
Cell metabolism assay.
Clone A cells (Apath, LLC) were seeded into a six-well plate in Dulbecco modified Eagle medium supplemented with 10% fetal bovine serum. After an overnight incubation to allow cell attachment, the cells were exposed to 5 μM [3H]PSI-6130 (21.3 Ci/mmol; 4000 dpm/pmol) in the presence or the absence of different concentration of zebularine for 48 h at 37°C in a 5% CO2 atmosphere. The cell culture medium was then removed, and the cell layer was washed three times with cold phosphate-buffered saline. After trypsinization, the cells were counted, resuspended in 1 ml of cold 60% methanol, and incubated overnight at −20°C. The samples were centrifuged at 14,000 rpm for 5 min, and the supernatant was collected and dried under a gentle filtered nitrogen flow and then stored at −20°C. The residues were resuspended in 100 μl of water, and 50-μl aliquots were injected into the HPLC column. PSI-6130 and its metabolites were separated by ion-exchange HPLC with a Whatman 10-μm SAX column (Whatman, Maidstone, England) and a series 200 HPLC system (Perkin-Elmer, Waltham, MA). The mobile phase consisted of buffer A and buffer B. Elution was performed by using a linear gradient of buffer B from 0 to 100% for 90 min. The radioactivity was analyzed by using a 610TR radiometric flow scintillation analyzer (Perkin-Elmer, Waltham, MA). All phosphorylated derivatives of PSI-6130 and RO2433 were detected (Fig. 1), and the retention time of each radiolabeled phosphate species corresponded to the retention times of the unlabeled reference compounds.
HCV replicon assay.
The HCV replicon assay was performed as previously described by Stuyver et al. (23). Briefly, 25 μl of threefold serial dilutions of PSI-6130 was added alone or in combination with 100 μM zebularine to each well of a 96-well plate. Clone A cells and clone A S282T cells, which were derived by sequential passaging in the presence of 2′-C-methyladenosine (22) and which contain the S282T mutation in the HCV RdRp, were added to the 96-well plate at 1,500 cells/well in 50 μl of medium without G418 (Geneticin; Invitrogen, Carlsbad, CA). After the plates were seeded, they were incubated at 37°C in a 5% CO2 atmosphere for 4 days. Replicon RNA was extracted and amplified by use of a single-step multiplex reverse transcription-PCR (RT-PCR) protocol as described previously (23). Antiviral activity was determined by subtracting the average threshold RT-PCR cycle (CT) of the test compound from the average CT of the no-drug control (ΔCT HCV). A ΔCT value of 3.3 equals a 1-log reduction (equal to the 90% effective concentration [EC90]) in replicon RNA levels. The cytotoxicities of the test compounds were also determined by calculating the ΔCT for rRNA (ΔCT RNA).
RESULTS
Deamination of PSI-6130 and PSI-6130-MP.
CDA catalyzes the hydrolytic deamination of various cytosine nucleosides to the corresponding uracil nucleosides. Our results showed that, compared to cytidine and deoxycytidine, PSI-6130 and 2′-C-methylcytidine were both poor substrates for human CDA. The Km value determined for PSI-6130 was approximately 100-fold higher than the values determined for cytidine and deoxycytidine, and the kcat value for PSI-6130 was more than 20-fold lower than the values for cytidine and deoxycytidine (Table 1). On the basis of the kcat/Km (catalytic efficiency) value, PSI-6130 was approximately 2,000-fold less efficient than cytidine and deoxycytidine as a substrate for human CDA. A similar difference was seen with 2′-C-methylcytidine. 2′-Deoxy-2′-fluorocytidine was also tested and was shown to be a substrate for human CDA (Table 1). The kcat/Km value for 2′-F-2′-deoxycytidine (0.14 μM−1 s−1) was very similar to that for cytidine and 2′deoxycytidine. The difference in catalytic efficiency between PSI-6130 and 2′-C-methylcytidine and cytidine, deoxycytidine, and 2′-deoxy-2′-fluorocytidine suggests that the 2′-C-methyl group was responsible for the decrease in enzyme efficiency.
TABLE 1.
Deamination of PSI-6130 and PSI-6130-MPa
| Enzyme | Substrate | kcat (s−1) | Km (μM) | kcat/Km (μM−1 s−1) |
|---|---|---|---|---|
| CDA | Cytidine | 5.4 ± 0.3 | 40 ± 5 | 0.14 |
| 2′-Deoxycytidine | 4.8 ± 0.2 | 41 ± 4 | 0.12 | |
| 2′-F-2′-deoxycytidine | 3.8 ± 0.4 | 27 ± 8 | 0.14 | |
| 2′-C-Methylcytidineb | 0.18 ± 0.02 | 3,100 ± 600 | 0.000058 | |
| PSI-6130b | 0.26 ± 0.02 | 4,000 ± 500 | 0.000065 | |
| DCTD | DCMP | 246 ± 16 | 87 ± 22 | 2.8 |
| PSI-6130-MP | 0.82 ± 0.07 | 2,000 ± 300 | 0.0004 |
All values were determined by spectrophotometric assay, except that an HPLC assay was used for the DCTD reactions with PSI-6130-MP.
The highest concentration tested was 4 mM.
In vitro metabolism studies revealed that, in addition to the formation of PSI-6130-TP in cells treated exogenously with the nucleoside, the 5′-triphosphate of the uridine congener, RO2433, was also formed (16) (Fig. 2). Since RO2433 was inactive in the replicon assay (5) (Table 2) and the compound was not a substrate for UCK-1, dCK, or thymidine kinase 1 or 2 at concentrations up to 1 mM (data not shown), we explored the possibility that deamination was occurring at the monophosphate level and was possibly catalyzed by DCTD. Deamination studies were performed with PSI-6130-MP and purified human DCTD, and the steady-state kinetic parameters for each are shown in Table 1. PSI-6130-MP was a weak substrate for DCTD; the compound had a significantly lower affinity compared to that of the natural substrate, dCMP, with Km values of 2.0 ± 0.3 mM and 0.087 ± 0.022 mM, respectively. The kcat for PSI-6130-MP was approximately 300-fold lower than the kcat for dCMP, and the overall catalytic efficiency (kcat/Km) for PSI-6130-MP was 6,900-fold lower than that for dCMP.
FIG. 2.
HPLC chromatograms of extracts of clone A replicon cells incubated for 48 h with PSI-6130 in the absence of zebularine (top panel) and in the presence of 10 μM (middle panel) and 50 μM (bottom panel) zebularine. Peaks were identified from the chromatograms of the standards. The PSI-6130-MP and RO2433-MP peaks were close to the background levels, with retention times of 8.57 min and 11.83 min, respectively.
TABLE 2.
Anti-HCV replicon activity of PSI-6130, RO2433, and PSI-7672
| Replicon | EC90 (μM)
|
||
|---|---|---|---|
| PSI-6130 | RO2433 | PSI-7672 | |
| Wild type | 4.9 ± 1.2 | >100 | 1.6 ± 0.5 |
| S282T | 23.8 ± 3.4 | >100 | 34.7 ± 14.5 |
Phosphorylation of RO2433 derivatives.
Since in vitro metabolism studies showed that RO2433 monophosphate (RO2433-MP), RO2433 diphosphate (RO2433-DP), and RO2433-TP were formed in cells treated with PSI-6130 (16) (Fig. 2), we examined the abilities of several kinases to use RO2433-MP and RO2433-DP as substrates. As shown in Table 3, RO2433-MP was phosphorylated to the diphosphate by YMPK at an approximately 200-fold lower catalytic efficiency than it was by the natural substrate, UMP. The next phosphorylation step, RO2433-DP to the RO2433-TP, was catalyzed by NDPK (Table 4). The kcat/Km value for RO2433-DP was 20-fold lower than that for UDP. Creatine kinase, 3-phosphoglycerate kinase, and PK are also candidates for the enzyme responsible for the phosphorylation of RO2433-DP, as these kinases are known to phosphorylate nucleoside diphosphates. The phosphorylation of RO2433-DP was tested with different isoforms of creatine kinase (MM, MB, and BB), 3-phosphoglycerate kinase, and PK. No activity was observed with the three isoforms of creatine kinase or 3-phosphoglycerate kinase at 200 μM RO2433-DP (data not shown). RO2433-DP was a weak substrate for PK. It was not possible to determine the Km and kcat values for RO2433-DP due to the very low affinity of RO2433-DP for PK. However, on the basis of the reaction rates with low concentrations of RO2433-DP, the kcat/Km ratio was estimated to be roughly 0.004, which was approximately 10-fold lower than that with NDPK.
TABLE 3.
Phosphorylation of RO2433-MP by YMPK
| Substrate | kcat (s−1) | Km (μM) | kcat/Km (μM−1 s−1) |
|---|---|---|---|
| UMPa | 81 ± 5 | 151 ± 31 | 0.54 |
| RO2433-MP | 7.9 ± 0.5 | 870 ± 100 | 0.0091 |
The values for UMP are taken from reference 19.
TABLE 4.
Phosphorylation of RO2433-DP by NDPK
| Substrate | kcat (s−1) | Km (μM) | kcat/Km (μM−1 s−1) |
|---|---|---|---|
| UDP | 145 ± 7 | 156 ± 25 | 0.93 |
| RO2433-DP | 27 ± 2 | 585 ± 104 | 0.046 |
Effects of zebularine on the metabolism and activity of PSI-6130 in wild-type and S282T mutant replicon cells.
To provide further evidence that DCTD is involved in the metabolism of PSI-6130-MP to RO2433-MP, cell metabolism studies were performed with clone A replicon cells and the DCTD inhibitor zebularine. The clone A cells were incubated for 48 h with 5 μM [3H]PSI-6130 in the presence or absence of 10, 50, or 100 μM zebularine. Zebularine is phosphorylated to the corresponding 5′-monophosphate, which is a potent inhibitor of DCTD (10, 17). HPLC analysis of the extracts prepared from the clone A replicon cells showed that the levels of the 5′-mono-, -di-, and -triphosphate derivatives of RO2433 decreased in the zebularine-treated cells and that the levels of PSI-6130-MP, PSI-6130 5′-diphosphate (PSI-6130-DP), and PSI-6130-TP showed a marked increase compared to the levels in the untreated control cells (Fig. 2; Table 5). There was no effect on the levels of the natural ribonucleoside triphosphates in the zebularine-treated cells compared to those in the control cells treated with PSI-6130 in the absence of zebularine (data not shown).
TABLE 5.
Effects of zebularine on PSI-6130 metabolism
| Zebularine concn (μM) | Concn (pmol/106 cells)
|
||||||
|---|---|---|---|---|---|---|---|
| PSI-6130 nucleotide
|
RO2433 nucleotide
|
Total TP | |||||
| MP | DP | TP | MP | DP | TP | ||
| 0 | 0.65 ± 0.20 | 3.30 ± 0.97 | 11.53 ± 0.70 | 0.79 ± 0.34 | 4.79 ± 1.79 | 11.51 ± 0.25 | 23.04 |
| 50 | 1.04 ± 0.22 | 5.48 ± 1.01 | 28.62 ± 6.66 | 0.27 ± 0.07 | 1.33 ± 0.19 | 5.62 ± 0.06 | 34.24 |
| 100 | 1.26 ± 0.33 | 6.61 ± 0.24 | 31.74 ± 1.24 | 0.22 ± 0.05 | 0.88 ± 0.15 | 3.53 ± 0.38 | 35.27 |
In an attempt to ascertain the contribution that RO2433-TP makes to the activity of PSI-6130, the EC90 for PSI-6130 was determined in the replicon assay in the presence and absence of zebularine by using the wild-type clone A and the S282T mutant replicon. Because zebularine, at a concentration of 100 μM, significantly increased the amount of PSI-6130-TP in the clone A replicon cells, it was believed that zebularine might affect the activity of PSI-6130 in the replicon assay and therefore might provide some insight into the contribution that each metabolite might make to the overall activity of PSI-6130. In the absence of zebularine, the EC90 for PSI-6130 was 4.9 ± 1.2 μM. However, when PSI-6130 was assayed in the presence of zebularine, the EC90 for PSI-6130 was reduced 2.3-fold to 2.1 ± 0.4 μM. Similarly, when PSI-6130 was assayed with the S282T mutant replicon, the EC90 was 23.8 ± 3.4 μM, and in the presence of zebularine, the EC90 for PSI-6130 was 8.9 ± 1.9 μM. Zebularine itself showed no activity in the replicon assay (EC90, >400 μM) and was not cytotoxic (50% cytotoxic concentration, >400 μM).
Inhibition of HCV RdRp by PSI-6130-TP and RO2433-TP.
Because high levels of RO2433-TP were present in cells treated with PSI-6130, it was of interest to see if RO2433-TP inhibits the HCV RdRp. Steady-state competitive inhibition assays were performed with both the wild-type and the S282T mutant enzymes. The S282T mutation is known to confer reduced sensitivity to 2′-hydroxy-2′-C-methyl nucleosides (18). As shown in Table 6, RO2433-TP inhibited the wild-type and the S282T HCV RdRp enzymes, with Ki values of 0.42 and 22 μM, respectively. PSI-6130-TP was a more potent inhibitor of the wild-type and S282T mutant polymerases than RO2433-TP. In addition, the S282T mutation had a significantly greater impact on the activity of RO2433-TP (23.7-fold change) than on that of PSI-6130-TP (7.5-fold) against the HCV RdRp with the S282T mutation compared to that against the wild-type HCV RdRp.
TABLE 6.
Inhibition of wild-type and S282T HCV RdRp enzymes by PSI-6130-TP and RO2433-TP
| Substrate | Wild type
|
S282T
|
Fold resistancea | ||||
|---|---|---|---|---|---|---|---|
| Ki (μM) | Km (μM) | Ki/Km | Ki (μM) | Km (μM) | Ki/Km | ||
| PSI-6130-TP | 0.059 ± 0.011 | 0.074 ± 0.014 (CTP) | 0.80 | 0.31 ± 0.05 | 0.052 ± 0.008 (CTP) | 6.0 | 7.5 |
| RO2433-TP | 0.42 ± 0.04 | 0.068 ± 0.007 (UTP) | 6.2 | 22 ± 2 | 0.15 ± 0.01 (UTP) | 147 | 23.7 |
Fold resistance = (Ki/Km for mutant)/(Ki/Km for wild type).
Anti-HCV replicon activity of the phosphoramidate prodrug of RO2433.
As stated above, RO2433 was inactive in the replicon assay when clone A cells were treated with the compound. To demonstrate the intrinsic ability of RO2433 to inhibit HCV RNA replication in the replicon assay if RO2433 was delivered to cells as the monophosphate, we treated clone A cells with the phosphoramidate of RO2433, 2′-deoxy-2′-fluoro-2′-C-methyluridine 5′-(phenyl methoxyalanyl phosphate) (PSI-7672). As shown in Table 2, an EC90 value of 1.62 ± 0.54 μM was obtained for this compound. To demonstrate that the end product, RO2433-TP, was formed in clone A cells treated with PSI-7672, clone A cells were incubated with the compound for 24 h. Extracts were prepared and analyzed by liquid chromatography/mass spectrometry to detect the presence of RO2433-MP and RO2433-TP. We found that PSI-7672 was indeed converted to RO2433-MP and subsequently to RO2433-TP (data not shown). We then tested PSI-7672 for its activity against the S282T mutant replicon, which confers reduced sensitivity to 2′-hydroxy-2′-C-methyl nucleoside analogs. The S282T mutant replicon demonstrated an EC90 value of 34.77 ± 14.5 μM (Table 2).
DISCUSSION
PSI-6130 is a potent and specific inhibitor of HCV RNA synthesis in clone A replicon cells. We previously reported that the phosphorylation of PSI-6130 to the corresponding 5′-triphosphate was catalyzed by enzymes of the deoxycytidine salvage pathway (19) (Fig. 3). PSI-6130 was shown to be a substrate for dCK, and subsequent phosphorylation to the 5′-di- and -triphosphates was catalyzed by YMPK and NDPK, respectively. Both the wild-type and the S282T mutant HCV RdRp enzymes were inhibited by PSI-6130-TP in enzyme inhibition studies (19). However, RdRp containing the S282T mutation showed 7.5-fold reduced sensitivity to PSI-6130-TP compared to that of the wild-type enzyme, on the basis of their respective Ki/Km ratios.
FIG. 3.
Proposed metabolic pathway for PSI-6130. The kinetic values for the phosphorylation of PSI-6130, PSI-6130-MP, and PSI-6130-DP are from previously published data (19).
Cell-based metabolism studies performed by Ma et al. (16) with primary human hepatocytes showed that a second pathway is involved in the metabolism of PSI-6130. These studies revealed that the exposure of cells to PSI-6130 resulted not only in the formation of PSI-6130-TP but also in the formation of RO2433-TP (16). Ma et al. (16) demonstrated that RO2433-TP had a significantly longer half-life (38 h) than PSI-6130 (4.7 h). Here we show that, like primary hepatocytes, PSI-6130 is metabolized to both PSI-6130-TP and RO2433-TP in clone A replicon cells (Fig. 2; Table 5). Although PSI-6130 was a poor substrate for human CDA, it was nevertheless deaminated by the enzyme to the corresponding uridine congener. However, RO2433 was not found to be active in the replicon assay (5) (Table 2) unless it was delivered as a monophosphate prodrug (Table 2). This finding was consistent with the enzymatic data, which showed that RO2433 was not a substrate for any of the pyrimidine kinases and therefore would not be subsequently anabolized to the 5′-triphosphate. Since RO2433-MP, -DP, and -TP were detected in the cell-based metabolism studies (16) (Fig. 2; Table 5) and because there are no known deaminases that specifically deaminate CDP, CTP, dCDP, dCTP, or analogs thereof in mammalian cells, the route to the uridine nucleotides most likely involved the deamination of PSI-6130-MP by DCTD. Enzymatic studies with purified human DCTD showed that this enzyme is capable of deaminating PSI-6130-MP to RO2433-MP. This enzyme is believed to be involved in regulating the intracellular concentrations of dCTP and dTTP for DNA synthesis, as the enzyme is activated in the presence of dCTP and is inhibited by dTTP (13, 17). Our enzyme kinetic study was performed in the absence of any activators or inhibitors for comparison reasons. Additional studies will be required to determine if the deamination of PSI-6130-MP is affected by various triphosphates, including PSI-6130-TP and RO2433-TP. Further evidence that DCTD can convert PSI-6130-MP to RO2433-MP came from cell-based metabolism studies, which showed that the incubation of cells with PSI-6130 in the presence of zebularine, a potent inhibitor of DCTD (10, 17), resulted in a significant decrease in the level of RO2433-TP and a concomitant increase in the level of PSI-6130-TP. Nevertheless, these results do not preclude the possibility that another enzyme(s) that is sensitive to inhibition by zebularine may exist in cells and is capable of deaminating PSI-6130-MP.
YMPK, the enzyme responsible for phosphorylating UMP, CMP, and dCMP, phosphorylates PSI-6130-MP to PSI-6130-DP (19). Here we showed that YMPK also phosphorylates RO2433-MP to the corresponding diphosphate but with an approximately 200-fold lower catalytic efficiency than that of UMP. The last step in the activation pathway is the phosphorylation of RO2433-DP to the corresponding 5′-triphosphate. Our enzymatic studies suggest that NDPK catalyzes this step in the metabolic pathway. The phosphorylation of RO2433-DP to the triphosphate showed a 20-fold lower catalytic efficiency than that of UDP. Figure 3 summarizes the overall metabolic pathway for PSI-6130.
A steady-state kinetic analysis of the inhibition of the 21-amino acid C-terminal truncated wild-type NS5B and S282T mutant enzymes was performed with both PSI-6130-TP and RO2433-TP. The Ki value for RO2433-TP was significantly higher than the Ki value for PSI-6130-TP for the wild-type (7-fold) and S282T (71-fold) RdRp enzymes. These results were consistent with those in a recent report by Ma et al., in which RO2433-TP showed a sixfold higher Ki than PSI-6130-TP (16). The Ki/Km ratios were approximately 8- and 25-fold higher for RO2433-TP with the wild-type and the S282T mutant enzymes, respectively. Therefore, it is possible that the contribution to the inhibition of HCV replicon RNA synthesis provided by PSI-6130-TP is greater than that provided by RO2433-TP. There was a 24-fold difference in the Ki/Km ratios between the wild-type and mutant enzymes with RO2433-TP, where the difference was only 7.5-fold for PSI-6130-TP. This result suggests that the reduced sensitivity in the replicon assay, due to the S282T substitution, is the result of a greater contribution by RO2433-TP than by PSI-6130-TP. The kinetic values reported here are significantly different from those reported previously (19). This could be due to differences in the amino acid sequence compared to the consensus sequence and/or a change in our assay conditions. Our previous RdRp clone contained four different amino acids, Arg20, Phe182, Thr215, and Gly402, compared to the consensus wild-type sequence Lys20, Leu182, Met215, and Arg402. Thus, those residues were also changed back to the consensus sequence. The changes in these residues may have affected the activity of the enzyme, as the new enzyme showed significantly higher levels of activity than the previous one. Previously, our reaction buffer contained both 1 mM MgCl2 and 0.75 mM MnCl2, but in this study, we used a buffer that contained 5 mM MgCl2. It is known that divalent metal ions can significantly affect the kinetics of RdRp (1, 9, 15). Experiments were performed in the presence of either magnesium ion or manganese ion, and the results showed that the Km value for natural nucleoside triphosphate was significantly higher in a buffer containing manganese than in one containing magnesium (data not shown). In addition, we have used an enzyme from an improved expression system with a different cloning vector. Therefore, the difference in the kinetic values from the previous study may be due to the amino acid changes in our original clone from that of the consensus sequence and/or our changes in the reaction conditions.
Cell-based metabolism studies demonstrated that the formation of RO2433-TP can be inhibited when cells are incubated with PSI-6130 in the presence of the DCTD inhibitor zebularine. Since our polymerase studies showed that RO2433-TP was less active than PSI-6130-TP against wild-type RdRp and the S282T mutant RdRp, clone A replicon cells and S282T mutant replicon cells were incubated with PSI-6130 in the presence and absence of zebularine to determine what effect a reduction in the level of RO2433-TP and an increase in the level of PSI-6130-TP might have on the activity of PSI-6130 in these replicon assays. The results of these studies suggest that in the replicon assay the cytidine metabolite is a more potent inhibitor of HCV RNA synthesis than the uridine metabolite. This finding correlates well with the results from the steady-state kinetic studies with both the wild-type and the S282T mutant RdRp enzymes. Comparison of the Ki/Km ratios for PSI-6130-TP and RO2433-TP showed that PSI-6130-TP was an 8- and 29-fold better inhibitor of the wild-type and the S282T mutant enzymes, respectively, than the triphosphate of the uridine metabolite.
In conclusion, PSI-6130 is a unique compound that gives rise to two molecules that are potent inhibitors of the HCV RdRp. Given that the bases of the two active triphosphate metabolites are structurally different (cytidine versus uridine), it could be suggested that PSI-6130 provides a novel mechanism for delivering a combination therapy. To date, it has been difficult to select for replicons with reduced sensitivity to PSI-6130 in passaging experiments. Therefore, it is enticing to speculate that in the clinic, the provision of two structurally different, active metabolites may increase the genetic barrier which the virus must overcome to achieve resistance, thus preventing or slowing the emergence of resistant virus. We continue to study the mechanism of action of PSI-6130 to try to elucidate the role that this combination of metabolites plays in the activity of the compound and in the emergence of resistance.
Acknowledgments
We thank Veronique Zennou for helpful discussions.
Footnotes
Published ahead of print on 12 November 2007.
REFERENCES
- 1.Alaoui-Lsmaili, M. H., M. Hamel, L. L'Heureux, O. Nicolas, D. Bilimoria, P. Labonte, S. Mounir, and R. F. Rando. 2000. The hepatitis C virus NS5B RNA-dependent RNA polymerase activity and susceptibility to inhibitors is modulated by metal cations. J. Hum. Virol. 3:306-316. [PubMed] [Google Scholar]
- 2.Alter, M. J., D. Kruszon-Moran, O. V. Nainan, G. M. McQuillan, F. Gao, L. A. Moyer, R. A. Kaslow, and H. S. Margolis. 1999. The prevalence of hepatitis C virus infection in the United States, 1988 through 1994. N. Engl. J. Med. 341:556-562. [DOI] [PubMed] [Google Scholar]
- 3.Blight, K. J., A. A. Kolykhalov, and C. M. Rice. 2000. Efficient initiation of HCV RNA replication in cell culture. Science 290:1972-1974. [DOI] [PubMed] [Google Scholar]
- 4.Carroll, S. S., J. E. Tomassini, M. Bosserman, K. Getty, M. W. Stahlhut, A. B. Eldrup, B. Bhat, D. Hall, A. L. Simcoe, R. LaFemina, C. A. Rutkowski, B. Wolanski, Z. Yang, G. Migliaccio, R. De Francesco, L. C. Kuo, M. MacCoss, and D. B. Olsen. 2003. Inhibition of hepatitis C virus RNA replication by 2′-modified nucleoside analogs. J. Biol. Chem. 278:11979-11984. [DOI] [PubMed] [Google Scholar]
- 5.Clark, J. L., J. C. Mason, L. Hollecker, L. J. Stuyver, P. M. Tharnish, T. R. McBrayer, M. J. Otto, P. A. Furman, R. F. Schinazi, and K. A. Watanabe. 2006. Synthesis and antiviral activity of 2′-deoxy-2′-fluoro-2′-C-methyl purine nucleosides as inhibitors of hepatitis C virus RNA replication. Bioorg. Med. Chem. Lett. 16:1712-1715. [DOI] [PubMed] [Google Scholar]
- 6.Collier, J., and R. Chapman. 2001. Combination therapy with interferon-alpha and ribavirin for hepatitis C: practical treatment issues. BioDrugs 15:225-238. [DOI] [PubMed] [Google Scholar]
- 7.Darby, S. C., D. W. Ewart, P. L. Giangrande, R. J. Spooner, C. R. Rizza, G. M. Dusheiko, C. A. Lee, C. A. Ludlam, F. E. Preston, et al. 1997. Mortality from liver cancer and liver disease in haemophilic men and boys in UK given blood products contaminated with hepatitis C. Lancet 350:1425-1431. [DOI] [PubMed] [Google Scholar]
- 8.Di Bisceglie, A. M., J. McHutchison, and C. M. Rice. 2002. New therapeutic strategies for hepatitis C. Hepatology 35:224-231. [DOI] [PubMed] [Google Scholar]
- 9.Ferrari, E., J. Wright-Minogue, J. W. Fang, B. M. Baroudy, J. Y. Lau, and Z. Hong. 1999. Characterization of soluble hepatitis C virus RNA-dependent RNA polymerase expressed in Escherichia coli. J. Virol. 73:1649-1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gribaudo, G., L. Riera, P. Caposio, F. Maley, and S. Landolfo. 2003. Human cytomegalovirus requires cellular deoxycytidylate deaminase for replication in quiescent cells. J. Gen. Virol. 84:1437-1441. [DOI] [PubMed] [Google Scholar]
- 11.Ishii, K., Y. Tanaka, C. C. Yap, H. Aizaki, Y. Matsuura, and T. Miyamura. 1999. Expression of hepatitis C virus NS5B protein: characterization of its RNA polymerase activity and RNA binding. Hepatology 29:1227-1235. [DOI] [PubMed] [Google Scholar]
- 12.Klumpp, K., V. Leveque, S. Le Pogam, H. Ma, W. R. Jiang, H. Kang, C. Granycome, M. Singer, C. Laxton, J. Q. Hang, K. Sarma, D. B. Smith, D. Heindl, C. J. Hobbs, J. H. Merrett, J. Symons, N. Cammack, J. A. Martin, R. Devos, and I. Najera. 2006. The novel nucleoside analog R1479 (4′-azidocytidine) is a potent inhibitor of NS5B-dependent RNA synthesis and hepatitis C virus replication in cell culture. J. Biol. Chem. 281:3793-3799. [DOI] [PubMed] [Google Scholar]
- 13.Liou, J. Y., P. Krishnan, C. C. Hsieh, G. E. Dutschman, and Y. C. Cheng. 2003. Assessment of the effect of phosphorylated metabolites of anti-human immunodeficiency virus and anti-hepatitis B virus pyrimidine analogs on the behavior of human deoxycytidylate deaminase. Mol. Pharmacol. 63:105-110. [DOI] [PubMed] [Google Scholar]
- 14.Lohmann, V., F. Korner, U. Herian, and R. Bartenschlager. 1997. Biochemical properties of hepatitis C virus NS5B RNA-dependent RNA polymerase and identification of amino acid sequence motifs essential for enzymatic activity. J. Virol. 71:8416-8428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lohmann, V., A. Roos, F. Korner, J. O. Koch, and R. Bartenschlager. 1998. Biochemical and kinetic analyses of NS5B RNA-dependent RNA polymerase of the hepatitis C virus. Virology 249:108-118. [DOI] [PubMed] [Google Scholar]
- 16.Ma, H., W. R. Jiang, N. Robledo, V. Leveque, S. Ali, T. Lara-Jaime, M. Masjedizadeh, D. B. Smith, N. Cammack, K. Klumpp, and J. Symons. 2007. Characterization of the metabolic activation of hepatitis C virus nucleoside inhibitor beta-d-2′-deoxy-2′-fluoro-2′-C-methylcytidine (PSI-6130) and identification of a novel active 5′-triphosphate species. J. Biol. Chem. 282:29812-29820. [DOI] [PubMed] [Google Scholar]
- 17.Maley, G. F., A. P. Lobo, and F. Maley. 1993. Properties of an affinity-column-purified human deoxycytidylate deaminase. Biochim. Biophys. Acta 1162:161-170. [DOI] [PubMed] [Google Scholar]
- 18.Migliaccio, G., J. E. Tomassini, S. S. Carroll, L. Tomei, S. Altamura, B. Bhat, L. Bartholomew, M. R. Bosserman, A. Ceccacci, L. F. Colwell, R. Cortese, R. De Francesco, A. B. Eldrup, K. L. Getty, X. S. Hou, R. L. LaFemina, S. W. Ludmerer, M. MacCoss, D. R. McMasters, M. W. Stahlhut, D. B. Olsen, D. J. Hazuda, and O. A. Flores. 2003. Characterization of resistance to non-obligate chain-terminating ribonucleoside analogs that inhibit hepatitis C virus replication in vitro. J. Biol. Chem. 278:49164-49170. [DOI] [PubMed] [Google Scholar]
- 19.Murakami, E., H. Bao, M. Ramesh, T. R. McBrayer, T. Whitaker, H. M. Micolochick Steuer, R. F. Schinazi, L. J. Stuyver, A. Obikhod, M. J. Otto, and P. A. Furman. 2007. Mechanism of activation of β-d-2′-deoxy-2′-fluoro-2′-C-methylcytidine and inhibition of hepatitis C virus NS5B RNA polymerase. Antimicrob. Agents Chemother. 51:503-509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Poynard, T., V. Ratziu, Y. Benhamou, P. Opolon, P. Cacoub, and P. Bedossa. 2000. Natural history of HCV infection. Baillieres Best Pract. Res. Clin. Gastroenterol. 14:211-228. [DOI] [PubMed] [Google Scholar]
- 21.Poynard, T., M. F. Yuen, V. Ratziu, and C. L. Lai. 2003. Viral hepatitis C. Lancet 362:2095-2100. [DOI] [PubMed] [Google Scholar]
- 22.Stuyver, L. J., T. R. McBrayer, P. M. Tharnish, J. Clark, L. Hollecker, S. Lostia, T. Nachman, J. Grier, M. A. Bennett, M. Y. Xie, R. F. Schinazi, J. D. Morrey, J. L. Julander, P. A. Furman, and M. J. Otto. 2006. Inhibition of hepatitis C replicon RNA synthesis by beta-d-2′-deoxy-2′-fluoro-2′-C-methylcytidine: a specific inhibitor of hepatitis C virus replication. Antivir. Chem. Chemother. 17:79-87. [DOI] [PubMed] [Google Scholar]
- 23.Stuyver, L. J., T. Whitaker, T. R. McBrayer, B. I. Hernandez-Santiago, S. Lostia, P. M. Tharnish, M. Ramesh, C. K. Chu, R. Jordan, J. Shi, S. Rachakonda, K. A. Watanabe, M. J. Otto, and R. F. Schinazi. 2003. Ribonucleoside analogue that blocks replication of bovine viral diarrhea and hepatitis C viruses in culture. Antimicrob. Agents Chemother. 47:244-254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tong, M. J., N. S. el-Farra, A. R. Reikes, and R. L. Co. 1995. Clinical outcomes after transfusion-associated hepatitis C. N. Engl. J. Med. 332:1463-1466. [DOI] [PubMed] [Google Scholar]
- 25.Yamashita, T., S. Kaneko, Y. Shirota, W. Qin, T. Nomura, K. Kobayashi, and S. Murakami. 1998. RNA-dependent RNA polymerase activity of the soluble recombinant hepatitis C virus NS5B protein truncated at the C-terminal region. J. Biol. Chem. 273:15479-15486. [DOI] [PubMed] [Google Scholar]